Abstract
Enterovirus 71 (EV71), a member of the Picornaviridae family, may cause serious clinical manifestations associated with the central nervous system. Enterovirus 3C protease is required for virus replication and can trigger host cell apoptosis via cleaving viral polyprotein precursor and cellular proteins, respectively. Although the role of the 3C protease in processing viral and cellular proteins has been established, very little is known about the modulation of EV71 3C function by host cellular factors. Here, we show that sumoylation promotes EV71 3C protein ubiquitination for degradation, correlating with a decrease of EV71 in virus replication and cell apoptosis. SUMO E2-conjugating enzyme Ubc9 was identified as an EV71 3C-interacting protein. Further studies revealed that EV71 3C can be SUMO (small ubiquitin-like modifier)-modified at residue Lys-52. Sumoylation down-regulated 3C protease activity in vitro and also 3C protein stability in cells, in agreement with data suggesting 3C K52R protein induced greater substrate cleavage and apoptosis in cells. More importantly, the recombinant EV71 3C K52R virus infection conferred more apoptotic phenotype and increased virus levels in culture cells, which also correlated with a mouse model showing increased levels of viral VP1 protein in intestine and neuron loss in the spinal cord with EV71 3C K52R recombinant viral infection. Finally, we show that EV71 3C amino acid residues 45–52 involved in Ubc9 interaction determined the extent of 3C sumoylation and protein stability. Our results uncover a previously undescribed cellular regulatory event against EV71 virus replication and host cell apoptosis by sumoylation at 3C protease.
Keywords: Post-translational Modification, Protein Degradation, Sumoylation, Ubiquitination, Virus
Introduction
Enterovirus 71 (EV71),2 similar to poliovirus, belongs to the genus Enterovirus of the Picornaviridae family. EV71 infection usually causes mild symptoms in childhood, such as herpangina or exanthema also known as hand, foot, and mouth disease (1). However, during the 1998 epidemic in Taiwan, EV71 infection was associated with severe central nervous system (CNS) diseases and complications, including encephalitis, aseptic meningitis, brain stem encephalitis, and rapidly fatal pulmonary edema and hemorrhage (2–5). Very little is known about the molecular basis of EV71-elicited neuropathogenesis.
EV71 possesses a single-stranded RNA genome of ∼7500 nucleotides in length with positive polarity (6). Although the replication mechanism of EV71 is largely unknown, studies of other members in the Picornaviridae family revealed that virus RNA encodes a large polyprotein (∼200 kDa), which is processed by viral protease 2A and 3C into 27 cleavage intermediates and end products (7). Most of the proteolytic reactions are accomplished by viral 3C protease, whereas viral 2A protease catalyzes only two cleavages on the polyprotein. In addition to viral precursor protein processing, enterovirus 3C protease is also involved in the inhibition of essential host functions such as transcription, cytoskeletal integrity, and polyadenylation (8–11). For instance, poliovirus 3C protease can cleave a number of cellular proteins such as TBP, CREB, and the cytoskeletal protein MAP-4 (8, 12, 13). Recently, EV71 3C protease was reported to cleave CstF-64 to inhibit cellular pre-mRNA polyadenylation (11). Cleavage of cellular proteins by 3C protease is considered to be associated with EV71-induced cell apoptosis, as expression of enterovirus 3C protease alone is sufficient to trigger cell apoptosis, in part, via activation of caspase-3 and poly-ADP-ribose polymerase (PARP) cleavage (14–16). Interestingly, genomic sequencing analysis of EV71 viral strains collected in Taiwan during the 1998–2003 epidemic seasons revealed some sequence variations in 3C protease (17). It is essential to elucidate whether these sequence variations alter 3C function and EV71 virulence, given the importance of 3C protease for enterovirus replication and host cell apoptosis.
Sumoylation, a SUMO peptide conjugated on the lysine residue of the protein substrate, is an important posttranslational control for cellular protein functions (18, 19) as well as virus replication (20). The conjugation of mammalian SUMO isoforms (SUMO-1,-2, and -3) to protein substrates is first activated by E1-activating enzyme (SAE1/SAE2), then transferred from E1 to the E2 conjugase (Ubc9) (21). In general, Ubc9 catalyzes the formation of an isopeptide bond between the C terminus of SUMO and the amino group of the target lysine via a direct interaction with a consensus motif ψKX(E/D) (where ψ is a large hydrophobic residue, and X is any residue) present in protein substrates (22, 23). Several studies demonstrated that the PIAS family of proteins, RanBP2, Pc2, HDAC4, and HDAC7, can act as SUMO E3 ligases to enhance substrate sumoylation (24–28). Conversely, SENP (SUMO-specific protease) family proteins function as isopeptidases to remove conjugated SUMO from substrates (29). Sumoylation results in distinct consequences to protein substrates, including alteration of protein activity and subcellular localization or stability (19). Regarding protein stability, sumoylation either antagonizes substrate ubiquitination by competing with the same lysine residue for modification (30) or facilitates substrate ubiquitination via SUMO-targeted ubiquitin ligase (STUbL) family proteins such as RNF4 (31–33). Whether SUMO modification affects EV71 virus activity has not yet been elucidated.
Here we report that EV71 3C function can be attenuated by SUMO modification. Ubc9 was identified as an EV71 3C-interacting protein. Further study revealed that 3C was modified by SUMO-1 at the lysine 52 residue in vivo and in vitro. Sumoylation decreased 3C in substrate cleavage as well as 3C protein stability in cells. More importantly, sumoylation at Lys-52 promoted 3C protein ubiquitination and degradation, correlating with a reduction of 3C substrate cleavage, apoptosis induction, and EV71 virus production in cells as well as EV71-elicited neurovirulence in a mouse model. Furthermore, EV71 3C mutations found in clinically isolated strains, including K52R and within amino acid residues 45–52, reduced Ubc9 binding capacity and were associated with clinical implications ranging from mild illness to CNS involvement. We further showed that 3C residues 45–52 involved in Ubc9 interaction contributed to 3C sumoylation potential and protein stability regulation. Collectively, our findings suggest that SUMO modification at EV71 3C protease may serve as a host cell defense against virus replication and apoptosis induction during EV71 infection.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Antibodies
GFP- or RFP-fused EV71 or polio 3C constructs were constructed by inserting each 3C cDNA into the GFP-Puro2-IRES and RFP-Puro2-IRES vectors, respectively. The EV71 cDNA was also subcloned into indicated tag vectors, including pBTM116 (LexA), pcDNA3-HA (HA), pCMV-FLAG-2B (FLAG), and pGEX-4T1 (GST). The constructs expressing EV71 3C point mutations were created by site-directed mutagenesis (QuikChange, Stratagene). Ubiquitin, PIAS1, and Ubc9 cDNAs were amplified by PCR and subcloned into pCMV-FLAG-2B, pcDNA3-HA, and GEX-4T1, respectively. The mammalian or bacterial constructs expressing HA-SENP1, HA-SENP2, FLAG-SUMO-1, His-SUMO-1, and His-SUMO-2 were reported previously (34). The construct for producing bacterial His-SUMO-1 11KR was a gift from Dr. Jaulang Hwang (Academia Sinica, Taiwan) (35), and mammalian FLAG-SUMO-1 11KR was generated by inserting cDNA of SUMO-1 11KR into p3XFLAG-CMV7.1 vector. pDNA-FLAG-SNURF WT and CS constructs expressing RNF4 WT and catalytically inactive (CS) mutant, respectively, in mammalian cells were gifts from Dr. Jorma Palvimo (University of Kuopio, Finland). The following primary antibodies were used: anti-HA (Covance), anti-PARP (Roche Applied Science), anti-CREB (Epitomics), anti-GFP (Santa Cruz; MBL), anti-FLAG (Sigma), anti-ubiquitin (Biomol), anti-cleaved caspase-3 (Cell Signaling), anti-VP1 (Millipore), anti-actin (Millipore), anti-EV 71 3C (11), anti-SUMO-1 and anti-SUMO-2/3 (36), and anti-PML rabbit polyclonal antibody (37).
Yeast Two-hybrid Screen and β-Galactosidase Assays
A yeast two-hybrid array screen using LexA-EV71 3C as bait was conducted as described previously (38). Briefly, the L40 strain transformed with LexA-EV71 3C was mated with AMR70 strains, which had been transformed with each specific prey gene in the pACT2 vector using 96-well plates. The resulting mated yeast cells were selected on a medium lacking histidine, leucine, and tryptophan then followed by a β-galactosidase assay. Quantitative β-galactosidase assays of yeast containing pairs of indicated bait and prey plasmids were determined by liquid culture following the Galacto-light Plus kit (Tropix) instructions.
Cell Culture, Transient Transfection, Immunoprecipitation, and Western Analyses
293T, HeLa, or HeLa stably expressing His-tagged SUMO-1 (a gift from Dr. Peter O'Hare, Imperial College, London), and RD (rhabdomyosarcoma) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) and streptomycin. Transfection of DNA constructs into HeLa, HeLa-His-SUMO-1, or 293T cells was performed using Lipofectamine 2000 (Invitrogen). Immunoprecipitation and Western analyses were performed as described previously (34). In brief, transfected cells were lysed directly in radioimmune precipitation assay buffer (200 mm Tris, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 1 mm PMSF, 1 mm DTT, 0.5% SDS, and 0.5% deoxycholate) with the addition of complete protease inhibitor mixture (Roche Applied Science) and 20 mm N-ethylmaleimide (Sigma). Lysates were further incubated (1 h) with indicated antibody and subsequently reacted (overnight) with protein A/G. Immunoprecipitated beads were washed and subjected to Western blot analysis using specific antibody.
GST Pulldown, in Vitro Sumoylation, and 3C Substrate Cleavage Assays
GST pulldown assays were performed as described (34) using 2 μg of recombinant GST or GST-Ubc9 fusion proteins along with the indicated in vitro synthesized 35S-labeled 3C WT or mutant proteins. In vitro sumoylation assay was performed as described previously (34) with recombinant proteins or in vitro synthesized 35S-labeled proteins as indicated. The Δ3A-3B-3Cmut-Δ3D, consisting of partial 3A and 3D domains linked to 3B and 3C catalytically inactive mutant (39), served as a substrate for in vitro cleavage reaction with GST-3C with or without sumoylation. In brief, GST-3C recombinant protein (5 μg) was subjected to in vitro sumoylation on GST glutathione-agarose beads. After washing twice with cleavage buffer (20 mm HEPES, 20% glycerol, and 100 mm KCl, pH7.9), the sample with or without sumoylation was split into six portions; one was saved as input, and 2 μl of 35S-labeled Δ3A-3B-3Cmut-Δ3D in vitro in 20 μl of cleavage buffer were added to the remaining five portions for the indicated digestion time.
Virus Propagation, Infection, and Virus Titer Determination
The EV71 (TW/2231/98) strain described previously (40) was used for this study. The infectious clone of EV71 harboring 3C K52R was created by PCR site-directed mutagenesis (Stratagene) using the full-length genome of EV71 WT (TW/2231/98) in pCR-XL-Topo vector as a template. These two viral strains were amplified in RD cells. The virus titer of infected RD cells harvested from amplification or at different post-infection time points was determined by plaque assay as described previously (40). In brief, 5 × 105 RD cells seeded in 6-cm dishes were cultured (37 °C, 14 h) followed by the addition of serial 10-fold dilutions of EV71 recombinant virus. After viral adsorption (1 h), infected cells were washed with PBS containing 0.2% BSA and covered with an overlay of Dulbecco's modified Eagle's medium containing 2% FBS mixed with 2% methylcellulose. Cells were further incubated (37 °C, 3 days), and plaques were stained with 0.1% crystal violet in a 10% formaldehyde solution. For mouse virus inoculation, three 1-day-old ICR mice were orally inoculated using a feeding tube with control media or EV71 virus 2231 wild type or 3C K52R strain in a total volume of 100 μl (1 × 107 plaque-forming units). Animals were measured for body weight every other day. Seven days after oral inoculation, mice were sacrificed, and organ histopathology was examined. All protocols were approved by the Institute of Biomedical Sciences, Academia Sinica.
Cell Apoptosis, Immunohistochemical Staining, and TUNEL Assay
Cell apoptosis induced by viral infection or transiently expressing EV71 3C protein was quantified by Cell Death Detection ELISA (Roche Applied Science) according to the manufacturer's instructions. For immunohistochemistry analysis, intestinal and spinal cord tissues removed from infected mice (6 mice/each virus infection; n = 3 independent experiments) were paraformaldehyde-fixed and paraffin-embedded for tissue sections. The tissue slides were deparaffinized, dehydrated, and further treated with 3% H2O2 buffer to block endogenous peroxidase activity and reacted with 0.01 m sodium citrate buffer for antigen retrieval. After blocking with 10% normal goat serum, the intestine and thoracic spinal cord sections were incubated (4 °C, overnight) with anti-VP1 and cleaved caspase-3 antibodies, respectively, then incubated with Super Picture HRP polymer conjugate secondary antibody (Zymed Laboratories Inc.) and further stained with DAB chromagen. Sections were also stained with hematoxylin (DAKO). Tissue sections of the thoracic spinal cord were subjected to Nissl staining, and the number of Nissl stain-positive neural cell bodies in the ventral horn area of the thoracic spinal cord was quantified. Apoptotic cells in tissue sections were visualized using the in situ Death Detection kit TMR red (tetramethylrhodamine red) (Roche Applied Science) according the manufacturer's instructions.
Statistical Analysis
Data from β-galactosidase assay, cleaved products, cell apoptosis, viral titer, Nissl body quantification, and mouse body weight in this study are expressed as the mean ± S.D. Statistical significance between different experimental groups was ascertained using Student's t test.
RESULTS
SUMO Modification of EV71 3C
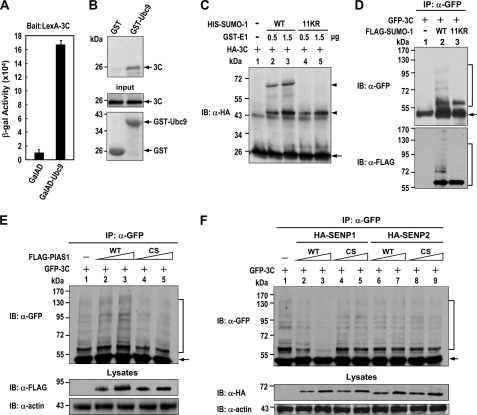
Using a yeast two-hybrid screen, we identified Ubc9 as a 3C-interacting protein. The interaction of 3C and Ubc9 was quantified using the β-galactosidase assay for yeast cells transformed with LexA-3C (bait) and GalAD-Ubc9 (prey) (Fig. 1A). The 3C-Ubc9 interaction was further evidenced by GST pulldown assays showing in vitro synthesized 35S-labeled 3C protein precipitated by GST-Ubc9 but not by the GST fusion protein alone (Fig. 1B). The results of the 3C-Ubc9 interaction led us to investigate whether 3C could be SUMO-modified. In vitro sumoylation reactions yielded two bands migrating more slowly than unmodified 3C (Fig. 1C, arrowhead). The level of these two slowly migrating bands was significantly elevated after increasing the amount of SUMO E1-activating enzyme in the reactions (lanes 2 and 3). To further differentiate bands derived from multiple mono-sumoylation or from mono-sumoylation utilizing poly-SUMO chain formation, a SUMO-1 mutant (11KR) incapable of forming a polychain (because all 11 Lys residues were converted to Arg residues) was subjected to in vitro sumoylation. SUMO-1 11KR mutant resulted in the generation of only one sumoylated band, suggesting that the slowly migrating band (below 72 kDa) missing from the SUMO-1 11KR samples was a poly-SUMO chain-conjugated 3C. Notably, 3C was modified by SUMO-1 more effectively than SUMO-2 in vitro compared with PML conjugated by SUMO-1 or SUMO-2 to a similar extent (supplemental Fig. 1). Furthermore, 3C also generated a major and several minor slowly migrating bands in cells co-expressing SUMO-1 (Fig. 1D, lane 2). Except for the major band, all other slowly migrating bands were abrogated in SUMO-1 11KR-transfected cells (lane 3), suggesting that 3C could be modified by mono-SUMO-1 as well as by the poly-SUMO-1 chain in cells. It should be noted that the pattern of these sumoylated bands was different from that observed in the in vitro sumoylation assays (Fig. 1C). This discrepancy could be the result of SUMO-1 overexpression, causing increased efficiency of 3C sumoylation and/or additional modifications in cells (see “Discussion”). Furthermore, the level of sumoylated bands was enhanced by PIAS1 and attenuated by SENP1 but not by its catalytically inactive mutant (CS) in HeLa cells stably expressing His-tagged SUMO-1 (Fig. 1, E and F). Furthermore, the inability of SENP2 to de-sumoylate 3C suggested a specificity of SENP1 for 3C de-sumoylation (Fig. 1F). Altogether, these data suggest that EV71 3C could be sumoylated both in vitro and in vivo.
FIGURE 1.
SUMO modification of the EV71 3C protease. A, shown are β-galactosidase assays of yeast cells transformed with LexA-3C and the indicated “prey.” Error bars represent the S.D. from three experiments performed in triplicate. B, an autoradiograph shows 35S-labeled 3C (top) pulled down by the GST-Ubc9 protein. input represents 20% of 35S-labeled protein subjected to binding assays. Coomassie Blue staining shows GST fusion proteins used for each binding reaction. C, Western blot analysis shows in vitro sumoylation of in vitro synthesized HA-tagged 3C proteins. The arrowhead and arrow indicate SUMO-1-modified and unmodified proteins, respectively. D–F, Western blot analysis shows GFP-tagged 3C modified by FLAG-tagged SUMO-1 in 293T cells (D) or HeLa cells stably expressing His-tagged SUMO-1 (E and F) with or without expressing increased amounts of the WT or catalytically inactive CS mutant of PIAS1 or SENPs as indicated. The SUMO-1-modified and unmodified GFP-3C is depicted by bracket and arrow, respectively.
3C Lys-52 Is the SUMO Conjugation Site
We next mapped the 3C sumoylation site(s) by replacing Lys residues (Fig. 2A, bold) with Arg residues and tested respective mutants in sumoylation assays. The results of the in vivo sumoylation assays showed that the K52R mutant eliminated the major and all other minor SUMO-modified bands compared with the WT and other Lys → Arg mutants (Fig. 2B). The importance of position Lys-52 for 3C sumoylation was further validated using in vitro sumoylation assays that demonstrated a loss of sumoylated bands in the K52R sample (Fig. 2C). These data suggest that Lys-52 was the conjugation site for mono- and poly-SUMO chain formation on EV71 3C. We further substantiated 3C sumoylation endogenously by infecting cells with recombinant EV71 virus. The 3C immunoprecipitation experiments revealed that the two slowly migrating bands detected by an anti-3C antibody were also recognized by an anti-SUMO-1 antibody from cell lysates harvested 8 h post-infection (Fig. 2D, asterisk). Notably, these two slowly migrating bands were modestly detected by the anti-SUMO-2/3 antibody (Fig. 2D, lane 14), consistent with data demonstrating that the EV71 3C was preferentially modified by SUMO-1 in vitro (supplemental Fig. 1). More importantly, these sumoylated bands were not observed in cells infected with recombinant EV71 virus harboring the 3C K52R mutation even though the unmodified 3C K52R protein was more abundant than the WT protein (Fig. 2D, lanes 2 and 5). Together, these results demonstrated that EV71 3C could be sumoylated and that sumoylation occurred at the Lys-52 residue. Notably, the extent of 3C sumoylated bands was attenuated at 12 h post-infection (lanes 3 and 9), suggesting additional regulation of 3C sumoylation during virus infection (see below). In addition, the migration positions of the 3C precursors 3BCD and 3CD, reported previously, are indicated (41).
FIGURE 2.
Lys-52 is the EV71 3C sumoylation site. A, amino acid residues of EV71 3C are shown. The Lys residues converted to Arg for sumoylation analysis are shown in bold. B, Western blot analysis shows GFP-3C WT or mutants modified by FLAG-tagged SUMO-1 in 293T cells as indicated. The SUMO-1-modified and unmodified GFP-3C are depicted by brackets and an arrow, respectively. C, Western blot analysis shows in vitro sumoylation of HA-tagged 3C WT or K52R proteins. The arrowhead and arrow indicate SUMO-1-modified and unmodified proteins, respectively. D, Western blot analysis shows endogenous sumoylated 3C from 1.5 × 107 RD cells infected (IF) with EV71 recombinant virus (10 m.o.i.) and harvested at the indicated post-infection time points. The arrows and asterisks indicate EV71 3C and its precursor proteins and sumoylated 3C, respectively.
EV71 3C Protease Function Is Down-regulated by Sumoylation
The finding of EV71 3C modified by SUMO-1 led us to test whether sumoylation regulates 3C substrate cleavage. To this end, a GST-3C fusion protein was subjected to an in vitro sumoylation reaction before measuring its capacity for substrate cleavage. The results of the in vitro sumoylation assay showed that about 25% of the GST-3C recombinant protein was sumoylated (Fig. 3A, top panel). A substrate Δ3A-3B-3Cmut-Δ3D previously used for analyzing 3C catalytic activity (39) was synthesized in vitro and subjected to cleavage by the GST-3C sample in the presence or absence of sumoylation reaction conditions. Notably, the sumoylated GST-3C protein sample yielded less Δ3A-3B-3Cmut-Δ3D cleavage product compared with the GST-3C sample incubated in the absence of sumoylation conditions (Fig. 3A, bottom panel, arrowhead, and the percent substrate cleavage further shown in graph format). These data suggest that sumoylation reduced 3C protease activity in vitro. We next examined whether sumoylation attenuates 3C substrate cleavage in cells. In line with the in vitro results, we observed that the 3C K52R mutant induced the cleavage product of ectopically expressed GFP-tagged TBP or HA-tagged CREB protein substrate in cells better than WT (Fig. 3, B and C). These data suggest that 3C function was down-regulated by sumoylation.
FIGURE 3.
Sumoylation down-regulates 3C substrate cleavage. A, a Western blot shows an in vitro sumoylation reaction of GST-3C proteins subjected to substrate cleavage (top). The bracket and arrow indicate SUMO-1-modified or unmodified GST-3C protein. − represents the GST-3C sample incubated with all components in the sumoylation reaction except ATP. An autoradiograph shows an in vitro cleavage reaction of the protein substrate Δ3A-3B-3Cmut-Δ3D incubated with the indicated GST-3C fusion proteins (bottom). The intact and cleaved products are marked by arrow and arrowheads, respectively. Error bars represent the mean ± S.D. (n = 3); *, p < 0.05; **, p < 0.01. B–D, immunoblots show ectopically expressed GFP-tagged TBP (B), HA-tagged CREB (C), or PARP (D) in HeLa cells transfected with the indicated GFP- or RFP-tagged 3C expression constructs. The intact and cleaved products are marked by an arrow and arrowhead, respectively. Error bars represent the mean ± S.D. (n = 3); **, p < 0.01. E, apoptosis of 5 × 104 HeLa cells transfected with the indicated GFP-tagged 3C constructs is shown. Error bars, mean ± S.D. (n = 3); **, p < 0.01). An immunoblot of the indicated GFP-3C proteins expressed in transfected-HeLa cells is shown.
Because 3C protease was reported to induce caspase-3 activation and cellular apoptosis (16), we further substantiated the effect of sumoylation on 3C protease function by examining caspase-3 substrate PARP cleavage and apoptosis in 3C-transfected cells. As expected, the 3C K52R mutant conferred greater levels of PARP cleavage product and apoptosis compared with WT (Fig. 3, D and E). Of note, the effects of the EV71 3C K52R mutant on TBP and PARP cleavage and apoptosis were similar to those observed for poliovirus 3C control (Fig. 3, B, D, and E). These results further support a role for the negative regulation of the 3C protease function by SUMO modification.
Sumoylation Modulates EV71 3C Stability
Because SUMO modification also regulates protein stability (18, 19), we further explored the stability of EV71 3C after sumoylation in cells. Cyclohexamide-chase experiments showed that the K52R protein half-life was greater than that of the WT, increasing from ∼3 to 4.8 h (Fig. 4A), indicating that the K52R mutation stabilized the 3C protein. Furthermore, treatment with the proteasome inhibitor MG132 attenuated 3C protein degradation (lane 8). Because proteasome-mediated protein degradation is generally associated with substrate ubiquitination, we next tested 3C ubiquitination and demonstrated that it was prevented by K52R (Fig. 4B). This observation was further substantiated by measuring 3C ubiquitination at an endogenous level in EV71 viral infection. Although the 3C WT protein could be ubiquitinated during viral infection, the 3C K52R mutant failed to do so (Fig. 4C, lanes 2 and 3 versus 5 and 6). In line with the failure of the K52R mutant, 3C K52R protein levels were higher than protein levels observed for WT 3C (Fig. 4C, middle panel). Notably, the 3C polyubiquitin chain was significantly increased 12 h post-infection (lane 3), correlating with a decrease in sumoylated 3C levels at this time point (Fig. 2D, lane 3). These results suggest that sumoylation may have facilitated 3C ubiquitination.
FIGURE 4.
Sumoylation promotes 3C protein degradation. A, shown is a Western blot of 3C WT or K52R expression in 293T cells treated with 50 μg/ml cyclohexamide (CHX) for the indicated periods of time in the presence or absence of 50 μg/ml MG132. The amount of 3C relative to levels in untreated cells is indicated at the bottom. The plot shows quantification of the 3C protein half-life from at least four independent experiments by nonlinear regression analysis. Significantly different levels of expression between 3C WT and K52R proteins were determined using Student's t test; **, p < 0.01. B and D, Western blot analysis shows 3C ubiquitination in 293T cells transfected with the indicated constructs. The ubiquitin-modified and unmodified GFP-3C proteins are depicted by brackets and an arrow, respectively. C, shown is a Western blot analysis of 3C ubiquitination from 1.5 × 107 RD cells infected with the indicated EV71 recombinant virus (10 m.o.i.) and harvested at the indicated time points of post-infection. The bracket indicates the ubiquitinated EV71 3C. E, a Western blot analysis shows 3C stability modulated by SENP1 and PIAS1. 5 × 106 293T cells transfected with 10 μg of empty vector (HA) or expression constructs corresponding to HA-SENP1 or HA-PIAS1 were cultured for 1 day then split into 12-well plates before transfection with 3C WT or the K52R mutant. Cells were further treated with 50 μg/ml cyclohexamide for the indicated periods of time. The amount of 3C relative to that in untreated cells is indicated at the bottom of each set.
If sumoylation facilitated 3C ubiquitination, removal of 3C sumoylation should abrogate its ubiquitination. To this end, SENP1 was tested in 3C ubiquitination experiments. As expected, coexpression of SENP1 reduced 3C sumoylation as well as ubiquitination (Fig. 4D, top and second panels, lanes 2). Notably, the effect of SENP1 in reducing 3C ubiquitination was specific because global ubiquitination was not significantly altered by SENP1 expression (bottom panel). Together, these results suggest that sumoylation at position Lys-52 promoted 3C ubiquitination and degradation. Consistently, coexpression of SENP1 or PIAS1 resulted in increased or decreased 3C protein stability in cyclohexamide-chase experiments, respectively (Fig. 4E). Conversely, SENP1 or PIAS1 expression failed to significantly alter the time course of K52R protein degradation (Fig. 4E), further supporting the involvement of sumoylation in 3C protein ubiquitination and degradation.
Because RNF4 was reported to facilitate ubiquitination of SUMO-modified proteins (42), we next tested whether 3C ubiquitination and degradation could be modulated by RNF4. Expression of RNF4 WT, but not a catalytically inactive mutant, modestly increased 3C ubiquitination levels (supplemental Fig. 2A), correlating with a marginal effect of RNF4 on 3C degradation (supplemental Fig. 2B). These results suggest the presence of other ubiquitin E3 ligase(s) or another molecular mechanism(s) operating the EV71 3C sumoylation-dependent ubiquitination and degradation pathway (see “Discussion”).
SUMO Modification of 3C Attenuates EV71-induced Apoptosis, Virus Production, and Neurovirulence
If Lys-52 sumoylation is important for 3C ubiquitination and degradation, it is conceivable that a recombinant EV71–3C K52R virus would trigger greater substrate cleavage and apoptosis than the WT virus. As expected, recombinant EV71–3C K52R virus infection resulted in more CREB cleavage in cells harvested at 9 and 12 h post-infections compared with WT virus (Fig. 5A). Likewise, more PARP cleavage products were observed in cells infected with the 3C K52R recombinant virus compared with WT virus-infected cells 8 and 12 h post-infection (Fig. 5B). Accordingly, recombinant 3C K52R recombinant virus-induced apoptosis was more pronounced 8 and 12 h post-infection than apoptosis observed after infection with WT virus (Fig. 5C), consistent with data observed for cells transiently expressing 3C protein only (Fig. 3E). These results suggest that sumoylation acted as a host cell defense mechanism resulting in EV71 3C attenuation. In line with this notion, we observed that WT recombinant virus produced reduced amounts of virus in RD cells compared with levels observed for the 3C K52R recombinant virus (Fig. 5D).
FIGURE 5.
Impairment of EV71 3C sumoylation potentiates viral replication and cell apoptosis. A and B, Western blot analysis of endogenous CREB (A) or PARP (B) cleavage in RD cells (5 × 105) infected with EV71 (10 m.o.i.) and harvested at the indicated time points of post-infection is shown. h.p.i., hours post-infection. The arrow and arrowhead indicate the intact or cleaved products corresponding to endogenous CREB or PARP, respectively. The percentage of CREB or PARP cleavage product over total PARP proteins in each sample is shown. Error bars, mean ± S.D. (n = 3); **, p < 0.01. C, apoptosis analysis of 6 × 104 RD cells infected with 10 m.o.i. each of either EV71 WT or 3C K52R mutant recombinant virus is shown. Apoptotic cells were determined using a Cell Death Detection ELISA kit (absorbance at 405 nm). Error bars, mean ± S.D. (n = 3); **, p < 0.01. D, a plot shows EV71 virus growth in RD cells. 5 × 105 cells were infected with the indicated EV71 recombinant virus (40 m.o.i.) and harvested at the indicated time points of post-infection, and virus titers were assessed using the plaque assay. Experiments were carried out in triplicate. Data represent the mean ± S.D. (n = 3); p < 0.05 (*) and p < 0.01 (**) indicate a statistical significant difference between EV71 3C WT- and K52R-infected cells harvested at the indicated time points. PFU, plaque-forming units. E, shown is a representative immunohistochemistry analysis of the small intestine and the ventral horns of the thoracic spinal cord sections from 1-day-old ICR mice orally inoculated with medium control (mock), EV71 2231 wild type, or with the 3C K52R strain at 1 × 107 plaque-forming units/mouse and scarified 7 days post-infection. Hematoxylin stained the nuclei (blue) in tissue sections in panels a–c and j–l. Scale bar, 100 μm. F, a bar graph shows the relative number of surviving neuron cells present in the thoracic spinal cord of EV71-infected mice observed in E. Error bars represent the S.D.; *, p < 0.05 and **, p < 0.01 (n = 3, in triplicate).
Previous studies reported that the EV71 virus could replicate in the intestinal tract and elicit central nervous system pathology in neonatal mice (43). We further demonstrated the importance of SUMO conjugation to 3C in the down-regulation of EV71-mediated pathogenesis in mice. Equal titers of EV71 WT or K52R recombinant virus were administrated orally to neonatal mice. After 7 days, mice were sacrificed, and organs were examined histologically. Mice infected with the EV71–3C K52R recombinant presented with more severe weight loss compared with WT-infected mice (supplemental Fig. 3), consistent with a recent report demonstrating that the degree of EV71 virulence correlated with weight loss in infected mice (44). In addition, characteristic lesions were observed in the mouse intestine and spinal cord after EV71 infection (Fig. 5E). The 3C K52R recombinant virus-infected mice presented with stronger VP-1 staining and severe villous blunting in the small intestine compared with WT virus-infected mice (panels a–c). These results correlated with the above finding showing that 3C K52R recombinant virus titers in infected cells were higher than WT titers (Fig. 5D). More importantly, Nissl body staining revealed that the number of neurons present in the ventral horn of the thoracic spinal cord were significantly diminished in 3C K52R-infected mice compared with WT-infected mice (Fig. 5, E, panels d–f and F), suggesting that 3C K52R viral infection resulted in increased neuron loss in this area. Consistent with this notion, 3C K52R viral infection resulted in elevated rates of apoptosis evidenced by TUNEL and cleaved caspase-3 staining (Fig. 5E, panels g–l). Together, these data support the notion that SUMO modification serves as a host cell defense mechanism designed to attenuate EV71-induced pathogenesis.
Ubc9 Binding Capacity Determines the Extent of EV71 3C Sumoylation and Stability Regulation
The results of the EV71–3C K52R infection mouse model led us to explore the clinical association of EV71 infections during the outbreaks in Taiwan. 3C sequence analysis of 395 EV71 strains isolated in Taiwan between 1998 and 2003 identified one patient with a 3C K52R mutation who presented with severe CNS involvement (supplemental Table 1).3 These results support a correlation between our EV71 mouse model and a clinical case. In addition, several isolated strains with mutations flanking the 3C Lys-52 residue were also identified, including mutations K45R, I47V, and V49I associated with the clinical diagnosis of more mild illness rather than CNS involvement (supplemental Table 1). To examine whether these mutations were also associated with 3C sumoylation, these 3C mutants were generated and tested for their sumoylation potential. The results of in vitro sumoylation assays revealed that K45R, I47V, and V49I mutations reduced 3C sumoylation to a similar extent (Fig. 6A). Similar observations were also made after analysis of in vivo sumoylation assays (Fig. 6B). The levels of in vitro and in vivo sumoylation of these 3C mutants are summarized in Fig. 6D. Although the incidence rates of the 3C sumoylation-defective mutants were not high in clinically associated EV71 strains,3 these results provide a potential correlation between EV71 3C sumoylation and disease severity.
FIGURE 6.
EV71 3C sumoylation correlates with Ubc9–3C binding. A, Western blot analysis shows in vitro sumoylation of HA-tagged 3C WT and mutants. The arrowheads and arrow indicate SUMO-1-modified and unmodified proteins, respectively. B, immunoblots show GFP-3C WT and mutant proteins modified by SUMO-1 in transfected 293T cells. The bracket and arrow indicate SUMO-1-modified and unmodified proteins, respectively. C, an autoradiograph shows 35S-labeled HA-tagged 3C WT or mutant proteins pulled down by GST (odd lanes) or by the GST-Ubc9 fusion protein (even lanes). input represents 10% of the amount of 35S-labeled protein subjected to binding assays. GST fusion proteins used for each binding reaction are shown. D, a summary of the EV71 3C mutants mediating Ubc9 binding, sumoylation, CREB cleavage, and protein stability is shown. The intensity of precipitated 3C protein by GST-Ubc9 was quantified by densitometry, and the relative binding of 3C to Ubc9 (mean ± S.D.) from four independent experiments is indicated after normalization to WT 3C binding (set to a value of 100). The intensity of SUMO-modified and -unmodified bands from both in vitro and in vivo sumoylation assays was quantified by densitometry, and the percentage of modified to unmodified 3C protein (mean ± S.D.; n = 4) is indicated after normalization to WT sumoylation (set to a value of 100). The CREB cleavage potential and protein half-life of each 3C mutant was quantified from four independent experiments (mean ± S.D.). Representative blots are shown in supplemental Fig. 5.
Given that mutation of the Lys-45, Ile-47 and Lys-49 residues altered the 3C sumoylation potential, we further examined whether other residues proximal to Lys-52 also affect 3C sumoylation, including residues Thr-46, Trp-48, Glu-50, His-51, and Leu-53. The results of in vitro and in vivo sumoylation analyses revealed that Ala substitution of Trp-48, Glu-50, or His-51 significantly reduced 3C sumoylation and that T46A and L53A mutants had slightly reduced sumoylation activity compared with the WT (Fig. 6, A, B, and D). These results suggest the importance of the Lys-52 flanking residues in mediating EV71 3C sumoylation. Consistent with this notion, we observed that poliovirus 3C, consisting of distinct residues surrounding the Lys-52 (supplemental Fig. 4A), failed to be sumoylated in cells (supplemental Fig. 4B). Replacement of the poliovirus 3C region with amino acids corresponding to the EV71 3C region, however, restored sumoylation effectively (supplemental Fig. 4B).
We next tested if the different EV71 3C mutants (from residues Lys-45 to Leu-53) mediated sumoylation based on differential Ubc9 interactions. To this end, various 3C mutant proteins synthesized in vitro were subjected to GST pulldown assays with GST-Ubc9. As expected, the levels of these 3C mutants precipitated by the GST-Ubc9 protein correlated with the extent of SUMO modification in these mutants (Fig. 6C). Further characterization of these 3C mutants revealed that the CREB cleavage potential and the protein stability of these mutants inversely correlated with their sumoylation levels (supplemental Fig. 5 and Fig. 6D). Collectively, these results suggest that the Ubc9 binding ability defined the EV71 3C sumoylation potential and conferred protein stability.
DISCUSSION
Many viruses interact with host proteins as a means of interfering with or usurping cellular machinery for creating a suitable replication environment. Sumoylation is one host cellular post-translational modification system previously reported to facilitate viral replication and/or to be inhibited by viral proteins during viral infection. For instance, sumoylation of influenza A viral NS1 protein and human immunodeficiency virus type 1 (HIV-1) p6 Gag regulates influenza virus rapid growth and HIV-1 infectivity (45, 46), respectively. Epstein-Barr virus BZLF1 protein, human cytomegalovirus IE1 protein, and herpes simplex virus ICP0 protein can disrupt PML nuclear bodies by inhibiting PML sumoylation (47–50). Likewise, avian adenoviral protein Gam1 abrogates cellular sumoylation events by targeting the SUMO E1 enzyme (51, 52). Although many studies have reported that the sumoylation system is inhibited or utilized by viral proteins for viral replication, very little is known regarding whether sumoylation can also act as a host cell defense system resulting in the attenuation of viral replication. In this study we demonstrated that the EV71 3C sumoylation-deficient virus triggered higher levels of apoptosis and elevated virus titer compared with WT virus, correlating with observations in virus-infected mice that presented with significantly increased apoptosis of neuronal cells in spinal cord and increased levels of viral VP1 protein in the intestine (Fig. 5). These results strongly implicate that sumoylation functions as a host cell defense mechanism against EV71 3C to reduce EV71 viral replication and viral infection-induced apoptosis. Our findings may serve as a template for studying sumoylation in the context of a defense mechanism against viral replication.
In addition, we identified Ubc9 as an EV71 3C-interacting protein and identified a 3C sumoylation site at the Lys-52 residue. Ubc9 generally binds to a protein substrate via the sumoylation ψKX(E/D) consensus motif (22, 23). Interestingly, EV71 3C Lys-52 and its flanking residues did not match this consensus motif. Instead, these residues fit the inverted sumoylation consensus motif (E/DXKψ), recently described for other sumoylated substrates (53). Although the binding of Ubc9 to the inverted sumoylation motif has not been characterized, our results from GST pulldown assay using mutated 3C proteins (Fig. 6C) indicated the involvement of this inverted sumoylation motif in mediating the Ubc9 interaction. In addition, the 3C residues between Lys-45 and Lys-49 also contributed to Ubc9 interactions to a certain extent that correlated with distinct sumoylation levels between these mutants (Fig. 6D). Interestingly, clinical EV71 isolates harboring 3C mutations in the Ubc9 interaction (supplemental Table 1) suggest that the EV71 virus may have evolved an escape mechanism from the host by reducing the 3C-Ubc9 interaction.
EV71 3C can be modified by mono-SUMO-1 as well as by the poly-SUMO-1 chain at the Lys-52 residue both in vitro and in vivo. The sumoylation pattern, the relative abundance of mono- versus poly-SUMO-1 chain formation on 3C seen in the in vitro assays (Fig. 1C) and during EV71 viral infection (Fig. 2D), was similar. However, when SUMO-1 was coexpressed with 3C in cells (Fig. 1D), the level of mono- versus poly-SUMO chain formation differed dramatically. This discrepancy could be due to SUMO-1 overexpression that increased the formation of different poly-SUMO chains on 3C. In addition, sumoylation-elicited 3C ubiquitination may also have contributed to this distinct pattern of poly-SUMO chain formation on 3C in cells as supported by the observation that treatment of GFP-3C proteins precipitated from SUMO-1-coexpressing cells, with a recombinant de-ubiquitinating enzyme, significantly reduced some of the slowly migrating bands.4 In line with the observation that SUMO was present in ubiquitinated 3C (Fig. 4D), these data support the notion that sumoylation is associated with 3C ubiquitination.
Sumoylation can either promote or antagonize protein substrate ubiquitination (30, 42). Initial observations that 3C WT but not K52R conferred robust ubiquitination (Fig. 4B) raised the possibility that Lys-52 may also have acted as a 3C ubiquitination site. In this scenario, sumoylation may have antagonized 3C ubiquitination. However, the results that SENP1 expression effectively removed 3C sumoylation also abrogated 3C ubiquitination excluded this possibility (Fig. 4D). In addition, the protein stability of 3C WT, but not K52R mutant, was positively and negatively regulated by SENP1 and PIAS1 (Fig. 4E), respectively. These results suggest that sumoylation of EV71 3C at position Lys-52 promotes 3C degradation by ubiquitination in cells. In agreement with this notion, EV71 3C sumoylation was detected early post-infection (8 h) followed by significantly reduced levels 12 h post-infection, correlating with a significantly increased level of 3C ubiquitination detected at the 12-h time point (Figs. 2D and 4C). The molecular mechanism of how sumoylation enhances EV71 3C ubiquitination is currently unclear. One possibility is that the SUMO moiety of sumoylated EV71 3C mediates a direct interaction with an E3 ubiquitin ligase, resulting in ubiquitin modification. For instance, RNF4 could preferentially mediate SUMO-2/3 polychain-conjugated substrates for ubiquitination via direct interaction between the SUMO-2/3 polychain and 4 copies of the SUMO-interacting motif in RNF4 (32). Because EV71 3C can be modified by SUMO-1 more effectively than SUMO-2 (Fig. 2D and supplemental Fig. 1), our observation that RNF4 ineffectively regulated 3C ubiquitination and protein stability was expected, suggesting that an other E3 ubiquitin ligase(s) likely exists for the ubiquitination of sumoylated 3C.
Alternatively, sumoylation may not mediate the direct interaction of EV71 3C protein with an E3 ubiquitin ligase. A previous study examining NEMO sumoylation revealed that sumoylation was responsible for retention of NEMO in the nuclear compartment where NEMO was phosphorylated and then ubiquitinated before translocation back to the cytosol (54). Thus, it is possible that sumoylation may alter EV71 3C subcellular localization for further ubiquitination and degradation. However, additional studies will be required to substantiate this hypothesis. In addition to sumoylation-mediated modulation of 3C stability in cells, we also showed that sumoylation could down-regulate 3C substrate cleavage activity in vitro. Although the Lys-52 residue is not within the 3C catalytic domain, sumoylation-reduced 3C substrate cleavage could result from blocking 3C interaction with its substrate or from a conformational change to its catalytic domain, which dampened catalytic activity.
In summary, our findings showed that EV71 3C sumoylation depended on the interaction between 3C and Ubc9. Sumoylation-attenuated EV71 3C function correlated with a reduction in viral replication and apoptosis, implying that EV71 strains with different sumoylation potential may cause infections presenting with distinct clinical severity.
Supplementary Material
Acknowledgments
We thank Drs. Peter O'Hare, Jaulang Hwang, and Jorma Palvimo for the HeLa-His-SUMO-1 stable cell line, the SUMO-1 11KR, and the FLAG-RNF4 constructs, respectively. We also thank Dr. Pang-Hsien Tu for analyzing histologic results and Dr. Che-Chang Chang, Dr. Jen-Chong Jeng, Chun-Chen Ho, and Huei-Ling Tang for generating constructs and reagents for the in vitro sumoylation assays.
This work was supported by National Science Council Grant NSC99-2321-B-002-025, Frontier Grant NSC99-2321-B-001-007, and an Academia Sinica Investigator Award (to H.-M. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1–S5.
L. Y. Chang, unpublished data.
S. C. Chen and H. M. Shih, unpublished data.
- EV71
- enterovirus 71
- TBP
- TATA box-binding protein
- PARP
- poly-ADP-ribose polymerase
- RD
- rhabdomyosarcoma
- m.o.i.
- multiplicity of infection
- CREB
- cAMP-response element-binding protein
- SUMO
- small ubiquitin-like modifier
- PML
- promyelocytic leukemia protein
- IB
- immunoblot
- IP
- immunoprecipitate.
REFERENCES
- 1. AbuBakar S., Chee H. Y., Al-Kobaisi M. F., Xiaoshan J., Chua K. B., Lam S. K. (1999) Virus Res. 61, 1–9 [DOI] [PubMed] [Google Scholar]
- 2. Cardosa M. J., Perera D., Brown B. A., Cheon D., Chan H. M., Chan K. P., Cho H., McMinn P. (2003) Emerg. Infect. Dis. 9, 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kehle J., Roth B., Metzger C., Pfitzner A., Enders G. (2003) J. Neurovirol. 9, 126–128 [DOI] [PubMed] [Google Scholar]
- 4. Ho M., Chen E. R., Hsu K. H., Twu S. J., Chen K. T., Tsai S. F., Wang J. R., Shih S. R. (1999) N. Engl. J. Med. 341, 929–935 [DOI] [PubMed] [Google Scholar]
- 5. Huang C. C., Liu C. C., Chang Y. C., Chen C. Y., Wang S. T., Yeh T. F. (1999) N. Engl. J. Med. 341, 936–942 [DOI] [PubMed] [Google Scholar]
- 6. Brown B. A., Pallansch M. A. (1995) Virus Res. 39, 195–205 [DOI] [PubMed] [Google Scholar]
- 7. Wimmer E., Hellen C. U., Cao X. (1993) Annu. Rev. Genet. 27, 353–436 [DOI] [PubMed] [Google Scholar]
- 8. Clark M. E., Lieberman P. M., Berk A. J., Dasgupta A. (1993) Mol. Cell. Biol. 13, 1232–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Armer H., Moffat K., Wileman T., Belsham G. J., Jackson T., Duprex W. P., Ryan M., Monaghan P. (2008) J. Virol. 82, 10556–10566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rubinstein S. J., Hammerle T., Wimmer E., Dasgupta A. (1992) J. Virol. 66, 3062–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weng K. F., Li M. L., Hung C. T., Shih S. R. (2009) PLoS Pathog. 5, e1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yalamanchili P., Datta U., Dasgupta A. (1997) J. Virol. 71, 1220–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joachims M., Harris K. S., Etchison D. (1995) Virology 211, 451–461 [DOI] [PubMed] [Google Scholar]
- 14. Li M. L., Hsu T. A., Chen T. C., Chang S. C., Lee J. C., Chen C. C., Stollar V., Shih S. R. (2002) Virology 293, 386–395 [DOI] [PubMed] [Google Scholar]
- 15. Calandria C., Irurzun A., Barco A., Carrasco L. (2004) Virus Res. 104, 39–49 [DOI] [PubMed] [Google Scholar]
- 16. Chang S. C., Lin J. Y., Lo L. Y., Li M. L., Shih S. R. (2004) J. Neurovirol. 10, 338–349 [DOI] [PubMed] [Google Scholar]
- 17. Wang H. Y., Tsao K. C., Hsieh C. H., Huang L. M., Lin T. Y., Chen G. W., Shih S. R., Chang L. Y. (2010) BMC Evol. Biol. 10, 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gareau J. R., Lima C. D. (2010) Nat. Rev. Mol. Cell Biol. 11, 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Geiss-Friedlander R., Melchior F. (2007) Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 20. Boggio R., Chiocca S. (2006) Curr. Opin. Microbiol. 9, 430–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson E. S. (2004) Annu. Rev. Biochem. 73, 355–382 [DOI] [PubMed] [Google Scholar]
- 22. Rodriguez M. S., Dargemont C., Hay R. T. (2001) J. Biol. Chem. 276, 12654–12659 [DOI] [PubMed] [Google Scholar]
- 23. Sampson D. A., Wang M., Matunis M. J. (2001) J. Biol. Chem. 276, 21664–21669 [DOI] [PubMed] [Google Scholar]
- 24. Nishida T., Yasuda H. (2002) J. Biol. Chem. 277, 41311–41317 [DOI] [PubMed] [Google Scholar]
- 25. Pichler A., Gast A., Seeler J. S., Dejean A., Melchior F. (2002) Cell 108, 109–120 [DOI] [PubMed] [Google Scholar]
- 26. Wotton D., Merrill J. C. (2007) Biochem. Soc. Trans. 35, 1401–1404 [DOI] [PubMed] [Google Scholar]
- 27. Gao C., Ho C. C., Reineke E., Lam M., Cheng X., Stanya K. J., Liu Y., Chakraborty S., Shih H. M., Kao H. Y. (2008) Mol. Cell. Biol. 28, 5658–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao X., Sternsdorf T., Bolger T. A., Evans R. M., Yao T. P. (2005) Mol. Cell. Biol. 25, 8456–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yeh E. T. (2009) J. Biol. Chem. 284, 8223–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ulrich H. D. (2005) Trends Cell Biol. 15, 525–532 [DOI] [PubMed] [Google Scholar]
- 31. Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., de Thé H. (2008) Nat. Cell Biol. 10, 547–555 [DOI] [PubMed] [Google Scholar]
- 32. Tatham M. H., Geoffroy M. C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., Palvimo J. J., Hay R. T. (2008) Nat. Cell Biol. 10, 538–546 [DOI] [PubMed] [Google Scholar]
- 33. van Hagen M., Overmeer R. M., Abolvardi S. S., Vertegaal A. C. (2010) Nucleic Acids Res. 38, 1922–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin D. Y., Huang Y. S., Jeng J. C., Kuo H. Y., Chang C. C., Chao T. T., Ho C. C., Chen Y. C., Lin T. P., Fang H. I., Hung C. C., Suen C. S., Hwang M. J., Chang K. S., Maul G. G., Shih H. M. (2006) Mol. Cell 24, 341–354 [DOI] [PubMed] [Google Scholar]
- 35. Yang M., Hsu C. T., Ting C. Y., Liu L. F., Hwang J. (2006) J. Biol. Chem. 281, 8264–8274 [DOI] [PubMed] [Google Scholar]
- 36. Chang C. C., Naik M. T., Huang Y. S., Jeng J. C., Liao P. H., Kuo H. Y., Ho C. C., Hsieh Y. L., Lin C. H., Huang N. J., Naik N. M., Kung C. C., Lin S. Y., Chen R. H., Chang K. S., Huang T. H., Shih H. M. (2011) Mol. Cell 42, 62–74 [DOI] [PubMed] [Google Scholar]
- 37. Liu J. H., Mu Z. M., Chang K. S. (1995) J. Exp. Med. 181, 1965–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li Z., Day C. P., Yang J. Y., Tsai W. B., Lozano G., Shih H. M., Hung M. C. (2004) Cancer Res. 64, 9080–9085 [DOI] [PubMed] [Google Scholar]
- 39. Shih S. R., Chiang C., Chen T. C., Wu C. N., Hsu J. T., Lee J. C., Hwang M. J., Li M. L., Chen G. W., Ho M. S. (2004) J. Biomed. Sci. 11, 239–248 [DOI] [PubMed] [Google Scholar]
- 40. Shih S. R., Tsai M. C., Tseng S. N., Won K. F., Shia K. S., Li W. T., Chern J. H., Chen G. W., Lee C. C., Lee Y. C., Peng K. C., Chao Y. S. (2004) Antimicrob. Agents Chemother. 48, 3523–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shih S. R., Weng K. F., Stollar V., Li M. L. (2008) J. Neurovirol. 14, 53–61 [DOI] [PubMed] [Google Scholar]
- 42. Hunter T., Sun H. (2008) Ernst. Schering Found. Symp. Proc. 1, 1–16 [DOI] [PubMed] [Google Scholar]
- 43. Chen Y. C., Yu C. K., Wang Y. F., Liu C. C., Su I. J., Lei H. Y. (2004) J. Gen. Virol. 85, 69–77 [DOI] [PubMed] [Google Scholar]
- 44. Wong T. W., Huang H. J., Wang Y. F., Lee Y. P., Huang C. C., Yu C. K. (2010) J. Antimicrob. Chemother. 65, 2176–2182 [DOI] [PubMed] [Google Scholar]
- 45. Xu K., Klenk C., Liu B., Keiner B., Cheng J., Zheng B. J., Li L., Han Q., Wang C., Li T., Chen Z., Shu Y., Liu J., Klenk H. D., Sun B. (2011) J. Virol. 85, 1086–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Votteler J., Neumann L., Hahn S., Hahn F., Rauch P., Schmidt K., Studtrucker N., Solbak S. M., Fossen T., Henklein P., Ott D. E., Holland G., Bannert N., Schubert U. (2011) Retrovirology 8, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Adamson A. L., Kenney S. (2001) J. Virol. 75, 2388–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Spengler M. L., Kurapatwinski K., Black A. R., Azizkhan-Clifford J. (2002) J. Virol. 76, 2990–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Müller S., Dejean A. (1999) J. Virol. 73, 5137–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bailey D., O'Hare P. (2002) J. Gen. Virol. 83, 2951–2964 [DOI] [PubMed] [Google Scholar]
- 51. Boggio R., Colombo R., Hay R. T., Draetta G. F., Chiocca S. (2004) Mol. Cell 16, 549–561 [DOI] [PubMed] [Google Scholar]
- 52. Chiocca S. (2007) Biochem. Soc. Trans. 35, 1419–1421 [DOI] [PubMed] [Google Scholar]
- 53. Matic I., Schimmel J., Hendriks I. A., van Santen M. A., van de Rijke F., van Dam H., Gnad F., Mann M., Vertegaal A. C. (2010) Mol. Cell 39, 641–652 [DOI] [PubMed] [Google Scholar]
- 54. Huang T. T., Wuerzberger-Davis S. M., Wu Z. H., Miyamoto S. (2003) Cell 115, 565–576 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.