Abstract
The retinoic acid-related orphan nuclear receptor γt (RORγt)/RORγ2 is well known as a master regulator of interleukin 17 (IL-17)-producing helper T (Th17) cell development. To develop a therapeutic agent against Th17-mediated autoimmune diseases, we screened chemical compounds and successfully found that digoxin inhibited IL-17 production. Further studies revealed that digoxin bound to the ligand binding domain of RORγt and suppressed Th17 differentiation without affecting Th1 differentiation. To better understand the structural basis for the inhibitory activity of digoxin, we determined the crystal structure of the RORγt ligand-binding domain in complex with digoxin at 2.2 Å resolution. The structure reveals that digoxin binds to the ligand-binding pocket protruding between helices H3 and H11 from the pocket. In addition, digoxin disrupts the key interaction important for the agonistic activity, resulting in preventing the positioning of helix H12 in the active conformation, thus antagonizing coactivator interaction. Functional studies demonstrated that digoxin inhibited RORγt activity and decreased IL-17 production but not RORα activity. Digoxin inhibited IL-17 production in CD4+ T cells from experimental autoimmune encephalomyelitis mice. Our data indicates that RORγt is a promising therapeutic target for Th17-derived autoimmune diseases and our structural data will help to design novel RORγt antagonists.
Keywords: Crystal Structure, Drug Action, Interleukin, Nuclear Receptors, Transcription Factors
Introduction
Interleukin 17 (IL-172 also known as IL-17A) is a pro-inflammatory cytokine that promotes inflammation, bone loss, and tissue damage. Many reports have implicated IL-17 in the pathogenesis of autoimmune disease: elevated levels of IL-17 in cerebrospinal fluid of multiple sclerosis (MS) patients (1, 2) and synovial fluid of rheumatoid arthritis (RA) patients (3). In the experimental autoimmune encephalomyelitis (EAE) mice model, suppression of IL-17 activity reduced severity of the symptom (4, 5). Thus, inhibiting IL-17 production has been regarded as a good strategy to treat autoimmune diseases. CD4+ T cells have been well known as the major source of IL-17 production, although the distinct subset of IL-17 producing CD4+ T cells has long been undefined. CD4+ T cells, which produce IL-17 arise directly from naive CD4+ T cells (6, 7), and these cells are widely known as IL-17-producing helper T (Th17) cells (6–9).
In 2006, Ivanov et al. (10) reported that retinoic acid-related orphan nuclear receptor γt (RORγt) was essential for Th17 differentiation in mice. The mice lacking RORγ were less susceptible to EAE (10). CD4+ splenocytes from RORγ−/− mice could not induce EAE (10). Other reports showed that human Th17 cells also express RORγt (11–15). Furthermore, recent two reports showed inhibitory effects of digoxin toward RORγt, and SR1001 against both RORγt and RORα which reduced the severity of EAE (16, 17). Given these reports, RORγt is thought to be a novel, promising therapeutic target for autoimmune diseases.
Nuclear receptors are ligand-regulated transcription factors and composed of several functional domains (18). The N-terminal domain is variable in both length and homology, and contains a constitutively active transactivation function known as activation function-1 (AF-1). Highly conserved DNA-binding domain at the central region recognizes specific DNA sequences in the promoter region of the target genes. The C-terminal ligand binding domain (LBD) is moderately conserved and contains the ligand-dependent transactivation function (termed AF-2). Activation of gene transcription occurs after binding of ligand to the LBD, leading to binding of coactivator to the LBD.
Structural studies have indicated that ligands regulate AF-2 activity by modulating the conformation of LBDs (18). Therefore, the three-dimensional structure of the LBD in complex with ligand is critical for understanding the structural mechanism of ligand action. Nuclear receptor LBDs adopt a canonical antiparallel α-helical sandwich fold generally composed of 12 α-helices (H1-H12) and a small β-sheet. Agonist ligands trigger the LBD activation function AF-2 by stabilizing a defined conformation in which the helix H12 packs against the LBD and generates a hydrophobic coactivator binding surface. Antagonist ligands interfere with the formation of an active LBD conformation and coactivator recruitment. To date, hydroxycholesterols (19–21) and T0901317 (22) have been reported as a ligand for RORγ. The analyses for x-ray crystal structures of the RORγ LBD in complex with hydroxycholesterols and the coactivator peptide have revealed the active conformation of the LBD (19). However, the structural basis of antagonism for RORγt remains unclear.
To develop a therapeutic agent for autoimmune diseases such as MS that inhibit IL-17 production, we screened chemical compounds that inhibit IL-17 production without affecting IL-2 production in EL-4 cells. In this screening, we identified digoxin as a suppressor of IL-17 production. Further studies have revealed that digoxin suppresses RORγt through binding and antagonizing RORγt-LBD, the same as reported recently by Litterman's group (16). We successfully determined the crystal structure of the RORγt LBD in complex with digoxin at 2.2 Å resolution and revealed the structural basis for antagonizing the RORγt activity by digoxin.
EXPERIMENTAL PROCEDURES
Cell Lines and Transfection
Mouse lymphoma EL-4 (23) cells (a gift from Dr. Tsuru, National Defense Medical College) were maintained in DMEM supplemented with 10% FBS. EL-4 cells were transfected with Nucleofector II (Lonza) according to the manufacturer's instructions.
Drug Library Screening
3 × 104 EL-4 cells were seeded in each well of 384 well plate and incubated with the chemical compounds from our library or vehicle (dimethyl sulfoxide; Sigma) for 2 h. Cells were then stimulated with phorbol 12-myristate 13-acetate (PMA) (25 ng/ml; Sigma) and ionomycin (125 ng/ml; Sigma) for 20 h. Subsequently, supernatants were collected to determine the concentrations of IL-17A by HTRF (Cisbio) and IL-2 by AlphaLISA (PerkinElmer) according to the manufacturer's protocols. The compounds which inhibited IL-17 production without affecting IL-2 production were selected.
Mice
C57BL/6 mice were purchased from Charles River Japan (Tokyo, Japan). 6–12-week-old mice were used. All experimental procedures were performed in accordance with the in-house guideline of the Institutional Animal Care and Use Committee in Daiichi Sankyo Co., Ltd.
T Cell Differentiation
CD4+ CD62L+ CD44low T cells were isolated from spleens by negative selection (R&D systems). Cells were stimulated with 10 μg/ml plate-bound anti-CD3e antibody (Ab) and 10 μg/ml soluble anti-CD28 Ab (BioLegend) in RPMI supplemented with 10% FBS. CD4+ T cells were isolated by positive selection with magnetic beads (Miltenyi Biotec) and stimulated with 2 μg/ml soluble anti-CD3e Ab and 1 μg/ml soluble anti-CD28 Ab (R&D systems). For Th17 differentiation, cells were cultured for 3 days in the presence of 5 ng/ml recombinant human TGF-β1 and 10 ng/ml recombinant mouse IL-6 (R&D systems), 10 μg/ml anti-interferon-γ (IFN-γ) Ab, and 10 μg/ml anti-IL-4 Ab (BD Biosciences). For Th1 differentiation, cells were cultured for 3 days in the presence of 10 ng/ml recombinant mouse IL-12 (R&D systems or Peprotech) and 10 μg/ml anti-IL-4 Ab. Also, we added recombinant mouse IL-2 (Roche Applied Science) at 50 ng/ml (naive T cells) or 5 ng/ml (CD4+ T cells).
Flow Cytometry
For intracellular cytokine staining, cells were stimulated with PMA (50 ng/ml) and ionomycin (750 ng/ml) for 6 h. After 1 h of incubation, BD Golgiplug (BD Biosciences) was added and incubated for 5 h. Then, cells were fixed, permeabilized, and stained with PE-conjugated anti-mouse IL-17A Ab and FITC-conjugated anti-mouse IFN-γ Ab (BD Biosciences). Cell sorting was performed using FACSCantoII (BD Biosciences).
Real-time PCR
RNA was extracted with RNeasy mini kit using DNase I (Qiagen), then cDNA was synthesized with high capacity cDNA Reverse Transcription kits (Applied Biosystems). TaqMan Gene Expression Assays and TaqMan Universal PCR Master Mix with ABI prism 7700 Sequence Detector (Applied Biosystems) were used for analysis of cDNA by real-time PCR. The expression of each gene was normalized to the expression of 18 S rRNA by the standard curve method. The assay ID of primer sets were as follows: RORγt Mm01261022_m1; RORα Mm01173773_m1; AhR Mm00478932_m1; IRF4 Mn00516431_m1; IL-17A; Mm00439619_m1; IL-17F Mm00521423_m1; IL-23R Mm00519942_m1; 18 S rRNA 4319413.
Vector Construction
Full-length wild type mouse RORγt (mRORγt) and mouse RORα (mRORα) were amplified from cDNA of mouse splenocytes and cloned into pUNO vector (Invivogen) or pSG5 vector (Agilent Technologies). 12 × RORE-Luc was constructed by inserting twelve repeats of the consensus RORγ-response element (GGTAAGTAGGTCAC) (24) into PGVP2 vector (TOYO-B-Net). The insert was obtained by primer ligation.
Cytokine Detection Assays
First, we transiently transfected EL-4 cells with plasmids encoding mRORγt, mRORα, or mock. These cells were incubated with digoxin for 1–4 h, then stimulated with PMA (50 ng/ml) and ionomycin (250 ng/ml) for 20–24 h. CD4+ T cells from EAE-induced mice were incubated with digoxin for 1–4 h, then stimulated with 1 μg/ml soluble anti-CD3e Ab and 1 μg/ml soluble anti-CD28 Ab for 24 h. Naive CD4+ T cells were incubated in Th17- or Th1- polarizing conditions in the presence or absence of digoxin for 72 h. Subsequently, supernatants were collected to determine the concentrations of IL-17A by HTRF and IFN-γ by AlphaLISA (PerkinElmer) according to the manufacturer's protocols.
Reporter Assay
EL-4 cells were co-transfected with PGV-P2-RORE vector and plasmids encoding mRORγt, mRORγ or mock. Cells were incubated with digoxin and incubated for 24 h. Luciferase activity was measured using Bright-Glo Luciferase Assay System (Promega).
EAE Induction
Male C57BL/6 mice was immunized subcutaneous with 200 μg of MOG35–55 peptide emulsified in a total of 100 μl of incomplete Freund's adjuvant containing 450 μg of Mycobacterium tuberculosis H37 Ra on day 0, supplemented with intravenous injection of 100 ng of pertussis toxin. The spleen of EAE-induced mice was collected on day 9.
Expression and Purification of the RORγt LBD Region
The human RORγt LBD (residues 265–507) was expressed with an N-terminal histidine tag and a thrombin cleavage site in E. coli BL21 (DE3) by using pET-15b vector (Novagen). The cells were grown in 2xYT at 37 °C up to 1.0 OD, then induced with 0.1 mm isopropyl-β-d-thiogalactopyranoside (IPTG). The incubation was continued overnight at 16 °C. Cells were resuspended in 20 mm Tris-HCl (pH 7.0), 0.5 m NaCl, 25 mm imidazole, 5 mm DTT, and Roche EDTA-free protease inhibitor mixture, followed by sonication. The lysate was centrifuged at 40,000 × g. for 30 min at 4 °C, and the supernatant was loaded on a HisTrap nickel affinity column (GE Healthcare). The column was washed with 20 mm Tris-HCl (pH 7.0), 0.5 m NaCl, 25 mm imidazole, 5 mm DTT, and the protein was eluted with a gradient of 25–500 mm imidazole. Fractions containing RORγt were pooled and the His6 tag was removed by thrombin digestion overnight at 4 °C, dialyzed against 20 mm Tris-HCl (pH 7.0), 0.5 m NaCl, 25 mm imidazole, 5 mm DTT and followed by a second nickel affinity column chromatography. Flow-through fractions were pooled and dialyzed against 20 mm Tris-HCl (pH 7.0), 0.1 m NaCl, 5 mm DTT. The RORγt LBD without His tag was further separated by ion exchange chromatography using a HiTrap SP HP column (GE Healthcare). Subsequent gel filtration was performed on a HiLoad 16/60 Superdex 75 column (GE Healthcare) using elution buffer 10 mm HEPES (pH 7.5), 0.15 m NaCl, 5 mm DTT. The purified protein was concentrated to 9 mg/ml and stored at −80 °C until used for binding assay or crystallization.
Binding Assay
5 nm Fluorescein-labeled LXR agonist T0901317, also known as the RORγ inverse agonist recently (22), was incubated with the RORγ LBD for 4 h at 4 °C in the presence of various concentration of testing compounds. Fluorescent polarization signals (mP) were measured with EnVision (PerkinElmer). Each value of fluorescent polarization samples in the presence of testing compounds were normalized by subtracting the value in the absence of testing compounds, and shown as ΔmP.
Statistical Analysis
All analyses for statistically significant differences were performed with the 2-tailed paired Student's t test. p-Values less than 0.05 were considered significant. All error bars shown in the figures in this article are S.D.
Crystallization, Data Collection, Structure Determination, and Refinement
Crystals were obtained at 22 °C with the vapor-diffusion method in sitting-drops containing 2 μl of protein-digoxin solution and 1 μl of reservoir solution (1.2 m sodium formate, 3% (v/v) 2-methyl-2,4-pentanediol, 0.1 m HEPES (pH 7.5)). Crystals were cryoprotected by equilibration in the reservoir solution containing 20% glycerol, then flash-cooled into the liquid N2 stream at 100 K. X-ray diffraction data were collected at the beamline AR-NW12 at Photon Factory, Tsukuba, Japan, then integrated and scaled using the program HKL-2000 (25). The structure was solved by molecular replacement with the program PHASER (26) using the crystal structure of the RORγ LBD (Protein Data Bank (PDB) accession code 3L0L) (19) without the ligand, peptide, and water molecules as a search model. The model was rebuilt manually with the program COOT (27) and refined with the program REFMAC (28). The stereochemical quality of the model was assessed with the program PROCHECK (29). Structure analysis was performed using the programs in the CCP4 suite (30). Figures were prepared with the program PyMol (31). The statistics of the diffraction data and structure refinement are summarized in Table 1. The atomic coordinates have been deposited in the PDB (accession code: 3B0W).
TABLE 1.
Data collection and refinement statistics
| Data collection statistics | |
| Wave length (Å) | 1.000 |
| Space group | P61 |
| Cell dimension | |
| a = b (Å) | 98.7 |
| c (Å) | 129.2 |
| Resolution range | 40.00–2.2 (2.28–2.2)a |
| Observed reflections | 413,006 |
| Unique reflections | 36,249 |
| Average redundancy | 11.4 (11.5) |
| Average I/σ(I) | 44.9 (4.6) |
| Completeness (%) | 99.8 (100.0) |
| Rmerge | 0.056 (0.470) |
| Refinement statistics | |
| Resolution range | 20.00–2.2 (2.26–2.2) |
| Rfactor | 0.212 (0.238) |
| Free R factor | 0.249 (0.302) |
| No. of atoms | |
| Protein | 3,516 |
| Ligand | 110 |
| Waters | 73 |
| R.m.s. deviations | |
| Bonds (Å) | 0.011 |
| Angles (°) | 1.258 |
| Average B-factors (Å2) | |
| Protein | 55.1 |
| Ligand | 60.2 |
| Waters | 41.4 |
| Ramachandran plot | |
| Most favored | 383 (94.8%) |
| Allowed | 19 (4.7%) |
| Generous allowed | 0 (0%) |
| Disallowed | 2 (0.5%) |
a Values in parentheses are for the highest-resolution shell.
RESULTS
Digoxin Inhibited the Differentiation into Th17 Cells in Vitro
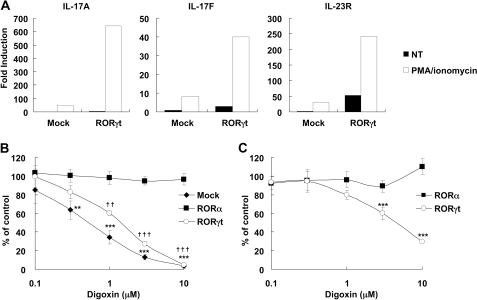
EL-4 cells produce IL-17 and IL-2 in response to PMA plus ionomycin treatment. To identify the small molecules that could inhibit IL-17 production, we have screened more than 700 compounds whose targets are already known and found digoxin as a candidate compound. Digoxin inhibited IL-17 production without affecting IL-2 production (data not shown). We next isolated CD4+ CD62L+ CD44low naive T cells and cultured them under Th17- or Th1-polarizing conditions to evaluate the effect of digoxin on the differentiation toward Th17 cells. Digoxin suppressed the generation of Th17 cells at the concentration of 10 μm. The IL-17 production during differentiation was also suppressed (Fig. 1). Interestingly, the generation of Th1 cells and the IFN-γ production during differentiation were not suppressed by digoxin, indicating that the inhibitory activity of digoxin is specific to Th17-differentiation. We obtained similar results using CD4+ T cells (supplemental Fig. S1A). Expression levels of IL-17A, IL-17F, and IL-23 receptor (IL-23R) mRNAs were kept much lower in digoxin-treated cells (supplemental Fig. S1B). The expression levels of RORγt (10), RORα (32), IRF-4 (33), and AhR (34) were up-regulated and no substantial difference in mRNA expression levels have been observed regardless with or without digoxin treatment (supplemental Fig. S1C). These results suggest that digoxin had an inhibitory activity on Th17 differentiation without affecting the expression levels of RORγt, RORα, IRF-4, and AhR in vitro.
FIGURE 1.
Digoxin suppressed differentiation to Th17 cells in vitro. Flow cytometry of intracellular IL-17 and IFN-γ (left panels) and cytokine production (right panels) during differentiation. Naive CD4+ T cells isolated from the spleens of C57BL/6 mice were cultured under Th1- (left upper panels) or Th17- (left lower panels) polarizing conditions in the presence or absence of digoxin. After 3 days in culture, supernatants were collected and cells were restimulated with PMA and ionomycin. Cytokine productions were assessed by intracellular staining (left panels, one representative of three data sets.). Concentrations of IL-17 and IFN-γ in supernatants were also determined by HTRF (for IL-17) and AlphaLISA (for IFN-γ) (right panels, data were shown as the mean ± S.D. of triplicate samples. ***, p < 0.001). All experiments were repeated at least once.
Binding Assay and Crystal Structure Determination
RORγt is known as a key regulator of Th17 differentiation. To determine whether digoxin binds and inhibits RORγt activity, we performed a competitive binding assay. As shown in the supplemental Fig. S2, digoxin competed with labeled T0901317 in a dose-dependent manner indicating that digoxin binds to RORγ LBD directly.
To gain structural insights into the mechanism of action, we determined the crystal structure of the RORγt LBD in complex with digoxin at 2.2 Å resolution (Rcryst = 0.212, Rfree = 0.249) (Fig. 2, A and B). The results of the crystallographic refinement are summarized in Table 1. The nomenclature of the secondary structure elements is based on the RXR LBD crystal structure (35). The final model in the asymmetric unit contains two protein molecules (monomer A: residues 265–479 and monomer B: residues 265–481), two digoxin molecules, and 73 water molecules. Both LBDs are very similar with root mean square deviations (rmsd) of 0.27 Å for 212 Cα atoms and were treated identically in this report except where noted otherwise.
FIGURE 2.
Structure of the RORγt LBD in complex with digoxin. A, chemical structure of digoxin with numbering. B, σA-weighted 2Fo-Fc electron density map shown as a dark blue mesh around digoxin. The protein is drawn as a stick model and digoxin is depicted as a ball-and-stick model. Contour level is 1.2 σ. C, schematic representation of the RORγt LBD (dark blue) with digoxin. The secondary structure assignment, and the N and C termini are labeled. Bound digoxin is depicted as a CPK model with carbon atoms in cyan and oxygen atoms in red. D, superposition of two RORγt LBDs in the asymmetric unit of the crystal. The protein is drawn as a cartoon model with molecule A and molecule B colored in pink and dark blue. Digoxin is shown as a stick model with carbons in orange for monomer A and carbons in cyan for monomer B. The structures of a lactone group, central steroidal core and digitoxose X of digoxin are superposed well. The conformation of digitoxose Y and Z are significantly different.
Overall Structure
The RORγt-LBD presents the canonical fold for the nuclear receptor LBDs (Fig. 2C) (36). Whereas the electron densities are excellent quality from residue 265 to residue 479 or residue 481, residues from a C-terminal part of H11 to H12 could not be modeled because no clear electron density was observed for this region. There are residual electron densities in the region around helices H3 and H4 (supplemental Fig. S3), which is the binding site for coactivator or corepressor peptides as well as helix H12 in some nuclear receptor structures (37). This region with residual densities is located between two LBDs in the asymmetric unit, related by 2-fold noncrystallographic symmetry. Residues from the C-terminal part of H11 to H12 of the RORγt LBD in complex with digoxin are most likely to be highly mobile and randomly oriented. In addition, it is probable that a small part of helix H12 from molecules in the crystal is located in the region near helices H3 and H4.
Digoxin in the Ligand-binding Pocket
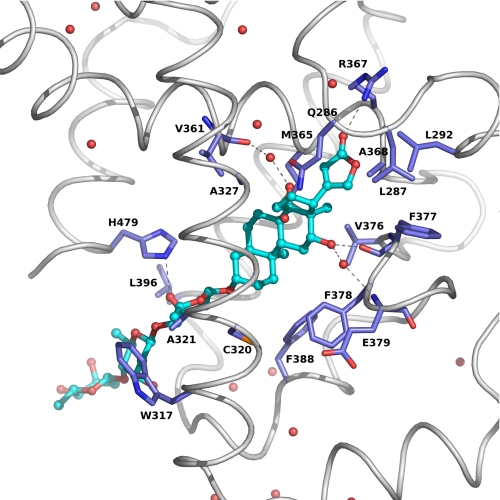
The crystal structure has showed that digoxin is bound in the ligand-binding pocket of the RORγt LBD (Fig. 2C). The average B values for digoxin (61.2 Å2 for the A chain and 59.3 Å2 for the B chain) are higher than the average B values for the protein (54.2 Å2 for the A chain and 56.1 5 Å2 for the B chain). The structures of a lactone group, central steroidal core, and digitoxose X of digoxin between A and B chains have almost the same conformation and are superposed well. However, the conformations of digitoxose Y and Z are significantly different (Fig. 2D) because digitoxose Y and Z protrudes between helices H3 and H11 and interacts the neighboring molecule in the crystal, suggesting that digitoxose Y and Z of digoxin are flexible in the complex structure. The average B values of the atoms of digitoxose X, Y, and Z are 57.0 Å2, 77.4 Å2 and 91.3 Å2 for the A chain, and 59.0 Å2, 75.2 Å2 and 80.0 Å2 for the B chain, respectively. However, both average B values of the lactone and steroidal core from the A and B chains are 47.6 Å2, which is the smaller value compared with those of the protein. Therefore, it is likely that the lactone group, central steroidal core and digitoxose X of digoxin bind to RORγt with specific interaction in the cavity of the protein, whereas both digitoxose Y and Z are flexible outside the protein. Inside the ligand pocket, digoxin forms extensive hydrophobic and hydrophilic interactions with RORγt, including three direct hydrogen bonds, two hydrogen bonds though water molecules, and 43 van der Waals contacts (Fig. 3). The O12, O23, and O3X atoms of digoxin make direct hydrogen bond interaction with O-Phe377 in β-strand 1 (s1), NH1-Arg367 in helix H5 and NE2-His479 in helix H11, respectively. The O12 and O14 atoms of digoxin make hydrogen bond interaction mediated through water molecules with N-Glu379 in loop s1-s2 and O-Val361 in H5, respectively. The following amino acid residues have a non-hydrogen atom closer than 4 Å to digoxin: Gln-286, Leu-287, Leu-292, Trp-317, Cys-320, Ala-321, Ala-327, Val-361, Arg-364, Met-365, Arg-367, Ala-368, Val-376, Phe-377, Phe-378, Phe-388, Leu-396, His-479.
FIGURE 3.
Details of the binding model of digoxin to the RORγt LBD. Residues of the RORγt involved in the binding of digoxin are shown as a stick model and digoxin is drawn as ball-and-stick. Hydrogen bonds are marked as dotted lines.
Comparison of the Digoxin-bound and the Agonist-bound LBDs
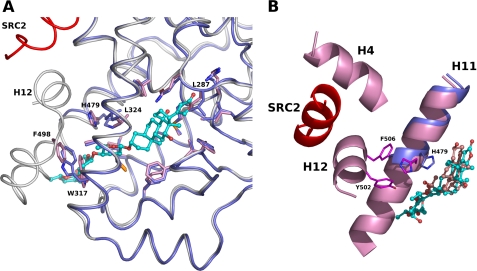
To better understand the structural basis of the digoxin antagonism, we compared the structures of the RORγt LBD with digoxin and with the agonist 25-hydroxycholesterol (PDB accession code: 3L0L) (19) (Fig. 4A). The overall structures of the digoxin-bound and hydroxylcholesterol-bound LBDs are similar from H1 to H11 with rmsd of 0.67 Å for 214 Cα atoms. Within the ligand-binding pocket, digoxin induces the conformational changes of the side chain atoms of Leu-287 in loop H1-H2 to create necessary space for the lactone group of digoxin. In addition, Leu-324 has a different rotamer for the accommodation of the steroidal core of digoxin. More dramatic differences caused by the digoxin binding to the RORγt LBD occurs in the C-terminal part of the LBD. Digoxin induces the conformational change of the side chain of His-479 in H11, forming a hydrogen bond between the O3X atom of digitoxose X in digoxin and NE2-His479. As a result, digoxin disrupts polar interaction observed in the agonist-bound RORγt LBD, involving His-479 in H11, Tyr-502 in H11′ and Phe-506 in H12, which would be important to stabilize the active conformation of helix H12 (Fig. 4B). Furthermore, digitoxose X and Y of digoxin points toward helix H12 and exits the binding pocket between H3 and H11. The superposition of these two structures has showed that there is a steric clash between digitoxose Z in the digoxin-bound LBD and helix H11 in the hydroxycholesterol-bound LBD structures. In addition, digoxin binding induces a different side chain conformation of Trp-317 in H3. As a consequence, a displacement of Phe-498 in loop H11′-H12 occurs to avoid a steric clash with Trp-317, which would also cause the large movement of H12. Thus, digoxin prevents the positioning of helix H12 in the active conformation as observed in the RORγt LBD complexes with the agonists.
FIGURE 4.
Superposition of the RORγt LBDs in complex with digoxin and with the agonist. A, digoxin induces a conformational change of Trp-317. The LBD (dark blue) with digoxin and the LBD (white) with the agonist hydroxycholesterol are depicted as a cartoon model. The coactivator SRC2 peptide is colored in red. Residues of RORγt involved in the binding of digoxin are shown as a stick model, and the corresponding residues in the agonist-bound LBD are also depicted. B, digoxin disrupts cation-π interaction observed in the agonist-bound RORγt LBD. Cartoon representation of the helix H11 (dark blue) of the digoxin-bound RORγt LBD and helices H4, H11, and H12 (pink) of the hydroxycholesterol-bound RORγt LBD are shown. The SRC2 peptide is colored in red. Residues His-479, Tyr-502, and Phe-506 involved in the cation-π interaction in the agonist-bound RORγt LBD (carbon, magenta) are shown as a stick model. The side chain atoms of His-479 (carbon, dark blue) are moved and make a hydrogen bond with digoxin. Digoxin (carbon, cyan) and hydroxycholesterol (carbon, pink) are shown as a ball-and-stick model.
Digoxin Inhibited the Activity of RORγt but Not RORα in T Cells
We next examined whether digoxin could inhibit the activity of RORγt in cells. As there was no report regarding cell lines that produce IL-17 in RORγt-dependent manner, we initially screened cell lines that could be used to monitor the activity of RORγt. We transfected mRORγt expression vector in several T cell lines and found that EL-4 cells responded to mRORγt overexpression for up-regulation of IL-17A and IL-23R mRNA expression. When cells were stimulated with PMA and ionomycin, significant levels of IL-17A, IL-17F, and IL-23R were observed in EL-4 cells transfected with mRORγt expression vector, while a smaller effect was seen without the stimulation (Fig. 5A).
FIGURE 5.
RORγt overexpression induces the expression of IL-17A, IL-17F, and IL-23R in EL-4 cells. A, EL-4 cells were transfected with control (Mock) or mRORγt expression vector (RORγt) followed by stimulation with PMA and ionomycin (PMA/ionomycin) or no stimulation (NT). mRNA expression of the indicated genes were analyzed by real-time PCR. Repeated experiments gave similar results. B, EL-4 cells were transiently transfected with control (Mock), mRORγt (RORγt), or mRORα expression vector (RORα), then stimulated with PMA/ionomycin in the presence of indicated concentrations of digoxin for 24 h. IL-17 concentrations of supernatants were determined by HTRF. Data were shown as the mean ± S.D. of quadruplicate samples measuring % of DMSO-treated control. ** and ††, p < 0.01, *** and †††, p < 0.001 compared with each DMSO-treated control. Repeated experiments gave similar results. C, EL-4 cells were transiently transfected with mRORγt or mRORα together with 12 × RORE-Luc luciferase construct. Then, cells were incubated in the presence of the indicated concentrations of digoxin for 24 h. Luciferase activity was analyzed. Data were shown as the mean ± S.D. for triplicate samples measuring % of DMSO-treated control. ***, p < 0.001 compared with each DMSO-treated control. All experiments were repeated at least once.
Then, we investigated the effect of digoxin toward IL-17 production using EL-4 cells transfected with mock, mRORγt, or mRORα plasmid. While mRORγt or mRORα expressing EL-4 cells produced a large amount of IL-17 compared with mock transfectants (data not shown), digoxin suppressed RORγt-induced IL-17 production in a dose-dependent manner (Fig. 5B), but not IL-2 production (data not shown). Interestingly, digoxin had no effect on RORα-induced expression of IL-17 (Fig. 5B), suggesting that digoxin specifically inhibited RORγt activity in cells. The IL-17 expression of mock-transfected EL-4 cells was also inhibited by digoxin (Fig. 5B), indicating that digoxin inhibited the activity of endogenous RORγt in EL-4 cells. Indeed, the production of IL-17 by EL-4 cells in response to PMA plus ionomycin or anti-CD3e Ab plus anti-CD28 Ab were decreased when the expression of RORγt was down-regulated with small interfering RNA (data not shown).
This prompted us to investigate whether digoxin could inhibit the transcriptional activity of RORγt. We performed reporter assays with 12 × RORE-Luc that contained twelve repeats of ROR binding elements at the promoter region (24). Consistent with the results above, the expression of exogenous mRORγt or mRORα enhanced the luciferase reporter activity in EL-4 cells (data not shown). Again, digoxin suppressed the luciferase activity in a dose-dependent fashion in cells transfected with mRORγt but not RORα (Fig. 5C). Together, these results indicate that digoxin inhibits the transcriptional activity of RORγt but not RORα in T cells, resulting in the suppression of Th17 differentiation and IL-17 production.
Digoxin Inhibited IL-17 Production from Differentiated Th17 Cells
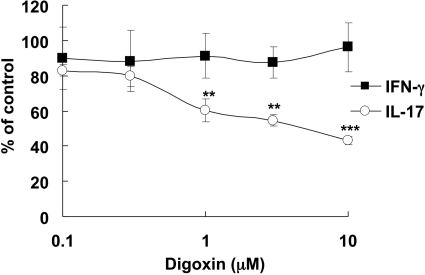
Differentiated Th17 cells express RORγt together with RORα that has similar DNA-binding specificity. As digoxin only inhibited the activity of RORγt, we next examined the ability of digoxin to modulate IL-17 production, the function of differentiated Th17 cells. CD4+ T cells in spleens from MOG-immunized EAE-induced mice were incubated with various concentrations of digoxin, and then stimulated with anti-CD3e Ab and anti-CD28 Ab. Digoxin suppressed IL-17 production in the stimulated cells in a dose-dependent manner, but not IFN-γ production at all (Fig. 6). Digoxin also inhibited MOG-mediated IL-17 production from CD4+ T cells in spleens of MOG-immunized mice co-cultured with γ -irradiated antigen presenting cells (data not shown). These data suggest that the suppression of RORγt-activity leads to inhibiting the IL-17 production from differentiated Th17 cells in response to stimulus, as well as to inhibit the differentiation toward Th17 cells.
FIGURE 6.
Digoxin inhibits IL-17 production but not IFN-γ production from CD4+ T cells derived from EAE mice. CD4+ T cells from MOG-immunized mice (Day 9) were stimulated with 1 μg/ml of soluble anti-CD3e Ab and 1 μg/ml of soluble anti-CD28 Ab for 24 h. Cytokine concentrations of the supernatants were determined by HTRF (for IL-17, open circles) and AlphaLISA (for IFN-γ, closed squares). Data were shown as the mean ± S.D. for quadruplicate samples measuring % of DMSO-treated control. **, p < 0.01, ***, p < 0.001 compared with each DMSO-treated control. All experiments were repeated at least once.
DISCUSSION
RORγt has been known as a master regulator in Th17 differentiation, and thought to be a promising therapeutic target for autoimmune diseases. We have screened chemical compounds for the inhibitors of IL-17 production and found that digoxin had an activity to inhibit RORγt. Recently, Littman's group have also reported that digoxin is an antagonist for RORγt (16). Consistent with the report, we found that digoxin inhibited Th17 differentiation and the RORγt-reporter activity by directly binding to RORγt but not inhibited RORα activity. In addition to these findings, we determined the crystal structure of the RORγt LBD in complex with digoxin, which revealed the structural basis of digoxin for the antagonizing RORγt activity. Moreover, our functional assay showed that digoxin inhibited IL-17 production in T cells from EAE mice.
Transcriptional activity of nuclear receptors is regulated by their interactions with co-activators such as steroid receptor coactivator (SRC) family of proteins. Coactivator contains an LXXLL motif that forms α-helix and binds into a hydrophobic cleft on the surface of the receptor, which is stabilized by the complex formation with agonist in the active LBD structure. In the RORγt LBD, when complexed with hydroxycholesterol and the coactivator SRC2 peptide, the SRC2 peptide binds to the cleft formed by helices H3, H4, and H12 (19). Some ligands, such as rogiglitazone, directly contact and stabilize helix H12 of the respective receptor, placing it in the agonist conformation (38). However, hydroxycholesterols stabilize the RORγt helix H12 through indirect cation-π interaction, involving residues His-479, Tyr-502, and Phe-506, as similarly observed in LXR (39) and VDR (40). The crystal structure analysis in this study has revealed that digoxin binds to the same cavity as agonist and disrupts this key cation-π interaction important for the active conformation (Fig. 4B). Mutational studies on RORα have described that the mutations of H484W in H11 (corresponding to His-479 in RORγt) and Y507F in H12 (corresponding to Tyr-502 in RORγt) significantly reduced the transcriptional activity of RORα (41). Moreover, the volume of digoxin is too large to be contained within the buried binding site. As a consequence, Helix-12 is displaced to allow the bulky extension to protrude between helices H3 and H11 from the cavity (Fig. 2C). In the antagonist-bound LBD structure of ERα and of RARα, the helix H12 packs on the groove formed by H3 and H4, due to the amphipathic character of H12 (42–44). In contrast, the binding of the antiestrogen ICI 164384 to the ERβ LBD completely abolishes the association between H12 and the remainder of the LBD thus H12 cannot adopt a defined position (45). The helix H12 of RORγt in complex digoxin appears to exist as a dynamic ensemble of conformations and/or in the position near helices H3 and H4, resulting in the incomplete formation of the binding surface for the LXXL motifs of the coactivator. Consequently, it is likely that antagonism of digoxin originates from the disruption of the key interaction critical for the agonistic activity and the presence of a large extension of digoxin that sterically prevents the proper LBD-coactivator interface being formed.
One of the reasons why it had been difficult to demonstrate the role of RORγt in IL-17 production is that there was no cell line that produce IL-17 in RORγt-dependent manner. Although ectopic expression of RORγt in mouse and human CD4+ naive T cells induced IL-17 expression (15, 46–48), no direct evidence has been reported due to the difficulty to distinguish its effect on the IL-17 production from the Th17 differentiation. In this study, we found that EL-4 cells transfected with RORγt produced elevated levels of IL-17 compared with mock transfectants. Similar findings were also recently reported by Solt et al. (17) during preparation of our manuscript. In our results using EL-4 cells, RORα as well as RORγt promoted IL-17 production. However, digoxin only inhibited RORγt-mediated IL-17 production but not in RORα-expressing cells (Fig. 5B).
RORα is a closely related family member of RORγt which is also expressed in differentiated Th17 cells. Unlike RORγt and RORγ, the expression of RORα is ubiquitous with various roles including lipid metabolism, smooth muscle differentiation, bone metabolism, cerebellar development, and smooth muscle differentiation. Actually, Solt et al. revealed that SR1001, a dual antagonist against RORγt and RORα, reduced severity of EAE (17). Thus, from the drug discovery point of view, it is an earnest issue whether inhibiting RORγt alone would be sufficient for suppressing IL-17 production. Indeed, our data using CD4+ T cells from EAE mice showed that RORγt supression was effective in blocking IL-17 expression (Fig. 6).
The expression level of IL-23R was also enhanced during Th17 differentiation. This enhancement was also suppressed completely by digoxin, indicating the importance of RORγt in IL-23R expression (supplemental Fig. S1B). Signals from IL-23R are thought to be essential to maintain Th17 cells (48). Thus, RORγt-antagonist could inhibit a Th17-mediated response completely by suppressing multiple steps such as Th17-differentiation, IL-17 production from differentiated Th17 cells and the maintenance of Th17 cells.
In summary, we identified digoxin as a RORγt antagonist and showed that RORγt-specific antagonists could be a promising therapeutic agent against Th17-derived autoimmune diseases. The crystal structure of the RORγt LBD in complex with digoxin also provides the basis for drug design to obtain more potent molecules of this potential new drug target.
Supplementary Material
Acknowledgments
We thank the staff of the Photon Factory NW-12 and BL-5A at Tsukuba for support with diffraction data collection, Dr. Sumiaki Tsuru for providing EL-4 cells, Atsushi Satoh for his support with screening, Mariko Kobayashi and Rinko Toyama for their assistance, Dr. Keiko Tamai for helpful discussions.
The atomic coordinates and structure factors (code 3B0W) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- IL
- interleukin
- RORγ
- retinoic acid-related orphan nuclear receptor γ
- LBD
- ligand-binding domain.
REFERENCES
- 1. Matusevicius D., Kivisäkk P., He B., Kostulas N., Ozenci V., Fredrikson S., Link H. (1999) Mult. Scler. 5, 101–104 [DOI] [PubMed] [Google Scholar]
- 2. Lock C., Hermans G., Pedotti R., Brendolan A., Schadt E., Garren H., Langer-Gould A., Strober S., Cannella B., Allard J., Klonowski P., Austin A., Lad N., Kaminski N., Galli S. J., Oksenberg J. R., Raine C. S., Heller R., Steinman L. (2002) Nat. Med. 8, 500–508 [DOI] [PubMed] [Google Scholar]
- 3. Kotake S., Udagawa N., Takahashi N., Matsuzaki K., Itoh K., Ishiyama S., Saito S., Inoue K., Kamatani N., Gillespie M. T., Martin T. J., Suda T. (1999) J. Clin. Invest. 103, 1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Komiyama Y., Nakae S., Matsuki T., Nambu A., Ishigame H., Kakuta S., Sudo K., Iwakura Y. (2006) J. Immunol. 177, 566–573 [DOI] [PubMed] [Google Scholar]
- 5. Hofstetter H. H., Ibrahim S. M., Koczan D., Kruse N., Weishaupt A., Toyka K. V., Gold R. (2005) Cell Immunol. 237, 123–130 [DOI] [PubMed] [Google Scholar]
- 6. Harrington L. E., Hatton R. D., Mangan P. R., Turner H., Murphy T. L., Murphy K. M., Weaver C. T. (2005) Nat. Immunol. 6, 1123–1132 [DOI] [PubMed] [Google Scholar]
- 7. Park H., Li Z., Yang X. O., Chang S. H., Nurieva R., Wang Y. H., Wang Y., Hood L., Zhu Z., Tian Q., Dong C. (2005) Nat. Immunol. 6, 1133–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dong C. (2006) Nat. Rev. Immunol. 6, 329–333 [DOI] [PubMed] [Google Scholar]
- 9. Harrington L. E., Mangan P. R., Weaver C. T. (2006) Curr. Opin. Immunol. 18, 349–356 [DOI] [PubMed] [Google Scholar]
- 10. Ivanov I. I., McKenzie B. S., Zhou L., Tadokoro C. E., Lepelley A., Lafaille J. J., Cua D. J., Littman D. R. (2006) Cell 126, 1121–1133 [DOI] [PubMed] [Google Scholar]
- 11. Annunziato F., Cosmi L., Santarlasci V., Maggi L., Liotta F., Mazzinghi B., Parente E., Filì L., Ferri S., Frosali F., Giudici F., Romagnani P., Parronchi P., Tonelli F., Maggi E., Romagnani S. (2007) J. Exp. Med. 204, 1849–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Acosta-Rodriguez E. V., Rivino L., Geginat J., Jarrossay D., Gattorno M., Lanzavecchia A., Sallusto F., Napolitani G. (2007) Nat. Immunol. 8, 639–646 [DOI] [PubMed] [Google Scholar]
- 13. Wilson N. J., Boniface K., Chan J. R., McKenzie B. S., Blumenschein W. M., Mattson J. D., Basham B., Smith K., Chen T., Morel F., Lecron J. C., Kastelein R. A., Cua D. J., McClanahan T. K., Bowman E. P., de Waal Malefyt R. (2007) Nat. Immunol. 8, 950–957 [DOI] [PubMed] [Google Scholar]
- 14. Yang L., Anderson D. E., Baecher-Allan C., Hastings W. D., Bettelli E., Oukka M., Kuchroo V. K., Hafler D. A. (2008) Nature 454, 350–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Unutmaz D. (2009) Eur. J. Immunol. 39, 1452–1455 [DOI] [PubMed] [Google Scholar]
- 16. Huh J. R., Leung M. W., Huang P., Ryan D. A., Krout M. R., Malapaka R. R., Chow J., Manel N., Ciofani M., Kim S. V., Cuesta A., Santori F. R., Lafaille J. J., Xu H. E., Gin D. Y., Rastinejad F., Littman D. R. (2011) Nature 472, 486–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Solt L. A., Kumar N., Nuhant P., Wang Y., Lauer J. L., Liu J., Istrate M. A., Kamenecka T. M., Roush W. R., Vidović D., Schürer S. C., Xu J., Wagoner G., Drew P. D., Griffin P. R., Burris T. P. (2011) Nature 472, 491–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Renaud J. P., Moras D. (2000) Cell Mol. Life Sci. 57, 1748–1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jin L., Martynowski D., Zheng S., Wada T., Xie W., Li Y. (2010) Mol. Endocrinol. 24, 923–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y., Kumar N., Solt L. A., Richardson T. I., Helvering L. M., Crumbley C., Garcia-Ordonez R. D., Stayrook K. R., Zhang X., Novick S., Chalmers M. J., Griffin P. R., Burris T. P. (2010) J. Biol. Chem. 285, 5013–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Y., Kumar N., Crumbley C., Griffin P. R., Burris T. P. (2010) Biochim. Biophys. Acta 1801, 917–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar N., Solt L. A., Conkright J. J., Wang Y., Istrate M. A., Busby S. A., Garcia-Ordonez R. D., Burris T. P., Griffin P. R. (2010) Mol. Pharmacol. 77, 228–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maeda H., Tsuru S., Shiraishi A. (1994) Jpn. J. Cancer Res. 85, 1137–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Medvedev A., Yan Z. H., Hirose T., Giguère V., Jetten A. M. (1996) Gene 181, 199–206 [DOI] [PubMed] [Google Scholar]
- 25. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 28. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 30. CCP4 (1994) Acta Crystallogr. D. Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 31. DeLano W. L. (2002) The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA [Google Scholar]
- 32. Yang X. O., Pappu B. P., Nurieva R., Akimzhanov A., Kang H. S., Chung Y., Ma L., Shah B., Panopoulos A. D., Schluns K. S., Watowich S. S., Tian Q., Jetten A. M., Dong C. (2008) Immunity 28, 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brüstle A., Heink S., Huber M., Rosenplänter C., Stadelmann C., Yu P., Arpaia E., Mak T. W., Kamradt T., Lohoff M. (2007) Nat. Immunol. 8, 958–966 [DOI] [PubMed] [Google Scholar]
- 34. Veldhoen M., Hirota K., Westendorf A. M., Buer J., Dumoutier L., Renauld J. C., Stockinger B. (2008) Nature 453, 106–109 [DOI] [PubMed] [Google Scholar]
- 35. Bourguet W., Ruff M., Chambon P., Gronemeyer H., Moras D. (1995) Nature 375, 377–382 [DOI] [PubMed] [Google Scholar]
- 36. Wurtz J. M., Bourguet W., Renaud J. P., Vivat V., Chambon P., Moras D., Gronemeyer H. (1996) Nat. Struct. Biol. 3, 87–94 [DOI] [PubMed] [Google Scholar]
- 37. Gronemeyer H., Gustafsson J. A., Laudet V. (2004) Nat. Rev. Drug Discov. 3, 950–964 [DOI] [PubMed] [Google Scholar]
- 38. Nolte R. T., Wisely G. B., Westin S., Cobb J. E., Lambert M. H., Kurokawa R., Rosenfeld M. G., Willson T. M., Glass C. K., Milburn M. V. (1998) Nature 395, 137–143 [DOI] [PubMed] [Google Scholar]
- 39. Williams S., Bledsoe R. K., Collins J. L., Boggs S., Lambert M. H., Miller A. B., Moore J., McKee D. D., Moore L., Nichols J., Parks D., Watson M., Wisely B., Willson T. M. (2003) J. Biol. Chem. 278, 27138–27143 [DOI] [PubMed] [Google Scholar]
- 40. Rochel N., Wurtz J. M., Mitschler A., Klaholz B., Moras D. (2000) Mol. Cell 5, 173–179 [DOI] [PubMed] [Google Scholar]
- 41. Kallen J. A., Schlaeppi J. M., Bitsch F., Geisse S., Geiser M., Delhon I., Fournier B. (2002) Structure 10, 1697–1707 [DOI] [PubMed] [Google Scholar]
- 42. Brzozowski A. M., Pike A. C., Dauter Z., Hubbard R. E., Bonn T., Engström O., Ohman L., Greene G. L., Gustafsson J. A., Carlquist M. (1997) Nature 389, 753–758 [DOI] [PubMed] [Google Scholar]
- 43. Shiau A. K., Barstad D., Loria P. M., Cheng L., Kushner P. J., Agard D. A., Greene G. L. (1998) Cell 95, 927–937 [DOI] [PubMed] [Google Scholar]
- 44. Bourguet W., Vivat V., Wurtz J. M., Chambon P., Gronemeyer H., Moras D. (2000) Cell 5, 289–298 [DOI] [PubMed] [Google Scholar]
- 45. Pike A. C., Brzozowski A. M., Walton J., Hubbard R. E., Thorsell A. G., Li Y. L., Gustafsson J. A., Carlquist M. (2001) Structure 9, 145–153 [DOI] [PubMed] [Google Scholar]
- 46. Manel N., Unutmaz D., Littman D. R. (2008) Nat. Immunol. 9, 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Crome S. Q., Wang A. Y., Kang C. Y., Levings M. K. (2009) Eur. J. Immunol. 39, 1480–1493 [DOI] [PubMed] [Google Scholar]
- 48. Ivanov I. I., Zhou L., Littman D. R. (2007) Semin. Immunol. 19, 409–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.