Abstract
There have been significant advances in techniques for the detection of biomarker signals in the oral cavity (e.g., ELISAs for proteins, PCR for RNA and DNA) as well as the engineering and development of microfluidic approaches to make oral-based point-of-care (POC) methods for the diagnosis for both local and systemic conditions a reality. In this section, we focus on three such approaches, namely, periodontal disease management, early markers for systemic diseases, and salivary markers useful for pharmacogenomic studies. Novel approaches using non-invasive, salivary samples and user-friendly devices offer results that are as sensitive and specific as laboratory-based analyses using blood or urine.
Keywords: biomarkers, saliva, cardiovascular disease, pharmacogenomics, periodontal diseases, lab-on-a-chip
Salivary Biomarkers For Screening Risk Assessment And Response To Periodontal Therapy
Periodontal disease is a common oral infectious disease that is also a leading cause of tooth loss in adults (Pihlstrom et al., 2005). Periodontal infection is initiated by tooth-associated microbial biofilms that stimulate a host response, leading to soft tissue destruction and alveolar bone loss (Darveau, 2010). Periodontal infections are implicated in a variety of other polygenic diseases, such as cardiovascular disease, stroke, and osteoporosis (Fig. 1). The bacterial biofilm serves as a chronic exposure of oral micro-organisms adhering to teeth, leading to a repeated microbial challenge and downstream effects of an altered host response (Garlet, 2010; Kebschull et al., 2010). Diagnostic methods in clinical practice today lack the ability both to detect the onset of inflammation and to identify those patients at greatest risk for periodontal disease progression (Zhang et al., 2009).
Figure 1.
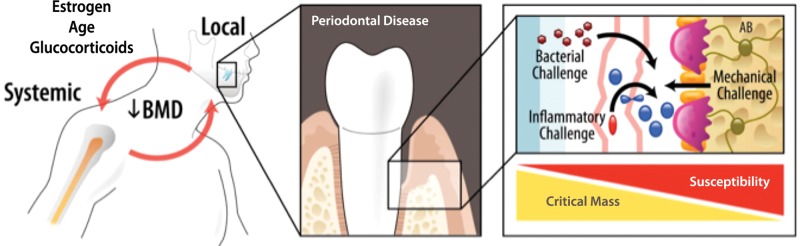
Inter-relationship between changes in the skeleton during bone metabolic diseases such as periodontitis and osteoporosis. The release of oral-fluid- and saliva-based biomarkers of disease may predict both tooth-site and skeletal bone alterations affected by pathogens, bone turnover, or biomechanical influences (Rios and Giannobile, 2010).
The development of new POC devices for periodontal surveillance will likely require less training and fewer resources than current diagnostic tests, could lead to better utilization by properly trained practitioners for simpler and less intensive treatment, and could result in cost-effective health-care delivery (Ramseier et al., 2008). In the future, patients will be screened for periodontal disease in settings other than the dental practice, such as at other health care clinics or at home, allowing them to be directed for more personalized treatments. Periodontal oral diagnostic devices will also enable large populations to be screened. In particular, underserved communities and resource-limited areas may be accessed more efficiently than by current cumbersome and poorly utilized screening programs (Yager et al., 2006). The potential to identify and monitor unique patient populations will help foster better identification of at-risk groups and increase access to treatment for those most in need, improving public health in periodontology and the oral health field in general.
Emerging clinical applications of lab-on-a-chip (LOC) technologies as point-of-care (POC) diagnostics developed for systemic diseases are now being readily applied to periodontology (Christodoulides et al., 2007; Herr et al., 2007a,b; Miller et al., 2010). The use of proteomic or multi-analyte approaches for the identification of periodontal diseases offers significant potential for providing periodontal disease “signatures” for risk (Ramseier et al., 2009; Gonçalves Lda et al., 2010; Haigh et al., 2010) (Fig. 2).
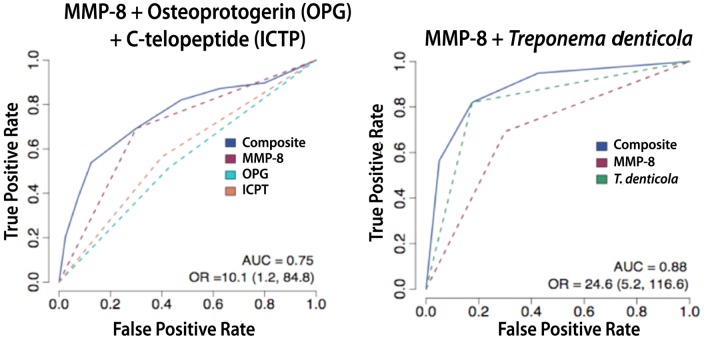
Figure 2.
Multi-analyte detection of salivary biomarkers predicts periodontal disease status. Receiver operator curves (ROC) of combinatorial permutations of salivary biomarkers [matrix metalloproteinase-8 (MMP-8), C-telopeptide (ICTP), osteoprotegerin (OPG)] coupled with biofilm subgingival pathogens such as Treponema denticola measured by quantitative PCR. AUC = area under the curve; OR = odds ratio; numbers in brackets are 95% confidence intervals. From Ramseier et al. (2009).
The use of saliva has successfully demonstrated its ease of use for POC applications for multiple disease entities (Malamud, 2006). However, for periodontal disease determination, a large body of research previously focused on gingival crevicular fluid biomarkers that provide local disease status, but represent a technically difficult approach for implementation in the clinical arena (Giannobile et al., 2009; Zhang et al., 2009). The field of periodontology is now able to detect a panel of salivary biomarkers to predict disease, including matrix metalloproteinase-8 (MMP-8) (Miller et al., 2006; Costa et al., 2010; Gursoy et al., 2010; Heikkinen et al., 2010), microbial factors (Iwano et al., 2010), and pro-inflammatory cytokines such as IL-1 beta (Miller et al., 2006; Ng et al., 2007; Fine et al., 2009; Suh et al., 2009).
Salivary biomarkers of disease have been evaluated in patients with periodontitis co-morbidities including rheumatoid arthritis (Mirrielees et al., 2010) and diabetes (Gumus et al., 2009). Further, oral-fluid-based biomarkers in local oral wound fluids have been used as assessments to predict the response to therapies such as periodontal surgery combined with MMP inhibition (Gapski et al., 2009) or tissue engineering constructs (Cooke et al., 2006; Sarment et al., 2006; Gapski et al., 2009). These wound repair biomarkers have also been used to determine the tissue healing responses of intra-oral soft tissue transplantation procedures (Morelli et al., 2011).
Periodontal disease is a multi-factorial disease involving infection, inflammation, and subsequent alveolar bone loss. Thus, biologic phenotypes may be of value, since they capture the microbial and inflammatory burden at the individual patient level. This will be important for the development of disease classifications with implications for targeted therapeutics (Casanova and Abel, 2004; Offenbacher et al., 2007). Analysis of recent data regarding the use of genetic, microbial, and protein saliva-based biomarkers supports the prediction for a propensity for gingival inflammation (Lee et al., 2011) or periodontal bony destruction (Kinney et al., 2011).
While the future of periodontal diagnostics by saliva-based techniques is promising, formidable obstacles need to be addressed prior to widespread use in a clinical setting. Validation of periodontal diagnostics will need to be benchmarked with existing gold standards of disease, including alveolar bone height and clinical attachment levels (Giannobile et al., 2009). One of the greatest challenges is not from bench to chair-side, but from chair-side to clinical practice. Acceptance by oral healthcare providers is necessary and may prove difficult. The dental community is not generally familiar with mass screening of populations for oral and systemic diseases (Ramseier et al., 2008). If more efficient periodontal therapy can be delivered, clinicians will be more likely to utilize new diagnostic approaches. A greater emphasis must be placed on clinician education in diagnostics, disease risk, and disease prevention through the public health sector before diagnostics will be integrated into routine clinical practice (Tabak, 2001). Although much needs to be done, the use of saliva-based oral fluid diagnostics offers a promising future for the diagnosis and monitoring of periodontal treatment outcomes.
Case Studies Of Salivary Diagnostics For Systemic Diseases
New developments in the area of medical microdevices can lead to medical results at POC, including bedside, ambulance, or other remote locations. Medical costs consume a staggeringly large fraction of the US and global economy, now accounting for 16.5% of the total US gross domestic product and growing at 7 to 8% each year. These expenses highlight the urgent need for the development of new tools for affordable healthcare. The electronics industry, with 5 decades of ~50% per year cost reductions, serves as an excellent model for what is possible in the healthcare industry should steps be taken now to create an effective bridge between healthcare and microelectronics industries. Despite remarkable progress toward POC clinical assay systems, few complete working prototypes have emerged. Although promising starts have been made with microfluidic LOC approaches, and important goals have been defined with the micro-total analysis system (µTAS) paradigm, the broad-scale release of workable devices has yet to be achieved. While their analysis core is substantially smaller than that of benchtop alternatives, the network of macroscopic laboratory-based infrastructure required for sample processing, analyte detection, data processing, and reagent handling implies that these platforms are best described as “chips-in-a-lab” rather than true “labs-on-a-chip”. The absence of a standard and modular analysis technology that spans multiple analyte classes motivates work toward universal mini-detection ensembles amenable to rapid prototyping with easy inclusion of newly validated biomarkers.
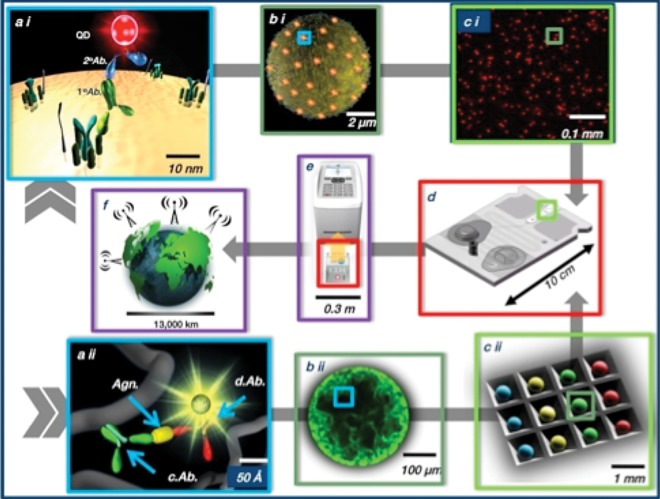
The POC analysis solution described here (Goodey et al., 2001; Goodey and McDevitt, 2003; Rodriguez et al., 2005; Weigum et al., 2007; Floriano et al., 2009; Jokerst et al., 2010; Jokerst and McDevitt, 2010) resulted in the programmablebio-nano-chip (PBNC). This device synergizes components and achievements from nanotechnology, clinical chemistry, bioinformatics, microfluidics, optics, image analysis, and pattern recognition to create a powerful new integrated measurement approach in a small device footprint. The PBNC system (Fig. 3) uses ~300-µm-diameter bead sensors composed of agarose “nano-nets” that populate a microelectro-mechanical (MEMS) support structure with integrated microfluidic elements. The beads are an efficient and selective protein capture medium suitable for the analysis of complex fluid samples. This work uses microscopy and computational studies to probe the 3D interior of the beads.
Figure 3.
Schematic illustration showing the main elements of the programmable bio-nano-chip system (PBNC). The universal detection system completes both cell-based (top image sequence, a i to c i) as well as soluble analyte tests (bottom image sequence, a ii to c ii).
The PBNC features a flexible assay design and has a diverse collection of validated analytes, and its modular design allows for rapid deployment for new biomarker signatures. Assays for nucleic acids, proteins, and cells are arranged in the PBNC to create analytical test modalities specific to different disease types. Collectively, the modularity, flexibility, and ability to process and learn new biomarker signatures represent a biologic form of “programmability”. Six major clinical trials are now active, involving the PBNC for major diseases in the areas of cardiac heart disease, oral cancer, ovarian cancer, and prostate cancer.
Saliva Testing For Pharmacogenomic Studies
The field of pharmacogenomics seeks to define five primary outcomes:
clinical response and differentiation;
risk identification;
dose selection guidance;
susceptibility, resistance, and differential disease diagnosis; and
polymorphic drug targets.
A pharmacogenomic test result, therefore, is much more informative than traditional therapeutic drug monitoring. A pharmacogenomic test result can inform physicians on the best therapeutic selection for an individual, including dose adjustment based upon a metabolic profile. Thus, the pharmacogenomic test has the potential to reduce adverse reactions or even death through accidental overdose. In many cases, the accidental overdose is the result of an individual’s genetically defined ability to metabolize particular compounds.
An excellent example of this is warfarin, which is used as an anticoagulant to protect against heart attack or stroke. Warfarin is taken by 42 million in the US each year, and dose adjustment is historically made by physicians using prothrombin (Rettie and Tai, 2006; Gak and Halkin, 2008; Babic et al., 2009; Daly, 2009; Tan et al., 2010). The dose is adjusted up or down as dictated by weekly or monthly tests to maintain optimal blood levels, since a suboptimal dose will not prevent the formation of embolisms, while an overdose can cause excessive bleeding. Pharmacogenomic testing has shown that human response to warfarin is dictated by several single nuclear polymorphisms (SNPs) located in the CYP2C9 gene. These SNPs have been defined and clinically validated. Currently, manufacturers marketing the drug include genetic information in the product labeling.
In preliminary studies utilizing genomic DNA obtained from oral fluids with a combination of custom buffers and commercially available membranes, we were able to amplify regions of DNA involved in sensitivity to warafin. In a locked nucleic acid format, the CYP2C9*2 and CYP2C9*3 mutations were distinguished from control wild-type sequences (Organtini K, Gonzalez JM, and Niedbala RS, unpublished observations), thus demonstrating proof-of-concept.
If the goal is to expand the use of oral-based diagnostics, then pharmacogenomics is a natural area for expansion. Test collection with an oral sample is as easy and non-invasive as dreamed of decades ago by some investigators. The key difference is that pharmacogenomics is based upon the collection of cells from which DNA can then be extracted, amplified, and analyzed. The mouth routinely sheds cells, or they may be easily loosened and collected by gentle brushing. The stability of DNA during collection and storage is the first challenge. Some commercial kits have been introduced, but they are expensive and not user-friendly. It is anticipated that the commercial opportunity for the collection of DNA will drive innovation, and new collectors are already being developed and introduced.
The second component to facilitate the use of oral fluid pharmacogenomics is the development of standard procedures to isolate the DNA from the collector and to amplify relevant genes efficiently. Thus far, there is only limited information outlining thoroughly evaluated procedures. Finally, the outcomes from the testing must be shown to be equivalent to current practices. Since DNA can be obtained from any cell in the body, there is theoretically no difference between samples collected from blood and those collected from the mouth. Oral-based pharmacogenomic testing is a natural extension of existing techniques. Additionally, there is a great deal of support for pharmacogenomic testing among regulatory bodies in the US. The successful use of oral fluids in this arena would expand the market for testing to millions of new opportunities.
There are several key milestones that will improve the chance for oral testing to become a standard in this rapidly developing field:
The literature shows that blood or oral fluids are viable for pharmacogenomics.
Oral fluids collection and processing costs are competitive with those for blood.
Oral fluids are included in pharmacogenomic regulations or guidance documents.
The last point above is perhaps the most important. Scientists are free to publish debate and define the parameters that will ensure quality scientific results. These results are vetted in the peer review process. However, the potential value of oral fluids included in guidance documents or regulations cannot be underestimated. Non-scientists will often look to such documents to avoid potential legal problems. Additionally, healthcare systems must also have a defined way to pay for the sampling and testing. Thus, successful use of oral testing for pharmacogenomics is technically feasible but still requires additional carefully controlled studies to create the body of evidence needed to obtain regulatory approval.
Footnotes
This work has been supported by NIH/NIDCR U01-DE014961, NCRR UL1RR024986, NIDCR/NIH UO1 DE15017, NIH U01 DE017793, UO1-DE 14964, and U19-DE 18385. We thank the American Association for Dental Research for hosting and sponsoring the Fall Focused Symposium.
WVG discloses that he holds intellectual property related to the material presented in this article.
References
- Babic N, Haverfield EV, Burrus JA, Lozada A, Das S, Yeo KT. (2009). Comparison of performance of three commercial platforms for warfarin sensitivity genotyping. Clin Chim Acta 406:143-147 [DOI] [PubMed] [Google Scholar]
- Casanova JL, Abel L. (2004). The human model: a genetic dissection of immunity to infection in natural conditions. Nat Rev Immunol 4:55-66 [DOI] [PubMed] [Google Scholar]
- Christodoulides N, Floriano PN, Miller CS, Ebersole JL, Mohanty S, Dharshan P, et al. (2007). Lab-on-a-chip methods for point-of-care measurements of salivary biomarkers of periodontitis. Ann NY Acad Sci 1098:411-428 [DOI] [PubMed] [Google Scholar]
- Cooke JW, Sarment DP, Whitesman LA, Miller SE, Jin Q, Lynch SE, et al. (2006). Effect of rhPDGF-BB delivery on mediators of periodontal wound repair. Tissue Eng 12:1441-1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa PP, Trevisan GL, Macedo GO, Palioto DB, Souza SL, Grisi MF, et al. (2010). Salivary interleukin-6, matrix metalloproteinase-8, and osteoprotegerin in patients with periodontitis and diabetes. J Periodontol 81:384-391 [DOI] [PubMed] [Google Scholar]
- Daly AK. (2009). Pharmacogenomics of anticoagulants: steps toward personal dosage. Genome Med 1:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darveau RP. (2010). Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol 8:481-490 [DOI] [PubMed] [Google Scholar]
- Fine DH, Markowitz K, Furgang D, Fairlie K, Ferrandiz J, Nasri C, et al. (2009). Macrophage inflammatory protein-1alpha: a salivary biomarker of bone loss in a longitudinal cohort study of children at risk for aggressive periodontal disease? J Periodontol 80:106-113 [DOI] [PubMed] [Google Scholar]
- Floriano PN, Christodoulides N, Miller CS, Ebersole JL, Spertus J, Rose BG, et al. (2009). Use of saliva-based nano-biochip tests for acute myocardial infarction at the point of care: a feasibility study. Clin Chem 55:1530-1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gak E, Halkin H. (2008). Shifting paradigms in the pharmacogenetics of warfarin. Pharmacogenomics 9:1373-1375 [DOI] [PubMed] [Google Scholar]
- Gapski R, Hasturk H, Van Dyke TE, Oringer RJ, Wang S, Braun TM, et al. (2009). Systemic MMP inhibition for periodontal wound repair: results of a multi-centre randomized-controlled clinical trial. J Clin Periodontol 36:149-156 [DOI] [PubMed] [Google Scholar]
- Garlet GP. (2010). Destructive and protective roles of cytokines in periodontitis: a reappraisal from host defense and tissue destruction viewpoints. J Dent Res 89:1349-1363 [DOI] [PubMed] [Google Scholar]
- Giannobile WV, Beikler T, Kinney JS, Ramseier CA, Morelli T, Wong DT. (2009). Saliva as a diagnostic tool for periodontal disease: current state and future directions. Periodontol 2000 50:52-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves Lda R, Soares MR, Nogueira FC, Garcia C, Camisasca DR, Domont G, et al. (2010). Comparative proteomic analysis of whole saliva from chronic periodontitis patients. J Proteomics 73:1334-1341 [DOI] [PubMed] [Google Scholar]
- Goodey A, Lavigne JJ, Savoy SM, Rodriguez MD, Curey T, Tsao A, et al. (2001). Development of multianalyte sensor arrays composed of chemically derivatized polymeric microspheres localized in micromachined cavities. J Am Chem Soc 123:2559-2570 [DOI] [PubMed] [Google Scholar]
- Goodey AP, McDevitt JT. (2003). Multishell microspheres with integrated chromatographic and detection layers for use in array sensors. J Am Chem Soc 125:2870-2871 [DOI] [PubMed] [Google Scholar]
- Gumus P, Buduneli N, Cetinkalp S, Hawkins SI, Renaud D, Kinane DF, et al. (2009). Salivary antioxidants in patients with type 1 or 2 diabetes and inflammatory periodontal disease: a case-control study. J Periodontol 80:1440-1446 [DOI] [PubMed] [Google Scholar]
- Gursoy UK, Kononen E, Pradhan-Palikhe P, Tervahartiala T, Pussinen PJ, Suominen-Taipale L, et al. (2010). Salivary MMP-8, TIMP-1, and ICTP as markers of advanced periodontitis. J Clin Periodontol 37:487-493 [DOI] [PubMed] [Google Scholar]
- Haigh BJ, Stewart KW, Whelan JR, Barnett MP, Smolenski GA, Wheeler TT. (2010). Alterations in the salivary proteome associated with periodontitis. J Clin Periodontol 37:241-247 [DOI] [PubMed] [Google Scholar]
- Heikkinen AM, Sorsa T, Pitkaniemi J, Tervahartiala T, Kari K, Broms U, et al. (2010). Smoking affects diagnostic salivary periodontal disease biomarker levels in adolescents. J Periodontol 81:1299-1307 [DOI] [PubMed] [Google Scholar]
- Herr AE, Hatch AV, Giannobile WV, Throckmorton DJ, Tran HM, Brennan JS, et al. (2007a). Integrated microfluidic platform for oral diagnostics. Ann NY Acad Sci 1098:362-374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AE, Hatch AV, Throckmorton DJ, Tran HM, Brennan JS, Giannobile WV, et al. (2007b). Microfluidic immunoassays as rapid saliva-based clinical diagnostics. Proc Natl Acad Sci USA 104:5268-5273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwano Y, Sugano N, Matsumoto K, Nishihara R, Iizuka T, Yoshinuma N, et al. (2010). Salivary microbial levels in relation to periodontal status and caries development. J Periodontal Res 45:165-169 [DOI] [PubMed] [Google Scholar]
- Jokerst JV, McDevitt JT. (2010). Programmable nano-bio-chips: multifunctional clinical tools for use at the point-of-care. Nanomedicine (Lond) 5:143-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokerst JV, Jacobson JW, Bhagwandin BD, Floriano PN, Christodoulides N, McDevitt JT. (2010). Programmable nano-bio-chip sensors: analytical meets clinical. Anal Chem 82:1571-1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebschull M, Demmer RT, Papapanou PN. (2010). “Gum bug, leave my heart alone!”—epidemiologic and mechanistic evidence linking periodontal infections and atherosclerosis. J Dent Res 89:879-902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney J, Morelli T, Braun T, Ramseier CA, Herr AE, Sugai JV, et al. (2011). Saliva/pathogen biomarker signatures and periodontal disease progression. J Dent Res 90:752-758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A, Bigelow-Ghaname C, Braun T, Sugai JV, Teles RP, Loesche WJ, et al. (2011). Salivary and microbial biomarkers during experimental acute gingival inflammation. J Dent Res 90(Spec Iss A):Abstract #439 http://iadr.confex.com/iadr/2011sandiego/webprogram/Paper145485.html [Google Scholar]
- Malamud D. (2006). Salivary diagnostics: the future is now. J Am Dent Assoc 137:284-286 [DOI] [PubMed] [Google Scholar]
- Miller CS, King CP, Jr, Langub MC, Kryscio RJ, Thomas MV. (2006). Salivary biomarkers of existing periodontal disease: a cross-sectional study. J Am Dent Assoc 137:322-329 [DOI] [PubMed] [Google Scholar]
- Miller CS, Foley JD, Bailey AL, Campell CL, Humphries RL, Christodoulides N, et al. (2010). Current developments in salivary diagnostics. Biomark Med 4:171-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirrielees J, Crofford LJ, Lin Y, Kryscio RJ, Dawson DR, III, Ebersole JL, et al. (2010). Rheumatoid arthritis and salivary biomarkers of periodontal disease. J Clin Periodontol 37:1068-1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli T, Neiva R, Oh TJ, Scheyer ET, Nevins ML, McGuire MK, et al. (2011). Angiogenic biomarkers and healing of living cellular constructs. J Dent Res 90:456-462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PY, Donley M, Hausmann E, Hutson AD, Rossomando EF, Scannapieco FA. (2007). Candidate salivary biomarkers associated with alveolar bone loss: cross-sectional and in vitro studies. FEMS Immunol Med Microbiol 49:252-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offenbacher S, Barros SP, Singer RE, Moss K, Williams RC, Beck JD. (2007). Periodontal disease at the biofilm-gingival interface. J Periodontol 78:1911-1925 [DOI] [PubMed] [Google Scholar]
- Pihlstrom BL, Michalowicz BS, Johnson NW. (2005). Periodontal diseases. Lancet 366:1809-1820 [DOI] [PubMed] [Google Scholar]
- Ramseier CA, Morelli T, Kinney JS, Dubois M, Rayburn LA, Giannobile WV. (2008). Periodontal disease: salivary diagnostics. In: Salivary diagnostics. Wong DT, editor. Ames, IA: Wiley Blackwell, pp.156-168 [Google Scholar]
- Ramseier CA, Kinney JS, Herr AE, Braun T, Sugai JV, Shelburne CA, et al. (2009). Identification of pathogen and host-response markers correlated with periodontal disease. J Periodontol 80:436-446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettie AE, Tai G. (2006). The pharmocogenomics of warfarin: closing in on personalized medicine. Mol Interv 6:223-227 [DOI] [PubMed] [Google Scholar]
- Rios HF, Giannobile WV. (2010). Osteoporosis and periodontal diseases. In: Oral manifestations of systemic diseases. Genco RJ, Williams RC, editors. New York: Aegis Press [Google Scholar]
- Rodriguez WR, Christodoulides N, Floriano PN, Graham S, Mohanty S, Dixon M, et al. (2005). A microchip CD4 counting method for HIV monitoring in resource-poor settings. PLoS Med 2:e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarment DP, Cooke JW, Miller SE, Jin Q, McGuire MK, Kao RT, et al. (2006). Effect of rhPDGF-BB on bone turnover during periodontal repair. J Clin Periodontol 33:135-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh KI, Kim YK, Kho HS. (2009). Salivary levels of IL-1beta, IL-6, IL-8, and TNF-alpha in patients with burning mouth syndrome. Arch Oral Biol 54:797-802 [DOI] [PubMed] [Google Scholar]
- Tabak LA. (2001). A revolution in biomedical assessment: the development of salivary diagnostics. J Dent Educ 65:1335-1339 [PubMed] [Google Scholar]
- Tan GM, Wu E, Lam YY, Yan BP. (2010). Role of warfarin pharmacogenetic testing in clinical practice. Pharmacogenomics 11:439-448 [DOI] [PubMed] [Google Scholar]
- Weigum SE, Floriano PN, Christodoulides N, McDevitt JT. (2007). Cell-based sensor for analysis of EGFR biomarker expression in oral cancer. Lab Chip 7:995-1003 [DOI] [PubMed] [Google Scholar]
- Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, et al. (2006). Microfluidic diagnostic technologies for global public health. Nature 442:412-418 [DOI] [PubMed] [Google Scholar]
- Zhang L, Henson BS, Camargo PM, Wong DT. (2009). The clinical value of salivary biomarkers for periodontal disease. Periodontol 2000 51:25-37 [DOI] [PubMed] [Google Scholar]