Abstract
Carma1, a caspase recruitment domain-containing membrane-associated guanylate kinase, initiates a unique signaling cascade via Bcl10 and Malt1 in NK cells. Carma1 deficiency results in reduced phosphorylation of JNK1/2 and activation of NF-κB that lead to impaired NK cell-mediated cytotoxicity and cytokine production. However, the precise identities of the downstream signaling molecules that link Carma1 to these effector functions were not defined. Here we show that transforming growth factor-β (TGF-β)-activated kinase 1 (TAK1) is abundantly present in NK cells, and activation via NKG2D results in its phosphorylation. Lack of Carma1 considerably reduced TAK1 phosphorylation, demonstrating the dependence of TAK1 on Carma1 in NKG2D-mediated NK cell activations. Pharmacological inhibitor to TAK1 significantly reduced NK-mediated cytotoxicity and its potential to generate IFN-γ, GM-CSF, MIP-1α, MIP-1β, and RANTES. Conditional in vivo knockdown of TAK1 in NK cells from Mx1Cre+TAK1fx/fx mice resulted in impaired NKG2D-mediated cytotoxicity and cytokine/chemokine production. Inhibition or conditional knockdown of TAK1 severely impaired the NKG2D-mediated phosphorylation of ERK1/2 and JNK1/2 and activation of NF-κB and AP1. Our results show that TAK1 links Carma1 to NK cell-mediated effector functions.
Keywords: Cytokines Induction, Immunology, Jun N-terminal Kinase (JNK), Receptors, Signal Transduction, Carma1, IFN-χ, NK Cells, NKG2D, TAK1
Introduction
TCR-, BCR-, and NKG2D-mediated stimulation triggers the activation and nuclear translocation of NF-κB and JNK transcription factors. Several genes that are regulated by NF-κB and JNK are crucial for lymphocyte development, proliferation, survival, migration, and effector functions (1). Carma1 is an adaptor protein that contains an N-terminal caspase recruitment domain and a C-terminal serine/threonine-rich domain. Carma1 is critical for antigen receptor-induced nuclear translocation of NF-κB via ubiquitination-mediated activation of the IκB kinase (IKK)4 complex (2–4). Carma1-deficient (Carma1−/−) mice displayed severely impaired NF-κB activation, T-cell development, IL-2 production, and TCR-induced proliferation (5, 6). Carma1 deficiency resulted in a significant reduction in follicular, marginal zone, and peritoneal B1 B cells (6). In addition, Carma1 deficiency impaired BCR-induced NF-κB-dependent proliferation of B cells and led to a reduction in the basal levels of serum immunoglobulins (5, 6). Carma1−/− mice failed to generate antibody responses to the T cell-dependent and -independent antigens (7).
Recently, Carma1 deficiency has been shown to affect NK-mediated cytokine generation (8, 9). However, a mechanistic understanding of how Carma1 transduces the receptor-mediated signaling into NK-mediated effector functions has yet to be determined. Multiple downstream effector molecules for Carma1 have been defined. TAK1 plays a critical role in the induction of transcription factors, NF-κB and AP1. TAK1 is a mitogen-activated protein kinase kinase kinase (MAPKKK) (10). Irrespective of these progresses, the role of TAK1 in regulating effector functions and its dependence on Carma1 in NK cells has yet to be defined.
In this study, we show that Carma1−/− NK cells were less effective in lysing tumor cells and have significantly impaired NCR1, CD244, NKG2D, Ly49D, and NK1.1-mediated cytokine and chemokine production. TAK1 phosphorylation was considerably reduced in the absence of Carma1. Pharmacological inhibition of TAK1 resulted in reduction of cytotoxicity and cytokine/chemokine production in NK cells. Conditional knockdown of TAK1 significantly impaired NKG2D-mediated cytokine and chemokine production in NK cells. Additionally, knockdown of TAK1 moderately reduced NKG2D-mediated tumor lysis by NK cells. Collectively, our results provide important insights into TAK1-regulated signaling events in NK cells.
MATERIALS AND METHODS
Mice and Cell Lines
Carma1−/− and WT mice have been backcrossed to C57BL/6 for eleven generations and were described earlier (5). Interferon-inducible TAK1 knockdown mice were generated (11) by crossing Mx1Cre mice (Jackson Laboratory, Bar Harbor, MN) with TAK1fx/fx mice (12) and backcrossing the resultant offspring. All mice used in this study were maintained in pathogen-free conditions at the Biological Resource Center at the Medical College of Wisconsin (Milwaukee, WI) or at the Loyola University Medical Center (Maywood, IL) and were used between 6 and 12 weeks of age. All the animal protocols used were approved by the animal facilities of the respective institutions. Interferon-inducible TAK1 knockdown Mx1Cre+TAK1fx/fx mice and control TAK1fx/fx mice were injected with 5 μg/g body weight of poly(I:C) on days 1 and 3 to induce TAK1 knockdown. Spleens of these treated mice were collected on day 4 (11). EL4, EL4H60-Low, EL4H60-High, RMA, RMA/S, and YAC1 cells and their culture conditions were as described previously (13, 14).
NK Cell Preparation
NK cells were purified as previously described (15). Briefly, single cell suspensions from different organs were passed through nylon wool columns to deplete adherent populations consisting of B cells and macrophages. Cells non-adherent to nylon wool were cultured with 1000 units/ml IL-2 (NCI-BRB-Preclinical Repository, Maryland, MD). The purity of the NK cultures was checked, and preparations with >90% of NK1.1+ cells were used.
Flow Cytometry
Single cell preparations were stained with fluorescent-labeled mAbs as described before (13). Antibodies for NK1.1 (PK136), CD3ϵ (145–2C11), NKG2D (A10), anti-CD244 (244F4), and anti-granzyme B (16G6) were obtained from e-Bioscience (San Diego, CA). Anti-H60a (205326) was obtained from R&D Systems (Minneapolis, MN). Anti-Ly49D was obtained from BD Pharmingen (San Jose, CA). An anti-NK1.1-secreting hybridoma clone (PK136) was obtained from ATCC and used. NK cells were stained in 1% FCS-PBS with appropriate antibodies (13). One million events were analyzed for each sample. Standard flow cytometric analyses were performed in LSR-II and analyzed with FACSDiva software (BD Biosciences).
NK Cell Effector Functions following Poly(I:C)-mediated Activation in Vivo
Poly(I:C)-mediated activation of NK cells in vivo. Wild-type and Carma1−/− mice were injected with Poly(I:C) (250 μg) or PBS, intravenously. After 18 h, a single cell suspension of the spleen was prepared. Splenocytes were co-cultured with EL4, EL4H60-High, or YAC1 at the rate of 1 million effector cells to 0.5 million target cells. Intracellular IFN-γ staining was performed after 12 h of co-culture. CD107a surface staining was performed after 4 h of co-culture. CD107a surface expression was also determined following plate-bound anti-NKG2D antibody-mediated activation.
Cytotoxicity Assays
NK-mediated cytotoxicity was quantified using chromium-51 (51Cr)-labeled target cells (16) at varied effector to target (E:T) ratios. Percent specific lysis was calculated using amounts of absolute, spontaneous, and experimental 51Cr release from target cells.
Quantification of Cytokines, Chemokines, and LAMP1 Expression
IL-2-cultured, Fc-blocked NK cells were activated with 5 μg/ml plate-bound mitogenic antibodies, and their culture supernatants were analyzed in a Bioplex assay (Bio-Rad). Intracellular IFN-γ was quantified as previously described (17). Briefly, Fc-blocked NK cells were activated with plate-bound mAbs. After 16 h cells were stained for surface NK1.1, fixed, permeabilized, and stained for intracellular IFN-γ using phycoerythrin-conjugated anti-IFN-γ mAb. For inhibitor assays, NK cells were incubated for 1 h with varying concentrations of 5Z-7-oxo-zeaenol, washed, and added into anti-NKG2D mAb-coated plates. After 18 h, cytokines and chemokines were quantified. IL-2-cultured NK cells were treated with IL-12, IL-18, or both for 18 h, and the supernatants were analyzed for the presence of indicated cytokines and chemokines. For quantifying IFN-γ-encoding mRNA, NK cells were activated for 6 h and harvested. RNA was extracted using an RNeasy Mini Kit (Qiagen, Valencia, CA). Real-time PCR was performed by using a previously published SYBR green protocol with an ABI7900 HT thermal cycler (18, 19). For quantifying granule release, NK cells were activated with an anti-NKG2D mAb and cultured in the presence of monensin (1:1000) and Alexa Fluor 488-conjugated anti-CD107a for 4 h. Cells were washed and stained for surface CD3 and NK1.1. CD107a (LAMP1) surface expression was analyzed by flow cytometry.
Western Blotting
10 μg of whole cell lysate was resolved using 10% SDS-PAGE gels, transferred to PVDF membranes, and probed with indicated antibodies (20). Antibodies against TAK1 (Upstate, Lake Placid, NY), phospho-TAK1 (Thr-184/187), total JNK1/2 (clone 56G8), phospho-JNK 1/2 (Thr-183/Tyr-185, clone 98-F2), total ERK1/2 and phospho-ERK1/2 (Thr-202/Tyr-204, clone D13.14.4E), total p38 and phospho-p38 (Thr-180/Tyr-182, clone 3D7, Cell Signaling, Boston, MA), and anti-actin (Roche Applied Science) were used, and signals were detected using an ECL kit (GE Healthcare, Piscataway, NJ). The -fold changes in the MAPK phosphorylation following NKG2D-mediated activation were calculated and compared. The band intensity of phospho-protein was normalized against the respective total protein. The -fold change in phosphorylation following 5, 20, or 60 min of activation was calculated using these normalized values by comparing it to that of unstimulated lanes.
Confocal Microscopy
NK cells were activated with plate-bound anti-NKG2D mAbs for 30 min at 37 °C. Activated NK cells were harvested and seeded in poly-l-lysine-coated chamber slides. Unstimulated NK cells were used as controls. After 1-h incubation at room temperature, cells were stained with Hoechst 33342. The cells were washed and fixed with ice-cold methanol and permeabilized with 0.1% Triton X-100 in 3% BSA. After blocking with 70% normal goat serum, cells were incubated overnight with anti-phospho JNK1/2 (Thr-183/Tyr-185, clone 98-F2), anti-phospho ERK1/2 (Thr-202/Tyr-204, clone D13.14.4E), total JNK1/2 (Clone 56G8), and total ERK1/2 antibodies (Cell Signaling Technology, Danvers, MA). The cells were washed and incubated with donkey anti-rabbit Alexa Fluor 488 antibody (Invitrogen) for 1 h. Phosphorylation of JNK1/2 and ERK1/2 were analyzed using confocal microscopy.
Electrophoretic Mobility Shift Assays
IL-2-cultured NK cells were activated with plate-bound anti-NKG2D mAb (A10). NK cells (1 × 106) were lysed, and nuclear protein extracts were generated using NE-PER reagent (Pierce). 2.5 μg of the nuclear protein extract was used in a binding reaction with 1× binding buffer, 2.5% glycerol, 5 mm MgCl2, 50 ng/μl of poly(dI:dC), and 0.05% Nonidet P-40 (LightShift EMSA optimization and control kit, Pierce). Biotin end-labeled duplex oligonucleotide probes for NF-κB (forward, 5′-AGT TGA GGG GAC TTT CCC AGG C/3Bio-3′ and reverse, 5′-GCC TGG GAA AGT CCC CTC AACT/3Bio/-3′ were added to the reaction mix at a final concentration of 2.0 ng/20-μl reaction mix. After 20 min, 5 μl of 5× loading buffer was added to the reaction mix, and the samples were resolved in a 6% polyacrylamide gel and transferred to nylon membrane. The protein-DNA probe complexes were cross-linked with a UV lamp (254 nm, 120 mJ/cm2). NF-κB-specific bands were detected using streptavidin-horseradish peroxidase conjugate in a chemiluminescence assay (LightShift chemiluminescent EMSA kit, Pierce). A similar assay was performed to detect the transcription factor, AP1 using biotin end-labeled duplex oligonucleotide probes (forward, 5′-/5Biosg/CGC TTG ATG ACT CAG CCG GAA-3′ and reverse, 5′-/5Biosg/TTC CGG CTG AGT CAT CAA GCG-3′).
Statistical Analyses
Statistical analyses were performed by using a two-tailed, unpaired, Student's t test, and p values of ≤0.05 were considered significant.
RESULTS
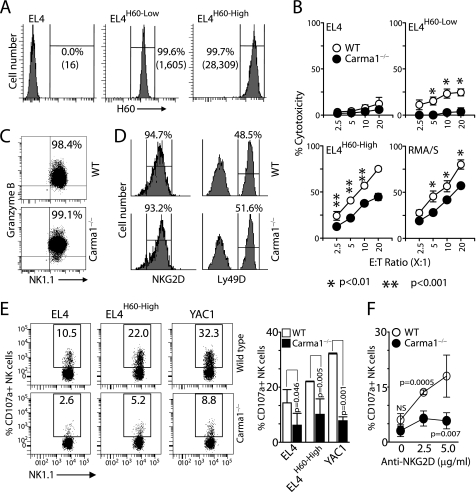
Lack of Carma1 Moderately Reduces the Natural Cytotoxicity of NK Cells
Carma1 expression is critical for antigen receptor-mediated signaling in T and B cells (20, 21). NKG2D is ubiquitously expressed on NK cells, and the activation through NKG2D results in cytotoxicity against ligand-expressing target cells (22). Earlier studies from us and others have shown that ectopic expression of H60 on tumor cells renders them susceptible to NKG2D-mediated cytotoxicity (13, 23, 24). To assess the ability of Carma1−/− NK cells in mediating cytotoxicity through NKG2D receptor, we tested EL4 cell lines that stably express H60 (EL4H60) in 51Cr-release assays (13). These stable cell lines express either physiological (EL4H60-Low) or pathological (EL4H60-High) (Fig. 1A) levels of H60 (25). Parental EL4 cells were used as negative controls. We tested IL-2-cultured splenic NK cells in cytotoxicity assays. Carma1−/− NK cells consistently showed reduced abilities to lyse both EL4H60-Low and EL4H60-High compared with WT (Fig. 1B). Cells that lack or have reduced expression of self MHC Class I molecules are susceptible to NK cell-mediated cytotoxicity (26). However, the signaling events that positively regulate this cytotoxicity are not fully understood. To test whether Carma1 plays a role in “missing-self” recognition, we measured the cytotoxic potential of IL-2-activated splenic NK cells against RMA/S, which expresses significantly lower levels of MHC Class I and its parental RMA tumor cells. Similar to EL4H60, RMA/S cells were also lysed with lower efficiency by Carma1−/− NK cells compared with that of WT (Fig. 1B). A moderate but significant reduction in cytotoxicity could have been due to a defect in the Carma1-mediated signaling pathway or due to a reduction in granzyme B. To exclude the later, we analyzed the level of granzyme B in WT and Carma1−/− NK cells by flow cytometry. Results presented in Fig. 1C indicate comparable levels of granzyme B between the WT and the knock-out-derived NK cells. Thus, we conclude that the moderate but significant defect in cytotoxicity in Carma1−/− NK cells is due to a failure in the successful mobilization of cytotoxic granules. The impaired cytotoxicity was not due to a reduction in NKG2D expression levels, because it was comparable between WT and Carma1−/− NK cells (Fig. 1D). To further confirm this defect in cytotoxicity, we challenged the WT and Carma1−/− mice with double-stranded, RNA-like polyinosine-polycytidylic acid polymer (poly(I:C)) that has been demonstrated to induce the production of IFN-α and IFN-β (27). Total splenic cells from the poly(I:C) or PBS-treated mice were co-cultured with EL4, EL4-H60High, or YAC-1 target cells for 4 h. We used the cell surface expression of CD107a as a measure of cytotoxicity in CD3−NK1.1+ NK cells. Results show a significant reduction in the levels of CD107a expression in Carma1−/− NK cells compared with that of WT (Fig. 1E). In addition, we also stimulated the splenic NK cells with plate-bound anti-NKG2D mAb and further confirmed the requirement of Carma1 in NK cell-mediated cytotoxicity (Fig. 1F). The increase in CD107a expression in NK cells following co-culture of splenocytes from PBS-treated mice was negligible compared with poly(I:C)-treated mice (data not shown).
FIGURE 1.
NK cell-mediated cytotoxicity is impaired in Carma1−/− NK cells. (A) Levels of H60 expression in stable EL4 transfectants as tested by anti-H60 antibody. (B) IL-2-activated splenic NK cells were tested against 51Cr-labeled EL4, EL4H60-Low, EL4H60-High and RMA/S target cells at the indicated E:T ratios. (C) Expression of granzyme B in IL-2-activated WT and Carma1−/− NK cells. NK cells were stained for NK1.1 and CD3, fixed, permeabilized and stained for intracellular granzyme B. CD3−NK1.1+ cells were gated and shown. (D) Expression levels of NKG2D or Ly49D activation receptors in WT and Carma1−/− NK cells. (E) Ex vivo NK cell cytotoxicity against EL4, EL4H60-High and YAC1 after in vivo poly(I:C) treatment was measured by surface staining for CD107a (LAMP1) in CD3−NK1.1+ cells following co-culture with the target cells. Mean ± S.D. of the percentage CD107a+ NK cells are shown. (F) Surface expression of CD107a in anti-NKG2D mAb activated NK cells following in vivo poly(I:C) treatment. Mean ± S.D. of the percentage CD107a+ NK cells are shown. Data shown in A-D are representative or averages of 6–8 mice in each category and representatives of three independent experiments. Data shown in E and F are representative of two independent experiments.
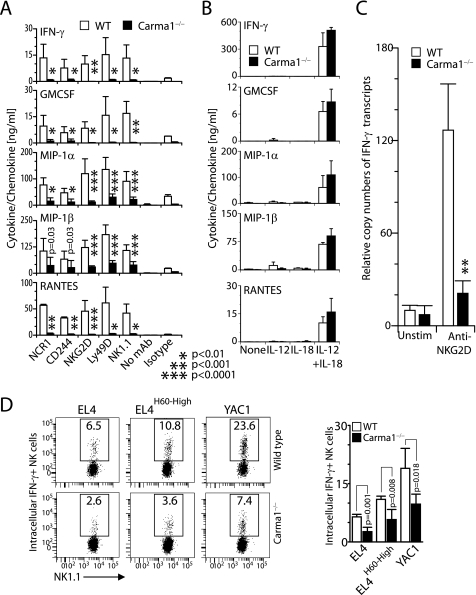
Lack of Carma1 Significantly Impairs Cytokine and Chemokine Production in NK Cells
Gene transcription and production of inflammatory cytokines are regulated by NF-κB and c-Jun/AP1 transcription factors. As part of their innate immune response, NK cells generate substantial quantities of cytokines such as IFN-γ, GM-CSF, and chemokines MIP-1α, MIP-1β, and RANTES (28). To determine the role of Carma1 in the generation of cytokines and chemokines, IL-2-cultured splenic NK cells were activated with titrated concentrations of plate-bound anti-NCR1 (goat polyclonal), anti-CD244 (2B4), anti-NKG2D (A10), anti-Ly49D (4E5), or anti-NK1.1 (PK136) mAbs. Supernatants were collected, and the levels of IFN-γ, GM-CSF, MIP-1α, MIP-1β, and RANTES were measured by using Bioplex assays. WT NK cells produced large amounts of IFN-γ and GM-CSF (Fig. 2A). In contrast, Carma1−/− NK cells were significantly impaired in their ability to produce IFN-γ and GM-CSF (Fig. 2A). Similar to these results, Carma1−/− NK cells also produced significantly lower amounts of chemokines, MIP-1α, MIP-1β, and RANTES compared with the WT. Thus, Carma1 is a critical regulator of signaling events that are initiated via NCR1·CD3, CD244·SAP, NKG2D·DAP12, Ly49D·DAP12, or NK1.1·FcRγ complexes. Activation by the mitogenic antibodies were specific, because neither no stimulation nor respective isotype antibody controls failed to elicit significant levels of these cytokines or chemokines (Fig. 2A). To determine whether this is a generalized hypo-responsiveness or exclusive defect associated with Immuno Tyrosine-based Activation Motif-containing activation receptors, we stimulated NK cells with IL-12, IL-18, or both. IL-12- and IL-18-mediated activation utilized the JAK/STAT pathway and did not involve Carma1 for downstream signaling. WT and Carma1−/−-derived NK cells responded equally well to IL-12- and IL-18-mediated activation (Fig. 2B). This demonstrated that Carma1−/− NK cells are fully capable of responding through their cytokine receptors. The substantial reduction could be due to the inabilities of the Carma1−/− NK cells to produce cytokines or due to a defect in cytokine secretion. To distinguish between these two possibilities, we quantified the amounts of IFN-γ-encoding mRNA before and after plate-bound anti-NKG2D mAb activation. Fig. 2C shows a significantly less copy number of IFN-γ-encoding mRNA in spleen-derived Carma1−/− NK cells. One of the potential reasons for the defect in the production of cytokine or chemokine from Carma1−/− NK cells may be a lower responsiveness to IL-2 during the in vitro culture. To further analyze this possibility, we co-cultured the total splenic cells from poly(I:C)-treated mice with EL4, EL4-H60High, or YAC-1 target cells for 12 h and quantified the levels of intracellular IFN-γ in CD3−NK1.1+ NK cells. Results presented in Fig. 2D show a significant reduction in the percentages of IFN-γ NK cells in Carma1−/− compared with that of WT. Thus, we conclude that Carma1 plays an obligatory role in the transcription of cytokine and chemokine genes, and its absence significantly reduces the ability of NK cells to produce these soluble mediators.
FIGURE 2.
Quantification of IFN-γ, GM-CSF, MIP-1α, MIP-1β and RANTES in culture supernatants following (A) receptor- or (B) IL-12- and IL-18-mediated activations, in Bioplex assays. Activation with isotype antibodies was performed and one representative control (Hamster IgG1) is shown. (C) IFN-γ-encoding transcripts in anti-NKG2D mAb-stimulated NK cells. Data shown in A-C are representative or averages ± S.D. of 6–8 mice in each category and representatives of three independent experiments. (D) Ex vivo IFN-γ production in NK cells from mice treated with poly(I:C) were measured after co-culturing with EL4, EL4H60-High and YAC1 cells. Cytokine production was measured by intracellular staining for IFN-γ in CD3−NK1.1+ cells. Data shown in D is representative of two independent experiments. Mean ± S.D. of the percentage IFN-γ+ NK cells are shown.
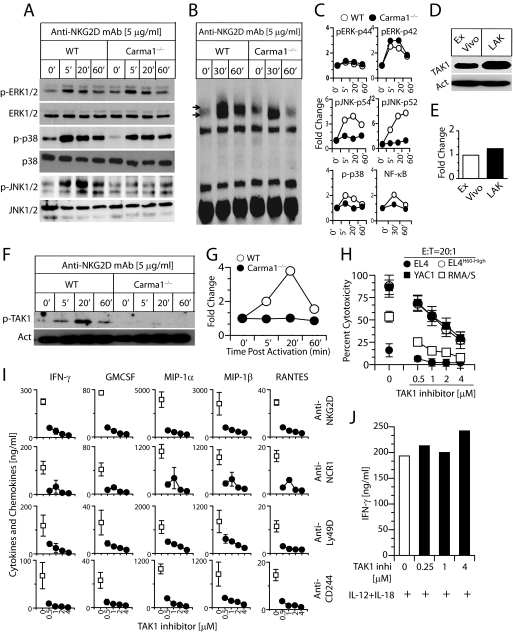
Lack of Carma1 Reduces NKG2D-mediated TAK1 Phosphorylation, Which Leads to Impaired JNK1/2 and NF-κB Activation
Earlier studies have shown that activation of MAPK is required for cytotoxic granule release and cytokine production in murine and human NK cells (29–32). To determine the role of Carma1 in NKG2D-mediated activation, we quantified the levels of MAPK phosphorylation and NF-κB activation in NK cells. Phosphorylation levels of ERK1/2 and p38 were comparable between WT and Carma1−/− NK cells (Fig. 3, A and C). However, the level of JNK1/2 phosphorylation was considerably reduced in Carma1−/− NK cells (Fig. 3, A and C). Cells were harvested at different time points of activation, and their nuclear protein extracts were analyzed for the level of NF-κB activation in electrophoretic mobility shift assays (EMSAs). After anti-NKG2D mAb-mediated activation, NF-κB was abundantly translocated into the nucleus in the WT but not in the Carma1−/− NK cells (Fig. 3, B and C).
FIGURE 3.
TAK1 is a critical intermediate in Carma1-mediated NK cell activation and function. IL-2-activated splenic NK cells were derived from WT and Carma1−/− mice. These NK cells were activated with plate-bound anti-NKG2D and analyzed for various signaling events downstream of NKG2D. (A) Phosphorylations of ERK1/2, JNK1/2 and p38. (B) Quantification of NF-κB in the nuclear extract after NKG2D-mediated activation. (C) Fold change in MAP kinase phosphorylation and nuclear translocation of NF-κB were compared between WT and Carma1−/− NK cells. (D) TAK1 protein expression in ex vivo and IL-2-cultured NK cells. (E) Fold change in TAK1 expression levels in ex vivo and IL-2-cultured NK cells. TAK1 expression was normalized with actin expression. (F) Phosphorylation of TAK1 after NKG2D-mediated activation. (G) Fold change in TAK1 phosphorylation were compared between WT and Carma1−/− NK cells. One representative experiment out of 3 is shown in A-G. (H) NK-mediated cytotoxicity after 5Z-7-oxo-zeaenol treatment. Wild type NK cells were treated with 5Z-7-oxo-zeaenol that specifically inhibits the kinase activity of TAK1. (I) Cytokine and chemokine production in WT NK cells that were pretreated with TAK1 inhibitor. (J) IFN-γ production following IL-12- and IL-18-mediated activation in WT NK cells pretreated with TAK1 inhibitor. Data presented are averages obtained from 3 mice and a representative of three independent experiments.
TAK1 is a member of the MAPKKK family and a key modulator of NF-κB and JNK that regulates pro-inflammatory signaling in multiple immune cell types (33). TAK1 regulates the activation of p38 and JNK1/2 through the phosphorylation of MKK3/6 (34) and MKK4 (35), respectively. Activation of c-Jun/AP1 is regulated by JNK1/2 (36). Because JNK1/2 is known to play an important role in the cytotoxic granule release, we hypothesized that the lack of Carma1 reduces TAK1 phosphorylation, which could be primarily responsible for the impairment in NK cytotoxicity. Activation and phosphorylation of TAK1 downstream of NKG2D receptor in NK cells is yet to be defined. Both fresh and IL-2-cultured NK cells contained ample amounts of TAK1 (Fig. 3, D and E). Earlier studies have shown that phosphorylation of Carma1 in the PRD domain by PKCβ and PKCθ allows Carma1 to form a signalosome with Bcl10, MALT1, TRAF2, TAK1, and IKKγ (NEMO) (37–39). Other studies have indicated that TAK1 can be activated via TRAF2 or TRAF6, independent of Carma1 (40). TAK1 also regulates the NF-κB pathway through its interaction with TRAF6 and by phosphorylating the NF-κB-inducing kinase (41). Autophosphorylation at threonines 184 and 187 with the help of TAB1 is critical for the kinase function of TAK1. To determine the role of Carma1 in TAK1 activation, we stimulated IL-2-cultured NK cells from WT and Carma1−/− mice with plate-bound anti-NKG2D mAb. Fig. 3 (F and G) indicates abundant phosphorylation of TAK1 in the WT NK cells, which peaked at 20 min after activation. However, its phosphorylation in Carma1−/− NK cells was considerably reduced. Thus, our results indicate that TAK1 is a critical link between Carma1 and JNK1/2 (c-Jun/AP1) or NF-κB activations downstream of NKG2D in NK cells.
Inhibition of the Kinase Activity of TAK1 Reduces NK Cell Effector Functions
To establish a functional role of TAK1 in NK cells, we used a pharmacological compound to inhibit the kinase activity of TAK1. IL-2-cultured NK cells were pretreated with titrating concentrations of a naturally occurring fungal resorcylic acid lactone, 5Z-7-oxo-zeaenol for 1 h at 37 °C (42). 5Z-7-Oxo-zeaenol specifically inhibits the catalytic activity of TAK1 by interacting with the ATP-binding site. Pretreated NK cells were tested for their ability to mediate cytotoxicity against EL4, EL4H60-High, RMA/S, and YAC1 cells. The E:T ratio was kept at 20:1. Our results presented in Fig. 3H demonstrate that inhibition of TAK1 activation with 5Z-7-oxo-zeaenol significantly affected the ability of NK cells to lyse EL4H60-High, YAC-1, and RMA/S target cells. Thus, our results implicate TAK1 in the cytotoxic granule release by NK cells. Next, we analyzed the role of TAK1 in NK cell-mediated cytokine or chemokine production. 5Z-7-Oxo-zeaenol-pretreated WT NK cells were stimulated with plate-bound mAbs to NKG2D, NCR1, Ly49D, and CD244. NKG2D, NCR1, and CD244 are ubiquitously expressed on all murine NK cells. Ly49D is expressed in ∼50% of the NK cells. Pretreatment of NK cells with TAK1 inhibitor significantly reduced their ability to generate cytokines and chemokines (Fig. 3I). 5Z-7-Oxo-zeaenol was tested between 4 and 0.5 μm. Even at the lowest concentrations of the inhibitor, generation of cytokines and chemokines was significantly reduced. Analyses of the viability of 5Z-7-oxo-zeaenol-pretreated NK cells revealed that the highest concentration used (4 μm) in these assays did not cause any detectable levels of cell death. Further, analysis of IFN-γ production in TAK1 inhibitor-pretreated NK cells following IL-12- and IL-18-mediated activation revealed that TAK1 inhibition did not affect the cytokine production, thus precluding the nonspecific effects of this drug (Fig. 3J).
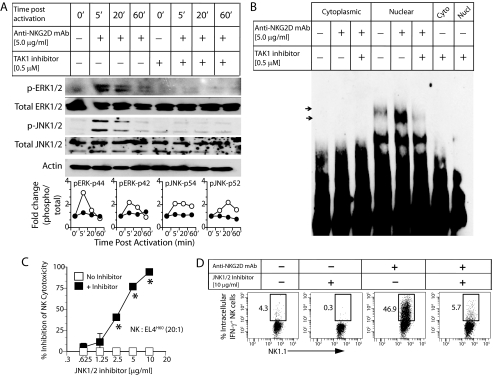
TAK1 Is Essential for NKG2D-mediated ERK1/2, JNK1/2 Phosphorylation, and NF-κB Activation
To further confirm the molecular mechanism by which TAK1 regulates the NK cell effector functions, we analyzed the status of ERK1/2, JNK1/2, and NF-κB activations in 5Z-7-oxo-zeaenol-pretreated (0.5 μm) NK cells. Pretreatment with this inhibitor considerably reduced both the ERK1/2 and JNK1/2 phosphorylation compared with that of untreated NK cells (Fig. 4A). The mechanism by which ERK1/2 phosphorylation is reduced after 5Z-7-oxo-zeaenol treatment in NK cells is not clear and requires additional investigations. Activation of NK cells through NKG2D receptor resulted in the nuclear translocation of NF-κB in the untreated NK cells (Fig. 4B). However, pretreatment with 5Z-7-oxo-zeaenol reduced the activation and nuclear translocation of NF-κB. To precisely determine the role of JNK1/2 in NKG2D-mediated effector functions, NK cells were pretreated with SP600125, a pharmacological inhibitor for JNK1/2, and tested for their ability to kill EL4H60-High tumor cells. JNK1/2 inhibition significantly reduced the cytotoxicity of NK cells against EL4H60-High tumor cells (Fig. 4C) at an E:T ratio of 20:1. Previously, it has been shown that, in addition to ERK1/2, JNK1/2 activation downstream of NKG2D is also critical for NK cell-mediated cytotoxicity (31). Additionally, JNK1/2 inhibitor also reduced the percentage of intracellular IFN-γ+ NK cells following anti-NKG2D-mediated activation (Fig. 4D). Based on these results, we conclude that TAK1 is an essential intermediate downstream of Carma1, and the functional impairment in Carma1−/− NK cells is due to a reduction in the phosphorylation levels of TAK1 that led to a decrease in JNK1/2 and NF-κB activations.
FIGURE 4.
TAK1 regulates signaling events and effector functions downstream of NKG2D in NK cells. WT NK cells were treated with 5Z-7-oxo-zeaenol that specifically inhibits the kinase activity of TAK1. Pretreated and untreated NK cells were activated with plate-bound anti-NKG2D mAb and analyzed for downstream signaling events. (A) Phosphorylation of ERK1/2 and JNK1/2. Fold change in MAP kinase phosphorylation were compared between WT and Carma1−/− NK cells and are shown in the bottom panel. (B) Quantification of NF-κB in the nuclear extract after NKG2D-mediated activation. WT NK cells were treated with SP600125, a specific inhibitor for JNK1/2. Untreated and inhibitor pretreated WT NK cells were analyzed for NKG2D-mediated (C) cytotoxicity and (D) IFN-γ production.
Conditional Knockdown of TAK1 Significantly Reduces NKG2D-mediated Effector Functions
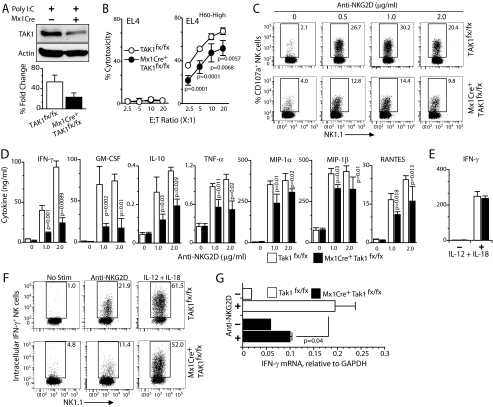
To confirm that TAK1 indeed regulates NKG2D-mediated effector functions via JNK1/2 and NF-κB/AP1, we used a mouse model where TAK1 could be conditionally knocked down. By crossing Mx1Cre mice with TAK1fx/fx mice (12), interferon-inducible TAK1 knockdown mice were generated (11). Splenic NK cells from poly(I:C)-treated TAK1fx/fx or Mx1Cre+TAK1fx/fx mice were cultured with IL-2. As expected, TAK1 expression was considerably reduced in poly(I:C)-treated NK cells obtained from Mx1Cre+TAK1fx/fx compared with TAK1fx/fx mice (Fig. 5A). Functional analyses of TAK1fx/fx and Mx1Cre+TAK1fx/fx NK cells revealed that cytotoxicity against EL4H60-High tumor cells was moderately but significantly reduced in the Mx1Cre+ TAK1fx/fx NK cells (Fig. 5B). Further, analyses of LAMP1 (CD107a) surface expression as an independent measure of cytotoxicity also revealed a reduction in the TAK1-deficient NK cells (Fig. 5C). Based on these observations, we conclude that the NK cell-mediated cytotoxicity can be in part regulated by TAK1.
FIGURE 5.
Conditional knockdown of TAK1 gene impairs NKG2D-mediated NK cell effector functions. A, poly(I:C) treatment induces the conditional knockdown of TAK1 gene in Mx1Cre+TAK1fx/fx- but not in TAK1fx/fx-derived NK cells. IL-2-cultured NK cells were analyzed for TAK1 expression by Western blot. Poly(I:C)-treated IL-2-activated NK cells from TAK1fx/fx and Mx1Cre+TAK1fx/fx were analyzed for their effector functions. B, NK cell-mediated cytotoxicity was tested against 51Cr-labeled EL4 and EL4H60-High target cells at the indicated E:T ratios. C, CD107a (LAMP1) surface expression in NK cells following plate-bound anti-NKG2D mAb-mediated activation. D, cytokine and chemokine production by NK cells following plate-bound anti-NKG2D mAb-mediated activation. E, IL-12- and IL-18-mediated IFN-γ production in TAK1fx/fx- and Mx1Cre+TAK1fx/fx-derived NK cells. F, IFN-γ production by NK cells was determined by intracellular staining. Following anti-NKG2D mAb activation, NK cells were stained for surface expression of CD3 and NK1.1, fixed, and permeabilized. These cells were stained for intracellular IFN-γ, gated for CD3−NK1.1+ cells and analyzed. G, anti-NKG2D-mediated IFN-γ gene transcription in TAK1fx/fx- and Mx1Cre+TAK1fx/fx-derived NK cells. Data presented in A–G are representative of a minimum of three independent experiments.
Next, we analyzed the ability of Mx1Cre+TAK1fx/fx NK cells to produce cytokines and chemokines through NKG2D-mediated activation. Similar to Carma1 deficiency, knockdown of TAK1 significantly reduced the production of IFN-γ, GM-CSF, IL-10, TNF-α, MIP-1α, MIP-1β, and RANTES (Fig. 5D). Our data presented in Fig. 2B indicated that IL-12- and IL-18-mediated cytokine and chemokine production did not require Carma1. To investigate the role of TAK1 in cytokine receptor-mediated activation, we stimulated NK cells from TAK1fx/fx and Mx1Cre+TAK1fx/fx mice with IL-12 and IL-18. Production of IFN-γ (Fig. 5E) was comparable between NK cells from TAK1fx/fx and Mx1Cre+TAK1fx/fx mice. Intracellular staining for IFN-γ revealed a similar pattern where NKG2D- but not IL-12- and IL-18-mediated IFN-γ production was defective in the Mx1Cre+TAK1fx/fx NK cells (Fig. 5F). Analyses of IFN-γ-encoding mRNA levels following NKG2D-mediated activation in TAK1fx/fx and Mx1Cre+TAK1fx/fx NK cells indicated that the reduction occurred at the transcriptional level (Fig. 5G). Collectively, these results demonstrate that TAK1 can regulate both the natural cytotoxicity and cytokine production in NK cells.
Lack of TAK1 Impairs NKG2D-mediated ERK1/2, JNK1/2, NF-κB, and AP1 Activation
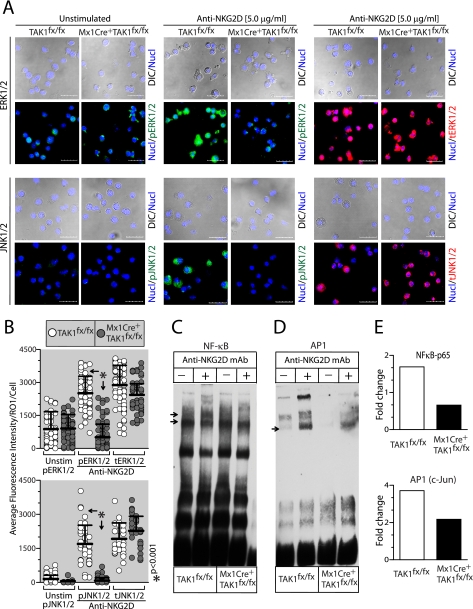
Next, we determined the downstream signaling events regulated by TAK1 in NKG2D-mediated activation. Toward this, we analyzed the ERK1/2 and JNK1/2 phosphorylation in NK cells from TAK1fx/fx and Mx1Cre+TAK1fx/fx. IL-2-cultured NK cells were stimulated with plate-bound anti-NKG2D antibody (5 μg/ml) for 30 min, transferred to chamber slides, and stained for phospho or total proteins. Activated cells were analyzed through confocal microscopy to determine the levels of ERK1/2 and JNK1/2 phosphorylations. Data presented in Fig. 6A demonstrate that activation of TAK1fx/fx mice-derived NK cells resulted in abundant phosphorylation of ERK1/2 and JNK1/2. However, a similar activation of NK cells from Mx1Cre+TAK1fx/fx mice resulted in reduced ERK1/2 or JNK1/2 protein phosphorylations. To obtain statistical validation of these observations, we quantified fluorescence intensity in individual NK cells. A single reference line was drawn across each NK cell, and an average of the fluorescence intensity was calculated and presented in Fig. 6B. These analyses further confirmed a significant reduction in ERK1/2 and JNK1/2 phosphorylation in NK cells from Mx1Cre+TAK1fx/fx mice. This is not due to a reduction in the total ERK1/2 or JNK1/2 proteins, because their fluorescence intensities were comparable between the TAK1fx/fx and Mx1Cre+TAK1fx/fx mice-derived NK cells (Fig. 6, A and B). Next, we analyzed the status of NF-κB and AP1 after NKG2D-mediated activation. Our results show NKG2D-mediated NF-κB activation was considerably reduced in NK cells from Mx1Cre+TAK1fx/fx mice (Fig. 6C). Additionally, the activation of AP1 (Fig. 6D) was also reduced in Mx1Cre+TAK1fx/fx NK cells following NKG2D cross-linking. These results reveal that TAK1 is critical for NKG2D-mediated activation of NF-κB and AP1 in NK cells. Quantification of these activated proteins further confirmed the obligatory role of TAK1 in NKG2D-mediated activation in NK cells (Fig. 6E). We conclude that TAK1 is a critical intermediate between Carma1 and NK-mediated effector functions.
FIGURE 6.
Conditional knockdown of TAK1 impairs signaling downstream of NKG2D receptor in NK cells. Poly(I:C)-treated NK cells from TAK1fx/fx and Mx1Cre+TAK1fx/fx were activated with plate-bound anti-NKG2D mAb and analyzed for signaling events downstream of NKG2D. A, phosphorylation of ERK1/2 and JNK1/2 following anti-NKG2D-mediated activation was analyzed through confocal microscopy. B, average fluorescence intensity of phosphorylated and total ERK1/2 and JNK1/2 in TAK1fx/fx- and Mx1Cre+TAK1fx/fx-derived NK cells were plotted and compared. EMSAs to detect NF-κB (C) and AP1 (D) following anti-NKG2D mAb-mediated activation are shown. E, the -fold changes in the nuclear translocation of NF-κB and AP1 in TAK1fx/fx- and Mx1Cre+TAK1fx/fx-derived NK cells. Data presented in A–E are representative of a minimum of three independent experiments.
DISCUSSION
In this study, we have determined the role of TAK1 in NK-mediated effector functions. Oligomerized Carma1 recruits multiple signaling components, including TAK1, to activate NF-κB and JNK1/2. By acting as both IKK kinase and a JNK kinase kinase, TAK1 links Carma1 to inducible transcription factors (33). TAK1 plays a critical role as a kinase for IKKβ, which leads to IκB phosphorylation (33). Independently, downstream of MKK6, TAK1 can also phosphorylate JNK1/2 leading to AP1 activation (c-Jun/AP1 heterodimer) (43). Evidence for this is provided from the AP1-dependent cytokine gene transcription in FcγRIIIA-stimulated human NK cells (44). In this study, using Carma1−/− and Mx1Cre+TAK1fx/fx mice, we defined that TAK1 is an essential molecular link between Carma1 and NK cell-mediated cytotoxicity and pro-inflammatory cytokine production. Because the development and maturation of NK cells in Carma1−/− mice were normal, we conclude that the functional inabilities were due to impairments in downstream signaling pathways and not due to inappropriate development, differentiation, or homeostasis of NK cells.
NK cells recognize tumor cells through multiple receptor-ligand interactions. Lack of Carma1 or knockdown of TAK1 moderately yet significantly reduced the NK cytotoxicity against tumor cells. Moderate but not a complete reduction in cytotoxic potentials of Carma1−/− and Mx1Cre+TAK1fx/fx NK cells indicate that these cells do not entirely depend on the Carma1/TAK1 pathway to mediate this effector function. As demonstrated by earlier studies, NK cytotoxicity may also depend on the Vav-1-mediated activation of JNK1/2, ERK1/2, and p38 (45, 46). Limited defects in the cytotoxic potentials of Carma1−/− or Mx1Cre+TAK1fx/fx mice-derived NK cells could be due to impairments in transport and release of cytotoxic granules. This notion is corroborated with the recent observations in NKG2D-expressing human NKL cell line, where inhibition of JNK1/2 activity resulted in the impaired movement of microtubule-organizing center, granzyme B, and paxillin to the immune synapse (31). Our present findings are different from our earlier observations with Bcl10−/− NK cells, where the NK-mediated cytotoxicity was not affected (47). One possible explanation is the ability of Carma1 to mediate Bcl10-independent functions, where Carma1 has been shown to recruit MALT1 in the absence of Bcl10 (48). Thus, we predict Carma1 and TAK1 but not Bcl10 to play an essential role in the cytotoxicity of NK cells.
NK cells generate inflammatory cytokines and chemokines as part of innate immunity. The absence of Carma1 in NK cells significantly reduced cytokine and chemokine generation mediated via NCR1, CD244, NKG2D, Ly49D, and NK1.1 receptors. In addition, conditional knockdown of TAK1 or the use of TAK1 inhibitor 5Z-7-oxo-zeaenol resulted in similar reductions in the production of IFN-γ, GM-CSF, IL-10, TNF-α, MIP-1α, MIP-1β, and RANTES. Reductions in the IFN-γ-encoding mRNA after NKG2D-mediated activation indicate that this defect occurs at the transcriptional level in both Carma1−/−- and Mx1Cre+TAK1fx/fx-derived NK cells. Nevertheless, these results also give credence that lack of Carma1 or TAK1 results in the generation of hypo-responsive NK cells. To investigate this hypothesis, we used pro-inflammatory cytokines IL-12 and IL-18 to stimulate NK cells. These cytokines mediate signaling events that are distinct from that of YINM- or ITAM-containing receptor-mediated activations. IL-12 and IL-18 utilize Tyk2/Jak2-mediated STAT4 phosphorylation to induce IFN-γ generation in NK cells (49, 50). Indeed, stimulation with IL-12 and IL-18 resulted in normal levels of GM-CSF, MIP-1α, MIP-1β, and RANTES from Carma1−/− and IFN-γ from Mx1Cre+TAK1fx/fx NK cells. Thus, YINM- or ITAM-independent pathways are fully operational in Carma1−/− and Mx1Cre+TAK1fx/fx NK cells refuting the possibility of global unresponsiveness. Additionally, in vivo stimulation of NK cells via type I IFNs (α and β) using poly(I:C), demonstrates that the defects in cytokine and chemokine production is not due to an inability of Carma1−/− NK cells to respond to IL-2 under the in vitro culture conditions that were used to generate LAK cells.
Molecular mechanisms by which the production of inflammatory cytokines are regulated in NK cells are emerging (31, 51). NKG2D and Ly49D recruit two distinct adaptor proteins to transduce activation signals (52–54). One of these adaptor proteins, DAP10, with a YINM motif recruits PI3K (55). The second adaptor molecule DAP12 with an ITAM motif associates with Syk and ZAP70 (56). NK1.1 recruits FcRγ to mediate its signal (57). NCR1 (NKp46) uses CD3ζ (58). CD244 belongs to the CD2 family and utilizes an immunoreceptor tyrosine-based switch motif to recruit signaling lymphocyte activation molecule-associated protein (59).
Following these receptor-mediated activations, TAK1 is recruited to the Carma1/Bcl10/Malt1/TRAF6 or Carma1/Malt1/TRAF6 signalosomes (48, 60). Reduction in the TAK1 phosphorylation in the Carma1−/− NK cells following NKG2D-mediated activation demonstrates an obligatory role for Carma1 in TAK1 recruitment to the signalosome and downstream signaling. Thus, an impaired phosphorylation of TAK1 in the Carma1−/− NK cells provides the mechanistic explanation for the reduced levels of JNK1/2 phosphorylation and NF-κB nuclear translocation in these cells. This notion is further confirmed by the fact that TAK1 is critical for JNK1/2 and NF-κB activation downstream of both TCR and BCR (10). Similarly, conditional knockdown of TAK1 in Mx1Cre+TAK1fx/fx NK cells (or use of 5Z-7-oxo-zeaenol) resulted in considerable reduction in the phosphorylation of JNK1/2 and NF-κB activation. Moreover, knockdown of TAK1 in NK cells resulted in the reduction of AP1 activation and ERK1/2 phosphorylation. Compared with NK cell cytotoxicity, a near complete reduction in cytokine and chemokine production following pretreatment with the TAK1 inhibitor 5Z-7-oxo-zeaenol or knockdown of TAK1 indicates Carma1 via TAK1 mediates NF-κB and JNK1/2 activations. MAPKs such as ERK1/2, JNK1/2, and p38 are potential downstream targets for multiple NK cell activation receptors (51). This indicates that JNK1 and -2 are critical downstream effectors of Carma1 when NK cells are activated through NKG2D. Our study also shows that lack of Carma1 did not affect the phosphorylation of either p38 or ERK1/2. However, inhibition or knockdown of TAK1 did affect the overall phosphorylation levels of ERK1/2. Presently, we do not have a mechanistic explanation for how ERK1/2 phosphorylations are regulated by TAK1. Together, we conclude that TAK1 is a critical molecular link between Carma1 and cytotoxic granule release or cytokine/chemokine gene transcriptions in NK cells.
Acknowledgment
We thank Dr. Dan Littman for providing the Carma1 knock-out mice.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 A1064826 (to S. M.).
- IKK
- IκB kinase
- NK
- natural killer cell
- Carma1
- caspase-recruitment domain membrane-associated guanylate kinase protein 1
- TAK1
- transforming growth factor-β-activated kinase 1
- Bcl10
- B-cell lymphoma-associated protein
- MAPK
- mitogen-activated protein kinase
- MAPKKK
- MAPK kinase kinase
- E:T
- effector to target ratio
- poly(I:C)
- polyinosine-polycytidylic acid polymer
- RANTES
- regulated on activation normal T cell expressed and secreted.
REFERENCES
- 1. Rawlings D. J., Sommer K., Moreno-García M. E. (2006) Nat. Rev. Immunol. 6, 799–812 [DOI] [PubMed] [Google Scholar]
- 2. Gaide O., Martinon F., Micheau O., Bonnet D., Thome M., Tschopp J. (2001) FEBS Lett. 496, 121–127 [DOI] [PubMed] [Google Scholar]
- 3. Wang D., You Y., Case S. M., McAllister-Lucas L. M., Wang L., DiStefano P. S., Nuñez G., Bertin J., Lin X. (2002) Nat. Immunol. 3, 830–835 [DOI] [PubMed] [Google Scholar]
- 4. Gaide O., Favier B., Legler D. F., Bonnet D., Brissoni B., Valitutti S., Bron C., Tschopp J., Thome M. (2002) Nat. Immunol. 3, 836–843 [DOI] [PubMed] [Google Scholar]
- 5. Egawa T., Albrecht B., Favier B., Sunshine M. J., Mirchandani K., O'Brien W., Thome M., Littman D. R. (2003) Curr. Biol. 13, 1252–1258 [DOI] [PubMed] [Google Scholar]
- 6. Newton K., Dixit V. M. (2003) Curr. Biol. 13, 1247–1251 [DOI] [PubMed] [Google Scholar]
- 7. Jun J. E., Wilson L. E., Vinuesa C. G., Lesage S., Blery M., Miosge L. A., Cook M. C., Kucharska E. M., Hara H., Penninger J. M., Domashenz H., Hong N. A., Glynne R. J., Nelms K. A., Goodnow C. C. (2003) Immunity 18, 751–762 [DOI] [PubMed] [Google Scholar]
- 8. Hara H., Ishihara C., Takeuchi A., Xue L., Morris S. W., Penninger J. M., Yoshida H., Saito T. (2008) J. Immunol. 181, 918–930 [DOI] [PubMed] [Google Scholar]
- 9. Gross O., Grupp C., Steinberg C., Zimmermann S., Strasser D., Hannesschläger N., Reindl W., Jonsson H., Huo H., Littman D. R., Peschel C., Yokoyama W. M., Krug A., Ruland J. (2008) Blood 112, 2421–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun L., Deng L., Ea C. K., Xia Z. P., Chen Z. J. (2004) Mol. Cell 14, 289–301 [DOI] [PubMed] [Google Scholar]
- 11. Tang M., Wei X., Guo Y., Breslin P., Zhang S., Zhang S., Wei W., Xia Z., Diaz M., Akira S., Zhang J. (2008) J. Exp. Med. 205, 1611–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sato S., Sanjo H., Takeda K., Ninomiya-Tsuji J., Yamamoto M., Kawai T., Matsumoto K., Takeuchi O., Akira S. (2005) Nat. Immunol. 6, 1087–1095 [DOI] [PubMed] [Google Scholar]
- 13. Regunathan J., Chen Y., Wang D., Malarkannan S. (2005) Blood 105, 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Malarkannan S., Shih P. P., Eden P. A., Horng T., Zuberi A. R., Christianson G., Roopenian D., Shastri N. (1998) J. Immunol. 161, 3501–3509 [PubMed] [Google Scholar]
- 15. Bennett M., Yu Y. Y., Stoneman E., Rembecki R. M., Mathew P. A., Lindahl K. F., Kumar V. (1995) Semin. Immunol. 7, 121–127 [DOI] [PubMed] [Google Scholar]
- 16. Mason L. H., Anderson S. K., Yokoyama W. M., Smith H. R., Winkler-Pickett R., Ortaldo J. R. (1996) J. Exp. Med. 184, 2119–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Malarkannan S., Horng T., Eden P., Gonzalez F., Shih P., Brouwenstijn N., Klinge H., Christianson G., Roopenian D., Shastri N. (2000) Immunity 13, 333–344 [DOI] [PubMed] [Google Scholar]
- 18. Fernandez-Sesma A., Marukian S., Ebersole B. J., Kaminski D., Park M. S., Yuen T., Sealfon S. C., García-Sastre A., Moran T. M. (2006) J. Virol. 80, 6295–6304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo H., Samarakoon A., Vanhaesebroeck B., Malarkannan S. (2008) J. Exp. Med. 205, 2419–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xue L., Morris S. W., Orihuela C., Tuomanen E., Cui X., Wen R., Wang D. (2003) Nat. Immunol. 4, 857–865 [DOI] [PubMed] [Google Scholar]
- 21. Ruland J., Duncan G. S., Elia A., del Barco Barrantes I., Nguyen L., Plyte S., Millar D. G., Bouchard D., Wakeham A., Ohashi P. S., Mak T. W. (2001) Cell 104, 33–42 [DOI] [PubMed] [Google Scholar]
- 22. Bauer S., Groh V., Wu J., Steinle A., Phillips J. H., Lanier L. L., Spies T. (1999) Science 285, 727–729 [DOI] [PubMed] [Google Scholar]
- 23. Cerwenka A., Bakker A. B., McClanahan T., Wagner J., Wu J., Phillips J. H., Lanier L. L. (2000) Immunity 12, 721–727 [DOI] [PubMed] [Google Scholar]
- 24. Diefenbach A., Jensen E. R., Jamieson A. M., Raulet D. H. (2001) Nature 413, 165–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Samarakoon A., Chu H., Malarkannan S. (2009) Mol. Immunol. 46, 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ljunggren H. G., Kärre K. (1985) J. Exp. Med. 162, 1745–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Djeu J. Y., Heinbaugh J. A., Holden H. T., Herberman R. B. (1979) J. Immunol. 122, 182–188 [PubMed] [Google Scholar]
- 28. Dorner B. G., Smith H. R., French A. R., Kim S., Poursine-Laurent J., Beckman D. L., Pingel J. T., Kroczek R. A., Yokoyama W. M. (2004) J. Immunol. 172, 3119–3131 [DOI] [PubMed] [Google Scholar]
- 29. Jiang K., Zhong B., Gilvary D. L., Corliss B. C., Hong-Geller E., Wei S., Djeu J. Y. (2000) Nat. Immunol. 1, 419–425 [DOI] [PubMed] [Google Scholar]
- 30. Wei S., Gilvary D. L., Corliss B. C., Sebti S., Sun J., Straus D. B., Leibson P. J., Trapani J. A., Hamilton A. D., Weber M. J., Djeu J. Y. (2000) J. Immunol. 165, 3811–3819 [DOI] [PubMed] [Google Scholar]
- 31. Li C., Ge B., Nicotra M., Stern J. N., Kopcow H. D., Chen X., Strominger J. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3017–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trotta R., Fettucciari K., Azzoni L., Abebe B., Puorro K. A., Eisenlohr L. C., Perussia B. (2000) J. Immunol. 165, 1782–1789 [DOI] [PubMed] [Google Scholar]
- 33. Ninomiya-Tsuji J., Kishimoto K., Hiyama A., Inoue J., Cao Z., Matsumoto K. (1999) Nature 398, 252–256 [DOI] [PubMed] [Google Scholar]
- 34. Greenblatt M. B., Shim J. H., Zou W., Sitara D., Schweitzer M., Hu D., Lotinun S., Sano Y., Baron R., Park J. M., Arthur S., Xie M., Schneider M. D., Zhai B., Gygi S., Davis R., Glimcher L. H. (2010) J. Clin. Invest. 120, 2457–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Geuking P., Narasimamurthy R., Lemaitre B., Basler K., Leulier F. (2009) PLoS One 4, e7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ventura J. J., Kennedy N. J., Lamb J. A., Flavell R. A., Davis R. J. (2003) Mol. Cell Biol. 23, 2871–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sommer K., Guo B., Pomerantz J. L., Bandaranayake A. D., Moreno-García M. E., Ovechkina Y. L., Rawlings D. J. (2005) Immunity 23, 561–574 [DOI] [PubMed] [Google Scholar]
- 38. Matsumoto R., Wang D., Blonska M., Li H., Kobayashi M., Pappu B., Chen Y., Wang D., Lin X. (2005) Immunity 23, 575–585 [DOI] [PubMed] [Google Scholar]
- 39. Shinohara H., Yasuda T., Aiba Y., Sanjo H., Hamadate M., Watarai H., Sakurai H., Kurosaki T. (2005) J. Exp. Med. 202, 1423–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baud V., Liu Z. G., Bennett B., Suzuki N., Xia Y., Karin M. (1999) Genes Dev. 13, 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schulze-Luehrmann J., Ghosh S. (2006) Immunity 25, 701–715 [DOI] [PubMed] [Google Scholar]
- 42. Ninomiya-Tsuji J., Kajino T., Ono K., Ohtomo T., Matsumoto M., Shiina M., Mihara M., Tsuchiya M., Matsumoto K. (2003) J. Biol. Chem. 278, 18485–18490 [DOI] [PubMed] [Google Scholar]
- 43. Huang H., Ryu J., Ha J., Chang E. J., Kim H. J., Kim H. M., Kitamura T., Lee Z. H., Kim H. H. (2006) Cell Death Differ. 13, 1879–1891 [DOI] [PubMed] [Google Scholar]
- 44. Aramburu J., Azzoni L., Rao A., Perussia B. (1995) J. Exp. Med. 182, 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Upshaw J. L., Leibson P. J. (2006) Semin. Immunol. 18, 167–175 [DOI] [PubMed] [Google Scholar]
- 46. Zompi S., Hamerman J. A., Ogasawara K., Schweighoffer E., Tybulewicz V. L., Di Santo J. P., Lanier L. L., Colucci F. (2003) Nat. Immunol. 4, 565–572 [DOI] [PubMed] [Google Scholar]
- 47. Malarkannan S., Regunathan J., Chu H., Kutlesa S., Chen Y., Zeng H., Wen R., Wang D. (2007) J. Immunol. 179, 3752–3762 [DOI] [PubMed] [Google Scholar]
- 48. Che T., You Y., Wang D., Tanner M. J., Dixit V. M., Lin X. (2004) J. Biol. Chem. 279, 15870–15876 [DOI] [PubMed] [Google Scholar]
- 49. Fehniger T. A., Shah M. H., Turner M. J., VanDeusen J. B., Whitman S. P., Cooper M. A., Suzuki K., Wechser M., Goodsaid F., Caligiuri M. A. (1999) J. Immunol. 162, 4511–4520 [PubMed] [Google Scholar]
- 50. Watford W. T., Hissong B. D., Bream J. H., Kanno Y., Muul L., O'Shea J. J. (2004) Immunol. Rev. 202, 139–156 [DOI] [PubMed] [Google Scholar]
- 51. Chen X., Trivedi P. P., Ge B., Krzewski K., Strominger J. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 6329–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Billadeau D. D., Upshaw J. L., Schoon R. A., Dick C. J., Leibson P. J. (2003) Nat. Immunol. 4, 557–564 [DOI] [PubMed] [Google Scholar]
- 53. Wu J., Song Y., Bakker A. B., Bauer S., Spies T., Lanier L. L., Phillips J. H. (1999) Science 285, 730–732 [DOI] [PubMed] [Google Scholar]
- 54. Wu J., Cherwinski H., Spies T., Phillips J. H., Lanier L. L. (2000) J. Exp. Med. 192, 1059–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gilfillan S., Ho E. L., Cella M., Yokoyama W. M., Colonna M. (2002) Nat. Immunol. 3, 1150–1155 [DOI] [PubMed] [Google Scholar]
- 56. McVicar D. W., Taylor L. S., Gosselin P., Willette-Brown J., Mikhael A. I., Geahlen R. L., Nakamura M. C., Linnemeyer P., Seaman W. E., Anderson S. K., Ortaldo J. R., Mason L. H. (1998) J. Biol. Chem. 273, 32934–32942 [DOI] [PubMed] [Google Scholar]
- 57. Arase N., Arase H., Park S. Y., Ohno H., Ra C., Saito T. (1997) J. Exp. Med. 186, 1957–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pende D., Parolini S., Pessino A., Sivori S., Augugliaro R., Morelli L., Marcenaro E., Accame L., Malaspina A., Biassoni R., Bottino C., Moretta L., Moretta A. (1999) J. Exp. Med. 190, 1505–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sayós J., Nguyen K. B., Wu C., Stepp S. E., Howie D., Schatzle J. D., Kumar V., Biron C. A., Terhorst C. (2000) Int. Immunol. 12, 1749–1757 [DOI] [PubMed] [Google Scholar]
- 60. Welteke V., Eitelhuber A., Düwel M., Schweitzer K., Naumann M., Krappmann D. (2009) EMBO Rep. 10, 642–648 [DOI] [PMC free article] [PubMed] [Google Scholar]