Abstract
p62/SQSTM1/A170 is a multimodular protein that is found in ubiquitin-positive inclusions associated with neurodegenerative diseases. Recent findings indicate that p62 mediates the interaction between ubiquitinated proteins and autophagosomes, leading these proteins to be degraded via the autophagy-lysosomal pathway. This ubiquitin-mediated selective autophagy is thought to begin with recognition of the ubiquitinated proteins by the C-terminal ubiquitin-associated (UBA) domain of p62. We present here the crystal structure of the UBA domain of mouse p62 and the solution structure of its ubiquitin-bound form. The p62 UBA domain adopts a novel dimeric structure in crystals, which is distinctive from those of other UBA domains. NMR analyses reveal that in solution the domain exists in equilibrium between the dimer and monomer forms, and binding ubiquitin shifts the equilibrium toward the monomer to form a 1:1 complex between the UBA domain and ubiquitin. The dimer-to-monomer transition is associated with a structural change of the very C-terminal end of the p62 UBA domain, although the UBA fold itself is essentially maintained. Our data illustrate that dimerization and ubiquitin binding of the p62 UBA domain are incompatible with each other. These observations reveal an autoinhibitory mechanism in the p62 UBA domain and suggest that autoinhibition plays a role in the function of p62.
Keywords: Autophagy, NMR, Protein Degradation, Protein Structure, Ubiquitin, X-ray Crystallography, UBA, p62
Introduction
Impairment of the ubiquitin-proteasome system is one of major causes of ubiquitin-positive inclusions found in various neurodegenerative diseases (1). Recent studies have identified the involvement of another degradation system, the autophagy-lysosomal pathway, in the formation of ubiquitin-positive inclusions as exemplified by the observation that autophagy-deficient mice exhibit substantial accumulation of such inclusions in tissues (2). p62/SQSTM1/A170, a multidomain protein found in ubiquitin-positive inclusions, has been shown to bind intracellular signaling factors (3–5). Accumulating evidence indicates that p62 is a receptor for ubiquitinated proteins that are targeted to the autophagosome for lysosomal degradation. Specifically, it is involved in autophagic elimination of damaged mitochondria, midbody rings, peroxisomes, and microbes (6–10).
The importance of p62 in autophagic degradation of proteins and organelles has been demonstrated by studies using tissue-specific autophagy-deficient mice. Elimination of Atg5 or Atg7, an essential gene in the formation of the autophagosome, in mouse neurons and hepatocytes resulted in toxicity accompanied by accumulation of ubiquitin-positive inclusions in the cells. In contrast, knock-out of both Atg7 and p62 (Atg7−/−/p62−/−) caused a dramatic reduction in the amount of inclusions in both types of cells (2, 11). A similar result was also reported in fruit flies (12). These observations indicate that p62 is critically involved in the development of ubiquitin-positive inclusions that should be degraded via autophagy. In addition to playing a role in the autophagy-lysosomal pathway, p62 itself was identified as a specific substrate degraded by the autophagy-lysosomal pathway (13–15), indicating that p62 levels in the cytosol might be tightly regulated by autophagy. p62 mediates cell signaling pathways related to cell stress, proliferation, cell death, and inflammation. The interaction of p62 with TRAF6 promotes its oligomerization and subsequent activation, which leads to Lys63 polyubiquitination of TRAF6 resulting in the activation of NF-κB activation. Death receptor ligation induces polyubiquitination of caspase-8 through the interaction of DISC with a cullin3-based ubiquitin ligase. p62 promotes aggregation of CUL3-modified caspase-8 within p62-dependent foci, leading to full activation and processing of the enzyme and driving commitment to cell death (5). Because p62 levels can be regulated through degradation by autophagy, cellular signals interacting with p62 may also be affected by autophagy.
A likely scenario of ubiquitin-mediated “selective autophagy” includes the following: p62 interacts with ubiquitinated proteins, leading to the formation of protein aggregates that are recruited to autophagosomes via interaction with LC3 (microtubule-associated protein 1 light chain 3), a mammalian homologue of Atg8. LC3 has been shown to localize to the isolation membrane/phagophore and mediate binding of protein aggregates to the autophagosome. The autophagosome then fuses with the lysosome/vacuole, and the ubiquitinated cargo is degraded. One functional domain identified in p62, the ubiquitin-associated (UBA)3 domain at its C terminus, plays a pivotal role in the incorporation of ubiquitinated substrates into the autophagy-lysosomal pathway. In the accepted mechanism, the UBA domain captures ubiquitinated proteins, and the PB1 (Phox and Bem1p) domain at the N terminus of p62 induces formation of protein aggregates through its self-oligomerizing nature. The aggregates are subsequently recruited to the autophagosome for degradation via the interaction between the LC3-interacting region/LC3 recognition sequence of p62 and LC3 at the isolation membrane (14, 16). The interaction between the p62 UBA domain and ubiquitin, which may underlie the initial step of the degradation pathway, has been extensively studied (11, 17, 18). Nevertheless, molecular details of this interaction are still unclear and remain to be addressed.
We present here the crystal structure of the UBA domain of mouse p62 and the solution structure of its ubiquitin-bound form together with biochemical and biophysical data, which, as a whole, reveals a novel ubiquitin-interaction mode of the UBA domain. The p62 UBA domain crystallized as a dimer and exists in a monomer-dimer equilibrium in solution. Although several UBA dimers have been reported (19–21), the p62 UBA dimer differs substantially from these, revealing a novel dimerization mode of the UBA domain. NMR data reveal that the binding mode between the p62 UBA domain and ubiquitin is similar to that seen in canonical UBA domains. However, only the monomeric p62 UBA domain is observed to bind ubiquitin. Structural comparison of the ubiquitin-free dimer and the ubiquitin-bound monomer of the p62 UBA domain indicates that the ubiquitin-bound form is unable to dimerize due to steric hindrance between the C-terminal tail of the p62 UBA domain and ubiquitin. The C-terminal tail is essential for dimer formation but dispensable for ubiquitin binding. Truncation of the tail dramatically enhances the apparent affinity of the p62 UBA domain for ubiquitin, presumably because of the absence of the monomer-dimer equilibrium that competes with ubiquitin binding. The existence of the monomer-dimer equilibrium may provide a regulatory process for the p62/ubiquitin interaction.
MATERIALS AND METHODS
Construction of Expression Vectors
The expression vector pGEX-6P1-p62 UBA (residues 391–438 of mouse p62 protein) was constructed using PCR. An oligonucleotide coding for the UBA domain of p62 with 5′ BamHI and 3′ XhoI restriction sites was ligated to the corresponding restriction sites in pGEX-6P1. All mutants of the p62 UBA domain, pGEX-6P1-p62 PB1 (residues 1–100) and PB1 mutants (DDD: D69A, D71A, and D73A and KRK: K7A, R68A, and K91A) were constructed using the QuikChange mutagenesis kit from the vectors pGEX-6P1-p62 UBA or pGEX-6P1-full-length p62 according to the manufacturer's instructions (Stratagene, La Jolla, CA).
Protein Sample Preparation
All the protein samples of the UBA domain and the PB1 domain, including mutants of mouse p62, were expressed as fusion proteins with GST in Escherichia coli. After purification by glutathione-Sepharose 4FF (GE Healthcare) column chromatography, the affinity tag was removed from the proteins using PreScission protease (GE Healthcare). The UBA and PB1 domains were further purified using HiLoad Superdex 75 pg 16/60 (GE Healthcare) size exclusion chromatography. The 15N- and 13C/15N-labeled samples were obtained by growing cells in M9 minimal media containing 15NH4Cl and [13C]glucose/15NH4Cl, respectively. The selenomethionine derivative of the p62 UBA domain was obtained using Le master medium.
p62 UBA Domain Crystallization, Data Collection, and Structural Refinement
Crystals were obtained using the a vapor diffusion hanging drop method with 100 mm sodium citrate, pH 5.0, and 1.8 m ammonium sulfate. This crystallization condition was applied to obtain crystals of the selenomethionine derivative of the p62 UBA domain. Protein stock concentration was 20 mg/ml for native crystal and 2 mg/ml for the selenomethionine derivative. Crystals grew in 1–3 days at 20 °C. For diffraction data collection, crystals were cryoprotected by addition of 20% (v/v) glycerol. Diffraction data of the p62 UBA were collected on an ADSC Quantum 315 detector (Area Detector System Corp., Poway, CA) at the beamline BL-5 at Photon Factory, Tsukuba, Japan. Data processing and scaling were performed with the program HKL2000 (22). Phases were obtained by the multiple-wavelength anomalous diffraction phasing technique using the selenomethionine derivative and the program Phaser (23). The initial model was constructed using the program Arp/Warp (24) and further refined using the program CNS (25) and the program REFMAC (26) in conjunction with the CCP4 suite (27). Diffraction data and refinement data statistics are shown in Table 1. All the structural figures were generated with the program PyMol. Data collection and structural refinement statistics are shown in Table 1.
TABLE 1.
Statistics of diffraction data and structure refinement
The highest resolution shell is shown in parentheses.
| Edge | Peak | High remote | Low remote | Native | |
|---|---|---|---|---|---|
| Data collection | |||||
| Source | Photon Factory-BL5 | ||||
| Detector | ADSC Quantum 315 | ||||
| Wavelength | 0.97928 Å | 0.97909 Å | 0.96405 Å | 0.98317 Å | 1.00000 Å |
| Space group | P43212 | ||||
| Cell dimensions | |||||
| a = b | 72.940 Å | 72.904 Å | 72.964 Å | 72.985 Å | 72.968 Å |
| c | 32.004 Å | 31.981 Å | 32.022 Å | 32.038 Å | 32.561 Å |
| Resolution | 50.0-1.7 Å | 50.0-1.4 Å | |||
| Total observations | 122,025 | 122,954 | 122,696 | 122,010 | 226,728 |
| Unique reflections | 9340 | 9386 | 9393 | 9393 | 17,371 |
| Rmerge | 7.1% (27.8%) | 9.0% (27.8%) | 6.4% (26.6%) | 4.8% (26.2%) | 4.8% (35.6%) |
| I/σI | 14.7 | 15.0 | 14.8 | 14.6 | 13.1 |
| Completeness | 96.5% (75.9%) | 97.2% (80.3%) | 97.0% (78.1%) | 96.9% (77.7%) | 97.0% (82.5%) |
| Redundancy | 13.1 (9.9) | 13.1 (10.1) | 13.1 (10.0) | 13.0 (9.6) | 13.1 (11.0) |
| Refinement | |||||
| Resolution | 50.00 to 1.40 Å | ||||
| Rwork/Rfree | 17.0/20.1% | ||||
| No. of atoms | 920 | ||||
| Protein | 838 | ||||
| Sulfate | 5 | ||||
| Water | 77 | ||||
| B-factors | |||||
| Main chain | 8.8 Å2 | ||||
| Side chain | 10.8 Å2 | ||||
| Sulfate | 18.0 Å2 | ||||
| Water | 23.9 Å2 | ||||
| All Atoms | 11.0 Å2 | ||||
| r.m.s.d. | |||||
| Bond lengths | 0.016 Å | ||||
| Bond angles | 1.697° | ||||
| Ramachandran plot | |||||
| Most favored | 93.1% | ||||
| Additional allowed | 6.9% | ||||
| Generously allowed | 0% | ||||
NMR Spectra
1H-15N HSQC spectra of wild-type and mutant p62 UBA were acquired in 20 mm sodium phosphate buffer, pH 6.8, 5 mm potassium chloride, 1 mm EDTA, and 10% D2O. For the dilution experiment of the p62 UBA domain, the protein sample was sequentially diluted 2-fold from 200 to 3.1 μm. For all other 1H-15N HSQC spectra, 100 μm protein samples were used except for the ubiquitin-binding experiment at low concentrations (10 μm). The normalized chemical shift changes for each of the spectra were defined by (δH2 + (δN/5)2)1/2, where δH and δN represent the chemical shift differences in the 1H and 15N dimensions, respectively. For backbone chemical shift assignments of the p62 UBA domain in various states, the 1HN, 15N, 13Cα, and 13Cβ chemical shifts of the p62 UBA domain under normal (free form, dimer), 6-equimolar ubiquitin (bound form, monomer), and 35% DMSO (free monomer) conditions were assigned using three-dimensional HNCACB, CBCA(CO)NH, HNCO, and HN(CA)CO experiments. All spectra were acquired using either 1 mm (free form), 400 μm (bound form), or 400 μm (35% DMSO) 13C/15N-labeled p62 UBA. Data were processed using the program NMRPipe (28) and analyzed with the program CARA (29). All the spectra were acquired with a Bruker Avance II 700 MHz spectrometer equipped with a TCI Cryoprobe head at 292 K.
Structural Determination of the p62 UBA Domain in the Presence of 6-Equimolar Ubiquitin
We assigned the chemical shifts of the p62 UBA domain under the 6-equimolar amount of ubiquitin using three-dimensional HNCACB, CBCA(CO)NH, HNCO, HN(CA)CO, HBHA(CO)NH, H(CCCO)NH, CC(CO)NH, and HCCH-TOCSY spectra of a 400 μm 13C/15N-labeled sample that was dissolved in 20 mm sodium phosphate, pH 6.8, 5 mm potassium chloride, 1 mm EDTA, and 10% D2O. The inter-proton distances used for the structural calculation of the p62 UBA domain were derived from three-dimensional 15N- and 13C-edited NOESY spectra. Any intermolecular NOESY cross-peaks were not observed in the intermolecular 13C-filtered NOESY spectra. The structure was calculated using the program CYANA (30) based on NOESY cross-peaks either manually picked using the program SPARKY or automatically (iteratively with the structural calculation) picked by ATNOS/CANDID modules in the program UNIO '08 (31, 32). Essentially, the same conformation of the p62 UBA domain structure was obtained using both calculation methods (backbone r.m.s.d. 0.89 Å through residues 394–438 of the mean structures). The final structural ensemble was calculated using the manually picked NOESY cross-peaks, which resulted in more converged structures (backbone r.m.s.d. was 0.54 ± 0.10 Å for the UNIO '08 ATNOS/CYANA structures and 0.13 ± 0.03 Å for the SPARKY/CYANA structure). The statistics for the structural calculation are given in Table 2.
TABLE 2.
Structural statistics of ubiquitin-bound p62 UBA
No violations in NOE distance restrains (>0.3 Å) were observed.
| NMR restraints | |
| Total distance restraints | 1144 |
| Short range | 598 |
| Medium range | 290 |
| Long range | 256 |
| Ensemble statisticsa | |
| Backbone atoms r.m.s.d. | 0.13 ± 0.03 Å |
| All heavy atoms r.m.s.d. | 0.69 ± 0.13 Å |
| Ramachandran plota | |
| Most favored regions | 78.0% (546) |
| Additional allowed regions | 22.0% (154) |
| Generously allowed regions | 0.0% (0) |
| Disallowed regions | 0.0% (0) |
a Statistics were calculated for residues 393–435 of the 20 lowest-energy structures.
Residual Dipolar Coupling
RDC samples were prepared as described (33). The 13C, 15N-lableled wild-type p62 UBA complex with ubiquitin was soaked in a cylindrically shaped 4% polyacrylamide gel, initially 6 × 9 mm in size, which was subsequently radially compressed to fit within an NMR tube, thereby increasing its length to 18 mm. 1H-15N residual dipolar couplings were obtained from IPAP-[1H-15N]-HSQC experiments carried out with 13C decoupling (34) at 298 K. The measurement was repeated three times to calculate uncertainties in the coupling constants. The order parameter for each residue was set to 1.0, respectively. The coupling constants from the helical regions were used. RDC data analysis was performed using REDCAT (35).
Isothermal Titration Calorimetry
ITC measurements were performed at 25 °C using a MicroCal VP-ITC calorimeter. Protein samples were dissolved in buffer containing 25 mm Tris-HCl, pH 8.0, and 150 mm NaCl. ITC buffers and samples were filtered and degassed prior to the experiment. For determination of dimer dissociation constants, 10 μl of either 1 mm (wild type) or 500 μm (W414F and I433A) protein samples were injected at 5-min intervals to the blank buffer in the 1.4-ml cell. For the determination of ubiquitin-binding constants, 10 μl of 2 mm ubiquitin sample was titrated to the cell containing 30 μm p62 UBA (wild type, W414F, and I433A) with the same experimental setup as for the dimer dissociation experiments. Injections were performed until the heat generated became negligible; typically, 24 injections were required for dimer dissociation and ubiquitin binding. Acquired data were integrated, base-line corrected, and analyzed using software provided by the manufacturer (Origin 7, MicroCal Software, Inc.). For each experiment, at least two independent titration experiments were performed.
RESULTS
Crystal Structure of the p62 UBA Domain
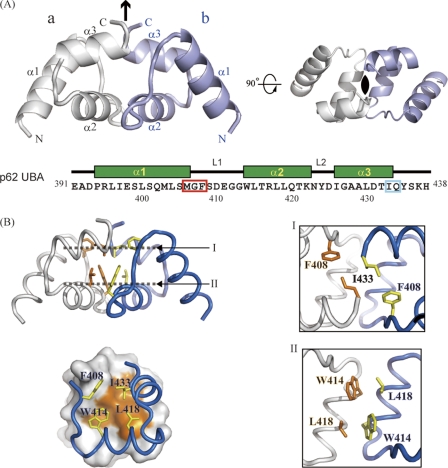
A polypeptide composed of residues 391–438 of mouse p62 was expressed, prepared, and crystallized. Using these crystals, the structure of the p62 UBA domain was determined at 1.4 Å resolution (Table 1). Two molecules of the p62 UBA domain formed a dimer in one crystallographic asymmetric unit with C2 symmetry (Fig. 1A). The structures of the subunits of an asymmetric unit were almost identical to each other (the r.m.s.d. value for backbone Cα atoms was 0.56 Å). The dimer interface was analyzed by visual inspection and using the program LIGPLOT (36). The two p62 UBA subunits are arranged so that helix-2 runs in an anti-parallel manner, and the aromatic ring of Trp414 at the beginning of helix-2 in one unit is involved in a hydrophobic interaction with the side chain of Leu418 in the other unit (Fig. 1B, panel II). The C-terminal part of helix-3 is also involved in the dimer interface, with Ile433 from one unit making hydrophobic contacts with Ile433 from the opposite unit (Fig. 1B, panel I). Helix-1 is not involved in the dimer interface. The surface made of hydrophobic residues in helix-2 and -3 creates a large dimer interface of 667 Å2. Auxiliary electrostatic interactions exist between residue pairs of Met406–Gln434, Arg417–Gly412, and Arg417–Gly413. The electron density terminated at Tyr435 and was not observed for three C-terminal residues, because of structural disorder.
FIGURE 1.
Crystal structure of p62 UBA dimer. A, ribbon representation of the crystal structure of the p62 UBA dimer. Noncrystallographic C2 symmetry axis is shown as a black arrow and a black oval. Each monomer unit is colored in white or light blue. The amino acid sequence with secondary structure indicated is shown below the structures. In the sequence, the MGF motif and the residues corresponding to di-leucine motif in canonical UBA domains are indicated by the red and cyan box, respectively. B, residues at the dimerization interface of the p62 UBA dimer are shown as stick models. Interactions in the dimer interface are shown in the right panel. The locations of planes I and II are indicated in the left panel, top. Each plane is the top view of the structure.
p62 UBA Domain Exists in a Dimer-Monomer Equilibrium in Solution
The considerable hydrophobicity and complementarity of the dimer interface in the crystal structure imply that the dimer is stable in aqueous solution. To test this assumption, we conducted a series of hydrodynamic studies on the p62 UBA domain (supplemental Table S1). Analytical gel filtration indicates that the estimated apparent molecular weight of wild-type (WT) UBA is two times the predicted molecular weight of monomeric UBA (∼5,800). This result is consistent with those from analytical ultracentrifugation, in which both sedimentation velocity and sedimentation equilibrium experiments indicate that WT UBA exists as a dimer with an apparent molecular weight of 10,800 ± 600 at a protein concentration of >17 μm, which is approximately two times that of monomeric UBA. To further characterize the p62 UBA domain in solution, we conducted solution NMR experiments. In the 1H-15N correlation spectrum of the 15N-labeled p62 UBA domain (∼1 mm), the number of cross-peaks from main chain amide groups roughly agreed with the number of amino acid residues in the p62 UBA domain. At this concentration, the p62 UBA domain exists mainly as a dimer. Hence, in the spectrum the cross-peaks from each monomer unit are degenerate, indicating that each subunit is in an equivalent environment in a symmetric dimer. These observations are consistent with the crystal structure of the p62 UBA domain, in which the two UBA molecules have a C2 axis of symmetry.
To test whether the solution dimer of the UBA domain is the same as that observed in the crystal, we mutated residues that form the dimeric interface in the crystal and observed their effects on dimer formation in solution. Seven mutants were prepared (W414A, W414K, W414F, L418A, L418V, I433A, and I433V). According to analytical gel filtration, the elution volumes indicate that all the mutants except I433V are monomeric at a concentration of 0.1 mm (supplemental Table S1). Analytical ultracentrifugation experiments showed that the apparent molecular weights of W414F and I433A mutants are 6,500. NMR results combined with results from the hydrodynamic studies suggest that the dimeric structure of the p62 UBA domain in solution is identical to that in the crystal.
NMR Analysis of the Dimer-Monomer Equilibrium in Solution
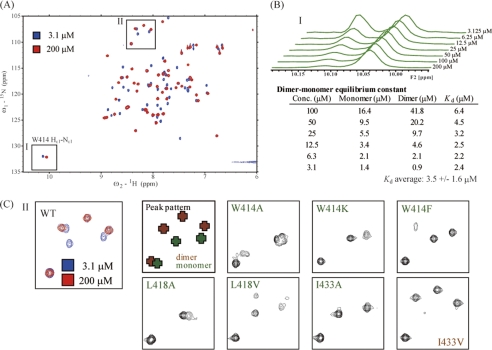
Interestingly, the 1H-15N correlation spectrum measured at a concentration of ∼1 mm p62 UBA domain contained an additional set of cross-peaks with intensities much weaker than the aforementioned main peak set. We assumed that these weak cross-peaks were due to monomeric p62 UBA domain. To test this assumption, we examined the concentration dependence of the 1H-15N correlation spectrum (Fig. 2, A and B). As the protein concentration decreased, the minor peak set increased in intensity, and the major peak set decreased in intensity. This mutually adverse change in peak intensity upon dilution points to the existence of a monomer-dimer equilibrium for the p62 UBA domain. The increase in intensity of the minor peaks at lower concentrations indicates that this set of peaks belongs to the monomeric species because dilution shifts the equilibrium toward the monomer. 1H-15N correlation spectra of the six monomeric mutants were also recorded, and these spectra are essentially identical to the minor set of peaks in the WT p62 UBA domain. Comparison of spectral subregions around glycine cross-peaks showed a striking similarity in the spectral pattern between the monomeric mutants and the minor set of peaks of WT p62 UBA (Fig. 2C). These results lead to the conclusion that the minor set of peaks observed in the 1H-15N correlation spectrum of WT p62 UBA belongs to the monomeric form.
FIGURE 2.
NMR analysis of p62 UBA dimerization. A, 1H-15N HSQC spectra of 15N-labeled WT p62 UBA measured at high (200 μm) and low (3.1 μm) concentrations. B, one dimensional projections of spectral region I in A generated from the 1H-15N HSQC spectra of 15N-labeled WT p62 UBA at various concentrations. The dissociation constant of the p62 UBA dimer was calculated from the populations of monomer and dimer species of the p62 UBA domain at each concentration based on the volume of Trp414 Hϵ1-Nϵ1 side chain peaks of monomer and dimer. C, spectral subregion II in A is magnified, and the same regions of the spectra of various mutants are indicated. All mutants except I433V showed the characteristic monomer cross-peak pattern. I433V maintains the characteristic dimer cross-peak pattern.
The equilibrium constant of the dimer dissociation (Kd,dim) was estimated to be in the micromolar range (∼3 μm) using the concentration-dependent change of the cross-peak intensity of the side chain of Trp414 (1Hϵ-15Nϵ cross-peak) (Fig. 2B). The exchange rate between monomer and dimer is slow relative to the NMR time scale, as we did not see any changes in chemical shift as a result of dilution.
Monomer-Dimer Equilibrium Competes with Ubiquitin Binding
Previously reported dissociation constants of the complex between ubiquitin and canonical UBAs range from tens to hundreds of micromolars (37). Hence, a question arises as to whether the ubiquitin interaction affects the monomer-dimer equilibrium of the p62 UBA domain. To examine this, we performed an NMR titration experiment using 15N-labeled p62 UBA and unlabeled ubiquitin. 1H-15N correlation spectra were obtained for samples of 10 μm 15N p62 UBA in the presence of varying amounts of unlabeled ubiquitin. At this UBA concentration, it was possible to simultaneously monitor spectral changes in the dimer and monomer species of the UBA domain upon addition of ubiquitin (Fig. 2B). Monomer cross-peaks gradually shifted upon addition of ubiquitin, which indicates the system is in the fast exchange regime relative to the NMR time scale (Fig. 3A). No cross-peaks corresponding to dimer species shifted, but intensities of some cross-peaks decreased upon addition of ubiquitin, which is characteristic of systems in the slow exchange regime with respect to the NMR time scale. This observation raises a possibility that three states (monomeric UBA, dimeric UBA, and the complex between monomeric UBA and ubiquitin) coexist during the titration. The complex and monomeric forms of UBA are thought to be in the fast-exchange equilibrium, and monomeric and dimeric UBA are in the slow-exchange equilibrium. With an excess of ubiquitin, no amide group produced more than one signal, demonstrating that the UBA domain binds to ubiquitin in a uniform manner. As a whole, the data suggest that only monomeric UBA can bind to ubiquitin, and ubiquitin binding induces dissociation of the UBA dimer by shifting the equilibrium toward the monomer.
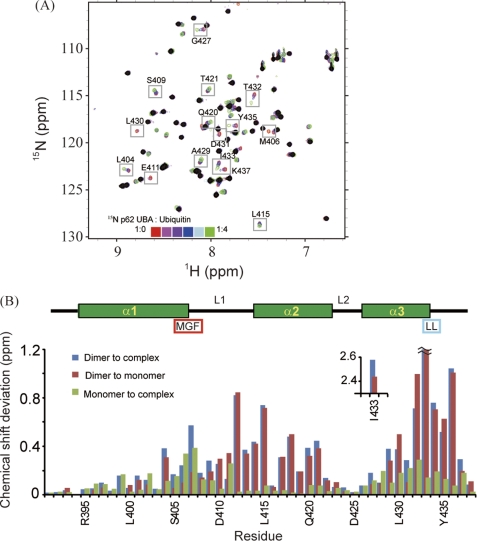
FIGURE 3.
Identification of p62 UBA dimerization interface and ubiquitin-binding site. A, overlay of 1H-15N HSQC spectra of 15N-labeled p62 UBA at low concentration (10 μm) in the absence and presence of different concentrations of ubiquitin. Peaks observed in the 1H-15N HSQC spectrum with a 200 μm sample are masked in black. B, normalized chemical shift changes of backbone amide groups are plotted in a function of amino acid residues. The secondary structures are shown schematically on the top. The position of the MGF motif and the di-leucine motif is also indicated. Normalized chemical shift changes are calculated by (δH2 + (δN/5)2)1/2, where δH and δN represent the chemical shift differences in the 1H and 15N dimensions.
This assumption is supported by titration experiments using 15N-labeled monomeric mutants of the p62 UBA domains, in which main chain resonances of the mutant UBA exhibited chemical shift changes in fast exchange upon titration of unlabeled ubiquitin. Titration of 15N-labeled ubiquitin with unlabeled WT p62 UBA showed fast-exchange binding kinetics, as observed for the monomer cross-peaks of WT 15N-labeled p62 UBA. Collectively, our NMR data indicate that it is the monomer, not the dimer, of the p62 UBA domain that binds to ubiquitin.
Titration experiments reported by Long et al. (17) on WT p62 UBA gave similar results. They suggested that a large conformational change takes place in monomeric UBA upon binding to ubiquitin. This interpretation seems to be due to the assumption that unliganded UBA dominantly exists as a monomer at 1 mm (17). However, our data clearly show that the majority of UBA is dimeric at concentrations >17 μm.
p62 UBA Domain Monomer Binds Ubiquitin Similar to Canonical UBA Domains
To further interpret the titration experiments, it was necessary to assign the chemical shifts for the amide 1H and 15N nuclei of monomeric and dimeric p62 UBA domains. Details of the assignment procedure are provided in supplemental Fig. S1. In brief, we first obtained the backbone assignments of 15N-labeled p62 UBA (0.4 mm) in a buffer containing 35% (v/v) dimethyl sulfoxide (DMSO). Under this condition, the p62 UBA domain exists as a monomer. The assignments were then transferred to the monomer of the p62 UBA domain in the standard buffer. The presence of DMSO did not perturb the structure of monomeric p62 UBA itself but destabilized the UBA dimer by weakening the hydrophobic interactions between the two UBA subunits (see supplemental material).
After obtaining unambiguous chemical shift assignments, we analyzed the chemical shift perturbations (CSPs) of the p62 UBA domain upon dimerization and ubiquitin binding (Fig. 3). Between monomer and dimer, moderate CSPs were identified in helix-2 and its preceding residues, and large CSPs (0.75–2.5 ppm) were observed in the C-terminal residues of helix-3 (Fig. 3B (red)). This observation is consistent with the crystal structure of dimeric p62 UBA, in which these regions make up the dimer interface. Canonical UBA domains share two common sequence patterns involved in ubiquitin binding as follows: the MGF motif at L1 loop and the di-leucine motif at the end of α3 (38). The MGF motif in the p62 UBA domain displays significant CSPs upon binding to ubiquitin (Fig. 3B (green)). Although the p62 UBA domain does not contain a di-leucine motif, significant CSPs were observed at the corresponding two residues (Fig. 3B (green)). Upon ubiquitin binding, CSPs were identified throughout helix-1 and -3, but helix-2 was unperturbed. These features agree with the conventional ubiquitin-binding mode of canonical UBA domains (37, 38), indicating that it is highly likely that the p62 UBA domain binds ubiquitin in a manner conserved among canonical UBA domains.
ITC Experiments, Dimerization of the p62 UBA Domain
To quantify the interaction between p62 UBA subunits, we examined dissociation of the dimer using ITC. The dimer-monomer equilibrium constant (Kd,dim) was determined to be 10 ± 2.9 μm (supplemental Fig. S2, A and C), which is slightly higher than that obtained by NMR (Fig. 2B and supplemental Fig. S2C). In agreement with experiments using the site-directed mutant experiments (NMR, gel filtration, and ultracentrifugation), heat generated in the ITC experiments using the mutants of W414F and I433A was negligible (supplemental Fig. S2C), confirming that those mutants are monomeric in solution.
ITC Experiments, Ubiquitin Binding of the p62 UBA Domain
The interaction between the p62 UBA domain and ubiquitin was also studied using ITC (supplemental Fig. S2, B and D). Less heat was generated upon titration of WT UBA with ubiquitin (ΔH = −1.0 kcal/mol) compared with the monomeric UBA mutants (ΔH = −5.5 and −3.9 kcal/mol for the W414F and the I433A mutant, respectively). Because dissociation of the UBA dimer is endothermic (ΔH = 7.8 kcal/mol), the heat generated during binding of ubiquitin by UBA was presumably counteracted by dissociation of the dimer. The dissociation constant of the interaction (Kd,bind) was calculated to be a few tens of micromolars (supplemental Fig. S2, B and D), which was similar to Kd,bind values determined by NMR. Thus, the p62 UBA domain is expected to experience both these processes while coexisting with ubiquitin.
Structure of p62 UBA Domain in Ubiquitin-UBA Complex Determined by Solution NMR
To gain more insight into the binding of the p62 UBA domain to ubiquitin, we attempted to determine the three-dimensional structure of the complex between the p62 UBA domain and ubiquitin. Efforts to determine a crystal or solution structure of the complex between the p62 UBA domain and ubiquitin have not yet been successful, but solution NMR experiments allowed determination of the three-dimensional structure of the p62 UBA domain in its ubiquitin-bound form.
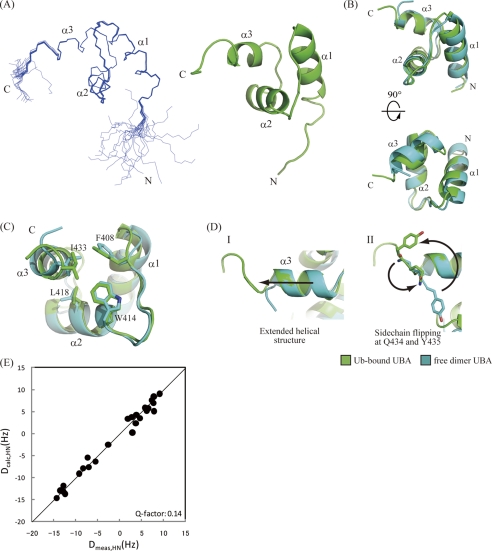
A 13C/15N-labeled p62 UBA sample was prepared with the inclusion of 6-equimolar amount of unlabeled ubiquitin. A series of triple resonance NMR experiments was carried out to obtain backbone and side chain assignments for 1H, 13C, and 15N nuclei, and three-dimensional 13C- and 15N-edited NOESY spectra were collected to obtain distance restraints. For structural calculations, the NOESY cross-peaks were assigned using automated assignment protocols to minimize human-derived artifacts. Overall statistics for the final calculated structures are given in Table 2. Residues 393–435 converged well, and the backbone r.m.s.d. of the 20 lowest energy structures was 0.13 ± 0.03 Å (Fig. 4). Two N-terminal and three C-terminal residues exhibited poor structural convergence because of the absence of long range NOEs. Because the {1H}-15N heteronuclear NOE values (see supplemental Fig. S3) of amide groups for these residues were low, both terminal residues are flexible in solution. Overall, the ubiquitin-bound structure of the p62 UBA domain can be almost perfectly superimposed upon its dimeric form in the crystal with a backbone r.m.s.d. value of ∼1 Å (Fig. 4B). To confirm this result, we also measured residual dipolar couplings (RDCs) of backbone N-HN pairs of p62 UBA in the complex (Fig. 4E). Overall, experimentally obtained RDCs for p62 UBA in complex with ubiquitin showed good agreement with the crystal structure of the p62 UBA dimer (Q-factor = 0.14), which demonstrates that the global fold of p62 UBA is essentially unchanged in the complex.
FIGURE 4.
NMR structure of p62 UBA domain in its ubiquitin-bound form. A, ensemble of the 20 lowest energy structures of the p62 UBA domain determined in a 6-equimolar amount of ubiquitin (left) and ribbon diagram of the energy-minimized averaged structure generated from the ensemble (right). B, superposition of the free-form crystal structure and the bound-form NMR structure of the p62 UBA domain. C, dimerization interface residues in both free- and bound-form structures of the p62 UBA domain are shown as stick models. D, structural comparison of the C terminus of the helix-3 between the free and the bound forms of the p62 UBA domain. The p62 UBA structure in its ubiquitin-bound form (NMR structure) is shown in green, and the free form (crystal structure) is shown in cyan. E, plot of calculated RDC constants of the free-form p62 UBA crystal structure (vertical axis) and measured RDC constants from the ubiquitin-bound p62 UBA sample (horizontal axis).
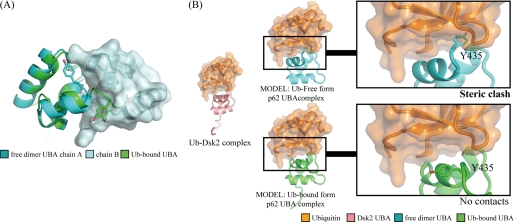
There are essentially no changes in the backbone conformation between the dimer and ubiquitin-bound form with the exception of helix-3 and the C-terminal tail. In the crystal structure of the dimer, helix-3 terminates at Ile433, but in the bound form, Gln434 and Tyr435 still retain a helical structure, making helix-3 one half-turn longer than in the dimer (Fig. 4D). A model of p62 UBA dimer suggests that the C-terminal extension of helix-3 would sterically interfere with the other subunit of the p62 UBA dimer (Fig. 5), indicating that the dimer and the ubiquitin-bound form are distinct, as predicted from the NMR titration experiments.
FIGURE 5.
Dimerization and ubiquitin binding of the p62 UBA domain. A, overlay of the free-form dimer crystal structure and the ubiquitin (Ub)-bound NMR structure of p62 UBA. The extended helix-3 that is formed upon binding to ubiquitin crashes to the other p62 UBA domain when p62 UBA is in a dimer configuration. B, model structures of the free-form crystal structure (cyan) and the ubiquitin-bound NMR structure (green) of p62 UBA complexed with ubiquitin (orange). The models were built based on the complex structure between Dsk2 UBA and ubiquitin (Protein Data Bank code 1WR1, pink and orange). Backbone atoms of the UBA domain from the complex were superimposed onto p62 UBA (cyan and green). The Dsk2 UBA domain from the original complexes was then removed. The C-terminal tail region, including the Tyr435 side chain, which has been shown to undergo a critical homomeric interaction for dimerization in this study, crashes with ubiquitin in the complex-form model (top), whereas no crashing is observed in the model generated from ubiquitin-bound form p62 UBA structure. These models suggest that the dimerization and ubiquitin binding are mutually exclusive.
C-terminal Tail of the p62 UBA Domain Is Required for Dimer Formation but Dispensable for Ubiquitin Binding
To study the importance of the C-terminal helical extension upon ubiquitin binding, a truncated mutant, p62 UBA (Y435*), which lacks Tyr435 and its C-terminal residues at positions 436–438, was prepared and examined by NMR. The 1H-15N correlation spectrum of p62 UBA (Y435*) displayed a cross-peak pattern similar to monomeric UBA at a concentration of 100 μm (supplemental Fig. S4), indicating that the mutant exists mainly as a monomer under this condition. Thus, the truncated residues play a key role in stabilizing the p62 UBA dimer. Because the C terminus of helix-3 from each subunit resides in proximity to the others in the dimer structure, it is assumed that residues 435–438 undergo intermolecular homomeric interactions with each other. However, the absence of electron density in the crystal and low heteronuclear NOE values (supplemental Fig. S3) for the C-terminal three residues in the dimer indicate that the conformation of the tail may not be uniform.
Interestingly, the Y435* mutant bound ubiquitin with a higher affinity (the dissociation constant was 7 μm) than WT p62 UBA, as indicated by NMR titration (supplemental Fig. S4C). This result confirms our assumption that the monomeric but not the dimeric form of the p62 UBA domain binds to ubiquitin and indicates that dimer formation of the UBA negatively regulates ubiquitin binding. Because the truncated UBA is capable of binding to ubiquitin, the C-terminal tail is not involved in the binding site but seemingly forms a critical dimer interface.
UBA Dimer Is Functional in Full-length p62
As p62 consists of multiple functional domains, the significance of the UBA dimerization needed to be clarified in the context of full-length protein. Therefore, a GST pulldown assay was conducted using GST-fused p62 and Lys48-linked or Lys63-linked polyubiquitin (supplemental Fig. S5). To clarify the effect of dimerization of the UBA domain, two derivatives, GST-W414F p62 and GST- Y435* p62, were tested. The former has a substitution of Trp414 by phenylalanine, and the latter lacks the C-terminal region. Either of these changes in p62 disabled the UBA dimerization when tested on the isolated UBA domain (see Fig. 2 and supplemental Fig. S4). Consistent with these data, both derivatives of GST-p62 bound more strongly to the tetraubiquitins than GST-WT p62, presumably because the UBA domains dominantly exist as active monomers.
DISCUSSION
Structural Determinants of Dimerization and Ubiquitin Binding
In this work, we revealed that p62 UBA could form a stable dimer in aqueous solution. Although a few other UBA dimers have been reported, they are structurally unrelated to the p62 UBA dimer, because it is apparent from the structures that these UBA dimers can bind ubiquitin without dissociating to monomers (17). The dimerization of p62 UBA was recently reported by others (39), in which a docking model was presented for p62 UBA dimers. When compared with our crystal structures, significant differences were found, for example, in the relative position and orientation of two UBA units in the dimer. These differences likely reflect limited information used for the model building in the report.
The comparison of two p62 UBA structures determined in our study reveals several critical determinants of UBA dimerization and ubiquitin binding. The supplemental Fig. S6 shows the structure of a representative UBA from Dsk2 and p62 UBA in free form. In Dsk2 UBA, Leu368, the first leucine in the consensus LL motif, contributes to forming a hydrophobic core via the interaction with Phe344, and the second leucine, Leu369, is outside the core to interact with ubiquitin. In contrast, p62 UBA does not have the LL motif; as an alternative, it has Ile433 and Gln434 at the equivalent position. The side chain of Ile433 is in the core as is Leu368 of Dsk2, but the Gln434 side chain is completely outside the core; its side chain amide hydrogen forms a hydrogen bond to the backbone amide oxygen of Met406 (supplemental Fig. S6C), which is a member of the MGF motif in the other subunit, stabilizing the dimer structure. This substitution from consensus Leu to Gln in p62 UBA is likely a determinant of the dimeric structure.
The Tyr435 side chain of p62 interacts with the hydrophobic residues on the top of the molecule making a protrusion, compared with Dsk2 UBA. This protrusion presumably makes a steric clash if ubiquitin binds to the UBA in the canonical manner (Fig. 5). Therefore, the conformation of Tyr435 likely inhibits ubiquitin binding. This interaction of Tyr435 appears to be important to stabilize the dimer, because p62 UBA becomes monomeric once Tyr435 and its C-terminal residues are deleted (supplemental Fig. S4). This deletion seems to have a negligible effect on the stability of the UBA folding itself judging from its 1H-15N correlation NMR spectrum, which exhibited mostly uniform intensities and line shapes of cross-peaks. The deletion mutant can bind to ubiquitin, probably because of the lack of the protrusion of Tyr435.
In the complex form of p62 UBA, residues at the C terminus of α3 gained a helical structure, and Tyr435 side chain flipped out so that steric clash with ubiquitin is avoided. The remaining part of the UBA domain does not represent significant changes upon complex formation.
Structure of p62 UBA in Complex with Ub
The UBA domain of p62 has previously been reported to undergo a drastic conformational change upon binding ubiquitin, where a repacking of the three-helix bundle has been proposed to occur (17). In light of our data, such a drastic conformational change does not appear to occur. Rather, our structural determination revealed that both the dimer and the ubiquitin-bound form of the p62 UBA domain adopt the same three-helix bundle structure and are almost superimposable. This binding mode is typical of other canonical UBA domains. Furthermore, because of the small magnitude of chemical shift differences (<0.4 pm) between the ubiquitin-bound and ubiquitin-free monomeric forms of the p62 UBA domain, the structure of the p62 UBA domain in its free monomeric state is most likely similar to that of the ubiquitin-bound state. The pattern of chemical shift differences between the monomer and the complex indicates that helices-1 and -3 are involved in ubiquitin binding, whereas helix-2 does not appear to interact with ubiquitin (Fig. 3B). This result is consistent with complex structures between other UBA domains and ubiquitin, in which helix-1 and helix-3 form major interfaces with ubiquitin (38, 40). Thus, our data indicate that the p62 UBA domain in its monomeric state possesses the same fold as the canonical UBA domains and binds to ubiquitin in the conserved manner. This conclusion is also supported by our RDC data collected for backbone amide groups of p62 UBA in complex with ubiquitin (Fig. 4E). The RDC data showed good agreement with the crystal structure of the UBA dimer in the free form (Q-factor = 0.14). This result demonstrated that the global fold of p62 UBA does not change upon the complex formation, and it essentially keeps the canonical UBA fold.
Full-length p62
p62 possesses a PB1 domain that is a ubiquitin-like (UbL) domain at its N terminus. Members of a class of proteins called UbL-UBA proteins have both an N-terminal UbL and a C-terminal UBA domain. Some of these proteins can adopt an autoinhibited form in which the N-terminal UbL domain noncovalently interacts with the C-terminal UBA domain (41). Thus, intramolecular interaction between the PB1 domain and the UBA domain in full-length p62 is possible. However, we observed no interaction between the isolated PB1 domain and the isolated UBA domain of p62 (supplemental Fig. S7), suggesting that such an intramolecular interaction is unlikely to occur.
It has been shown that C-terminal UBA domains of UbL-UBA proteins protect these proteins from proteasomal destruction so that these shuttle proteins can be reused after delivering polyubiquitylated proteins to the proteasome (42). Intriguingly, proteasomal degradation of green fluorescent protein (GFP)-fused p62 was promoted when the C-terminal eight residues of its UBA domain were deleted (ΔC8 in supplemental Fig. S8). Because p62 UBA lacking these residues is unable to form dimers (supplemental Fig. S4), the UBA dimerization may have some role in regulating the lifetime of p62 in cells. For example, the protein might be isolated from the ubiquitin-proteasomal system when it is in the dimer form and awaiting a signal that breaks the dimer for taking part in the system.
Conclusion
Our study demonstrated that dimerization and ubiquitin binding of the p62 UBA domain are mutually exclusive due to the potential steric clash between the C-terminal tail of p62 UBA and ubiquitin. This feature provides the domain with an autoinhibitory mechanism for ubiquitin binding, which may endow the domain with a regulatory process for its function, although self-oligomerization caused by the N-terminal PB1 domain of p62 may complicate matters. The biological relevance of this autoinhibitory mechanism of the p62 UBA domain needs to be clarified in future studies.
Supplementary Material
Acknowledgments
We thank Dr. A. Ojida and Dr. I. Hamachi for their technical help in performing ITC experiments.
This work was supported by grants-in-aid for scientific research and the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Science, and Culture of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8, Table S1, and “Methods”.
The atomic coordinates and structure factors (codes 3B0F and 2RRU) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- UBA
- ubiquitin-associated domain
- ITC
- isothermal titration calorimetry
- r.m.s.d.
- root mean square displacement
- RDC
- residual dipolar coupling
- HSQC
- heteronuclear single quantum coherence
- UbL
- ubiquitin-like
- CSP
- chemical shift perturbation.
REFERENCES
- 1. Goldberg A. L. (2003) Nature 426, 895–899 [DOI] [PubMed] [Google Scholar]
- 2. Komatsu M., Waguri S., Koike M., Sou Y. S., Ueno T., Hara T., Mizushima N., Iwata J., Ezaki J., Murata S., Hamazaki J., Nishito Y., Iemura S., Natsume T., Yanagawa T., Uwayama J., Warabi E., Yoshida H., Ishii T., Kobayashi A., Yamamoto M., Yue Z., Uchiyama Y., Kominami E., Tanaka K. (2007) Cell 131, 1149–1163 [DOI] [PubMed] [Google Scholar]
- 3. Kirkin V., McEwan D. G., Novak I., Dikic I. (2009) Mol. Cell 34, 259–269 [DOI] [PubMed] [Google Scholar]
- 4. Komatsu M., Ichimura Y. (2010) FEBS Lett. 584, 1374–1378 [DOI] [PubMed] [Google Scholar]
- 5. Moscat J., Diaz-Meco M. T. (2009) Cell 137, 1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim P. K., Hailey D. W., Mullen R. T., Lippincott-Schwartz J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20567–20574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 8. Pohl C., Jentsch S. (2009) Nat. Cell Biol. 11, 65–70 [DOI] [PubMed] [Google Scholar]
- 9. Yoshikawa Y., Ogawa M., Hain T., Yoshida M., Fukumatsu M., Kim M., Mimuro H., Nakagawa I., Yanagawa T., Ishii T., Kakizuka A., Sztul E., Chakraborty T., Sasakawa C. (2009) Nat. Cell Biol. 11, 1233–1240 [DOI] [PubMed] [Google Scholar]
- 10. Zheng Y. T., Shahnazari S., Brech A., Lamark T., Johansen T., Brumell J. H. (2009) J. Immunol. 183, 5909–5916 [DOI] [PubMed] [Google Scholar]
- 11. Wooten M. W., Geetha T., Babu J. R., Seibenhener M. L., Peng J., Cox N., Diaz-Meco M. T., Moscat J. (2008) J. Biol. Chem. 283, 6783–6789 [DOI] [PubMed] [Google Scholar]
- 12. Nezis I. P., Simonsen A., Sagona A. P., Finley K., Gaumer S., Contamine D., Rusten T. E., Stenmark H., Brech A. (2008) J. Cell Biol. 180, 1065–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 15. Ichimura Y., Kumanomidou T., Sou Y. S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., Komatsu M. (2008) J. Biol. Chem. 283, 22847–22857 [DOI] [PubMed] [Google Scholar]
- 16. Noda N. N., Kumeta H., Nakatogawa H., Satoo K., Adachi W., Ishii J., Fujioka Y., Ohsumi Y., Inagaki F. (2008) Genes Cells 13, 1211–1218 [DOI] [PubMed] [Google Scholar]
- 17. Long J., Gallagher T. R., Cavey J. R., Sheppard P. W., Ralston S. H., Layfield R., Searle M. S. (2008) J. Biol. Chem. 283, 5427–5440 [DOI] [PubMed] [Google Scholar]
- 18. Tan J. M., Wong E. S., Dawson V. L., Dawson T. M., Lim K. L. (2008) Autophagy 4, 251–253 [Google Scholar]
- 19. Prag G., Misra S., Jones E. A., Ghirlando R., Davies B. A., Horazdovsky B. F., Hurley J. H. (2003) Cell 113, 609–620 [DOI] [PubMed] [Google Scholar]
- 20. Peschard P., Kozlov G., Lin T., Mirza I. A., Berghuis A. M., Lipkowitz S., Park M., Gehring K. (2007) Mol. Cell 27, 474–485 [DOI] [PubMed] [Google Scholar]
- 21. Trempe J. F., Brown N. R., Lowe E. D., Gordon C., Campbell I. D., Noble M. E., Endicott J. A. (2005) EMBO J. 24, 3178–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Otwinowski Z., Minor W. (1997) Macromol. Crystallogr. A 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 23. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 25. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 26. Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 247–255 [DOI] [PubMed] [Google Scholar]
- 27. Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 28. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 29. Rochus K. (2004) The Computer Aided Resonance Assignment Tutorial, Cantina Verlag, Goldau, Switzerland [Google Scholar]
- 30. Güntert P. (2004) Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 31. Herrmann T., Güntert P., Wüthrich K. (2002) J. Biomol. NMR 24, 171–189 [DOI] [PubMed] [Google Scholar]
- 32. Herrmann T., Güntert P., Wüthrich K. (2002) J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 33. Chou J. J., Gaemers S., Howder B., Louis J. M., Bax A. (2001) J. Biomol. NMR 21, 377–382 [DOI] [PubMed] [Google Scholar]
- 34. Wang Y. X., Marquardt J. L., Wingfield P., Stahl S. J., Lee-Huang S., Torchia D., Bax A. (1998) J. Am. Chem. Soc. 120, 7385–7386 [Google Scholar]
- 35. Valafar H., Prestegard J. H. (2004) J. Magn. Reson. 167, 228–241 [DOI] [PubMed] [Google Scholar]
- 36. Wallace A. C., Laskowski R. A., Thornton J. M. (1995) Protein Eng. 8, 127–134 [DOI] [PubMed] [Google Scholar]
- 37. Hurley J. H., Lee S., Prag G. (2006) Biochem. J. 399, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang D., Raasi S., Fushman D. (2008) J. Mol. Biol. 377, 162–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Long J., Garner T. P., Pandya M. J., Craven C. J., Chen P., Shaw B., Williamson M. P., Layfield R., Searle M. S. (2010) J. Mol. Biol. 396, 178–194 [DOI] [PubMed] [Google Scholar]
- 40. Ohno A., Jee J., Fujiwara K., Tenno T., Goda N., Tochio H., Kobayashi H., Hiroaki H., Shirakawa M. (2005) Structure 13, 521–532 [DOI] [PubMed] [Google Scholar]
- 41. Su V., Lau A. F. (2009) Cell. Mol. Life Sci. 66, 2819–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Heinen C., Acs K., Hoogstraten D., Dantuma N. P. (2011) Nat. Commun. 2, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.