Abstract
Menin is a tumor suppressor protein that is encoded by the MEN1 (multiple endocrine neoplasia 1) gene and controls cell growth in endocrine tissues. Importantly, menin also serves as a critical oncogenic cofactor of MLL (mixed lineage leukemia) fusion proteins in acute leukemias. Direct association of menin with MLL fusion proteins is required for MLL fusion protein-mediated leukemogenesis in vivo, and this interaction has been validated as a new potential therapeutic target for development of novel anti-leukemia agents. Here, we report the first crystal structure of menin homolog from Nematostella vectensis. Due to a very high sequence similarity, the Nematostella menin is a close homolog of human menin, and these two proteins likely have very similar structures. Menin is predominantly an α-helical protein with the protein core comprising three tetratricopeptide motifs that are flanked by two α-helical bundles and covered by a β-sheet motif. A very interesting feature of menin structure is the presence of a large central cavity that is highly conserved between Nematostella and human menin. By employing site-directed mutagenesis, we have demonstrated that this cavity constitutes the binding site for MLL. Our data provide a structural basis for understanding the role of menin as a tumor suppressor protein and as an oncogenic co-factor of MLL fusion proteins. It also provides essential structural information for development of inhibitors targeting the menin-MLL interaction as a novel therapeutic strategy in MLL-related leukemias.
Keywords: Protein Motifs, Protein Structure, Protein-Protein Interactions, Tumor Suppressor Gene, X-ray Crystallography, Mixed Lineage Leukemia, Multiple Endocrine Neoplasia 1
Introduction
Menin is a tumor suppressor protein encoded by the MEN1 (multiple endocrine neoplasia 1) gene (1) that controls cell growth in endocrine tissues. Mutations in Men1 occur with an estimated frequency of one in 30,000 individuals and are associated with MEN1 tumors of the parathyroid glands, pancreatic islet cells, and anterior pituitary gland (2). Menin is an ubiquitously expressed nuclear protein (3) that is engaged in a complex network of interactions with diverse proteins, including transcription factors such as JunD (4), NF-κB (5), and SMAD3 (6); chromatin-associated proteins such as mSin3A (7), MLL (mixed lineage leukemia) (8, 9), and lens epithelium-derived growth factor (10); DNA repair proteins such as the DNA damage repair protein FANCD2 (11); and the replication protein A subunit RPA2 (12). The diversity of interacting partners suggests a role of menin in multiple biological pathways, including cell growth regulation, cell cycle control, genome stability, bone development, and hematopoiesis (13, 14). Despite its importance in many physiological and pathological processes, no structural information about menin or menin complexes with protein partners are currently available.
Menin also functions as a critical oncogenic co-factor of MLL fusion proteins required for their leukemogenic activity (15). Translocations of the MLL gene frequently occur in aggressive human acute leukemias, both in children and adults (16, 17). Patients with leukemias harboring MLL translocations have very unfavorable prognoses and respond poorly to currently available treatments. Menin is a highly specific binding partner for MLL and MLL fusion proteins and is required to regulate expression of MLL target genes, including Hoxa9 and Meis1 (9, 15, 18, 19). Importantly, loss of menin binding by MLL fusion proteins abolishes their oncogenic potential in vitro and in vivo (15, 19). Disruption of the menin-MLL interaction using genetic methods blocks development of acute leukemia in mice (15). Therefore, the menin-MLL interaction represents an attractive therapeutic target for development of novel drugs for acute leukemias with MLL rearrangements (15, 18, 19).
Lack of a menin structure significantly limits the understanding of menin function as a tumor suppressor protein (20) and its role as a co-factor of leukemogenic MLL fusion proteins. We have recently characterized in detail the menin-MLL interaction by employing biophysical and biochemical methods (21). As a next step toward revealing the molecular mechanism of the MLL-mediated leukemogenesis, we have determined the first three-dimensional structure of menin and mapped the MLL binding site. Because human menin was recalcitrant to crystallization efforts, we crystallized a menin homolog from N. vectensis, which is highly conserved with the human protein and is expected to have a very similar structure. The structure of menin revealed the presence of a very large central cavity that comprises many hydrophobic and negatively charged amino acids. By site-directed mutagenesis, we have shown that this cavity represents a binding site for MLL. Furthermore, we have expressed several MEN1 missense mutations and characterized their effect on the thermodynamic stability of the protein and its interactions with MLL. Overall, our work provides an important structural insight into understanding the function of menin and constitutes the essential molecular basis for development of small molecules to inhibit the menin-MLL interaction in acute leukemias related to MLL translocations.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
The synthetic gene encoding N. vectensis menin was ordered from GenScript and subcloned into pET32a vector (Novagen). Site-directed mutagenesis was performed to introduce a stop codon at residue 487 and internal deletion of residues 426–442.
Nematostella menin was expressed in Rosetta2(DE3) cells (Novagen) and purified using affinity chromatography column HisTrap HP (GE Healthcare) followed by ion exchange employing Q Sepharose FF (GE Healthcare). To remove the thioredoxin-His6 tag, the protein was cleaved by 3C protease O/N and loaded onto nickel-nitrilotriacetic acid superflow resin (Qiagen). At the final step protein was purified by size exclusion chromatography using column HiLoad 16/60 Superdex 75 pg (GE Healthcare). Selenomethionine (SeMet) protein was obtained by growing Rosetta2(DE3) cells in M9 minimal media supplemented with 50 mg/liter l(+)-selenomethionine 99+% (Acros Organics). SeMet protein was purified according to the above protocol established for unlabeled protein.
Purification of full-length human menin was described elsewhere (21). We have performed two sets of point mutations using site-directed mutagenesis; mutations designed to abolish MLL binding (S155K, M278K, Y323K, E359K, E363K) and MEN1 point mutations (P12L, H139D, A242V, and A309P). Expressions and purifications were carried out using similar protocol as for the wild type protein.
MLL Binding Experiments
Dissociation constants for binding of MLL MBM1 to human menin and menin mutants were determined by fluorescence polarization method using previously published protocol (21). Briefly, the fluorescein-labeled MLL-derived peptide, FITC-MBM1 (MLL4–15) at 50 nm, was titrated with a range of menin concentrations in the FP buffer (50 mm TRIS, pH 7.5, 50 mm NaCl, 1 mm DTT). After 1-h incubation of the protein-peptide complexes, change in fluorescence polarization and anisotropy were monitored at 525 nm after excitations at 495 nm using PHERAstar microplate reader (BMG Labtech, Inc.). Results were used to calculate binding affinity (Kd) for the MLL-derived peptide with wild type menin or menin mutants using the Origin program (version 7.0).
Stability Studies
Thermodynamic stabilities of human menin and mutants were measured by employing thermal shift assay using Thermofluor 384 ELS system. Protein unfolding was examined by monitoring the fluorescence of 1-anilinonaphthalene-8-sulfonic acid by increasing the temperature from 20 to 60 °C. All samples were prepared in quadruplicates and contained proteins at 5 μm concentrations in 50 mm Tris, pH 7.5, 50 mm NaCl buffer, and 50 μm 1-anilinonaphthalene-8-sulfonic acid. To limit evaporation, the samples were covered using mineral oil.
Crystallization
Initial crystals of Nematostella menin were obtained using the sitting drop technique at 10 °C in 20% PEG 3350, 0.1 m Bis-Tris, pH 6.0, 0.2 m ammonium sulfate. High quality crystals (40 × 40 × 100 μm) used for diffraction experiments were obtained from microseeding and formed after 2 days. Prior to data collection, crystals were transferred into a cryosolution containing 20% glycerol and flash-frozen in liquid nitrogen.
Structure Determination
Data were collected at a temperature of 100 K on beamlines 19-BM of the Structural Biology Center (22) and 21-ID-F of the Life Science Collaborative Access Team at the Advanced Photon Source. HKL-2000 (23) was used for data processing. Structure determination was performed using single-wavelength anomalous diffraction and HKL-3000 (24), which is integrated with SHELXD, SHELXE (25), MLPHARE (26), DM (27), ARP/wARP (28), CCP4 (29), and SOLVE and RESOLVE (30). The initial low resolution model obtained from data collected for a crystal containing SeMet-substituted protein was improved by manual rebuilding in COOT (31). The low resolution model was later improved in HKL-3000 using higher resolution data collected for crystal of the native protein. Final adjustment of the model and refinement was done with COOT and REFMAC (32). MolProbity (33) and ADIT (34) were used for model validation. The coordinates and structure factors of Nematostella menin structure were deposited in the Protein Data Bank with the accession code 3RE2. The statistics of data collection and structure refinement are summarized in Table 1. Electrostatic potential was calculated using APBS (35). Figures were prepared in PyMOL (version 1.4). Model of human menin was built using Modeler 9 (version 8) (36), and analysis of sequence conservation was carried out employing ConSurf (37) based on multiple sequence alignment of 21 menin homologs.
TABLE 1.
Data collection, structure solution, and refinement statistics
Ramachandran plots were calculated using MolProbity software. Numbers in parentheses refer to the highest resolution shell. FOM, figure of merit; r.m.s.d., root mean square deviation.
| SeMet | Native | |
|---|---|---|
| Data collection | ||
| Beamline | 21-ID-F | 19-BM |
| Wavelength (Å) | 0.9787 | 0.9791 |
| Unit cell (Å) | a = 91.5, c = 114.5 | a = 89.9, c = 115.6 |
| Space group | P43211 | P43212 |
| Solvent content (%) | 46 | 44 |
| Resolution range (Å) | 50.00–3.25 | 50.00–1.95 |
| Highest resolution shell (Å) | 3.31-3.25 | 1.98-1.95 |
| Unique reflections | 8061 (380) | 35,267 (1725) |
| Redundancy | 11.6 (12.1) | 13.8 (13.5) |
| Completeness (%) | 100.0 (100.0) | 99.9 (100.0) |
| Rmerge (%) | 8.9 (64.1) | 9.1 (78.0) |
| Average I/σ(I) | 42.9 (5.0) | 34.8 (3.8) |
| B from Wilson plot (Å2) | 87.2 | 23.6 |
| Structure solution | ||
| Se sites (localized/possible) | 9/11 | |
| FOMMLPHARE | 0.20 | |
| FOMDM | 0.74 | |
| Refinement | ||
| R (%) | 16.1 | |
| Rfree (%) | 21.1 | |
| Mean B value (Å2) | 34.5 | |
| r.m.s.d. bond lengths (Å) | 0.019 | |
| r.m.s.d. bond angles | 1.6° | |
| No. of amino acid residues | 446 | |
| No. of water molecules | 339 | |
| Ramachandran plot | ||
| Most favored regions (%) | 98.6 | |
| Additional allowed regions (%) | 1.4 | |
RESULTS
Determination of Crystal Structure of Menin Homolog from N. vectensis
Human menin is a 610-amino acid-long protein with a strongly conserved sequence among all species (1). We carried out extensive bioinformatic analysis of the menin sequence and failed to find any protein of known structure with significant sequence similarity. Our initial efforts to crystallize human menin were also not successful. Sequence analysis identified several internal fragments predicted to be disordered, and we hypothesized that the presence of such unstructured regions hinders the crystallization of human menin. Based on the sequence alignment, we identified a homolog from N. vectensis, which has one of the shortest sequences among all menin homologs (Fig. 1A). N. vectensis is the starlet sea anemone, a primitive animal whose complete genome has been sequenced (38). Importantly, the sequence identity between human menin and the Nematostella homolog exceeds 50% for the structural domain, strongly indicating that their three-dimensional structures should be very similar. Based on the prediction of disordered regions, we generated a construct of the Nematostella menin homolog with the deletion of one loop (residues 426–442) and truncation of the C terminus (residues 487–539). The truncated protein crystallized readily, and the crystals diffracted to 1.95Å resolution.
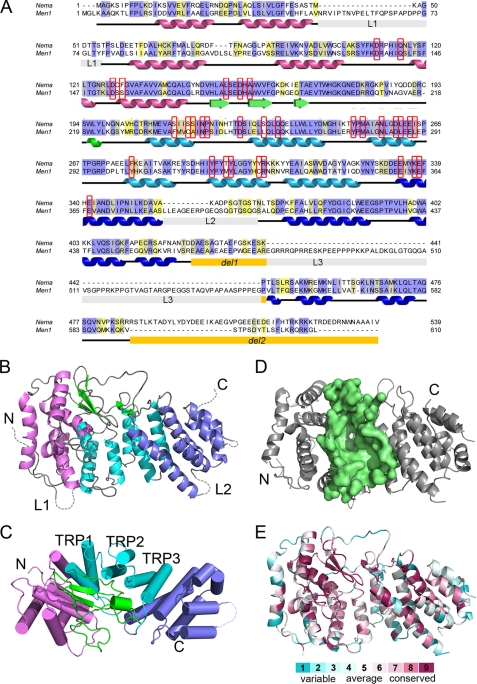
FIGURE 1.
Crystal structure of the menin homolog from N. vectensis. A, sequence alignment of Nematostella (Nema) and human (Men1) menins. Color coding highlights identical (blue) and conserved (yellow) residues. Positions of secondary structure elements derived from the crystal structure are shown and colored following the schemes in B and C. The regions deleted in Nematostella menin for crystallization are labeled del1 and del2; the three long loops are labeled L1, L2, and L3. Residues lining putative MLL binding site are in red boxes. B, the overall fold of menin is composed of three TRP motifs (cyan), which are flanked by N-terminal (purple) and C-terminal (blue) helical bundles and covered by a three-stranded antiparallel β-sheet (green). C, the structure of Nematostella menin rotated by 90°. Helices are shown as cylinders, and the TRP motifs are numbered. D, the central cavity in Nematostella menin. The residues lining the cavity are shown in surface representation (pale green). E, representation of conserved residues based on multiple sequence alignment of 21 menin homologs prepared using the ConSurf server (37).
We determined the crystal structure of Nematostella menin by single-wavelength anomalous diffraction with the HKL-3000 program (Table 1) (24). The structure has a predominantly α-helical fold (Fig. 1, B and C), and the core of the protein comprises three tetratricopeptide repeat (TRP1, -2, and -3) motifs that are flanked on their N and C termini by two α-helical bundles and covered by a three-stranded antiparallel β-sheet (Fig. 1, B and C). TRP2 is a structural motif composed of a pair of antiparallel α-helices, which might serve as a scaffold for protein-protein interactions and often mediates the assembly of multiprotein complexes (39). Analysis of structural similarities using FATCAT (40) revealed that the menin fold is reminiscent of several other TRP-containing proteins such as bacterial RapH phosphatase (41) and anaphase-promoting complex protein APC6 (42). Interestingly, the β-sheet motif is absent in these TRP proteins and represents a novel element specific to menin.
The most striking feature of the three-dimensional structure of menin is the presence of a very large central cavity that is enclosed by all four subdomains (the TPR motifs, two α-helical bundles, and a β-sheet) (Fig. 1D). Analysis of this cavity with the CASTp program (43) revealed an approximate volume of 3700 Å3. The large size and elongated shape of this site strongly suggests that it serves as a binding site for protein-protein interactions (see below).
Homology Model of Human Menin
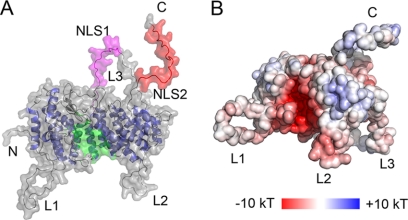
Because of the high sequence identity (>50%), we built a homology model of human menin (Fig. 2) using the Nematostella menin structure as a template. As expected the highest degree of sequence conservation is found within the protein core, which strongly indicates that the protein structure, is strictly preserved between human menin and its Nematostella homolog. A very interesting feature of human menin is the presence of long loops, L1 (residues 51–77), L2 (residues 382–401), and L3 (residues 461–548, the longest), as well as an unstructured C-terminal tail (residues 593–610) (Fig. 2A). With the exception of the C terminus, these loops are substantially shorter in the Nematostella homolog. Several phosphorylation sites (e.g. Ser-487 and Ser-543 in loop L3 and Ser-583 in the C terminus) (44) and two nuclear localization sequences (NLS1 in L3 and NSL2 in the C terminus) (3) have been mapped to these fragments, indicating that these regions most likely play an important role in regulation of menin function and localization.
FIGURE 2.
Homology model of human menin. A, a model of full-length human menin shown in surface representation. Loops are labeled L1, L2, and L3; NLS1 is shown in magenta, NLS2 is shown in red, and the central cavity is shown in green. B, electrostatic potential calculated for the homology model of human menin.
Central Cavity in Menin Constitutes MLL Binding Site
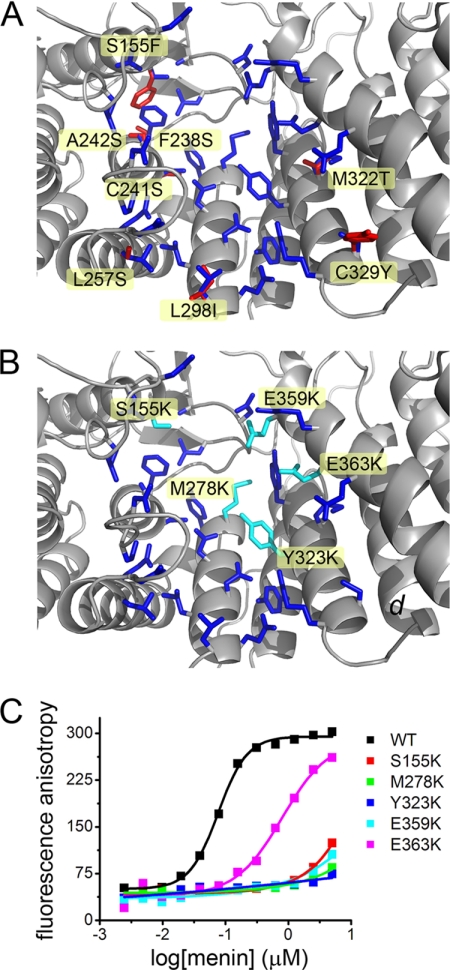
Menin exerts its biological function by binding to multiple protein partners (13, 14). Based on the menin structure, we concluded that the large central cavity in the protein most likely constitutes the binding site for some of the protein partners. This cavity is formed by several α-helices from the three TRP motifs and the N- and C-terminal α-helical bundle domains (Fig. 3). The character of this site is defined by a number of hydrophobic residues (Leu-177, Pro-245, Leu-257, Leu-286, Tyr-276, Met-278, Tyr-319, Met-322, Tyr-323, and Phe-328) and several acidic side chains (Asp-136, Asp-153, Asp-180, Asp-285, Glu-288, Glu-359, Glu-363, and Glu-366) (Fig. 3), which gives rise to its strong negative surface potential (Fig. 2B). Importantly, relatively few residues lining the cavity differ in identity between the human and Nematostella menin, indicating that the structure of this site is highly preserved (Fig. 3A).
FIGURE 3.
The central cavity in menin constitutes the MLL MBM1 binding site. A, side chains forming the central cavity in human menin are shown in blue. Residues that differ between the human and Nematostella homologs are labeled and shown in red. B, residues mutated in human menin to disrupt MLL binding are labeled and shown in cyan. C, fluorescence polarization binding assay demonstrating decrease of MLL MBM1 binding affinity to menin mutants. Kd value for wild type menin is 77 ± 38 nm and 812 ± 305 nm for E363K mutant. Four remaining menin mutants bind MBM1 very weakly with Kd > 10 μm.
We have recently characterized the menin-MLL interaction and found that two short fragments of MLL (MBM1 and MBM2) interact with menin, and MBM1 (MLL residues 4–15) represents the high affinity binding motif (21). We found that MBM1 binds to menin in an extended conformation and that the three hydrophobic residues (Phe-7, Pro-8, and Pro-11) have the most significant contribution to the binding affinity. In addition, the MBM1 contains three positively charged residues (Arg-6, Arg-8, and Arg-12), which further contribute to the interaction with menin (21). To test whether the central cavity in human menin constitutes the binding site for MBM1, we have made several point mutations in this site. The mutations were designed to cause electrostatic repulsion with MLL by introducing positively charged lysine residues (S155K, M278K, Y323K, E359K, and E363K; Fig. 3B). We have also verified that such mutations did not impair the thermodynamic stability of menin (supplemental Table S1). As expected, all mutations affected binding of MBM1 to menin (Fig. 3C). Four of the five mutations (S155K, M278K, Y323K, and E359K) significantly impaired binding of MBM1 to menin by >100-fold (Fig. 3C). Substitution of E363K had weaker effect and decreased the affinity of MBM1 to menin by ∼10-fold. Overall, these experiments clearly indicate that the MBM1 motif of MLL binds to the central cavity in menin, validating that it represents a protein-protein interaction site.
Sequence analysis of the N. vectensis genome indicates a lack of an MLL homolog (45). However, because of very high sequence similarity of residues lining the binding site, we expected that Nematostella menin binds the MLL MBM1 motif. Indeed, we observed that Nematostella menin interacts with the MBM1 fragment, although the affinity is approximately three orders of magnitude weaker than that of the human menin (supplemental Fig. S1).
MEN1 Mutations Impair Menin Stability and Protein-Protein Interactions
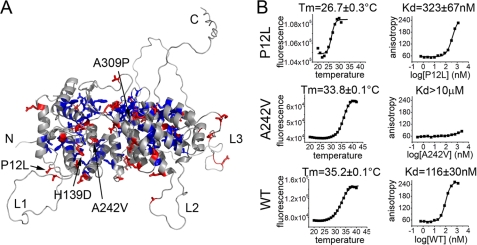
To date, >1300 germline and somatic mutations have been discovered in Men1 (13), resulting in MEN1 tumors. However, the comparison of clinical features in patients with Men1 mutations reveals no phenotype-genotype correlations (13). The majority of mutations discovered in Men1 are nonsense mutations leading to truncation of the protein or frameshift mutations (13). The three-dimensional structure revealed that menin is a single domain protein, and therefore, the truncation of its amino acid sequence will inevitably result in disruption of the menin fold leading to loss of menin function. Approximately 20% of MEN1 mutations are missense mutations that result in single residue substitutions in menin. We analyzed 159 unique missense mutations and found that 66% of these mutations occur in buried residues (Fig. 4A). Considering a very high degree of sequence conservation among menin homologs (Fig. 1E), these mutations most likely destabilize the protein structure. The remaining 34% of MEN1 missense mutations occur at solvent exposed sites and might impair protein-protein interactions (Fig. 4A). Interestingly, eight mutations occur in the long unstructured loop L3 and will likely leave the menin structure unaffected. The occurrence of such mutations in MEN1 further emphasizes the important role of the flexible regions in regulation of menin function.
FIGURE 4.
Characterization of MEN1 missense mutations. A, distribution of MEN1 missense mutations mapped onto the homology model of human menin. Buried and solvent exposed residues mutated in MEN1 are shown in blue and red, respectively. B, comparison of thermodynamic stability and MLL binding affinity for P12L, A242V, and wild type human menin.
To experimentally test the effects of disease-related mutations in human menin, we expressed four MEN1 mutants: P12L, H139D, A242V, and A309P. Two of these mutations (H139D and A309P), which replace strongly conserved residues, did not yield soluble protein, most likely due to destabilization of menin structure. For the two remaining mutants, we have obtained soluble proteins, tested their stability, and MLL binding affinity, and compared them to the wild type menin (Fig. 4B). The P12L mutation, which affects partially exposed residue, results in a relatively small, 3-fold decrease in affinity toward MLL. However, this mutation significantly decreased the thermodynamic stability of the protein, reducing the Tm by >8 °C (Fig. 4B). This effect corroborates previous studies demonstrating enhanced proteolytic degradation of the P12L mutant leading to the loss of menin (46). Interestingly, the mutation of A242V, which affects the residue located in the central cavity in menin, does not affect the stability of the protein (Fig. 4B). However, it does abolish binding of MLL (Fig. 4B) and potentially other menin binding partners.
DISCUSSION
Menin functions as an adapter protein that is involved in the interactions with multiple protein partners (13, 14). Here, we report the first three-dimensional structure of menin, which represents a significant step toward understanding the role of menin as a tumor suppressor and as an oncogenic co-factor of MLL fusion proteins. We have determined the crystal structure of menin from N. vectensis, which is a very close homolog of human protein. The high level of sequence conservation between the two proteins (>50% identical residues for structure regions) strongly indicates very similar three-dimensional structures. A rationale for selection of Nematostella protein for crystallization experiments was based on significantly shorter fragments that are predicted to be disordered as compared with human menin. The deletion of a single loop and truncation of the C terminus in Nematostella menin yielded a protein that easily crystallized.
Menin directly interacts with MLL and functions as a critical oncogenic co-factor of MLL fusion proteins required for their leukemogenic activity (15, 18, 19). Based on the structure of Nematostella menin, we identified the presence of a very large cavity in the center of the menin structure, and by mutagenesis, we demonstrated that in human menin, it represents the MLL binding site. Importantly, the residues forming this cavity are highly conserved between human and Nematostella menin, strongly indicating that this structure is preserved in both proteins. Furthermore, the size and the character of the central cavity in menin are highly complementary to the MLL MBM1 motif. Interestingly, when menin structure is superimposed onto the structures of other TRP-containing proteins, the location of the MLL binding site in menin overlaps with the peptide binding sites found in the TRP proteins (47). Previously, we found that MBM1 binds to menin in an extended conformation (21), and this binding mode might be similar to the peptide complexes with the TRP proteins (47). In addition to MLL binding, menin interacts with many other proteins and the central cavity in menin structure might constitute the binding site for other protein-protein interactions, such as JunD (4).
The interaction between menin and MLL fusion proteins has been emphasized as a valid drug target for acute leukemias with MLL translocations (15, 19). Knowledge of the three-dimensional structure of menin and mapping the binding site for MLL provides an essential structural basis for development of small molecule inhibitors targeting the menin-MLL interaction. Importantly, the very large size of this cavity and a well defined structure strongly emphasizes that this site should be amenable to targeting by small molecule inhibitors.
The function of menin as a tumor suppressor is less understood and might be in part related to the cell cycle regulation in endocrine tissues. For example, the interaction of menin with MLL is required for regulation of expression of the cyclin-dependent kinase inhibitors p27kip1 and p18ink4c (48). However, menin function as a tumor suppressor is likely more complex and might involve transcriptional repression via association with transcription factors such as JunD (4), NF-κB (5), or general co-repressor Sin3A (7). Furthermore, menin interacts with proteins involved in DNA replication, recombination, and repair such as replication protein A (RPA) (12) and FANCD2 (Fanconi anemia complementation group D2 protein) (11), which implicates menin function in maintenance of genome integrity. Overall, menin might play a role as an adapter protein that tethers multiple proteins, and the knowledge of its three-dimensional structure will likely have an invaluable impact on understanding which interactions are essential to the function of menin as an oncogenic co-factor and as a tumor suppressor.
To date, no structural information for menin complexes is available, and therefore, the structure of menin presented here should prove valuable for mapping menin interactions with protein partners. For example, we have found that patient mutation A242V yields a protein variant that is incapable of interacting with MLL and possibly with other menin partners. Furthermore, the structure of menin provides a better understanding of the effect of MEN1 mutations. Because the majority of disease-related missense mutations change buried residues, they most likely result in destabilization of the menin structure and loss of protein function (46). We have expressed several MEN1 mutations and found that two substitutions (H139D and A309P) result in protein that is insoluble and that the P12L mutation destabilizes the protein. Importantly, knowledge of the menin structure might also serve to distinguish causative from benign mutations in the Men1 gene to potentially facilitate diagnosis of the MEN1 tumors.
Supplementary Material
Acknowledgment
We thank Dr. Matthew D. Zimmerman for critical reading of the manuscript.
This work was supported by Research Scholar Grant RSG-11-082-01-DMC from the American Chemical Society (to T. C.) and a Leukemia and Lymphoma Society Translational Research Program grant (to J. G.).
The atomic coordinates and structure factors (code 3RE2) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
- TRP
- tetratricopeptide repeat
- SeMet
- selenomethionine.
REFERENCES
- 1. Chandrasekharappa S. C., Guru S. C., Manickam P., Olufemi S. E., Collins F. S., Emmert-Buck M. R., Debelenko L. V., Zhuang Z., Lubensky I. A., Liotta L. A., Crabtree J. S., Wang Y., Roe B. A., Weisemann J., Boguski M. S., Agarwal S. K., Kester M. B., Kim Y. S., Heppner C., Dong Q., Spiegel A. M., Burns A. L., Marx S. J. (1997) Science 276, 404–407 [DOI] [PubMed] [Google Scholar]
- 2. Marx S. J. (2005) Nat. Rev. Cancer 5, 367–375 [DOI] [PubMed] [Google Scholar]
- 3. Guru S. C., Goldsmith P. K., Burns A. L., Marx S. J., Spiegel A. M., Collins F. S., Chandrasekharappa S. C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 1630–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Agarwal S. K., Guru S. C., Heppner C., Erdos M. R., Collins R. M., Park S. Y., Saggar S., Chandrasekharappa S. C., Collins F. S., Spiegel A. M., Marx S. J., Burns A. L. (1999) Cell 96, 143–152 [DOI] [PubMed] [Google Scholar]
- 5. Heppner C., Bilimoria K. Y., Agarwal S. K., Kester M., Whitty L. J., Guru S. C., Chandrasekharappa S. C., Collins F. S., Spiegel A. M., Marx S. J., Burns A. L. (2001) Oncogene 20, 4917–4925 [DOI] [PubMed] [Google Scholar]
- 6. Kaji H., Canaff L., Lebrun J. J., Goltzman D., Hendy G. N. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3837–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim H., Lee J. E., Cho E. J., Liu J. O., Youn H. D. (2003) Cancer Res. 63, 6135–6139 [PubMed] [Google Scholar]
- 8. Yokoyama A., Wang Z., Wysocka J., Sanyal M., Aufiero D. J., Kitabayashi I., Herr W., Cleary M. L. (2004) Mol. Cell Biol. 24, 5639–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hughes C. M., Rozenblatt-Rosen O., Milne T. A., Copeland T. D., Levine S. S., Lee J. C., Hayes D. N., Shanmugam K. S., Bhattacharjee A., Biondi C. A., Kay G. F., Hayward N. K., Hess J. L., Meyerson M. (2004) Mol. Cell 13, 587–597 [DOI] [PubMed] [Google Scholar]
- 10. Yokoyama A., Cleary M. L. (2008) Cancer Cell 14, 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jin S., Mao H., Schnepp R. W., Sykes S. M., Silva A. C., D'Andrea A. D., Hua X. (2003) Cancer Res. 63, 4204–4210 [PubMed] [Google Scholar]
- 12. Sukhodolets K. E., Hickman A. B., Agarwal S. K., Sukhodolets M. V., Obungu V. H., Novotny E. A., Crabtree J. S., Chandrasekharappa S. C., Collins F. S., Spiegel A. M., Burns A. L., Marx S. J. (2003) Mol. Cell Biol. 23, 493–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lemos M. C., Thakker R. V. (2008) Hum. Mutat. 29, 22–32 [DOI] [PubMed] [Google Scholar]
- 14. Balogh K., Patócs A., Hunyady L., Rácz K. (2010) Mol. Cell Endocrinol. 326, 80–84 [DOI] [PubMed] [Google Scholar]
- 15. Yokoyama A., Somervaille T. C., Smith K. S., Rozenblatt-Rosen O., Meyerson M., Cleary M. L. (2005) Cell 123, 207–218 [DOI] [PubMed] [Google Scholar]
- 16. Sorensen P. H., Chen C. S., Smith F. O., Arthur D. C., Domer P. H., Bernstein I. D., Korsmeyer S. J., Hammond G. D., Kersey J. H. (1994) J. Clin. Invest. 93, 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cox M. C., Panetta P., Lo-Coco F., Del Poeta G., Venditti A., Maurillo L., Del Principe M. I., Mauriello A., Anemona L., Bruno A., Mazzone C., Palombo P., Amadori S. (2004) Am. J. Clin. Pathol. 122, 298–306 [DOI] [PubMed] [Google Scholar]
- 18. Chen Y. X., Yan J., Keeshan K., Tubbs A. T., Wang H., Silva A., Brown E. J., Hess J. L., Pear W. S., Hua X. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1018–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Caslini C., Yang Z., El-Osta M., Milne T. A., Slany R. K., Hess J. L. (2007) Cancer Res. 67, 7275–7283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang Y., Hua X. (2007) Mol. Cell Endocrinol. 265–266, 34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grembecka J., Belcher A. M., Hartley T., Cierpicki T. (2010) J. Biol. Chem. 285, 40690–40698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rosenbaum G., Alkire R. W., Evans G., Rotella F. J., Lazarski K., Zhang R. G., Ginell S. L., Duke N., Naday I., Lazarz J., Molitsky M. J., Keefe L., Gonczy J., Rock L., Sanishvili R., Walsh M. A., Westbrook E., Joachimiak A. (2006) Journal of synchrotron radiation 13, 30–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Otwinowski Z., Minor W. (1997) in Methods in Enzymology: Macromolecular Crystallography, Vol. 276, pp. 307–326, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 24. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 25. Sheldrick G. M. (2008) Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 26. Otwinowski Z. (ed) (1991) Isomorphous Replacement and Anomalous Scattering, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 27. Cowtan K. D., Main P. (1993) Acta Crystallogr. D Biol. Crystallogr. 49, 148–157 [DOI] [PubMed] [Google Scholar]
- 28. Perrakis A., Morris R., Lamzin V. S. (1999) Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 29. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 30. Terwilliger T. (2004) Journal of synchrotron radiation 11, 49–52 [DOI] [PubMed] [Google Scholar]
- 31. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 33. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang H., Guranovic V., Dutta S., Feng Z., Berman H. M., Westbrook J. D. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1833–1839 [DOI] [PubMed] [Google Scholar]
- 35. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Eswar N., Webb B., Marti-Renom M. A., Madhusudhan M. S., Eramian D., Shen M. Y., Pieper U., Sali A. (2006) Curr. Protoc. Bioinformatics Chapter 5, Unit 5 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N. (2010) Nucleic Acids Res. 38, W529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Putnam N. H., Srivastava M., Hellsten U., Dirks B., Chapman J., Salamov A., Terry A., Shapiro H., Lindquist E., Kapitonov V. V., Jurka J., Genikhovich G., Grigoriev I. V., Lucas S. M., Steele R. E., Finnerty J. R., Technau U., Martindale M. Q., Rokhsar D. S. (2007) Science 317, 86–94 [DOI] [PubMed] [Google Scholar]
- 39. Blatch G. L., Lässle M. (1999) Bioessays 21, 932–939 [DOI] [PubMed] [Google Scholar]
- 40. Veeramalai M., Ye Y., Godzik A. (2008) BMC Bioinformatics 9, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Parashar V., Mirouze N., Dubnau D. A., Neiditch M. B. (2011) PLoS Biol. 9, e1000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang J., Dye B. T., Rajashankar K. R., Kurinov I., Schulman B. A. (2009) Nat. Struct. Mol. Biol. 16, 987–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J. (2006) Nucleic Acids Res. 34, W116–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. MacConaill L. E., Hughes C. M., Rozenblatt-Rosen O., Nannepaga S., Meyerson M. (2006) Mol. Cancer Res. 4, 793–801 [DOI] [PubMed] [Google Scholar]
- 45. Sullivan J. C., Ryan J. F., Watson J. A., Webb J., Mullikin J. C., Rokhsar D., Finnerty J. R. (2006) Nucleic Acids Res. 34, D495–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yaguchi H., Ohkura N., Takahashi M., Nagamura Y., Kitabayashi I., Tsukada T. (2004) Mol. Cell Biol. 24, 6569–6580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. (2000) Cell 101, 199–210 [DOI] [PubMed] [Google Scholar]
- 48. Milne T. A., Hughes C. M., Lloyd R., Yang Z., Rozenblatt-Rosen O., Dou Y., Schnepp R. W., Krankel C., Livolsi V. A., Gibbs D., Hua X., Roeder R. G., Meyerson M., Hess J. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 749–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.