Abstract
Pathogen proteins targeting the actin cytoskeleton often serve as model systems to understand their more complex eukaryotic analogs. We show that the strong actin filament nucleation activity of Vibrio VopL depends on its three W domains and dimerization through a unique VopL C-terminal domain (VCD). The VCD displays a novel all-helical fold and interacts with the pointed end of the actin nucleus, contributing to the nucleation activity directly and through duplication of the W domain repeat. VopL promotes rapid cycles of filament nucleation and detachment, but generally has no effect on elongation. Profilin inhibits VopL-induced nucleation by competing for actin binding to the W domains. Combined, the results suggest that VopL stabilizes a hexameric double-stranded pointed end nucleus. Analysis of hybrid constructs of VopL and the eukaryotic nucleator Spire suggest that Spire may also function as a dimer in cells.
Keywords: actin nucleation, W domain, VopL/VopF, Spire
Nucleation is rate-limiting during actin polymerization, and is catalyzed in cells by proteins called actin filament nucleators 1. Known filament nucleators in eukaryotic cells include the Arp2/3 complex together with its large family of Nucleation Promoting Factors (NPFs) 2, Formins 3,4, Spire 5, Cobl 6 and Lmod 7. Additionally, microbial pathogens often subvert the host cell actin cytoskeleton for infection, particularly by interfering with nucleation. Vibrios are Gram-negative bacteria that cause wound infection, gastrointestinal disease, and diarrhea 8. Vibrio parahaemolyticus and Vibrio cholerae produce the type III secretion system (T3SS) virulence factors VopL 9 and VopF 10. VopL and VopF share 57% sequence identity. Both proteins have strong actin nucleation activity, and through their ability to disrupt actin homeostasis are suggested to play a critical role in Vibrio host cell entry and infection 9,10. Recently, a VopF homolog, VopN, was identified in a divergent T3SS island of V. cholerae strains isolated from cholera outbreaks in Peru and Bangladesh 11. VopN is also a potent nucleator. It localizes to the host stress fibers, and is involved in the disruption of cell polarity and tight junctions and in eliciting the diarrheal response in a rabbit model 11.
Among filament nucleators, the WASP-homology 2 (WH2 or W) domain is emerging as the most common actin-binding motif 1, present in Spire 5, Cobl 6, VopL and VopF 9,10, Sca2 12, TARP 13, Lmod 7, and the NPFs of the Arp2/3 complex 2. The W domain consists of 17–27-aa, comprising an N-terminal helix that binds in the target-binding cleft between actin subdomains 1 and 3 and a C-terminal extended portion featuring the conserved 4-aa motif LKKT(V) 1. In Spire, Cobl, VopL and VopF, Sca2, and some NPFs, the W domain occurs in tandem repeats, with each W domain binding one actin monomer to stabilize a small nucleus of three to four actin subunits 5,6,9,10. However, the spacing between W domains varies considerably among these proteins, which appears to determine the configuration of the actin nuclei that they generate. Thus, the four W domains of Spire are connected by short inter-W linkers and, as a result, Spire is thought to stabilize a long-pitch (longitudinal) actin tetramer 5. This arrangement might not be optimal for nucleation, as Spire has low nucleation activity in vitro 5,14,15. In contrast, Cobl, with just three W domains, has strong nucleation activity. Cobl differs from Spire in that it contains a long linker between its second and third W domains. It has been proposed that this property enables Cobl to stabilize a short-pitch trimeric nucleus 6, which presumably is a more effective way to promote polymerization. This model appears plausible, but remains untested and cannot explain why VopL and VopF, which contain three W domains (like Cobl) but short inter-W linkers (like Spire), display strong nucleation activity 9,10. We set out to study the mechanism of actin filament nucleation by VopL as an important step to understand its role in Vibrio infection and because of its relevance for our understanding of other eukaryotic and bacterial nucleators. Our structure-function studies reveal among other findings that dimerization plays a critical role in VopL-mediated nucleation, by enabling the formation of a hexameric pointed end actin nucleus. The results further suggest that Spire, and possibly other eukaryotic nucleators, may also function as dimers or higher oligomers in cells.
RESULTS
Nucleation requires all three W domains and C-terminal VCD
To understand how VopL can achieve strong nucleation activity with just three W domains and short inter-W linkers, we analyzed the polymerization activity of VopL fragments designed according to its domain organization (Fig. 1a and Supplementary Fig. 1). VopL is a 484-aa protein. The three W domains are located toward the middle of the sequence (residues 130–227), positioned in between a short proline-rich (Pro-rich) sequence (residues 99–113) and a C-terminal 238-aa domain (residues 247–484) of unknown function and structure, referred to here as VCD (for VopL C-terminal domain).
Figure 1.

Nucleation activity of VopL constructs. (a) Domain organization of VopL, constructs used in this study and alignment of W domains illustrating the sites of point mutations. (b) Time course of polymerization of 2 μM Mg-ATP-actin (6% pyrene-labeled) alone or in the presence of 25 nM VopL constructs (color-coded). Polymerization rates are reported as mean and s.e.m. values (number of measurements ≥ 3). (c) Effect of VopL construct concentration on the polymerization rate of pyrene-actin (see also Supplementary Fig. 2). (d) Time course of actin polymerization by VopL constructs containing point mutations in the W domains (shown in part a). (e) Polymerization of 1.5 μM Mg-ATP-actin (33% Oregon Green-labeled) alone or in the presence of 0.1 nM VopL constructs visualized by TIRF microscopy on NEM-myosin II-coated coverslips, including time-lapse micrographs (see Supplementary Videos 1–4) of representative 19.5 μm2 fields (left) and plots of the growth of 20 filament barbed-ends over time (right). Circles and arrowheads indicate the pointed and barbed ends of representative filaments. The average number of filaments in a 133 μm2 field of view at 300 s from two independent experiments is indicated as f μm-2. Errors are ±s.d.
Bulk polymerization was measured using the pyrene-actin assay 16. While the polymerization activity of Spire was mapped to its four-W domain repeat 5, VopL construct 3W, comprising the three W domains, had negligible activity compared to the spontaneous polymerization of actin alone (Fig. 1b). In contrast, 3W-VCD, comprising in addition the VCD, had very strong activity (Fig. 1b), which increased in a dose-dependent manner (Fig. 1c and Supplementary Fig. 2a). This construct had even stronger activity than the Arp2/3 complex analyzed at the same concentration (25 nM) with activation by 250 nM N-WASP WCA (Supplementary Fig. 3). A fragment corresponding to the four W domain repeat of Drosophila Spire (residues 366–482) had negligible activity at this concentration (Supplementary Fig. 3), which is consistent with previous reports 5,14,15 (analyzed further below).
All three W domains of VopL contributed towards its strong nucleation activity (Fig. 1b). Thus, removing the first W domain (construct 2W-VCD) resulted in a ~95% drop in activity, and the polymerization activities of constructs 1W-VCD and VCD, lacking the first two and all three W domains, were very low at 25 nM and did not increase appreciably with concentration (Fig. 1b,c and Supplementary Fig. 2b,c). To further clarify the role of each W domain in nucleation, we analyzed the effect of point mutations of the W domains within construct 3W-VCD. Each W domain was independently mutated at four positions, with Ala substitutions of two consecutive hydrophobic residues of the N-terminal helix and the first two residues of the LKKT(V) motif (Fig. 1a). While the second and third W domain mutants showed a less severe ~60% decrease in activity, the first W domain mutant displayed a ~90% drop in activity compared to 3W-VCD (Fig. 1d). Thus, the activity of mutant 3W-VCD(W1*) was similar to that of construct 2W-VCD lacking the first W domain (Fig. 1b), suggesting that the combination of the four point mutations completely abolished actin binding to the first W domain. In contrast, mutants 3W-VCD(W2*) and 3W-VCD(W3*) presumably can still recruit actin subunits through their incapacitated W domains (albeit less efficiently), most likely due to additional longitudinal interactions with the adjacent actin subunits or VCD (see below). To some extent, these mutated W domains can be viewed as long inter-W domain linkers, similar to linker-2 in Cobl. It is thus interesting to note that shortening Cobl‘s linker-2 abolishes its nucleation activity, whereas replacing this linker with an unrelated sequence of similar length restores most of the endogenous activity 6, emphasizing the importance of linker length.
The pyrene-actin assay measures bulk polymerization, but cannot resolve potential changes in filament elongation rate that could occur if VopL were to stay bound to the barbed end of the filament after nucleation. We used TIRF (total internal reflection fluorescence) microscopy to directly observe actin assembly in the presence of VopL constructs by monitoring the polymerization of 1.5 μM Mg2+-ATP-actin of which 33% was labeled with Oregon Green at Cys374 for visualization 17,18 (Fig. 1e, Supplementary Videos 1–4). In the absence of VopL constructs, spontaneously assembled actin filaments elongated from their barbed ends at the rate of 8.4 sub s-1, and only 1.0 × 10-3 filament μm-2 were observed on average after 300 s. In the presence of 0.1 nM VopL constructs P-3W-VCD and 3W-VCD, filament nucleation was dramatically stimulated, resulting in the formation of 31 and 29 × 10-3 filament μm-2. Construct VCD had negligible activity under these conditions, producing only 1.3 × 10-3 filament μm-2. Filaments nucleated in the presence of VopL constructs elongated from their barbed ends approximately at the same rate as control filaments, in contrast to Formins that remain processively bound to barbed ends and regulate filament elongation 19.
VCD mediates dimerization and binds at the pointed end
We then addressed the mechanism by which the VCD contributes to the overall nucleation activity of VopL, while displaying negligible activity on its own (Fig. 1b,c). Sequence analysis predicted that the last 23 residues of VCD (462–484) formed a coiled coil (Supplementary Fig. 4), suggesting that it could mediate VopL dimerization. Consistent with this prediction, multi-angle light scattering combined with size exclusion chromatography (SEC-MALS) showed that all VopL constructs containing an intact VCD formed dimers (Fig. 2a and Supplementary Table 1).
Figure 2.

VCD mediates dimerization and binds to the pointed end of the polymerization nucleus. (a) SEC-MALS mass measurements of VopL constructs (theoretical masses in parenthesis). Supplementary Table 1 lists the masses of other constructs. (b) Time course of polymerization of 2 μM Mg-ATP-actin alone or induced by 25 nM VopL constructs. (c,d) Gel electrophoresis of supernatant (S) and pellet (P) fractions after co-sedimentation at 224,000 × g of F-actin with VCD or 1W-VCD. (e) Mass measurements of 1W-VCD and its complex with actin. (f) TIRF time-lapse micrographs of actin assembly induced by P-3W-VCD immobilized on a coverslip, showing a filament anchored to the coverslip by its pointed end that dissociates between frames 220 and 280 s (top) and a filament that remains anchored throughout (bottom) (Supplementary Videos 5 and 6). Circles and arrows indicate the pointed and barbed ends of the filaments. Shown on the right are growth plots of 20 filaments, including tethered and non-tethered (dashed lines). (g) TIRF time-lapse micrographs of actin (green) assembly by 3W-VCD bound to Qdots (red). Upper panels show a Qdot that nucleates 14 filaments (arrowheads and numbers, Supplementary Videos 7 and 8). Middle and lower panels show filaments whose pointed (circles) or barbed (arrows) ends appear to be bound to a Qdot (Supplementary Videos 9 and 10). (h) Plots of the growth of filaments, including non-bound, fast dissociated, pointed and barbed end-bound, and two filaments whose barbed ends detach from the Qdot and their elongation rates decrease during the experiment. Errors are ±s.d.
Simultaneously mutating five hydrophobic residues at heptad positions a and d of the coiled coil to glutamic acid (Supplementary Fig. 4) introduced repulsive forces that disrupted dimerization (Fig. 2a). Interestingly, however, the nucleation activity of the resulting monomeric construct 3W-VCD*, while substantially lower than that of 3W-VCD, was higher than that of 3W (Fig. 2b). This result suggested that VCD does not merely mediate dimerization, but additionally contributes to the recruitment of actin subunits during nucleation. To explore this possibility, we performed a high-speed (224,000 × g) co-sedimentation assay of VCD with F-actin. In the absence of actin filaments, VCD was found in the supernatant, whereas in the presence of F-actin a small fraction was also detected in the pellet (Fig. 2c), indicating that VCD binds F-actin. Interestingly, construct 1W-VCD, comprising in addition the third W domain, sequestered actin monomers and formed a stable complex with actin in the pelleting assay (Fig. 2d). The mass of this complex was 135,000 Da (Fig. 2e), consistent with the presence of two actin molecules and a 1W-VCD dimer in the complex. This complex formed relatively fast, after incubation of 1W-VCD with F-actin for only 1 h, and thus could not occur solely by sequestration of actin monomers dissociating spontaneously from the filament, suggesting that 1W-VCD also has severing or depolymerizing activity.
To investigate the location of VopL during nucleation, we used TIRF microscopy to observe filament assembly by VopL construct P-3W-VCD immobilized on a coverslip (Fig. 2f and Supplementary Videos 5 and 6). More than 96% of the filaments nucleated by immobilized P-3W-VCD were immediately detached from the coverslip, making it difficult to determine which end of the filament was associated with VopL during nucleation. A small fraction of the filaments (~3.8%) remained tethered to the coverslip, and appeared to be bound through their pointed ends. Filaments elongated at the control rate of ~8.3 sub s-1 regardless of whether they were free or attached to coverslips, and they did not buckle like filaments attached by their barbed ends to immobilized Formin 20.
To further clarify the location of VopL during nucleation, we labeled construct 3W-VCD with quantum dots (Qdots). The majority of the Qdots did not display nucleation events (Supplementary Table 2). Of the fraction of Qdots showing nucleation (3.9%), most appeared to assemble multiple filaments, undergoing very rapid cycles of nucleation and dissociation (Fig. 2g and Supplementary Videos 7 and 8). It was not possible to unambiguously determine which end of the filament was associated with the Qdot in these fast events. However, because the elongation rate of these filaments (~8 sub s-1) was unchanged compared to control filaments (Fig. 2h), it appeared likely that VopL was at the pointed end during nucleation but quickly dissociated as filaments started to grow. A relatively small fraction of the filaments (18.5%) remained associated with Qdots after nucleation for more than a minute. Of these, 10.9% were clearly associated through the pointed end, as indicated by brighter (less bleached) barbed ends, and displayed an elongation rate of 7.7 sub s-1, similar to that of free filaments (Fig. 2g,h and Supplementary Video 9). Unpredictably, ~7.6% of the filaments were unambiguously found to be associated through their barbed ends (Fig. 2g). Barbed end-associated filaments grew faster (Fig. 2h) and occasionally buckled (Supplementary Video 10), as observed with Formins 20. These rare events were only observed with Qdots, and it is unclear whether they represent a core function of VopL. Qdots can have multiple VopL molecules attached, artificially increasing the local concentration of actin monomers bound to W domains and the likelihood that newly formed filaments remain tethered (note that part of the binding interface of the W domain is exposed on the side of the actin filament 21). Nevertheless, this is the first observation of a W domain-based nucleator that can accelerate barbed end filament elongation. The closest parallel is with Eva/VASP proteins. Although not generally thought to participate in nucleation, Ena/VASP proteins interact with actin through W domain-related sequences and accelerate processive barbed end elongation when concentrated on a bead 22. A similar situation could occur in cells if the local concentration of secreted VopL is sufficiently high. Yet, the vast majority of our observations suggest that during nucleation VopL is at the pointed end of the polymerization nucleus, dissociates fast after nucleation, and is not involved in processive elongation.
Crystal structure of VCD
The crystal structure of VCD was determined ab initio, using data collected at a single X-ray wavelength from a selenomethionine derivatized crystal (Methods and Table 1). The structure revealed a novel dimeric fold (Fig. 3a and Supplementary Video 11). Each monomer consisted of three discrete domains: base (Arg247–Ala280; Ser395–Ser461), arm (Asp281–Leu394) and coiled coil (Tyr462–Ser484). Dimerization was mainly mediated by the coiled coil domain and preceding loop 460–462, where the side chains of Tyr460 and Tyr462 from the two monomers interlocked into each other. Also contributing to dimerization was the loop Gln418–Gly421 that formed a short antiparallel β-sheet at the dimer interface. The extended arm domain was the most flexible part of the structure, as reflected by higher temperature factors and large differences between the two subunits of the dimer (Supplementary Fig. 5a,b). Moreover, the crystals were rarely isomorphous, and differences among them resulted primarily from changes in the orientation of the arm domains, as illustrated by a comparison of two structures derived from two different crystals (Supplementary Fig. 5c). As a result, residues Ser324–Ser339 of the arm domain were not visualized in the structure.
Table 1.
Data collection and refinement statistics
| Data collection | |
| Space group | P 21 21 21 |
| Cell dimensions | |
| a, b, c (Å) | 56.93, 90.49, 101.86 |
| αβγ (°) | 90, 90, 90 |
| Wavelength (Å) | 0.9772 |
| Resolution (Å) | 3.1–67.7 (3.1–3.2)* |
| Rmerge | 12.2 (42.6) |
| I / σI | 20.56 (2.4) |
| Completeness (%) | 99.5 (99.9) |
| Redundancy | 32.8 (34.9) |
| Refinement | |
| Resolution (Å) | 3.1–67.7 (3.1–3.26)* |
| No. reflections | 9,773 |
| Rwork / Rfree | 24.0 / 29.3 |
| No. atoms | |
| Protein | 3,484 |
| Ligand/ion | - |
| Water | - |
| B-factors | |
| Protein | 149.12 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.512 |
Values in parentheses are for highest-resolution shell
Figure 3.

Crystal structure of VCD and SAXS structures of VCD and 1W-VCD+actin. (a) Two perpendicular views of the crystal structure of VCD and definition of domains and secondary structure shown along the sequence of this domain (Supplementary Video 11). Different colors highlight different domains: base (magenta, green), arm (pink), coiled coil (gold). (b) Experimental X-ray scattering pattern of VCD (red) and 1W-VCD+actin (blue) as a function of momentum transfer s=4πsin(θ)/λ, where 2θ is the scattering angle and λ=0.103 nm is the X-ray wavelength. The inset shows the normalized distance distribution functions computed from the scattering pattern with the program GNOM36. (c,d) Two perpendicular orientations of the average SAXS envelopes of VCD and 1W-VCD+actin (same orientations as for the crystal structure).
Solution structure by SAXS of VCD and 1W-VCD+actin
We used Small Angle X-ray Scattering (SAXS) to study the solution structures of VCD and 1W-VCD in complex with actin to determine the location of actin subunits in the polymerization nucleus. The molecular mass of VCD determined from the scattering intensity (Fig. 3b) extrapolated to zero angle, I(0), was 50.6 kDa, confirming the presence of a dimer in solution (theoretical mass: 53.7 kDa). The radius of gyration (Rg), characterizing the overall spread of the particle in solution, was 38 Å. The unconstrained (i.e. not imposing two-fold symmetry) surface envelope resulting from averaging of 50 ab initio structures computed with the program DAMMIF23 displayed pseudo two-fold symmetry, as expected for the VCD dimer (Fig. 3c). The low-resolution SAXS envelope bore obvious similarities with the crystal structure, particularly for the most stable portions of the structure, including the base and coiled coil domains (Supplementary Fig. 6). The flexible arm domains, however, were only partially resolved in the SAXS envelope and seemed to adopt a different orientation in solution than in the crystal structure, giving rise to a more extended structure.
Construct 1W-VCD forms a 2:2 complex with actin (Fig. 2d,e). The Rg of this complex measured by SAXS was 45 Å (Fig. 3b,d), and its molecular mass determined from the I(0) was 136.2 kDa which, consistent with the light scattering measurement (Fig. 2e), confirmed the presence of a 2:2 complex in solution. The average SAXS envelope clearly reproduced the VCD portion of the complex, but did not contain sufficient extra density to account for two actin molecules (Fig. 3d). This result indicates that at least one of the actin subunits is bound loosely, i.e. flexibly tethered to VCD although tightly bound to W3 (Kd =0.4 μM, Supplementary Fig. 7), such that averaging diminishes its contribution to the final envelope. The extra density observed was located at the interface between the base and arm domains. In the crystal structure of VCD, the N-termini of each monomer emerge near the interface between the base and arm domains (Fig. 3a), which combined with the location of the actin subunit in the SAXS envelope suggests a tentative model of the polymerization nucleus (see below).
Profilin inhibits VopL-induced nucleation
VopL has two Pro-rich sequences, one between W domains 2 and 3 (188PAPPPLP194) that could potentially bind one profilin-actin complex 24, and one N-terminal to the W domain repeat (99PPAPPLPGAIPPAPP113) that could bind two profilin-actin complexes. The latter sequence is reminiscent of the Formin-Homology 1 (FH1) domain, which is also positioned N-terminal to the actin-binding region and recruits profilin-actin complexes to stimulate filament elongation12,19. In contrast, profilin inhibited in a dose-dependent manner bulk polymerization induced by 5 nM VopL construct P-3W-VCD, carrying both Pro-rich sequences, and construct 3W-VCD, containing only the second Pro-rich sequence (Fig. 4a).
Figure 4.

Profilin inhibits polymerization induced by VopL. (a) Polymerization rates of 2 μM Mg-ATP-actin induced by 5 nM VopL constructs (color-coded) or 0.5 μM F-actin seeds (black) as a function of profilin concentration. (b) Visualization by TIRF microscopy of the effect of 2.5 μM profilin on the polymerization of 1.5 μM Mg-ATP-actin alone (Supplementary Video 12) or induced by 0.1 nM 3W-VCD and P-3W-VCD (Supplementary Videos 13 and 14) or mutants 3W-VCDP191E and P-3W-VCDP191E (Supplementary Videos 15—18). Plots of the growth of 20 individual filaments are shown on the right. (c,d) Comparison of the nucleation activities and elongation rates measured by TIRF with (blue) or without (red) profilin. Errors are ±s.d.
To further explore the effect of profilin we turned to direct observation of polymerization by TIRF microscopy (Fig. 4b and Supplementary Videos 12–14). In control experiments, the addition of 2.5 μM profilin reduced somewhat the nucleation efficiency and barbed end elongation rate of actin alone. However, in accord with the bulk polymerization assay, profilin strongly inhibited nucleation by P-3W-VCD and 3W-VCD (Fig. 4b,c). Filaments that did nucleate in the presence of profilin and VopL constructs grew from their barbed ends at similar rates as control filaments.
Profilin’s inhibitory effect on VopL-induced nucleation could be explained by at least two different mechanisms: 1) profilin could bind to the Pro-rich sequences of VopL and sterically block the formation of a polymerization nucleus, or 2) the W domain, whose binding site on actin partially overlaps with that of profilin in crystal structures 24,25, cannot compete effectively with profilin for actin binding. To distinguish between these two possibilities, we introduced the mutation P191E in the middle of the second Pro-rich sequence, which according to crystal structures 24 should abolish profilin binding to this site. This mutation was introduced in constructs P-3W-VCD and 3W-VCD, and was expected to partially or completely release the inhibitory effect of profilin if the first mechanism was responsible for it. In control experiments (without profilin), mutants P-3W-VCDP191E and 3W-VCDP191E displayed normal nucleation activity, but the addition of profilin inhibited their activities to the same extent as for the original constructs in bulk polymerization (Fig. 4a) and TIRF experiments (Fig. 4b,c and Supplementary Videos 15–18). Filament elongation rates were not affected by the mutation. In bulk polymerization the assay, maximum inhibition was attained at a profilin concentration of 2 μM, equal to the concentration of actin in these experiments.
Combined, these results suggest that profilin competes with the W domain for actin binding, probably due to its higher affinity for ATP-actin (KD~0.1 μM) 26 than that of most W domains. For instance, the affinity of the third W domain of VopL for ATP-actin measured here by isothermal titration calorimetry was 0.41 μM (Supplementary Fig. 7), consistent with previous measurements for other W domains 25,27. In our experiments, as it is probably the case in cells, the concentration of profilin was much higher than that of VopL, putting the W domain at a considerable disadvantage for binding to actin. In cells, where profilin-actin is the largest fraction of umpolymerized actin 28, the inhibitory effect of profilin could be substantial, implying that most W domain-based nucleation might proceed from the comparatively small pool of free actin. An analogous conclusion was reached for Formins, whose nucleation activity is also inhibited by profilin 29.
Analysis of hybrid constructs of VopL and Spire
In agreement with previous reports 5,30, we found that the W domain repeat of Spire (Fig. 5a) had low nucleation activity at 25 nM, but its activity increased appreciably at 250 nM (Fig. 5b). In contrast, VopL construct 3W had low nucleation activity, which did not increase with concentration. Spire linker-3 (between W domains 3 and 4) is known to play a critical role in its nucleation activity 5,30. Accordingly, a hybrid construct in which linker-2 of VopL construct 3W was replaced by Spire linker-3, 3W-sL3 (Fig. 5a), behaved similarly to Spire in that its activity increased with concentration (Fig. 5b). Thus, we find that the specific sequence and length (Spire linker-3 is seven residues shorter than VopL linker-2) of inter-W linkers plays a critical role in the nucleation activity of tandem W domain nucleators, possibly by contributing directly to the recruitment of actin subunits 5,30.
Figure 5.
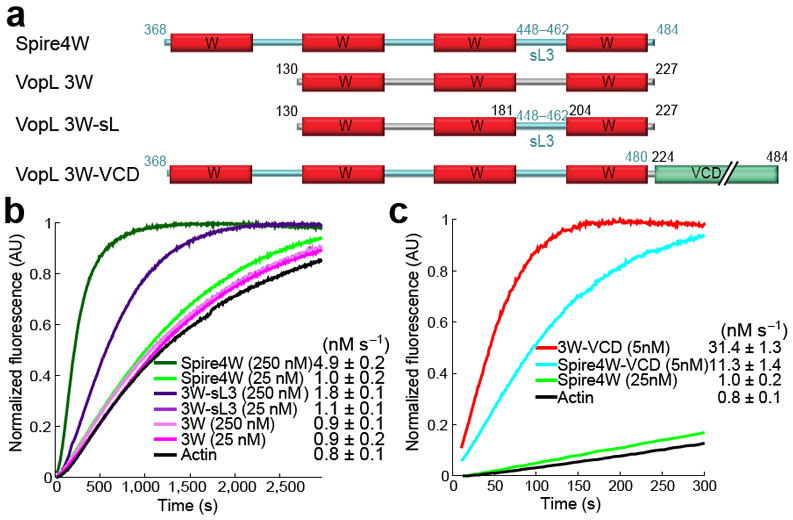
Role of dimerization and specific sequence of W domains and inter-W linkers in nucleation. (a) Design of hybrid constructs of VopL and Drosophila Spire’s repeat of four W domains (Spire4W). In construct 3W-sL3, VopL linker-2 was replaced with Spire linker-3. In construct Spire4W-VCD, VopL’s VCD was fused C-terminal to the last W domain of Spire, using the LKKT(V) motifs as reference for fusion. (b) Time courses of polymerization of 2 μM Mg-ATP-actin induced by two different concentrations (25 and 250 nM) of constructs Spire4W, 3W-sL3 and 3W. (c) Comparison of the time courses of actin polymerization induced by constructs VopL 3W-VCD and Spire4W-VCD and Spire4W at the indicated concentrations. Errors are ±s.d.
An important factor determining the strong nucleation activity of VopL seems to be dimerization. We asked whether dimerization of Spire’s repeat of four W domains would have a similar effect on its nucleation activity. We generated a hybrid construct of Spire’s W domain repeat and VopL’s VCD (Spire4W-VCD), using the LKKT(V) sequence of the last W domain as a reference for fusion (Fig. 5a). For a reliable comparison of the nucleation activities of constructs 3W-VCD and Spire4W-VCD, the concentration of both proteins was lowered to 5 nM in the pyrene-actin polymerization assay, resulting in polymerization rates of 31.4 nM s-1 and 11.3 nM s-1, respectively (Fig. 5c). This result confirmed that dimerization of tandem W domains and the contribution of a pointed end stabilizing factor such as VCD leads to dramatic increases in nucleation activity. In addition, this result established the importance of the specific sequence of the W domains and inter-W linkers, since even after dimerization the activity of VopL with six W domains was much higher than that of Spire with eight W domains.
DISCUSSION
Proposed mechanism of nucleation
Collectively, the results presented here suggest a structure-function model of VopL-mediated nucleation, some features of which are extendable to other filament nucleators (Fig. 6). Through dimerization, VopL stabilizes a hexameric pointed end nucleus and promotes extremely fast cycles of filament nucleation and detachment, but generally has no effect on elongation and is inhibited by profilin. VCD plays a dual role in this activity: duplication of the W domain repeat, to mediate the formation of the hexameric nucleus, and contributing directly to the recruitment of actin subunits by binding to the pointed end of the nucleus. In the crystal structure, the N-terminus of VCD emerges near the interface between the base and arm domains. N-terminal to it, and connected by a linker similar in length to the inter-W domain linkers, is the third W domain of VopL. We attempted to manually dock the crystal structures of VCD and that of a W–actin complex 25, guided by their relative positions in the SAXS envelope and constrained by the necessity to bring the C-terminus of the W domain within polypeptide bond distance to the N-terminus of VCD. This approach resulted in a model displaying nearly ideal shape and charge complementarity between VCD and the bound actin subunit (Fig. 6a,b and Supplementary Video 19). This model predicts that the first actin subunit of the polymerization nucleus contacts both subunits of the VopL dimer. But, where is the second actin? Because VopL has two-fold symmetry and the pointed end of the filament does not, we should consider two possibilities. A second actin subunit could be bound symmetrically on the other side of VCD, but this would produce substantial steric clashes between actin subunits. Moreover, this conformation is different from that of the pointed end of the filament where actin subunits are staggered by half a monomer length, and it is probably unfavorable for nucleation. More likely the second actin subunit will be positioned with respect to the first one similarly to subunits of the actin filament 31. The linker between the last W domain and VCD is sufficiently long to permit such an arrangement, which would not produce steric clashes but would imply that the second actin subunit is tethered loosely, consistent with the SAXS envelope. Detachment of VopL from the polymerization nucleus probably results from incompatibility (steric hindrance) between W–actin interactions and longitudinal actin–actin contacts in the filament. Indeed, the helix of the W domain binds in the cleft between actin subdomains 1 and 3 25, which also mediates inter-subunit contacts in the filament 31,32. Thus, in a recent structure of actin subunits bound longitudinally to tandem W domains, the actin subunits are separated by the insertion of the W domain helix at the actin-actin interface 21. It is therefore important to draw a clear distinction between polymerization nucleus (nucleation intermediate) and the actin filament. In the nucleus, the actin subunits are still bound to the W domains, which is incompatible with longitudinal inter-subunit contacts in the filament. As a result, VopL (and other tandem W domain nucleators) must dissociate when the nucleus begins to adopt a filament-like conformation (Fig. 6c), as observed in our experiments (Fig. 2g). Steric hindrance of the W domain with inter-actin contacts in the filament also suggests the possibility that during dissociation VopL could carry with it a few actin subunits bound to the W domains and VCD, as supported by the fact that construct 1W-VCD displays severing or depolymerizing activity (Fig. 2d).
Figure 6.
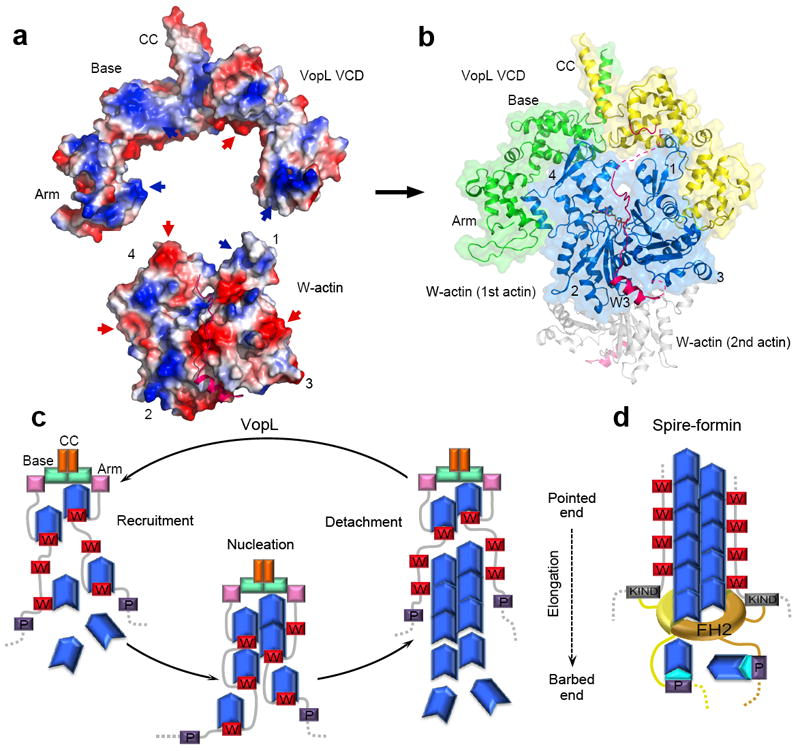
Proposed mechanisms of actin nucleation by VopL and Spire. (a) Electrostatic surface representation (blue, positively charged; red, negatively charged) of the structures of VCD and W–actin (PDB code: 2D1K) 25, illustrating the existing shape and charge complementarity (indicated by red and blue arrows) between the two structures. (b) Such complementarity and the necessity to connect the C-terminus of the third W domain to the N-terminus of VCD lead to a model of the complex in which the first actin subunit interacts with both subunits of the VopL dimer (see Supplementary Video 19). The second actin subunit (gray) might bind on the opposite side of VCD according to the actin filament model 31. (c) Model of nucleation by VopL. VCD plays a dual role, contributing directly to the recruitment of actin subunits by binding to the pointed end of the polymerization nucleus, and enabling the formation of a hexameric nucleus by duplication of the W domain repeat. VopL detaches fast after nucleation, probably due to steric hindrance of the W domains with longitudinal contacts between actin subunits in the filament 21. Upon detachment, VopL might carry with it actin subunits that become part of the nucleus in a new round of polymerization. (d) Spire’s activity is also enhanced by dimerization, which in cells is mediated by interaction of Spire’s KIND domain with formins 15,33,34.
General implications for tandem W domain-based nucleation
Our results support the notion that isolated W domain repeats are intrinsically weak nucleators; they mediate actin recruitment, but other factors, particularly oligomerization, contribute toward a strong nucleation activity. Thus, duplication of the W domain repeat of Spire, possibly aided by the pointed end binding activity of VCD, resulted in a dramatic increase of its nucleation activity. This result is not merely a curiosity, but might represent a general paradigm for how Spire (Fig. 6d) and other tandem W domain nucleators function. Thus, there is now strong evidence that Spire interacts directly with the Formin Cappuccino, and that the two proteins function together during Drosophila oogenesis 15,33,34. A similar interaction occurs among their mammalian counterparts, Spir-1, Spir-2, Formin-1 and Formin-2, which also function as collaborative regulators of oocyte development 15,33. The interaction involves the KIND domain of Spire and a short sequence immediately C-terminal to the actin-binding Formin-Homology 2 (FH2) domain 15. Because the FH2 domain forms a dimer, the interaction leads to indirect dimerization of Spire and a marked increase in nucleation activity 15 which, based on our findings, probably results from duplication of the W domain repeat. A somewhat similar situation occurs with the tumor suppressor protein APC, a recently described nucleator that while binding actin through sequences unrelated to the W domain, depends on dimerization for optimal activity and synergizes in vivo with the Formin mDia1 35. Pathogens frequently mimic eukaryotic functions, and not surprisingly a bacterial protein mimicking the Spire-Cappuccino interaction has now been discovered 12. Rickettsia Sca2 is an actin filament nucleator that combines an N-terminal FH2-like domain with a repeat of three W domains. While the FH2-like domain mediates processive elongation, the region C-terminal to the FH2 domain, which includes the W domains, appreciably enhances the nucleation activity of Sca2 12. Another example is the T3SS protein TARP, a filament nucleator from Chlamydia trachomatis. Although TARP contains a single W domain, oligomerization through its central Pro-rich domain enables it to recruit multiple actin subunits to promote polymerization 13. Finally, Cobl also contains a sequence (residues 550–600) predicted to form a coiled coil, but whether dimerization (direct or though partner interactions) contributes to its activity is still unknown. These examples suggest that the mechanism of nucleation uncovered here for Vibrio VopL may have general relevance for our understanding of other tandem W domain-based nucleators.
METHODS
Protein preparation
The Vibrio parahaemolyticus genomic DNA was a gift of Narjol González-Escalona. VopL fragments 90–484 (P-3W-VCD), 130–484 (3W-VCD), 160–484 (2W-VCD), 200–484 (1W-VCD), 247–484 (VCD) and 130–227(3W) were cloned into vector pTYB12 (New England BioLabs). The cDNA of Drosophila Spire was purchased from Open Biosystems (clone SD10157), and the fragment 366–482 (Spire4W) and hybrid constructs Spire4W-VCD (Spire366–480-VopL224–484) and 3W-sL (VopL130–181-Spire448–462-VopL204–227) were cloned as above. Human profilin-1 was obtained as described 24. Constructs were expressed in E. coli BL21(DE3) cells. Cells were resuspended in chitin-column equilibration buffer (25 mM Tris-HCl [pH 7.5], 500 mM NaCl, 1 mM EDTA, 100 μM PMSF) and lysed using a Microfluidizer (MicroFluidics Corp.). After purification on a chitin affinity column, proteins were purified by ion exchange chromatography and dialyzed against 25 mM Tris [pH 7.5], 50 mM NaCl and 0.2 mM CaCl2.
Actin polymerization assay
Actin polymerization was measured as the fluorescence increase resulting from the incorporation of pyrene-labeled actin into filaments, using a Cary Eclipse fluorescence spectrophotometer (Varian). Prior to data acquisition, 2 μM Mg-ATP-actin (6% pyrene-labeled) was mixed with different concentrations (see figures) of VopL or Spire constructs in F-buffer (10 mM Tris [pH 7.5], 1 mM MgCl2, 50 mM KCl, 1 mM EGTA, 0.1 mM NaN3, 0.02 mg ml-1 BSA, 0.2 mM ATP). Data acquisition started 10 s after mixing. Similar experiments were performed with addition of varying profilin concentrations. Polymerization experiments with the Arp2/3 complex were performed as described 37. Control experiments were carried out with addition of 10 × F-buffer. Polymerization rates were calculated as the slope at 50% polymerization and converted to nM s-1 (nM monomers adding to filaments s-1) assuming a total concentration of polymerizable actin of 1.9 μM as described 21.
TIRF microscopy
Total Internal Reflection Fluorescence (TIRF) microscopy images were collected at 10 s intervals with an iXon EMCCD camera (Andor Technology) using an Olympus IX-71 microscope fit with through-the-objective TIRF illumination. Mg-ATP-actin (33% Oregon Green-labeled) was mixed with 2 × TIRF buffer (10 mM Imidazole [pH 7.0], 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 50 mM DTT, 0.2 mM ATP, 50 μM CaCl2, 15 mM glucose, 20 μg ml-1 catalase, 100 μg ml-1 glucose oxidase, and 0.5% (w/v) methylcellulose 400 centipoise) and 0.1 nM VopL or VopL-Qdot constructs (with or without 2.5 μM profilin), and transferred to a flow cell for imaging at 23°C. For immobilized experiments, flow cells were pre-incubated with 10 nM P-3W-VCD for 5 min and washed with 60 μL 1 × TIRF buffer, before the addition of 1.5 μM Mg-ATP-actin. Filaments were considered tethered to the coverslip if the position of one of the ends did not change for five frames. In most experiments (except experiments with immobilized P-3W-VCD), the coverslip was coated with NEM-myosin II to capture the actin filaments.
Size exclusion chromatography-multi-angle light scattering (SEC-MALS)
Samples (100 μl at 1–2 mg ml-1) were fractionated by size exclusion chromatography using an TSK-gel Super SW2000 column (Tosoh Bioscience) coupled to an Agilent 1100 HPLC system (Agilent Technologies). The molecular species separated by the column were analyzed through a DAWN HELEOS multi-angle light scattering (MALS) detector and an Optilab rEX refractive index detector and their masses calculated with the Astra software (Wyatt Technology Corp.).
Sedimentation assay with F-actin
Actin (20 μM) in G-buffer (2 mM Tris [pH 7.4], 0.2 mM CaCl2, 0.2 mM ATP, 1 mM DTT, 1 mM NaN3) was polymerized with addition of 50 mM KCl, 2 mM MgCl2, and 1 mM ATP for 30 min at room temperature. VopL constructs 1W-VCD and VCD were centrifuged at 224,000 × g for 30 min to remove potential aggregates. F-actin (10 μM) was incubated with 10 μM VopL constructs for 1 h at room temperature. Samples were centrifuged at 224,000 × g for 30 min. Equal volumes of supernatant and pellet were analyzed by SDS-PAGE.
Protein labeling with Qdots
The AviTag 17-aa sequence GGGLNDIFEAQKIEWHE was added at the C-terminus of construct 3W-VCD. This sequence is biotinylated at Lys12 by E. coli biotin ligase during protein expression 38. Construct 3W-VCD-AviTag was expressed in BL21(DE3) cells, carrying plasmid pBirAcm (Avidity), for overexpression of E. coli biotin ligase 39. Protein expression and purification were conducted as above, in medium supplemented with 100 μM biotin. Conjugation with streptavidin-coupled quantum dot 625 (QD-625, Invitrogen) was carried out according to the manufacturer’s protocol, and confirmed by agarose gel electrophoresis.
Crystallization, data collection and structure determination
VCD was dialyzed against 25 mM HEPES [pH 7.5], 50 mM NaCl, 2 mM DTT and concentrated to 5 mg ml-1. Crystals of selenomethionine-substituted VCD were obtained by hanging drop vapor-diffusion at 4°C. Typical 2 μL drops consisted of a 1:1 (v/v) mixture of protein solution and a well solution containing 0.2 M sodium fluoride, 13–15% (w/v) PEG 3350. Crystals were flash-frozen in liquid nitrogen after a short passage through a solution containing 20% (w/v) PEG 400 added to the crystallization buffer. X-ray data was collected at CHESS beamline A1. Data indexation and scaling were performed with the program XDS 40. The structure was determined using the single-wavelength anomalous diffraction method. Selenium sites were found and phases were calculated with the program SHELXD 41. Density modification and non-crystallographic symmetry averaging were performed with the program DM 42. Model building and refinement were performed with the programs Coot 43 and Phenix 44 (Table 1). Illustrations of structures were prepared with the program PyMOL (Schrödinger).
Small Angle X-Ray Scattering (SAXS)
SAXS datasets were collected at 10°C from solutions of VCD and 1W-VCD+actin at BioCAT beamline 18-ID (APS, Argonne National Laboratory). VCD was measured at 2.5 mg ml-1 and 1W-VCD+actin was measured at two concentrations, 0.5 and 1.0 mg ml-1. Protein scattering profiles were obtained by subtracting the average of 20 buffer profiles from that of 20 protein profiles. The distance distribution function, p(r), radius of gyration, Rg, and maximum particle dimensions, Dmax, were computed with the indirect transform package GNOM36. Ab initio shape reconstructions were calculated with the programs DAMMIF23. For each reconstruction, 50 ab initio models were generated and averaged with the program DAMAVER45.
Supplementary Material
Acknowledgments
Supported by National Institutes of Health grant R01 GM073791 to RD and R01 GM079265 to DRK. We thank Narjol González-Escalona (Food and Drug Administration) for providing the Vibrio parahaemolyticus genomic DNA and Thomas Irving and Liang Guo for assistance during SAXS data collection at the BioCAT beamline. BioCAT is supported by NIH grant RR-08630 and APS by Department of Energy Contract W-31-109-Eng-38. Crystal data collection at CHESS was supported by NSF grant DMR-0936384 and NIH grant RR-01646.
Footnotes
AUTHOR CONTRIBUTIONS S.N. conducted the project, expressed proteins, performed sedimentation, crystallization, structure determination and SAXS experiments and participated in most of the other experiments; M.B. performed pyrene actin polymerization assays; G.R. performed ITC and SEC-MALS experiments; M.J.G., J.D.W. and D.R.K. performed and analyzed TIRF experiments; R.D. performed crystallographic studies, supervised the research and wrote the paper.
Accession codes. Protein Data Bank: The atomic coordinates and structure factors of the VCD have been deposited with accession code 3RYL.
References
- 1.Dominguez R. Structural insights into de novo actin polymerization. Curr Opin Struct Biol. 2010;20:217–25. doi: 10.1016/j.sbi.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–77. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- 3.Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- 4.Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30:342–53. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–8. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- 6.Ahuja R, et al. Cordon-Bleu Is an Actin Nucleation Factor and Controls Neuronal Morphology. Cell. 2007;131:337–350. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chereau D, et al. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320:239–43. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris JG., Jr Cholera and other types of vibriosis: a story of human pandemics and oysters on the half shell. Clin Infect Dis. 2003;37:272–80. doi: 10.1086/375600. [DOI] [PubMed] [Google Scholar]
- 9.Liverman AD, et al. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proc Natl Acad Sci U S A. 2007;104:17117–22. doi: 10.1073/pnas.0703196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tam VC, Serruto D, Dziejman M, Brieher W, Mekalanos JJ. A Type III Secretion System in Vibrio cholerae Translocates a Formin/Spire Hybrid-like Actin Nucleator to Promote Intestinal Colonization. Cell Host and Microbe. 2007;1:95–107. doi: 10.1016/j.chom.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Tam VC, et al. Functional Analysis of VopF Activity Required for Colonization in Vibrio cholerae. MBio. 2010;1 doi: 10.1128/mBio.00289-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haglund CM, Choe JE, Skau CT, Kovar DR, Welch MD. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat Cell Biol. 2010;12:1057–63. doi: 10.1038/ncb2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jewett TJ, Fischer ER, Mead DJ, Hackstadt T. Chlamydial TARP is a bacterial nucleator of actin. Proc Natl Acad Sci U S A. 2006;103:15599–604. doi: 10.1073/pnas.0603044103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosch M, et al. Analysis of the function of Spire in actin assembly and its synergy with formin and profilin. Mol Cell. 2007;28:555–68. doi: 10.1016/j.molcel.2007.09.018. [DOI] [PubMed] [Google Scholar]
- 15.Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol. 2007;179:117–28. doi: 10.1083/jcb.200706196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris ES, Higgs HN. Biochemical analysis of mammalian formin effects on actin dynamics. Methods Enzymol. 2006;406:190–214. doi: 10.1016/S0076-6879(06)06015-0. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88:1387–402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neidt EM, Skau CT, Kovar DR. The cytokinesis formins from the nematode worm and fission yeast differentially mediate actin filament assembly. J Biol Chem. 2008;283:23872–83. doi: 10.1074/jbc.M803734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124:423–35. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 20.Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci U S A. 2004;101:14725–30. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rebowski G, et al. Structure of a longitudinal actin dimer assembled by tandem w domains: implications for actin filament nucleation. J Mol Biol. 2010;403:11–23. doi: 10.1016/j.jmb.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Breitsprecher D, et al. Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. Embo J. 2008;27:2943–54. doi: 10.1038/emboj.2008.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Cryst. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferron F, Rebowski G, Lee SH, Dominguez R. Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. Embo J. 2007;26:4597–606. doi: 10.1038/sj.emboj.7601874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chereau D, et al. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci U S A. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perelroizen I, Marchand JB, Blanchoin L, Didry D, Carlier MF. Interaction of profilin with G-actin and poly(L-proline) Biochemistry. 1994;33:8472–8. doi: 10.1021/bi00194a011. [DOI] [PubMed] [Google Scholar]
- 27.Marchand JB, Kaiser DA, Pollard TD, Higgs HN. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
- 28.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–65. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 29.Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18:9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuchero JB, Coutts AS, Quinlan ME, Thangue NB, Mullins RD. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat Cell Biol. 2009;11:451–9. doi: 10.1038/ncb1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–5. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 32.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–8. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 33.Pechlivanis M, Samol A, Kerkhoff E. Identification of a short Spir interaction sequence at the C-terminal end of formin subgroup proteins. J Biol Chem. 2009;284:25324–33. doi: 10.1074/jbc.M109.030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dahlgaard K, Raposo AA, Niccoli T, St Johnston D. Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev Cell. 2007;13:539–53. doi: 10.1016/j.devcel.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okada K, et al. Adenomatous polyposis coli protein nucleates actin assembly and synergizes with the formin mDia1. J Cell Biol. 2010;189:1087–96. doi: 10.1083/jcb.201001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- 37.Boczkowska M, et al. X-ray scattering study of activated Arp2/3 complex with bound actin-WCA. Structure. 2008;16:695–704. doi: 10.1016/j.str.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–9. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith PA, et al. A plasmid expression system for quantitative in vivo biotinylation of thioredoxin fusion proteins in Escherichia coli. Nucleic Acids Res. 1998;26:1414–20. doi: 10.1093/nar/26.6.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–22. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 42.Cowtan K. Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:470–8. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallog. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


