Abstract
In the healthy subgingiva, oral treponemes account for a small percentage of the total bacteria. However, in diseased periodontal pockets, treponemes thrive and become a dominant component of the bacterial population. Oral treponemes are uniquely adept at capitalizing on the environmental conditions that develop with periodontal disease. The molecular basis of adaptive responses of oral treponemes is just beginning to be investigated and defined. The completion of several treponeme genome sequences and the characterization of global regulatory systems provide an important starting point in the analysis of signaling and adaptive responses. In this review, we discuss existing literature focused on the genetic regulatory mechanisms of Treponema denticola and present an overview of the possible roles of regulatory proteins identified through genome analyses. This information provides insight into the possible molecular mechanisms utilized by oral spirochetes to survive in the periodontal pocket and transition from a minor to a dominant organism.
Keywords: treponemes, two-component regulatory systems, AtcRS, Hpk2, Rrp2, c-di-GMP
Introduction
Periodontal disease, a common health problem of middle-aged adults, affects millions every year, and its costs to society are high (Dye et al., 2007). Periodontitis has been aptly referred to as a “polymicrobial disruption of host homeostasis,” the intensity of which is determined by variables including the composition of the oral flora, host genetic predisposition, and underlying disorders (Darveau, 2010). The bacteriology of periodontal disease is complex, due to the presence of nearly 700 bacterial species in the oral cavity, with ~ 400 found in association with subgingival plaque (Paster et al., 2006). As periodontal disease develops, a shift occurs in the relative abundance of specific species. Elevated numbers of Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola, which form the red-microbial complex, correlate with gingivitis, chronic periodontitis, acute necrotizing ulcerative gingivitis, endodontic lesions, and have been associated with cardiovascular disease, stroke, endometriosis, preterm delivery of low-birthweight infants, and diabetes mellitus (Socransky et al., 1998; Yuan et al., 2001; Kshirsagar et al., 2007; Chen et al., 2008; Makiura et al., 2008; Kavoussi et al., 2009; Inaba and Amano, 2010). While oral Treponemes constitute a low percentage of the bacterial population in gingival crevicular fluid of healthy individuals, they are abundant in periodontal pockets (Dewhirst et al., 2000; Ellen and Galimanas, 2005). Due to T. denticola’s abundance at diseased sites, its close association with other periopathogens (Porphyromonas gingivalis), its localization at the plaque-tissue interface, and its production of powerful proteases, it is considered a prime contributor to periodontal-disease-associated tissue destruction (Holt and Ebersole, 2005). T. denticola, which produces dentilisin (a chymotrypsin-like protease) (Fenno et al., 1998), disrupts epithelial cells in vitro in a dentilisin-dependent manner (Chi et al., 2003). A direct role in tissue destruction in vivo has not been demonstrated. However, T. denticola does reside within epithelial cells in individuals with chronic periodontitis (Colombo et al., 2007). Periodontal-disease-associated tissue destruction leads to drastic changes in the physiochemical environment of the subgingival crevice and periodontium. This review seeks to summarize recent progress that has been made in delineating the key regulatory proteins and molecules that mediate environmental sensing and adaptive responses of T. denticola, and to provide potential insight into the molecular mechanisms that allow this periopathogen to thrive in disease sites in the periodontium.
Spirochetes
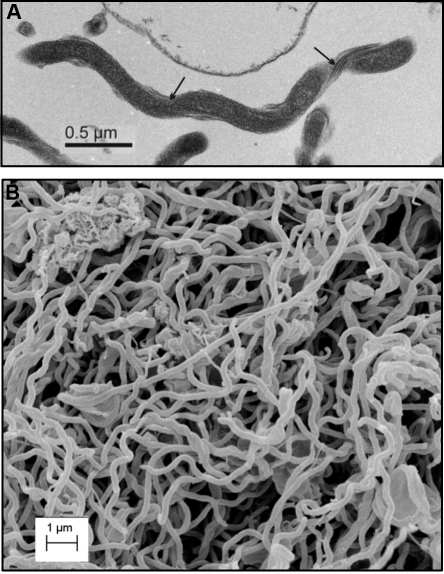
Spirochetes are a diverse group of bacteria that share a common spiral-shaped or flat-wave-form ultrastructure. The phylum Spirochaetes branched early from the evolutionary tree, dividing into 3 families (Spirochaetaceae, Brachyspiraceae, and Leptospiraceae) and 9 genera. Of these, the Treponema, Borrelia, and Leptospira are human health concerns. The genus Treponema includes the causative agents of syphilis and periopathogens. The Leptospira are causative agents of leptospirosis, and the Borrelia cause relapsing fever and Lyme disease. Spirochetes possess a distinctive ultrastructure (Fig. 1) and a unique mode of motility. They range from 5 to 20 µm in length and 0.1 to 0.5 µm in diameter. They are similar to Gram-negative bacteria, possessing both inner and outer membranes (Fig. 2), but most species lack lipopolysaccharide (LPS). LPS is replaced by lipoproteins and glycolipids (Radolf and Lukehart, 2006; Samuels and Radolf, 2010).
Figure 1.
Electron microscopic analysis of T. denticola. Transmission (A) and scanning (B) electron micrographs of T. denticola ATCC 35405. Scale bars are indicated. The arrows in panel A indicate the endoflagella bundles.
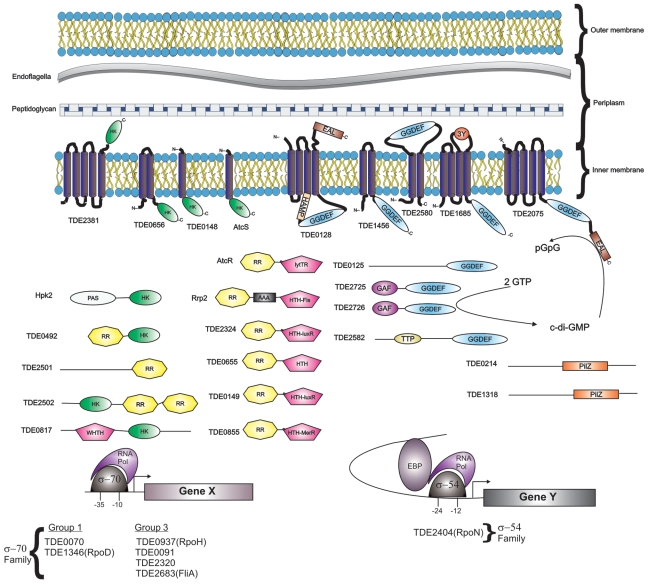
Figure 2.
Cellular architecture and genetic regulatory and signaling networks of T. denticola 35405. The T. denticola cellular architecture and predicted location of regulatory and signaling proteins are depicted. The membrane topology of the N- and C-termini of each protein is indicated. The functional domains of specific ORFs are indicated by color coding and abbreviations as follows: histidine kinase (HK-green), response regulator receiver (RR, yellow), GGDEF (blue), EAL (brown), and PilZ (orange). DNA-binding domains, which are color-coded pink, are labeled with their subfamily designation (HTH, helix-turn-helix; WHTH, winged helix-turn-helix; lytTR/AlgR/AgrA/LytR family). Other functional domains indicated include TCR 3Y motif (labeled as 3Y), tetratricopeptide repeat domain (TTP), HAMP dimerization domain (HAMP), AAA ATPase domain (AAA), GAF, and PAS domains. The sigma factors of T. denticola are listed at the bottom, with the schematic indicating the general way in which they interact with DNA and/or other regulatory proteins. Enhancer binding proteins are abbreviated as EBP.
Spirochete ultrastructure is defined by the physical influence of internal flagella (endoflagella). Endoflagella form two distinct bundles that are contained within the periplasmic space, with each inserting into the inner membrane at opposite ends of the cell [reviewed in Charon et al. (1992)]. The bundles extend two-thirds the length of the cell, wrapping around the inner membrane in a right-handed sense (refer to Fig. 1A). Intriguing questions remain as to how spirochetes regulate directionality of movement and chemotactic responses. Treponema denticola devotes nearly 6% of its genome to genes involved in motility and chemotaxis. The unusually high number of methyl-accepting chemotaxis proteins implies that it can respond to a wide range of chemoattractants (Seshadri et al., 2004). Known chemoattractants include serum, albumin, and glucose (Ruby et al., 2008). Much remains to be determined regarding the regulatory mechanisms of spirochetal motility and chemotaxis (Radolf and Lukehart, 2006; Samuels and Radolf, 2010). Recent studies indicate that the secondary messenger molecule, c-di-GMP, may be a key regulator of chemotaxis and motility in other spirochetes (Rogers et al., 2009; Sultan et al., 2010). The possible role of c-di-GMP in treponemal biology is discussed in detail below.
Phylogenetics and Genome Composition of the Oral Treponemes
The human oral microbiome harbors 76 treponemal phylotypes, with T. denticola being the most prevalent (Paster et al., 1991). T. denticola strain ATCC 35405, the most extensively characterized oral treponeme, possesses a 2.84-Mb circular chromosome (2786 ORFs) (Seshadri et al., 2004). T. vincentii ATCC 35580 and T. lecithinolyticum OMZ684T harbor 2.51-Mb (2559 ORFs) and 1.47-Mb (2059 ORFs) chromosomes, respectively (www.homd.com). Approximately 1200 of T. denticola’s ORFs have assigned functions. Consistent with their fastidious growth requirements, the genomes of oral treponemes carry only a limited set of genes that are associated with biosynthetic pathways (Seshadri et al., 2004). Plasmids are not a significant genomic component of oral treponemes (Chan et al., 1996).
Two-Component Regulatory (TCR) Systems
The mechanisms used by oral treponemes to sense and respond to the physiochemical changes associated with periodontal disease remain largely unknown. Microarray studies have demonstrated that the T. denticola transcriptome is responsive to heat shock, oxygen shock, and osmotic downshift (McHardy et al., 2010). In bacteria, two-component regulatory (TCR) systems sense and transmit signals in response to environmental stimuli (Galperin, 2006). TCR systems typically consist of a histidine kinase and a response regulator that can vary in domain architecture and output effector mechanisms (Galperin, 2006). Typically, signal transduction is initiated with the sensing of stimuli by histidine kinases. Histidine kinases autophosphorylate and then transfer the phosphate to the receiver domain of a response regulator protein, inducing conformational changes that activate the protein. In some hybrid kinases, which possess an integrated response regulator receiver domain, the phosphate may undergo intramolecular transfer steps before being transferred to its final aspartate residue. Hybrid kinases may use accessory proteins to complete the phosphotransfer process. The mechanistic and domain architecture diversity of TCR systems has been detailed in several reviews (Galperin et al., 2001; Galperin, 2006). Many response regulators act at the transcriptional level. However, some lack DNA-binding domains and regulate through protein-protein interactions or the production of secondary messenger molecules.
Most bacteria encode 20 to 30 histidine-kinase response-regulator pairs. This allows for the fine-tuned control of responses to environmental stimuli (Galperin et al., 2001). T. denticola strain 35405 encodes 6 histidine kinases, 7 response regulators, and 2 kinase-response regulator hybrids that have potential global regulatory ability (Table 1). T. lecithinolyticum and T. vincentii encode considerably fewer histidine kinases and response regulators. T. denticola TCR systems may offer a biological advantage to this periopathogen. Determination of additional genome sequences will provide insight into the potential adaptive capabilities of other treponemes. With information derived from the T. denticola strain 35405 genome sequence (Seshadri et al., 2004), a schematic depicting putative regulatory mechanisms and networks was generated (Fig. 2). This model is speculative. It is clear that our understanding of signaling mechanisms in oral treponemes as a whole is in its infancy and requires considerable investigation.
Table 1.
Histidine Kinases and Response Regulators of T. denticola Strain 35405
| ORF | Notes |
|---|---|
| Histidine Kinases | |
| TDE0032(AtcS) | forms a TCR system with the response regulator, AtcR; transcription is up-regulated during late-stage growth |
| TDE0148 | forms a TCR system with TDE0149; single transmembrane (TM) domain; co-transcribed withTDE149 and TDE0150 (a ribonuclease containing a cyclic- nucleotide-binding domain). |
| TDE0656 | forms a TCR system with TDE0655; 2 TM domains; may function as an intramembrane sensor; may be transcribed as part of an operon consisting of TDE0658-TDE0653; analogous locus is conserved in Bacillus |
| TDE1970(Hpk2) | no TM domain; N-terminal PAS domain; significant homology with Hpk2 of B. burgdorferi |
| TDE0817 | orphan kinase; no TM domain; N-terminal winged helix-turn-helix DNA-binding domain |
| TDE2381 | orphan kinase with a periplasmic kinase domain; 5 TM domains; adjacent to genes involved in cobalamine metabolism (TDE2382 and TDE2383). |
| Response Regulators | |
| TDE0033(AtcR) | forms a TCR system with AtcS; only spirochetal protein with a LytTR DNA-binding domain |
| TDE0149 | LuxR helix-turn-helix domain |
| TDE0655 | helix-turn-helix domain |
| TDE1969(Rrp2) | RpoN interaction domain and a Fis DNA-binding domain |
| TDE2501 | forms a TCR system with TDE2502; lacks an obvious effector domain |
| TDE0855 | orphan response regulator; excisionase DNA-binding domain; homologous protein (74% similar) found in Spirochaeta; may be co-transcribed with TDE0856-TDE0859 |
| TDE2324 | orphan response regulator; LuxR helix-turn-helix domain; located upstream of an eight-TM domain containing hypothetical protein (TDE2325) and a cobalamine synthesis protein cobQ (TDE2326) |
| Hybrids | |
| TDE0492 | homology with a TCR system of S. aureus that is involved in virulence and biofilm formation; no TM domain |
| TDE2502 | consists of a kinase domain and 2 receiver domains; possibly co- transcribed with TDE2501; no TM domain |
Two T. denticola strain 35405 TCR systems have been identified that appear to have potential global regulatory capability: AtcRS (Frederick et al., 2008) and Hpk2-Rrp2 (Sarkar et al., 2010) (Fig. 3). The AtcS sensor kinase and AtcR response regulator are encoded by TDE0032 and TDE0033, respectively. The Hpk2 sensor kinase and Rrp2 response regulator are encoded by TDE1970 and TDE1969, respectively. These genes are highly conserved among T. denticola isolates, and the putative functional residues are invariant in sequence. Interestingly, orthologs of AtcR and AtcS are not found in other spirochetes for which genome sequences are known. The AtcRS regulatory network may, in part, define the unique biological properties of T. denticola. In contrast, the Hpk2-Rrp2 system may have a more universal function, since it is present in several other spirochete species, including the Borrelia.
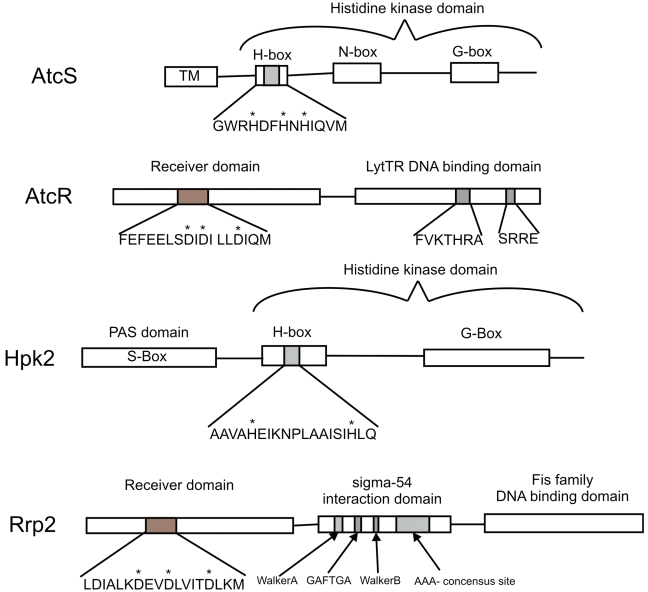
Figure 3.
Domain architecture of the AtcSR and Hpk2-Rrp2 systems. The putative functional domains of each protein are indicated. The specific sequences of regions that undergo phosphorylation are indicated, with the likely target residues designated by an asterisk. In addition, the residues required for LytTR functional activity are also shown.
AtcS is a 29-kDa inner membrane anchored protein with the characteristic H (his phosphorylation domain), N and G boxes (nucleotide-binding domains) of histidine kinases. The H box harbors 3 possible autophosphorylation sites (H57, H60, and H62). H57 and H60 are predicted to project into a solvent-accessible, putative nucleotide-binding pocket and be properly positioned to accept phosphate (Frederick et al., 2008). AtcS has 43 to 65% amino acid similarity with the CitA domain of the C4 dicarboxylate sensor kinases of Bacillus (Janausch et al., 2002). The highest degree of homology occurs within the kinase domains. The kinase domain interacts with its cognate response regulator, thus allowing for intermolecular phosphotransfer (Ohta and Newton, 2003). Hence, the homology between AtcS and CitA most likely reflects the structural conservation that is required for the kinase response-regulator interaction. The mechanism by which AtcS senses environmental signals, and the nature of those signals, remains to be determined, since the protein lacks an obvious sensing domain.
AtcR is predicted to be a 28-kDa cytoplasmic protein with 3 possible phosphor-accepting sites (D52, D54, and D59) (Frederick et al., 2008). AtcR displays 60% amino acid similarity with the Clostridium VirR response regulator (Ba-Thein et al., 1996). Both VirR and AtcR possess a LytTR domain. LytTR domains were originally identified in the AlgR/AgrA/LytR family of transcriptional regulators (Nikolskaya and Galperin, 2002). LytTR binding motifs have been mapped upstream of environmentally regulated genes, suggesting a role for AtcR in adaptive responses. Of the nearly 6000 sequenced response regulator proteins, only 5% harbor LytTR domains. The T. denticola AtcR protein is the only known oral spirochete protein to harbor a LytTR domain. The completion of additional treponemal genome sequences will further our understanding of the distribution of this regulatory domain and its role in oral spirochete biology.
Hpk2 is a conserved 46-kDa protein that is universally distributed among T. denticola strains (Sarkar et al., 2010). While T. vincentii encodes an Hpk2 ortholog (56% amino acid identity and 75% similarity), there is no obvious ortholog in T. lecithinolyticum. Hpk2 harbors an N-terminal PAS domain (acronym for Per-ARNT-Sim). PAS domains detect specific environmental stimuli, including oxygen (Moglich et al., 2009). It is noteworthy that the T. denticola Hpk2 PAS domain harbors a unique 15-aa insertion that is not found in any other PAS-domain-containing protein. This unique insertion may modulate Hpk2 signaling activity. The PAS domain is followed by an H-Box with 3 possible autophosphorylation sites (H185, H197, and H219) and an H-ATPase domain (ATP-Mg2+-binding sites) that spans the C-terminal third of the protein.
T. denticola Rrp2 is a conserved 52-kDa protein carried by several spirochete species, including the Borrelia (Sarkar et al., 2010). T. vincentii (68% amino acid identity and 84% similarity), but not T. lecithinolyticum, harbors an Rrp2 ortholog. Rrp2 possesses 3 putative phosphor-acceptor residues (D45, D48, and D53) in its receiver domain. The B. burgdorferi Rrp2 ortholog is a σ54-dependent response regulator that is an essential activator of the RpoN-RpoS regulatory pathway (Yang et al., 2003). RpoN up-regulates RpoS, which positively regulates genes involved in the transmission cycle (Ouyang et al., 2008). In B. burgdorferi, replacement of wild-type rrp2 with a site-directed mutant deficient in ATP binding abolished the transcriptional expression of several important, plasmid-encoded virulence factors that are also regulated by temperature and pH (Revel et al., 2002; Ojaimi et al., 2003). As in the Borrelia, T. denticola Rrp2 may regulate transcriptional responses to environmental changes. Interestingly, T. denticola lacks RpoS (Seshadri et al., 2004); hence, the mechanisms of Rrp2-mediated gene regulation may be distinctly different from those of other spirochetes.
The ability of AtcS and Hpk2 to autophosphorylate and transfer phosphate to their cognate response regulators has been demonstrated with recombinant proteins (Frederick et al., 2008; Sarkar et al., 2010). Neither TCR system uses accessory proteins, since transfer occurs with defined purified proteins. Interestingly, for both systems, phosphotransfer requires pre-loading of the kinase prior to incubation with the response regulator. Phosphotransfer requires kinase dimerization to generate a stable structure that can interact with the response regulator (Ohta and Newton, 2003). If the kinase is not pre-loaded with phosphate, unphosphorylated response regulator may interact with kinase monomers, preventing dimerization and inhibiting interactions required for phosphotransfer.
The PAS Domain of Hpk2 Influences Responsiveness to Environmental Stimuli
T. denticola is an obligate anaerobe, and, upon exposure to micro-aerophilic conditions, it produces hydrogen sulfide which serves to deplete localized oxygen, thus restoring an anaerobic microenvironment (Lai and Chu, 2008). The presence of a heme-binding pocket in the Hpk2 PAS domain suggests that this protein could play a role in sensing oxygen. In light of the significant differences in oxygen concentration that exist in specific microenvironments within periodontal pockets (Mettraux et al., 1984), the ability to respond to even small concentration changes could be a key aspect of T. denticola biology. Sarkar and co-workers tested the kinase activity of an N-terminal truncation variant lacking the PAS domain (Hpk2ΔPAS) (Sarkar et al., 2010). No significant difference in phosphate incorporation was observed for full-length Hpk2 and Hpk2ΔPAS (p > 0.05) when the assays were conducted under aerobic conditions. However, under anaerobic conditions, a significant reduction in phosphate incorporation was observed with Hpk2ΔPAS (relative to Hpk2; p < 0.05). Hpk2ΔPAS was also unable to transfer phosphate to Rrp2. Analysis of the data suggests that, under anaerobic conditions, signaling through the PAS domain is required for optimal autophosphorylation activity.
Transcriptional Analyses of atcR, atcS, hpk2, and rrp2
Consistent with that observed for most histidine-kinase response-regulator cognate pairs, atcR and atcS and hpk2 and rrp2 are co-transcribed (Frederick et al., 2008; Sarkar et al., 2010). Both operons are up-regulated during late-log-phase growth and can be expressed, at least in part, as larger operons (Frederick et al., 2008; Sarkar et al., 2010). AtcR-atcS is transcribed as an operon consisting of ORFs TDE0037(abrB)-TDE0030 and as a smaller bicistronic transcript. Hpk2-rrp2 is transcribed as an operon consisting of ORFs TDE1974-TDE1968. It remains to be determined if these large operons are the dominant transcriptional units during growth in the human host. Transcriptional start site analyses have revealed that the 2 atcS-atcR promoters are group 1 σ70 consensus-type promoters (Wosten, 1998; Frederick et al., 2008). The possible roles of alternative sigma factors in T. denticola biology are discussed below.
ORF TDE0037 (designated as AbrB), which can be co- transcribed with atcR, encodes a transition-state transcriptional regulatory protein (Frederick et al., 2008). AbrB family member proteins orchestrate gene expression patterns in response to environmental signals (Phillips and Strauch, 2002). Transcription of abrB is typically autoregulated and up-regulated during periods of growth transition. Analysis of the sequence upstream from T. denticola abrB identified a consensus AbrB-binding site, suggesting that AbrB may be a key player in the growth-phase-dependent transcriptional patterns of atcRS (Frederick et al., 2008).
Consideration of the putative functions of proteins encoded by genes co-transcribed with hpk2-rrp2 could provide insight as to how the up-regulation of this operon could contribute to the success of T. denticola in the periodontal pocket. TDE1968 (FtsJ), TDE1971 (DnaX), and TDE1974 (MurG) all encode proteins that carry out functions required for rapid growth. FtsJ, a 23S rRNA methyltransferase, stabilizes the 50S subunit of the 70S ribosome, allowing for efficient translation (Hager et al., 2002). In B. burgdorferi, inactivation of ftsJ results in impaired growth and morphological abnormalities (Morozova et al., 2005). DnaX (TDE1971) encodes the gamma/tau subunit of DNA polymerase III, a protein involved in DNA replication (Maki and Kornberg, 1988). MurG (TDE1974), a glycosyltransferase, catalyzes a terminal step in peptidoglycan synthesis. MurG is essential for cell viability and may be part of the divisome (Mohammadi et al., 2007). Last, ORFs TDE1972 and TDE1973 are annotated as a possible peptide toxin and colicin V production factor, respectively. While the specific role or activity of these particular proteins has not been directly demonstrated, it is possible that their production could inhibit the growth of competing organisms. The collective up-regulation of Hpk2-Rrp2, its adjacent genes, and other genes of the regulon could facilitate the rapid growth of T. denticola in the periodontal pocket.
Other Regulatory Networks Identified Through Genome Sequence Determination
C-di-GMP and Treponemal Biology
C-di-GMP, an important secondary-messenger molecule in bacteria, regulates cellular processes, including motility, expression of virulence genes, cell-to-cell signaling, exopolysaccharide production, and the transition between biofilm and planktonic modes of growth (Hengge, 2009). C-di-GMP is synthesized by diguanylate cyclases, which are defined by the presence of GGDEF domains (formerly referred to as the DUF1 domain). The intracellular pool of c-di-GMP, which can be highly localized within a cell, is controlled by the opposing activities of diguanylate cyclases and phosphodiesterases that possess an EAL or HD-GYP domain. GGDEF domains exist in several different contexts and have been identified in response-regulatory proteins (B. burgdorferi Rrp1) (Ryjenkov et al., 2005), in hybrid proteins that also contain an EAL domain, or as stand-alone enzymes (Romling et al., 2005). The effector mechanisms of c-di-GMP are just beginning to be defined. C-di-GMP has been shown to bind to PilZ domain-containing proteins, inducing conformational changes that modulate protein activity (Ryjenkov et al., 2006; Freedman et al., 2009). C-di-GMP has also been shown to bind to proteins that lack obvious PilZ domains (Chin et al., 2010). The determinants required for these non-PilZ-based interactions are not known. C-di-GMP also regulates at the mRNA level by binding to riboswitches (non-coding segments of mRNA that form specific secondary structures) (Sudarsan et al., 2008). The inability to identify conserved riboswitches in spirochetes could be reflective of sequence divergence consistent with the evolutionary separation of spirochetes from other organisms.
The importance of c-di-GMP in spirochetal biology is highlighted by recent studies of B. burgdorferi (Ryjenkov et al., 2005; Rogers et al., 2009; Freedman et al., 2009; Sultan et al., 2010). B. burgdorferi encodes a single diguanylate cyclase designated as response regulatory protein 1 (Rrp1). Microarray analyses of a B. burgdorferi rrp1 deletion mutant revealed that Rrp1, and by extension c-di-GMP, regulates the transcription of nearly 10% of the genome (Rogers et al., 2009). The genes regulated encode proteins with diverse functions. Genome sequence analyses suggest that c-di-GMP may also serve as an important regulatory molecule in T. denticola. Strain 35405 harbors proteins with GGDEF, EAL, and PilZ domains (Table 2). One of the PilZ domain proteins (TDE0214) has significant homology to the PlzA protein of B. burgdorferi (Freedman et al., 2009). C-di-GMP has the potential to regulate numerous processes that are central to the ability of T. denticola to survive and thrive in the subgingival crevice. The analysis of the role of c-di-GMP and of proteins that constitute the c-di-GMP regulatory network in the biology of oral treponemes represents a fertile area for future research.
Table 2.
C-di-GMP Regulatory Network Proteins of T. denticola 35405
| Family | ORF | Notes |
|---|---|---|
| GGDEF | TDE0125 | putative N-terminal ligand-binding domain |
| TDE1456 | terminal gene of a 5-gene operon (other genes are conserved hypotheticals); analogous locus is conserved in T. vincentii. | |
| TDE1685 | 3Y motif (occur in other sensor proteins) | |
| TDE2580 | possible operon with TDE2581 and TDE2582 | |
| TDE2581 | homologous to GGDEF proteins but does not have a GGDEF domain. | |
| TDE2582 | tetratricopeptide repeat domain (possible adhesion function) | |
| TDE2725 | cyclic-nucleotide-binding domain; may be co-transcribed with TDE2726 | |
| TDE2726 | cyclic-nucleotide-binding domain; possibly arose through gene duplication of 2725; T. vincentii contains 2 homologs in tandem but lacks TDE2725 | |
| GGDEF/EAL | TDE0128 | GGDEF and EAL domains are separated by a putative TM domain; HAMP domain |
| TDE2075 | possible operon with TDE2076 (solute binding protein). | |
| PilZ | TDE0214 | homolog of B. burgdorferi PlzA (c-di-GMP binding protein); possible operon with a tetratricopeptide repeat (TPR) protein |
| TDE1318 | only identifiable homolog is in Spirochaeta thermophila (34% identical); may form a 4-gene operon that includes genes involved in competence and recombination |
Sigma (σ) Factors and Anti-σ Factors of T. denticola 35405
The ability of bacteria to selectively regulate specific genes is dependent in part on the activity of sigma factors (σ). Sigma factors interact with RNA polymerase and direct binding to specific promoter sequences [reviewed in Wosten (1998)]. There are two main families of σ factors: σ70 and σ54. The σ70 family proteins bind to consensus -10 and -35 regions upstream of the transcriptional start site. The σ54 proteins bind to conserved -12 and -24 regions. The σ70 family is further divided into 3 main groups that bind to different nucleotide sequences. All eubacteria produce one or more σ70 family member proteins. Group 1 σ70 proteins direct the transcription of genes required for exponential growth. Group 2 σ70 proteins recognize DNA sequences similar to those recognized by group 1, but these sigma factors are not essential for exponential growth or survival. The stationary phase σ factor, RpoS (σ38 or σs), is an example of a σ70 group 2 sigma factor. The σ70 group 3 proteins consist of the alternative σ factors that control the expression of regulons in response to specific environmental stimuli or during a tightly defined window during a life/developmental/pathogenesis stage. The flagellar σ factor, σ28, extracytoplasmic function (ECF) σ factors, and heat-shock sigma factors are members of this group. T. denticola 35405 encodes 2 group 1 σ70 factors and 4 group 3 σ70 proteins (Table 3).
Table 3.
Sigma, Anti-sigma, and Anti-anti-sigma Factors of T. denticola 35405
| Family/Group | ORF | Notes and Comments |
|---|---|---|
| σ70 family | ||
| Group 1 | TDE0070 | lacks N-terminal domain required for open complex formation which could render the protein non-functional |
| TDE1346 (RpoD) | regulates housekeeping genes; conserved among spirochetes | |
| Group 2 | None detected | |
| Group 3 | TDE0937 (RpoH) | heat-shock-response sigma factor |
| TDE0091 | ECF family; part of operon containing the anti-σ factor TDE0090, cysteinyl-tRNA synthetase (CysS), and a transglutaminase; suggests possible role in cysteine and glutathione metabolism | |
| TDE2320 | ECF family; putative operon with TDE2319 and TDE2321 (hypotheticals) | |
| TDE2683 (FliA) | σ28; regulator of flagellar biosynthesis | |
| σ54 family | TDE2404 (RpoN) | Possible regulator of global transcriptional responses to environmental stimuli |
| Anti-σ factors | TDE00201 (FlgM) | Possible regulator of σ28 |
| TDE1121 | RsbW homolog; likely co-transcribed with its cognate anti-anti-σ factor (TDE1122) and the σ factor regulatory protein (TDE1123). | |
| TDE0090 | Possible regulator of TDE0091 (see above) | |
| TDE2319 | Possible regulator of TDE2320 (see above) | |
| Anti-anti-σ factors | TDE0632 | RsbV homolog; putative operon with TDE0630 and TDE0631 |
| TDE1122 | RsbV homolog; putative operon with TDE1123 | |
| TDE2083 | Contains a zinc finger DNA-binding domain. | |
| σ factor regulatory protein | TDE0630 | RsbU homolog; 2 TM domains with a cytoplasmic phosphatase domain; may regulate TDE0632 |
| TDE0631 | RsbU homolog; 10 TM domains with a periplasmic phosphatase domain; may regulate TDE0632 | |
| TDE1123 | RsbU homolog: 3 TM domains, a GAF domain and phosphatase domain; may regulate TDE1122 | |
The activity of σ70 family sigma factors can be regulated by anti-sigma factors, anti-anti-sigma factors, and σ factor regulatory proteins. Two examples of such regulation are the FliA-FlgM system (Aldridge and Hughes, 2002) and the RsbUVW system originally described in Gram-positive bacteria (Pane-Farre et al., 2009). Homologs of all of the key players of these systems are encoded by the T. denticola 35405 genome (Table 3), and would contribute to the signaling capacity of the organism.
Members of the σ54 family of σ factors are structurally distinct from the σ70 proteins and regulate by a different mechanism. They require a separate transcription factor, the enhancer binding protein (EBP), to direct transcription (Buck et al., 2000). The sole σ54 protein encoded by T. denticola is RpoN (TDE2404). The T. denticola response regulator, Rrp2, which is discussed in detail above, possesses a σ54 (i.e., RpoN) interaction domain (Sarkar et al., 2010). Rrp2 most likely serves as an EBP for RpoN. In Borrelia burgdorferi, the Rrp2-RpoN-RpoS system controls the transcription of several environmentally regulated virulence factors (Hubner et al., 2001; Smith et al., 2007). However, the lack of an RpoS (σ38) homolog in T. denticola suggests that RpoN may either act directly or utilize a yet-to-be-identified regulatory partner. T. denticola encodes at least 3 proteins that contain σ54 interaction domains (TDE2079, TDE2309, and TDE2593). TDE2309 and TDE2079 contain GAF domains which are associated with the binding of cyclic nucleotides and small molecules (Martinez et al., 2002). Interestingly, homologs of these σ54 interaction domain-containing proteins are not found in T. vincentii or T. lecithinolyticum.
Conclusions and Future Directions
As periodontal disease develops, dynamic changes occur in the bacterial population in the subgingival crevice. The physiochemical properties that develop favor specific species such as T. denticola, which transitions from a minor species in the healthy subgingiva to a dominant species in periodontal pockets. The ability to capitalize on the changing environment requires that bacteria be able to sense, read, and respond to nutrient availability, oxygen levels, metabolic by-product concentrations, blood and serum concentrations, pH, and numerous other variables. The molecular mechanisms used by the oral treponemes to drive these adaptive responses have, until recently, been largely undefined. It is our hope that this review will provide a resource that will assist investigators seeking to define the molecular basis of adaptive responses of the oral treponemes. The identification of the key molecular players and mechanisms involved in adaptive responses will open the door for translational studies seeking to develop preventive or treatment strategies for periodontal disease.
Footnotes
This work was supported by a grant to R.T. Marconi from NIH-NIDCR (5R01DE017401-04). Microscopy was performed at the VCU Department of Anatomy and Neurobiology Microscopy Facility and supported by funding from an NIH-NINDS Center Core Grant (5P30NS047463-02).
References
- Aldridge P, Hughes KT. (2002). Regulation of flagellar assembly. Curr Opin Microbiol 5:160-165 [DOI] [PubMed] [Google Scholar]
- Ba-Thein W, Lyristis M, Ohtani K, Nisbet IT, Hayashi H, Rood JI, et al. (1996). The virR/virS locus regulates the transcription of genes encoding extracellular toxin production in Clostridium perfringens. J Bacteriol 178:2514-2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M, Gallegos MT, Studholme DJ, Guo Y, Gralla JD. (2000). The bacterial enhancer-dependent sigma(54) (sigma(N)) transcription factor. J Bacteriol 182:4129-4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EC, Klitorinos A, Gharbia S, Caudry SD, Rahal MD, Siboo R. (1996). Characterization of a 4.2-kb plasmid isolated from periodontopathic spirochetes. Oral Microbiol Immunol 11:365-368 [DOI] [PubMed] [Google Scholar]
- Charon NW, Greenberg EP, Koopman MB, Limberger RJ. (1992). Spirochete chemotaxis, motility, and the structure of the spirochetal periplasmic flagella. Res Microbiol 143:597-603 [DOI] [PubMed] [Google Scholar]
- Chen YW, Umeda M, Nagasawa T, Takeuchi Y, Huang Y, Inoue Y, et al. (2008). Periodontitis may increase the risk of peripheral arterial disease. Eur J Vasc Endovasc Surg 35:153-158 [DOI] [PubMed] [Google Scholar]
- Chi B, Qi M, Kuramitsu HK. (2003). Role of dentilisin in Treponema denticola epithelial cell layer penetration. Res Microbiol 154:637-643 [DOI] [PubMed] [Google Scholar]
- Chin KH, Lee YC, Tu ZL, Chen CH, Tseng YH, Yang JM, et al. (2010). The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J Mol Biol 396:646-662 [DOI] [PubMed] [Google Scholar]
- Colombo AV, da Silva CM, Haffajee A, Colombo AP. (2007). Identification of intracellular oral species within human crevicular epithelial cells from subjects with chronic periodontitis by fluorescence in situ hybridization. J Periodontal Res 42:236-243 [DOI] [PubMed] [Google Scholar]
- Darveau RP. (2010). Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol 8:481-490 [DOI] [PubMed] [Google Scholar]
- Dewhirst FE, Tamer MA, Ericson RE, Lau CN, Levanos VA, Boches SK, et al. (2000). The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol Immunol 15:196-202 [DOI] [PubMed] [Google Scholar]
- Dye BA, Tan S, Smith V, Lewis BG, Barker LK, Thornton-Evans G, et al. (2007). Trends in oral health status: United States, 1988-1994 and 1999-2004. Vital Health Stat 11(248):1-92 [PubMed] [Google Scholar]
- Ellen RP, Galimanas VB. (2005). Spirochetes at the forefront of periodontal infections. Periodontol 2000 38:13-32 [DOI] [PubMed] [Google Scholar]
- Fenno JC, Hannam PM, Leung WK, Tamura M, Uitto VJ, McBride BC. (1998). Cytopathic effects of the major surface protein and the chymotrypsinlike protease of Treponema denticola. Infect Immun 66: 1869-1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick JR, Rogers EA, Marconi RT. (2008). Analysis of a growth-phase-regulated two-component regulatory system in the periodontal pathogen Treponema denticola. J Bacteriol 190:6162-6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman JC, Rogers EA, Kostick JL, Zhang H, Iyer R, Schwartz I, et al. (2009). Identification and molecular characterization of a cyclic-di-GMP effector protein, PlzA (BB0733): additional evidence for the existence of a functional cyclic-di-GMP regulatory network in the Lyme disease spirochete, Borrelia burgdorferi. FEMS Immunol Med Microbiol 58:285-294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. (2006). Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol 188:4169-4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Nikolskaya AN, Koonin EV. (2001). Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett 203:11-21 [DOI] [PubMed] [Google Scholar]
- Hager J, Staker BL, Bugl H, Jakob U. (2002). Active site in RrmJ, a heat shock-induced methyltransferase. J Biol Chem 277:41978-41986 [DOI] [PubMed] [Google Scholar]
- Hengge R. (2009). Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7:263-273 [DOI] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. (2005). Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000 38:72-122 [DOI] [PubMed] [Google Scholar]
- Hubner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV. (2001). Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc Natl Acad Sci USA 98:12724-12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba H, Amano A. (2010). Roles of oral bacteria in cardiovascular diseases—from molecular mechanisms to clinical cases: implication of periodontal diseases in development of systemic diseases. J Pharmacol Sci 113:103-109 [DOI] [PubMed] [Google Scholar]
- Janausch IG, Zientz E, Tran QH, Kroger A, Unden G. (2002). C4-dicarboxylate carriers and sensors in bacteria. Biochim Biophys Acta 1553:39-56 [DOI] [PubMed] [Google Scholar]
- Kavoussi SK, West BT, Taylor GW, Lebovic DI. (2009). Periodontal disease and endometriosis: analysis of the National Health and Nutrition Examination Survey. Fertil Steril 91:335-342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kshirsagar AV, Offenbacher S, Moss KL, Barros SP, Beck JD. (2007). Antibodies to periodontal organisms are associated with decreased kidney function. The Dental Atherosclerosis Risk In Communities study. Blood Purif 25:125-132 [DOI] [PubMed] [Google Scholar]
- Lai Y, Chu L. (2008). Novel mechanism for conditional aerobic growth of the anaerobic bacterium Treponema denticola. Appl Environ Microbiol 74:73-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki S, Kornberg A. (1988). DNA polymerase III holoenzyme of Escherichia coli. II. A novel complex including the gamma subunit essential for processive synthesis. J Biol Chem 263:6555-6560 [PubMed] [Google Scholar]
- Makiura N, Ojima M, Kou Y, Furuta N, Okahashi N, Shizukuishi S, et al. (2008). Relationship of Porphyromonas gingivalis with glycemic level in patients with type 2 diabetes following periodontal treatment. Oral Microbiol Immunol 23:348-351 [DOI] [PubMed] [Google Scholar]
- Martinez SE, Beavo JA, Hol WG. (2002). GAF domains: two-billion-year-old molecular switches that bind cyclic nucleotides. Mol Interv 2:317-323 [DOI] [PubMed] [Google Scholar]
- McHardy I, Keegan C, Sim JH, Shi W, Lux R. (2010). Transcriptional profiles of Treponema denticola in response to environmental conditions. PLoS One 5:e13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettraux GR, Gusberti FA, Graf H. (1984). Oxygen tension (pO2) in untreated human periodontal pockets. J Periodontol 55:516-521 [DOI] [PubMed] [Google Scholar]
- Moglich A, Ayers RA, Moffat K. (2009). Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17:1282-1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi T, Karczmarek A, Crouvoisier M, Bouhss A, Mengin-Lecreulx D, den Blaauwen T. (2007). The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol Microbiol 65:1106-1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova OV, Dubytska LP, Ivanova LB, Moreno CX, Bryksin AV, Sartakova ML, et al. (2005). Genetic and physiological characterization of 23S rRNA and ftsJ mutants of Borrelia burgdorferi isolated by mariner transposition. Gene 357:63-72 [DOI] [PubMed] [Google Scholar]
- Nikolskaya AN, Galperin MY. (2002). A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res 30:2453-2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta N, Newton A. (2003). The core dimerization domains of histidine kinases contain recognition specificity for the cognate response regulator. J Bacteriol 185:4424-4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojaimi C, Brooks C, Casjens S, Rosa P, Elias A, Barbour A, et al. (2003). Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect Immun 71:1689-1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Z, Blevins JS, Norgard MV. (2008). Transcriptional interplay among the regulators Rrp2, RpoN and RpoS in Borrelia burgdorferi. Microbiology 154(Pt 9):2641-2658 [DOI] [PubMed] [Google Scholar]
- Pane-Farre J, Jonas B, Hardwick SW, Gronau K, Lewis RJ, Hecker M, et al. (2009). Role of RsbU in controlling SigB activity in Staphylococcus aureus following alkaline stress. J Bacteriol 191:2561-2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Dewhirst FE, Weisburg WG, Tordoff LA, Fraser GJ, Hespell RB, et al. (1991). Phylogenetic analysis of the spirochetes. J Bacteriol 173:6101-6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Olsen I, Aas JA, Dewhirst FE. (2006). The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000 42:80-87 [DOI] [PubMed] [Google Scholar]
- Phillips ZE, Strauch MA. (2002). Bacillus subtilis sporulation and stationary phase gene expression. Cell Mol Life Sci 59:392-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radolf JD, Lukehart SA, editors (2006). Pathogenic Treponema: molecular and cellular biology. Norfolk, UK: Caister Academic Press [Google Scholar]
- Revel AT, Talaat AM, Norgard MV. (2002). DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc Natl Acad Sci USA 99:1562-1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers EA, Terekhova D, Zhang HM, Hovis KM, Schwartz I, Marconi RT. (2009). Rrp1, a cyclic-di-GMP-producing response regulator, is an important regulator of Borrelia burgdorferi core cellular functions. Mol Microbiol 71:1551-1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U, Gomelsky M, Galperin MY. (2005). C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol 57:629-639 [DOI] [PubMed] [Google Scholar]
- Ruby JD, Lux R, Shi W, Charon NW, Dasanayake A. (2008). Effect of glucose on Treponema denticola cell behavior. Oral Microbiol Immunol 23:234-238 [DOI] [PubMed] [Google Scholar]
- Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. (2005). Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J Bacteriol 187: 1792-1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryjenkov DA, Simm R, Romling U, Gomelsky M. (2006). The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem 281:30310-30314 [DOI] [PubMed] [Google Scholar]
- Samuels DS, Radolf JD, editors (2010). Borrelia: molecular biology, host interaction and pathogenesis. Norfolk, UK:Caister Academic Press [Google Scholar]
- Sarkar J, Frederick J, Marconi RT. (2010). The Hpk2-Rrp2 two-component regulatory system of Treponema denticola: a potential regulator of environmental and adaptive responses. Mol Oral Microbiol 25:241-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri R, Myers GS, Tettelin H, Eisen JA, Heidelberg JF, Dodson RJ, et al. (2004). Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc Natl Acad Sci USA 101:5646-5651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AH, Blevins JS, Bachlani GN, Yang XF, Norgard MV. (2007). Evidence that RpoS (sigmaS) in Borrelia burgdorferi is controlled directly by RpoN (sigma54/sigmaN). J Bacteriol 189:2139-2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL. (1998). Microbial complexes in subgingival plaque. J Clin Periodontol 25: 134-144 [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, et al. (2008). Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321:411-413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan SZ, Pitzer JE, Miller MR, Motaleb MA. (2010). Analysis of a Borrelia burgdorferi phosphodiesterase demonstrates a role for cyclic-di-guanosine monophosphate in motility and virulence. Mol Microbiol 77:128-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosten MM. (1998). Eubacterial sigma-factors. FEMS Microbiol Rev 22:127-150 [DOI] [PubMed] [Google Scholar]
- Yang XF, Alani SM, Norgard MV. (2003). The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc Natl Acad Sci USA 100:11001-11006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K, Chang CJ, Hsu PC, Sun HS, Tseng CC, Wang JR. (2001). Detection of putative periodontal pathogens in non-insulin-dependent diabetes mellitus and non-diabetes mellitus by polymerase chain reaction. J Periodontal Res 36:18-24 [DOI] [PubMed] [Google Scholar]