Abstract
Establishment of the microbiota of the gut has been shown to differ between infants delivered by Caesarian section (C-section) and those delivered vaginally. The aim of the present study was to compare the oral microbiota in infants delivered by these different routes. The oral biofilm was assayed by the Human Oral Microbe Identification Microarray (HOMIM) in healthy three-month-old infants, 38 infants born by C-section, and 25 infants delivered vaginally. Among over 300 bacterial taxa targeted by the HOMIM microarray, Slackia exigua was detected only in infants delivered by C-section. Further, significantly more bacterial taxa were detected in the infants delivered vaginally (79 species/species clusters) compared with infants delivered by C-section (54 species/species clusters). Multivariate modeling revealed a strong model that separated the microbiota of C-section and vaginally delivered infants into two distinct colonization patterns. In conclusion, our study indicated differences in the oral microbiota in infants due to mode of delivery, with vaginally delivered infants having a higher number of taxa detected by the HOMIM microarray.
Keywords: newborn, Caesarian section, vaginal delivery, bacterial taxa, HOMIM, Slackia exigua
Introduction
The first exposure to micro-organisms in vaginally delivered infants occurs during passage through the birth canal, whereas the first exposure to bacteria in infants born by Caesarian section (C-section) is from the skin of parents and health providers, and medical equipment. Different modes of delivery lead to differences in the intestinal microbiota in infants (Penders et al., 2006; Dominguez-Bello et al., 2010). Vaginally born children have been reported to have a more diverse gut microbiota, whereas children born by C-section had higher numbers of Clostridium difficile and delayed acquisition of bifidobacteria and Escherichia coli (Ahrné et al., 2005; Penders et al., 2006). In the oral cavity, mutans streptococci were detected more frequently and at a younger age in children delivered by C-section than in those delivered vaginally (Li et al., 2005). These authors hypothesized that C-section, compared with vaginal birth, lowered the exposure to commensal, protective bacteria from the mother during birth, reducing the natural barrier to colonization by oral pathogens.
Acquisition of oral bacteria in early childhood results mainly from transmission from the mother (Könönen, 2000; Tanner et al., 2002), but there is less information about other factors influencing establishment of the microbiota in the oral cavity than reported for the gut. Establishment of the gut microbiota was found not to be a predetermined species-by-species succession, but rather a coordinated interplay between external and internal factors (Fanaro et al., 2003; Penders et al., 2006). External factors for the gut microbiota included the environment during birth, the mother’s microbiota, and infant feeding method (Fallani et al., 2010). Internal factors included the developmental stage of the gastrointestinal tract and host factors (Benson et al., 2010).
The aim of the present study was to compare oral microbiota, seeking differences in colonization patterns in infants delivered vaginally or by C-section. The human Oral Microbe Identification Microarray was used to detect bacterial taxa.
Study Population & Methods
Study Population
All mothers living in a small inland town and a coastal university city in Northern Sweden who had delivered a healthy baby in the previous 3 mos were invited to consent for their infant to participate in the study. From 300 invited women, 207 accepted (69%), and all infants delivered by C-section (n = 41) and 26 randomly selected vaginally delivered infants were selected for microbial analyses. Phone interviews were conducted with the non-participants, and the only reason given for non-participation was lack of time. The study was approved by The Regional Ethical Review Board, Umeå, Sweden, and participating mothers signed informed consent at recruitment.
Mode of delivery (C-section or vaginal), intravenous treatment with antibiotics during delivery, and body weight and length were checked against medical records. The mothers completed a questionnaire on other possible confounders, such as health issues (allergy, infections, stomach problems), the infant’s use of antibiotics, feeding mode (breast- or bottle-fed), use of a pacifier, and the presence of teeth.
Microbiota by 16S rRNA Probes in HOMIM Microarray
We collected oral biofilm samples by carefully swabbing the cheeks, tongue, and alveolar ridges. DNA was purified from samples with the use of the Gen Elute Bacterial Genomic DNA kit (Sigma Aldrich, St. Louis, MO, USA) to obtain 60-1220 ng DNA, which exceeded the amount required for the microarray assay. Four samples were excluded because of low yield after DNA extraction, leaving 38 and 25 of the samples from C-section and vaginal delivery groups, respectively.
The purified DNA of samples was assayed with 422 oligonucleotide probes to the 16S rRNA gene targeting more than 300 bacterial taxa in the HOMIM microarray (http://mim.forsyth.org/homim.html). Samples were analyzed at the HOMIM microarray facility at The Forsyth Institute, Cambridge, MA, USA (Colombo et al., 2009). Hybridization signals were read on a six-level scale (0-5), with a lower limit of detection of 104 cells (Colombo et al., 2009).
Statistical Procedures
Body length and weight at birth and at 3 mos were averaged among infants delivered by the two birth delivery modes. Differences between means were tested by two-sided, independent t tests. Dichotomized scores from the HOMIM microarray analyses were used. Lack of signal was set to 0, and all signal levels ≥ 1 to 1. Differences in prevalence distribution between groups were tested with a Chi2 test. The False Discovery Rate method was used to identify a p-value with less than one false rejection of H0 when true (p < 0.005). Thus, a p-value < 0.005 was considered statistically significant to account for multiple comparisons.
Multivariate partial least-squares discriminant analysis (PLS-DA) modeling was performed (SIMCA P+, version 12.0, Umetrics AB, Umeå, Sweden) as described (Sjöström et al., 1986; Bylesjö et al., 2006). In contrast to traditional regression models, the PLS-DA technique, which defines the maximum separation between class members (here mode of delivery) in the data, is suitable for data where the number of observations is smaller than the number of variables, and where the independent variables co-vary. Dichotomous HOMIM signals, and the selected individual characteristics, gender, weight and length at birth and 3 mos, gestational wks at delivery, treatment with antibiotics during delivery, feeding mode (bottle- or breast-fed), use of pacifier, presence and number of teeth, and town of residence built the X-block, and mode of delivery the Y-block (outcome). An identical model including breast-fed infants born in or later than gestational wk 37 only was also run. Variables were autoscaled to unit variance, and cross-validated prediction of Y calculated (Wold, 1978). Cross-validation was done by a systematic prediction of 1/7th of the data by the remaining 6/7th of the data. The importance of each x-variable in the model is given by a variable importance in projection (VIP) value. VIP > 1.0 was considered influential and VIP ≥ 1.5 highly influential (Sjöström et al., 1986). The R2- and Q2-values give the capacity of the x-variables to explain (R2) and predict (Q2) the outcome.
Results
Cohort Description
There were no differences by gender or by other characteristics, including breast-feeding, between infants born vaginally and those born by C-section (Table 1). Two infants were born in gestational wk 35 (one delivered by C-section and one by the vaginal route), whereas all other infants were born in gestational wk 37 or later. All infants were healthy at birth and at 3 mos of age. None of the infants had ever received antibiotic treatment, and none was ever given supplements containing probiotic bacteria. With the exception of 15 mothers who received intravenous antibiotics in association with a C-section because of an acute clinical complication, none had antibiotics at delivery. There were no significant differences between participating and non-participating infants in length and weight at birth and at 3 mos of age, or in the socio-economic variables of their families (data not shown).
Table 1.
Gender Proportions, Body Weight and Length, and Feeding Method for Infants Delivered Vaginally or by Caesarian Section
| Vaginal Delivery n = 251 | Caesarian Section n = 381 | p-value | |
|---|---|---|---|
| Boys/Girls (numbers) | 16/10 | 19/21 | 0.264 |
| Length (cm)2 | |||
| at birth | 51.1 (50.0–52.2) | 49.2 (48.4–50.0) | 0.849 |
| at 3 mos of age | 61.3 (60.4–62.4) | 60.2 (59.2–60.8) | 0.100 |
| Weight (g)2 | |||
| at birth | 3603 (3394–3813) | 3425 (3190–3660) | 0.202 |
| at 3 mos of age | 6364 (5986–6742) | 6069 (5807–6331) | 0.221 |
| Breast-fed at 3 mos of age (%)3 | |||
| Exclusively or partially | 27 | 32 | 0.836 |
| Not at all | 73 | 68 | |
Numbers vary slightly for various analyses due to single missing values. Oral samples from four infants, three born by C-section and one vaginally, were not analyzed by microarray, because of low DNA yield.
Mean (95% CI limits). Differences of means were tested with Student’s independent t test after confirmation of a normal distribution.
Differences in proportions were tested with the Chi2-test.
Bacteria Detected by HOMIM Microarray
There was reactivity to 85 of the 300 taxa in the HOMIM microarray in oral biofilms of three-month-old infants (Appendix Table). Bacteria detected belonged to 6 phyla or divisions, and approximately half of the taxa detected belonged in Firmicutes, particularly Streptococcus species, which were detected in all infants (Table 2). Other genera detected in 80 to 99% of all children were Actinomyces, Gemella, and Veillonella. A smaller proportion of the infants (< 15% of the combined groups) had species in the genera Bacteroides, Selenomonas, Aggregatibacter, Kingella, Neisseria, and the TM7 division.
Table 2.
Proportions (%) of Three-month-old Infants with a Positive HOMIM Signal by Genus
| Phylogenetic Group and Genus | Vaginal Delivery(%) | Caesarian Section(%) | p-value |
|---|---|---|---|
| Actinobacteria | |||
| Actinomyces | 92.0 | 81.6 | 0.247 |
| Rothia | 68.0 | 65.8 | 0.856 |
| Bacteroidetes | |||
| Bacteroides | 8.0 | 0.0 | 0.076 |
| Capnocytophaga | 16.0 | 0.0 | 0.011 |
| Porphyromonas | 4.0 | 0.0 | 0.214 |
| Prevotella | 40.0 | 7.9 | 0.002 |
| Firmicutes | |||
| Eubacterium | 16.0 | 13.2 | 0.752 |
| Gemella | 96.0 | 100.0 | 0.214 |
| Granulicatella | 36.0 | 60.5 | 0.057 |
| Lactobacillus | 16.0 | 63.2 | 0.000 |
| Parvimonas | 4.0 | 18.4 | 0.093 |
| Selenomonas | 12.0 | 10.5 | 0.856 |
| Solobacterium | 20.0 | 10.5 | 0.293 |
| Streptococcus | 100 | 100 | 1.000 |
| Veillonella | 84.0 | 97.4 | 0.055 |
| Fusobacteria | |||
| Fusobacterium | 44.0 | 34.2 | 0.434 |
| Leptotrichia | 44.0 | 13.2 | 0.006 |
| Proteobacteria | |||
| Aggregatibacter | 4.0 | 0.0 | 0.214 |
| Campylobacter | 40.0 | 28.9 | 0.363 |
| Haemophilus | 28.0 | 7.9 | 0.033 |
| Kingella | 8.0 | 0.0 | 0.076 |
| Neisseria | 8.0 | 5.3 | 0.663 |
| TM7 Division | 12.0 | 0.0 | 0.029 |
Species or species clusters detected in all infants were Streptococcus Cluster II, Streptococcus Cluster III, Streptococcus anginosus/intermedius, and Streptococcus oralis (Appendix Table). Species detected in ≥ 80% of all children were Streptococcus Cluster I, Streptococcus mitis biovar 2, Streptococcus australis, Streptococcus parasanguinis I and II, Actinomyces gerensceriae, Gemella hemolysans, Veillonella atypical, and Veillonella parvula. Species detected in only a few (< 15%) infants included Streptococcus sanguinis and Streptococcus mutans, species of Neisseria, Aggregatibacter, and Kingella, and Actinomyces naeslundii genospecies1 and 2 (Actinomyces clusters I and II, respectively) (Appendix Table).
Species Distribution by Mode of Delivery
There were higher numbers of taxa detected by the microarray in swabs from infants delivered vaginally (79 species/clusters) than in infants delivered by C-section (54 species/clusters) (p = 0.001). Of the species or clusters detected, 31 were detected only in infants born vaginally, compared with 6 species or clusters that were detected only in C-section infants (Appendix Fig. 1).
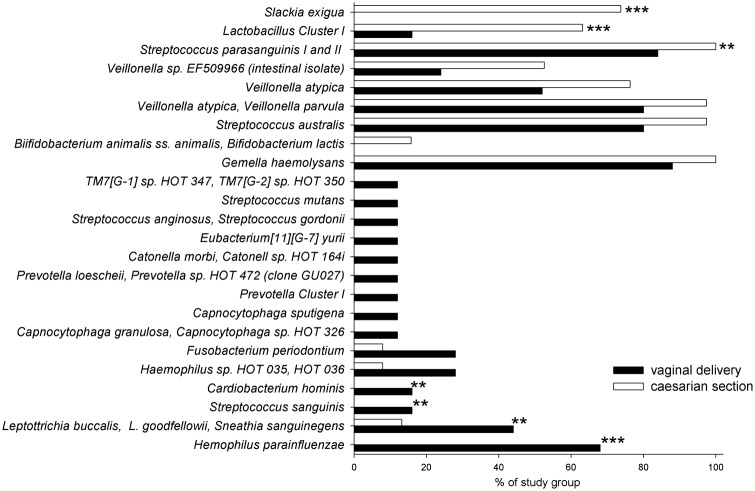
The detection frequencies of 22 species and 2 clusters differed between modes of infant delivery. Species or clusters detected more frequently from C-section compared with vaginally born infants were Slackia exigua (p < 0.001) and Lactobacillus Cluster I (p < 0.001) (Fig.). Haemophilus parainfluenzae (p = 0.005) was detected more frequently from vaginally delivered compared with C-section infants (Fig.). Additional species/clusters displayed marginal significance levels (p-values between 0.02 and 0.006; Fig.).
Figure.
Reactivity to 24 probes (out of 85 probe reactions) that differed significantly (p < 0.005) or marginally (p < 0.01) in three-month-old infants born vaginally or by Caesarian section. ***p ≤ 0.005, **p < 0.01 tested by Chi-square. No indication for p-values between 0.02 and 0.01.
PLS-DA Multivariate Modeling of HOMIM 16S rRNA-based Microarray Data
A model with two significant components was obtained by PLS-DA modeling with mode of delivery as outcome (y-variable) and dichotomized HOMIM microarray signals as the x-block, including selected individual characteristics (see statistics) as potential confounders. This model virtually clustered infants delivered by C-section separately from those delivered vaginally (Appendix Fig. 2). The multivariate model, which had an explanatory and predictive capacity of 62% (R2 = 0.618) and 44% (Q2 = 0.0.457), respectively, confirmed associations found in the univariate analyses. Species strongly associated (VIP ≥ 1.5) with being born by C-section were Slackia exigua, Lactobacillus Cluster I, Veillonella sp. EF509966, Veillonella atypical, and V. parvula, and those associated with being vaginally delivered were S. sanguinis, Streptococcus sp. HOT 058, and Cardiobacterium hominis (Table 3). The model remained strong (R2 = 0.693, Q2 = 0.496), and the same taxa remained strongly influential when the two pre-term (gestational wk 35) and all formula-fed infants were excluded.
Table 3.
Variable Importance (VIP) for Bacteria Associated with Mode of Delivery from PLS-DA Multivariate Modeling
| Associated with Vaginal Delivery | Associated with Caesarian Section Delivery | ||
|---|---|---|---|
| Bacterial Group1 | VIP | Bacterial Group1 | VIP |
| Streptococcus sanguinis | 1.50 | Slackia exigua | 3.26 |
| Cardiobacterium hominis | 1.50 | Lactobacillus Cluster I3 | 1.85 |
| Streptococcus anginosus, Streptococcus gordonii | 1.44 | Veillonella sp. EF509966 (intestinal isolate) | 1.54 |
| Prevotella Cluster I2 | 1.43 | Veillonella atypica, Veillonella parvula | 1.51 |
| Prevotella loescheii, Prevotella sp. HOT 472 | 1.43 | Streptococcus parasanguinis I and II | 1.34 |
| Eubacterium [11] [G-7] yurii | 1.43 | Veillonella parvula | 1.23 |
| Catonella morbi, Catonella sp. HOT 164 | 1.43 | Gemella haemolysans | 1.32 |
| TM7 [G-1] sp. HOT 347, TM7 [G-2] sp. HOT 350 | 1.43 | Gemella morbillorum | 1.26 |
| Haemophilus parainfluenzae | 1.41 | Streptococcus australis | 1.20 |
| Leptotrichia buccalis, Leptotrichia goodfellowii, Sneathia sanguinegens | 1.31 | Bifidobacterium animalis ss. animalis, Bifidobacterium lactis | 1.13 |
| Bacteroidetes phylum | 1.28 | Streptococcus cristatus | 1.08 |
| Campylobacter gracilis | 1.27 | Kingella oralis, Eikenella sp. HOT 009 | 1.03 |
| Capnocytophaga granulose, Capnocytophaga sp. HOT 326 | 1.26 | Selenomonas noxia | 1.03 |
| Haemophilus sp. HOT 035, HOT 036 | 1.25 | Selenomonas sputigena, Selemonas sp. HOT 143 | 1.03 |
| Kingella oralis | 1.18 | Aggregatibacter segnis, Aggregatibacter sp. HOT 512 | 1.03 |
| Prevotella nigrescens | 1.05 | Campylobacter concisus | 1.03 |
| Corynebacterium matruchotii | 1.05 | Streptococcus sp. HOT 070, 071 | 1.02 |
| Neisseria elongata | 1.05 | Granulicatella elegans | 1.02 |
| Fusobacterium periodontium | 1.05 | ||
| Capnocytophaga sputigena | 1.05 | ||
| Streptococcus mutans | 1.04 | ||
| Prevotella Cluster IV | 1.01 | ||
| Prevotella melaninogenica, Prevotella histicola | 1.01 | ||
Results are shown for a basic model including all infants with HOMIM microarray data (n = 63) and (in bold) a second model restricted to breast-fed infants and those born in gestational week 37 or later (n = 42). In the PLS-DA model including all 63 infants (model R2 = 0.620, Q2 = 0435), length at birth, gestational weeks, and town of residence were associated with mode of birth (VIP ≥ 1.0), whereas in the PLS-DA model restricted to breast-fed infants and infants born in or after gestational week 3 (n = 42) (model R2 = 0.693, Q2 = 0.496), only length at 3 mos was influential in addition to bacteria.
Unnamed taxa are identified by their Human Oral Taxon (HOT) number from HOMD (Dewhirst et al., 2010).
Prevotella cluster I (probe Y65) targets P. loescheii, Prevotella spp clone GU027, Prevotella spp strain C3MKM081, and Prevotella spp strain TFI B31FD (HOT numbers 317, 472, 658).
Lactobacillus cluster I (probe W94) targets L. casei, L. paracasei, and L. rhamnosus.
Discussion
The present study investigated the oral microbiota in infants delivered vaginally or by C-section to evaluate if there were differences associated by birth delivery mode. Higher numbers of taxa were detected among infants delivered vaginally, compared with those delivered by C-section, with probes to the 16S rRNA gene of cultivated and uncultivated oral bacteria in a microarray format (HOMIM; Paster and Dewhirst, 2009). Further, the results indicated differences in the microbiota depending on delivery method, including a novel finding that Slackia exigua was detected exclusively, and in high prevalence, in infants delivered by C-section. These findings indicate that there were differences in the microbiota of the oral cavity depending on birth delivery method, as has been reported for the microbiota of the lower gastrointestinal tract (Penders et al., 2006; Dominguez-Bello et al., 2010).
In the current study, 85 species or species clusters out of the approximately 300 taxa evaluated by the HOMIM microarray were detected in the three-month-old infants. This is fewer bacterial taxa than reported for adults in whom approximately 65 to 70% of the microarray species were detected by the same assay (Colombo et al., 2009; Preza et al., 2009). While there are no direct comparisons between the numbers of taxa detected by HOMIM in infants and those in adults, a lower species diversity of infants compared with individuals in later ages is consistent with separate reports for infants and adults (Könönen, 2000; Hao and Lee, 2004; Morelli, 2008). The lower number of taxa detected in oral biofilms from C-section compared with vaginally delivered infants is in accord with the lower diversity reported for the gut in samples taken immediately after C-section birth (Dominguez-Bello et al., 2010).
After birth, bacterial colonization of the gastrointestinal tract, including the mouth, is influenced by the transmission of bacteria from the environment and by genetic factors (Mandar and Mikelsaar, 1996; Dominguez-Bello et al., 2010). In the first few months of life, the major influences on the oral microbial succession are person-to-person transmission, composition of the infant’s saliva, mode of feeding, and microbial cross-talk. In the neonate, oral bacterial colonization starts with streptococci from the viridans group (Pearce et al., 1995; Könönen, 2000), whereas significant colonization of anaerobes was not detected in infants before 2 mos of age (Könönen, 2000). While there are no comparable data with HOMIM in infants, the present frequent detection of species in Firmicutes, and particularly within the genus Streptococcus, is consistent with previous reports of oral colonization by streptococci in infants (Pearce et al., 1995; Könönen, 2005). It is unlikely that these species are transients, considering the detection threshold of about 104 cells for the HOMIM microarray, indicating that species detection in this assay likely reflects colonization and growth (Paster and Dewhirst, 2009; Olson et al., 2011). Notably, treatment of the mothers with antibiotics during delivery was not influential on the oral microbiota in three-month-old infants.
The present dataset was characterized by a larger number of variables than study participants, and by the presence of species that might be interdependent based on shared environmental needs or inter-species co-aggregation. Under these conditions, the multivariate PLS-DA method is suitable to search for subject clustering and for identifying the variables characterizing clusters. With x-variables that were scaled to unit variance, and cross-validation of the explanatory capacity to account for over-interpretation, there was good power to discriminate between modes of delivery. The results were also stable after the model was restricted to infants born in wk 37 or later and those being breast-fed. With PLS-DA, several bacterial species differed between infants based on their delivery method. The biological significance of the species evaluated, and detection of fewer species in the mouth in early infancy of infants delivered by C-section, compared with those delivered vaginally, however, is unknown and will require longitudinal evaluation. It is nevertheless notable that a greater microbial diversity in the intestine and in the mouth has been reported to be associated with health (Marsh, 2006; Preza et al., 2008; Sjögren et al., 2009; Turnbaugh et al., 2009; Luoto et al., 2011).
A difference based on mode of delivery was detection of Slackia exigua in over 76% of the C-section infants (all with HOMIM scores ≥ 2) compared with non-detection in the vaginal delivery group. S. exigua is a Gram-positive, strictly anaerobic, asaccharolytic species that has been isolated in root canal infections, periodontitis, extra-oral surgical wounds, and intestinal abscesses (Abiko et al., 2010; Kim et al., 2010). While it is not known why S. exigua was detected in the C-section but not in the vaginal delivery group, it seems possible that the more diverse microbial biofilm of vaginally delivered infants could suppress establishment of this periodontitis-associated species. The association between S. exigua and mode of delivery requires further investigation.
In conclusion, the present study indicates a different colonization pattern in the oral cavity between three-month-old infants delivered vaginally and those delivered by Caesarian section. The reasons for the differences are unknown, as is whether these differences have long-term impact on the oral or general health of the child. Possible reasons for differences will likely include the relative influence of host receptor and mucosal and saliva immune phenotypes, and interactions with environmental exposures.
Acknowledgments
Dr. Conny Wikström, Umetrics, Sweden, is acknowledged for expertise support in PLS-DA.
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
The present study was supported by grants from Västerbotten County Council (TUA/FoU) and The Swedish Patent Revenue Foundation, and by a Public Health Service Grant DE-015847 (AT) from the National Institute of Dental and Craniofacial Research, USA, and by Henning and Johan Throne-Holst’s Foundation (PL).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Abiko Y, Sato T, Mayanagi G, Takahashi N. (2010). Profiling of subgingival plaque biofilm microflora from periodontally healthy subjects and from subjects with periodontitis using quantitative real-time PCR. J Periodontal Res 45:389-395 [DOI] [PubMed] [Google Scholar]
- Ahrné S, Lönnermark E, Wold AE, Aberg N, Hesselmar B, Saalman R, et al. (2005). Lactobacilli in the intestinal microbiota of Swedish infants. Microbes Infect 7:1256-1262 [DOI] [PubMed] [Google Scholar]
- Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, et al. (2010). Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA 107:18933-18938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylesjö M, Rantalainen M, Cloarec O, Nicholson JK, Holmes E, Trygg J. (2006). OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J Chemometrics 20:341-351 [Google Scholar]
- Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. (2009). Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis and periodontal health using the Human Oral Microbe Identification Microarray. J Periodontol 80:1421-1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Paster BJ, Tanner AC, Yu WH, Lakshmanan A, et al. (2010). The human oral microbiome. J Bacteriol 192:5002-5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA 107:11971-11975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R, et al. (2010). Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding and antibiotics. J Pediatr Gastroenterol Nutr 51:77-84 [DOI] [PubMed] [Google Scholar]
- Fanaro S, Chierici R, Guerrini P, Vigi V. (2003). Intestinal microflora in early infancy: composition and development. Acta Paediatr Suppl 91:48-55 [DOI] [PubMed] [Google Scholar]
- Hao WL, Lee YK. (2004). Microflora of the gastrointestinal tract: a review. Methods Mol Biol 268:491-502 [DOI] [PubMed] [Google Scholar]
- Kim KS, Rowlinson MC, Bennion R, Liu C, Talan D, Summanen P, et al. (2010). Characterization of Slackia exigua isolated from human wound infections including abscesses of intestinal origin. J Clin Microbiol 48:1070-1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könönen E. (2000). Development of oral bacterial flora in young children. Ann Med 32:107-112 [DOI] [PubMed] [Google Scholar]
- Könönen E. (2005). Anaerobes in the upper respiratory tract in infancy. Anaerobe 11:131-136 [DOI] [PubMed] [Google Scholar]
- Li Y, Caufield PW, Dasanayake AP, Wiener HW, Vermund SH. (2005). Mode of delivery and other maternal factors influence the acquisition of Streptococcus mutans in infants. J Dent Res 84:806-811 [DOI] [PubMed] [Google Scholar]
- Luoto R, Kalliomäki M, Laitinen K, Delzenne NM, Cani PD, Salminen S, et al. (2011). Initial dietary and microbiological environments deviate in normal-weight compared to overweight children at 10 years of age. J Pediatr Gastroenterol Nutr 52:90-95 [DOI] [PubMed] [Google Scholar]
- Mandar R, Mikelsaar M. (1996). Transmission of mother’s microflora to the newborn at birth. Biol Neonate 69:30-35 [DOI] [PubMed] [Google Scholar]
- Marsh PD. (2006). Dental plaque as a biofilm and a microbial community – implications for health and disease. BMC Oral Health 6(Suppl 1):S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli L. (2008). Postnatal development of intestinal microflora as influenced by infant nutrition. J Nutr 138:1791S-1795S [DOI] [PubMed] [Google Scholar]
- Olson JC, Cuff CF, Lukomski S, Lukomska E, Canizales Y, Wu B, et al. (2011). Use of 16S ribosomal RNA gene analyses to characterize the bacterial signature associated with poor oral health in West Virginia. BMC Oral Health 11:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paster BJ, Dewhirst FE. (2009). Molecular microbial diagnosis. Periodontol 2000 51:38-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce C, Bowden GH, Evans M, Fitzsimmons SP, Johnson J, Sheridan MJ, et al. (1995). Identification of pioneer viridans streptococci in the oral cavity of human neonates. J Med Microbiol 42:67-72 [DOI] [PubMed] [Google Scholar]
- Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. (2006). Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118:511-521 [DOI] [PubMed] [Google Scholar]
- Preza D, Olsen I, Aas JA, Willumsen T, Grinde B, Paster BJ. (2008). Bacterial profiles of root caries in elderly patients. J Clin Microbiol 46:2015-2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preza D, Olsen I, Willumsen T, Boches SK, Cotton SL, Grinde B, et al. (2009). Microarray analysis of the microflora of root caries in elderly. Eur J Clin Microbiol Infect Dis 28:509-517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren YM, Jenmalm MC, Böttcher MF, Björkstén B, Sverremark-Ekström E. (2009). Altered early infant gut microbiota in children developing allergy up to 5 years of age. Clin Exp Allergy 39:518-526 [DOI] [PubMed] [Google Scholar]
- Sjöström M, Wold S, Söderström B. (1986). PLS discriminant plots. In: Pattern recognition in practice II. Gelsema ES, Kanal LN, editors (p. 486). Elsevier: Amsterdam [Google Scholar]
- Tanner AC, Milgrom PM, Kent R, Jr, Mokeem SA, Page RC, Liao SI, et al. (2002). Similarity of the oral microbiota of pre-school children with that of their caregivers in a population-based study. Oral Microbiol Immunol 17:379-387; erratum in Oral Microbiol Immunol 18:338, 2003 [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. (2009). A core gut microbiome in obese and lean twins. Nature 457:480-484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S. (1978). Cross validatory estimation of the number of components in factor and principal component models. Technometrics 20(Pt 1): 397-405 [Google Scholar]