Abstract
MicroRNA-183 (miR-183), miR-96, and miR-182 comprising the miR-183/96/182 cluster are highly expressed in photoreceptor cells. Although in vitro data have indicated an important role for this cluster in the retina, details of its in vivo biological activity are still unknown. To observe the impact of the miR-183/96/182 cluster on retinal maintenance and light adaptation, we generated a sponge transgenic mouse model that disrupted the activities of the three-component microRNAs simultaneously and selectively in the retina. Although our morphological and functional studies showed no differences between transgenic and wild type mice under normal laboratory lighting conditions, sponge transgenic mice displayed severe retinal degeneration after 30 min of exposure to 10,000 lux light. Histological studies showed that the outer nuclear layer thickness was dramatically reduced in the superior retina of transgenic mice. Real time PCR experiments in both the sponge transgenic mouse model and different microRNA stable cell lines identified Arrdc3, Neurod4, and caspase-2 (Casp2) as probable downstream targets of this cluster, a result also supported by luciferase assay and immunoblotting analyses. Further studies indicated that expression of both the cluster and Casp2 increased in response to light exposure. Importantly, Casp2 expression was enhanced in transgenic mice, and inhibition of Casp2 partially rescued their light-induced retinal degeneration. By connecting the microRNA and apoptotic pathways, these findings imply an important role for the miR-183/96/182 cluster in acute light-induced retinal degeneration of mice. This study demonstrates a clear involvement of miRs in the physiology of postmitotic cells in vivo.
Keywords: MicroRNA, Neurodegeneration, Photoreceptors, Vision, Vitamin A
Introduction
MicroRNAs (miRs)2 are endogenous and short (about 22 nucleotides) noncoding RNA molecules that bind to complementary sequences in the 3′-UTR of multiple target mRNAs, usually inhibiting their translation or causing their destabilization (1, 2). miRs are well conserved in eukaryotic organisms and widely expressed in different tissues and cell types (3). To date, 885 and 689 miRs have been identified in human and mouse genomes, respectively (miRBASE Release 13.0, March, 2009). Each miR regulates about 200–800 target genes, and 60% of protein-encoding genes in humans are predicted to be regulated by miRs (4). Several miRs are tissue-specific, suggesting specialized roles in tissue differentiation and maintenance (5, 6). Progress toward identifying the in vivo functions of miRs has been limited, not only by the complex biology of these molecules but also by the experimental accessibility of the biological systems that they affect.
The retina is a layered tissue in the back of the eye that contains specialized photoreceptor cells called rods and cones as well as other cell types, including ganglion, bipolar, horizontal, and amacrine cells (7). Its development, maintenance, and light-sensitive visual functions are highly regulated. At least 78 miRs are preferentially expressed in vertebrate retina (6, 8, 9), indicating the importance of miRs in modulating gene expression profiles in retinal cells. Moreover, inactivation of Dicer (an essential RNase III endonuclease required for miR maturation) leads to progressive functional and structural degeneration of mouse retina (10, 11).
In 2007, Xu et al. (6) identified miR-183, miR-96, and miR-182 as a sensory organ-specific paralogous miR cluster called the miR-183/96/182 cluster. These miRs are located within 4 kb of each other on mouse chromosome 6qA3 and are transcribed in the same direction from telomere to centromere. This genomic arrangement is highly conserved in vertebrates (supplemental Fig. S1A), and the sequences of the three-component miRs are similar to each other. Real time PCR and in situ hybridization data reveal that the miR-183/96/182 cluster is highly expressed in photoreceptors and interneurons of the inner nuclear layer but is not detectable in brain (6, 12). Levels of the cluster are down-regulated during dark adaptation and up-regulated in light-adapted retina, with rapid decay and increased transcription being responsible for the respective changes (13). In the P347S rhodopsin transgenic mouse, a human retinitis pigmentosa model, and three other photoreceptor-linked mouse models of RP (Opsin−/−, Δ307 and rds) (12), expression of the miR-183/96/182 cluster was dramatically decreased (12), suggesting that such decreased expression is associated with retinal disease. However, another study showed that there were no significant transcriptional or phenotypic changes in miR-182 depleted mice (14). Similarly, a naturally occurring seed region substitution mutation in miR-96 of human patients did not produce any detectable visual impairment (15). The absence of retinal defects in these cases may stem from redundancy in the miR-183/96/182 cluster, i.e. because the three-component miRs are similar, they can functionally substitute for each other.
Exposure to excessive white light can induce retinal degeneration in a variety of animals. However, the molecular mechanism by which light induces retinal degeneration is presently unclear. Apoptosis has been implicated as a common final pathway in retinal degeneration (16), and several apoptosis-related genes have been shown to be activated during photoreceptor cell death. Casp1 and Casp2 were detected in the ONL of degenerating rat retina (17). Bid and its upstream gene, p38, were also activated in apoptotic photoreceptors (18). Bax and Bak double knock-out in mice protected photoreceptors from light damage (19, 20). Moreover, inhibition of Casp3 preserved the retina in rd1/rd1 mice (21), tubby mice (22), and S334ter rats (23). Casp2 is a cysteine-aspartate protease that mediates apoptosis induced by many types of cellular stress, including reactive oxygen species, DNA damage, and endoplasmic reticulum stress. Once activated, Casp2 cleaves Bid and causes mitochondrial outer membrane permeabilization and cytochrome c release, which in turn activate Casp9 and Casp3, resulting in apoptosis (24).
To clarify the roles of the miR-183/96/182 cluster in retinal maintenance and light adaptation, we generated a sponge transgenic mouse model to disrupt the activity of these three miRs simultaneously in the retina. Then we characterized the retinal defects in these transgenic mice and identified transcriptional changes in their retinas. Although there were no significant morphological and functional differences between wild type (WT; C57/BL6) and transgenic mice under laboratory cyclic lighting conditions, transgenic mice displayed severe retinal degeneration after acute bright light exposure.
MATERIALS AND METHODS
Generation of the Sponge Transgenic Mouse Model
To generate miR-183/96/182 cluster sponge elements, we introduced 10 copies each of complementary sequences to miR-183, miR-96, and miR-182, each with mismatches at positions 9–12 for improved stability (25), into the 3′-UTR of enhanced green fluorescent protein (EGFP) in a pNOVA expression vector. Sponge element expression was driven by an Opsin promoter and terminated with an SV40 polyadenylation element (supplemental Fig. S2A). Special primer pairs bridging the Opsin promoter and EGFP coding sequences were designed for PCR detection. To assess the specificity and efficiency of the Opsin promoter, the plasmid was sequenced and then transfected into Y79 cells (retinal carcinoma cells, American Type Culture Collection, Manassas, VA) and HEK293 cells (controls). Bright EGFP-positive cells were noted in the transfected Y79 cells, but not in the nontransfected cells or HEK293 cells, confirming the efficiency and specificity of the Opsin promoter (supplemental Fig. S2B). The 6.8-kb transgene fragment was cut out by NotI and MluI and then injected into fertilized eggs of BL6/SJL hybrid mice using the pronuclear injection method in the Transgenic and Targeting Facility at Case Western Reserve University. Founder mice were then back-crossed with C57/BL6 mice many times to obtain a clean background.
Genotyping Analyses
Genomic DNA from the tails of 4-week-old mice was digested with DirectPCR (VIAGEN Biotech Inc., Los Angeles, CA) plus proteinase K (final concentration, 50 μg/ml) at 55 °C overnight and then heat-inactivated at 85 °C for 45 min. Sequences of primer pairs used to screen the sponge transgenic mice were as follows: SpongeFor, 5′-ATATCTCGCGGATGCTGAAT-3′, and SpongeRev, 5′-GAACTTCAGGGTCAGCTTGC-3′. Rd_For/Pde_Rev and Pde_For/Pde_Rev primer pairs were designed to distinguish the Pde6brd1 mutant allele from the normal allele. β-Actin, detected by β-actin_For/β-actin_Rev primer pairs, was used as an internal control. Sequences for these primers were as follows: Rd_For, 5′-CCAGTGGCCTTCCAACCTAA-3′; Pde_For, 5′-CCAGTGGCCTTCCAACCTAC-3′, and Pde_Rev, 5′-GAAAGTTGAACATTTCATCAGCTG-3′; β-actin_For, 5′-AGTGTGACGTTGACATCCGTA-3′, and β-actin_Rev, 5′-GCCAGAGCAGTAATCTCCTTCT3′.
RT-PCR and Real Time PCR
Total RNAs were extracted from the mouse retina or cells with the MirVanaTM miR isolation kit (Ambion, Austin, TX). For miR real time PCR, all reagents, including the Universal cDNA synthesis kit, SYBR® Green master mix, LNATM PCR primer sets, and 5 S rRNA PCR primer set, were purchased from Exiqon (Woburn, MA). For mRNA RT-PCR and real time PCR, primer sets were designed within exon junction areas listed in supplemental Table S1. In brief, mRNAs were reverse-transcribed with a SuperScript® III first-strand synthesis kit (Invitrogen). Then 50 ng of cDNA was mixed with FS Universal SYBR Green master mix (Roche Applied Science) and the target gene primer set (final concentration, 1 μm for each primer) to produce a 20-μl reaction mixture. All real time PCR experiments were done in an Eppendorf Mastercycler® ep realplex real time PCR system.
Splinted Ligation
Total RNAs were extracted from mouse retina or cells with the MirVanaTM miR isolation kit, and 0.5–4 μg of total RNA was used for each assay. All procedures followed manufacturer's instructions for the miRtect-IT miRNA labeling and detection kit (United States Biochemical Corp., Cleveland, OH) (26). Twenty pmol of detection oligonucleotide was 5′-end-radiolabeled with 2.5 μm [γ-32P]ATP (150 mCi/ml, PerkinElmer Life Sciences). Designs of bridge oligonucleotides and mimic oligonucleotides were based on miR sequences registered in the miRBase sequence database (see supplemental Table S2). Ligated DNA-RNA complexes were separated by 16% mini urea-polyacrylamide gels and exposed to a phosphor screen after drying for 2 h. The radioisotope signal detected by the phosphor screen was recorded with Storm 820 (Amersham Biosciences). Because the miR-183, miR-96, and miR-182 sequences are similar, we tested the probe specificities before doing the experiments (supplemental Fig. S3).
Electroretinography (ERG)
Two- to 4-month-old female mice were dark-adapted for more than 24 h prior to an ERG and then anesthetized by intraperitoneal injection of a mixture (20 μl/g body weight) containing ketamine (6 mg/ml) and xylazine (0.44 mg/ml) in 10 mm sodium phosphate, pH 7.2, with 100 mm NaCl. To stabilize body temperature, mice were placed on a heated pad (<40 °C) in a Ganzfeld chamber, and positioning of the contact lens electrodes was adjusted. Then two needle electrodes were placed under the skin of the forehead and tail. Before the start of recording, the base line was checked to verify that the mouse was alive and the lens was well contacted. Single flash ERGs were recorded simultaneously from both eyes, with scotopic flash ERGs from −3.7 to 0.6 log candela·s/m2 (interstimulus intervals between 10 and 60 s). After 5 min of light adaptation (30 candela·s/m2), photopic flash ERGs from −2.1 to 0.6 log candela·s/m2 were recorded. Amplitudes were measured by EM software for Windows (LKC Technologies, Inc.) from base line to the negative peak for a-waves, and from the latter (or base line, if absent) to the positive peak for b-waves.
Ultra-high Resolution Optical Coherence Tomography (OCT)
Ultra-high resolution SD-OCT (Bioptigen) was employed for in vivo imaging of mouse retinas. Mice were anesthetized by intraperitoneal injection of a mixture (20 μl/g body weight) containing ketamine (6 mg/ml) and xylazine (0.44 mg/ml) in 10 mm sodium phosphate, pH 7.2, with 100 mm NaCl. Pupils were dilated with 1% tropicamide. Four pictures acquired in the B-scan mode were used to construct each final averaged SD-OCT image. The superior retina, inferior retina, and optical nerve head regions were imaged for each eye.
Retinal Histology and Immunocytochemistry
Mouse eyeballs were extracted immediately after euthanasia. After removal of the cornea and lens, eyecups were fixed in 4% neutral formalin overnight at 4 °C. Following 5, 10, and 20% sucrose dehydration, eyecups were embedded in 20% sucrose/OCT (1:1), and 12-μm thick sections were cut. Hematoxylin and eosin (H&E) staining was performed by the Pathology Core Facility at Case Western Reserve University. For immunocytochemistry, slides were incubated for 2 h at 55 °C, then blocked with 1.5% normal goat serum for 1 h, and incubated with primary antibody overnight. After washing three times in PBS (137 mm NaCl, 2.7 mm KCl, 4.3 mm Na2HPO4, 1.4 mm KH2PO4, pH 7.2–7.4), slides were incubated with fluorescent conjugated secondary antibody, peanut agglutinin lectin (PNA) (for detection of cones, 1:500 dilution), and DAPI (for counterstaining cell nuclei, 1:500 dilution). The primary antibody was anti-Opsin 1D4 antibody (1:200 dilution, a kind gift from R. S. Molday (Vancouver, British Columbia, Canada)).
Laser Capture Microscopy
Laser capture microscopy was done in the core facility at the Cleveland Clinic Foundation. Basically, 24 h after 30 min of 10,000 lux light exposure, eyeballs from 4-month-old mice were collected fresh and embedded in Tissue-Tech O.C.T. compound. All samples were cut at 12 μm thickness and mounted on Leica membrane slides (PET-Membrane, Wetzlar, Germany). Rapid HistoGene® staining (Applied Biosystems, Foster City, CA) was used to identify different regions of the retina. Briefly, the sections were fixed in 75% ethanol for 30 s and then immersed in HistoGene staining solution for 30 s, followed by dehydration and washing with graded alcohols (75, 95, and 100%) for 20 s each. Regions of the retina were isolated using an HC PLan Fluotar 10×/0.3NA lens on a Leica AS-LMD laser microdissection microscope (Leica Microsystems, Bannockburn, IL) equipped with a 337 nm nitrogen laser and a Hitachi HV-C20A 3-chip color CCD camera. Fragments were collected in lysis buffer from a MirVanaTM miR isolation kit for subsequent experiments.
Creation of Stable Cell Lines
Partial pri-miR sequences of miR-183, miR-96, and miR-182 were inserted into pMXs-IRES-EGFP retroviral vector (supplemental Fig. S4A), and the plasmids were transfected into Phoenix cells with calcium phosphate. Medium was replaced 12 h after transfection, and the transfected cells were incubated for 24 h at 32 °C. Viruses were harvested from the medium, diluted 1:6 with target cell medium, and then supplied with Polybrene (final concentration 5 μg/ml). NIH3T3 cells were incubated with this virus mixture at 32 °C supplemented with 5% CO2. After three cycles of viral infection, NIH3T3 cells were treated with 0.05% trypsin (Thermo Scientific, Logan, UT) for 2 min to dissociate them from the plates. The cells were then sorted to obtain those 5% with the greatest EGFP fluorescence intensity as assessed by flow cytometry at the Flow Cytometry Core Facility at Case Western Reserve University.
Immunoblotting
Cells or retinas were lysed in NTEN buffer (100 mm NaCl, 0.5% Nonidet P-40, 50 mm Tris-HCl, pH 7.6) with a proteinase inhibitor mixture (Roche Applied Science). Proteins were separated by 15% polyacrylamide gels and detected by anti-Casp2 (1:200 dilution, Biovision, Mountain View, CA), anti-Opsin 1D4 (1:1,000 dilution), anti-Bid (final concentration 1 μg/ml, R&D Systems, Minneapolis, MN), anti-middle wave-sensitive opsin (Mw-opsin, 1:500 dilution, EMD Millipore, Billerica, MA), and anti-β-tubulin (1:1,000 dilution, Santa Cruz Biotechnology, Santa Cruz, CA) antibodies.
Luciferase Reporter Assay
pGL3P-4×Neurod4-3′-UTR, pGL3P-4×Arrdc3-3′-UTR, and pGL3P-4×Casp2-3′-UTR plasmids were constructed for the luciferase reporter assay. Basically, four copies of targeted regions from the target gene 3′-UTR flanked by XbaI and FseI restriction sites were inserted between the luciferase coding sequence and SV40 polyadenylation element of the pGL3-promoter vector (pGL3P) (supplemental Fig. S5A). NIH3T3 cells were maintained in DMEM with 10% FBS, and transfections were carried out with the LipofectamineTM 2000 reagent (Invitrogen). After obtaining the engineered plasmids, we first tested their linear expression range (supplemental Fig. S5B). Plasmids overexpressing miR-183/96/182 clusters were constructed by inserting partial pri-miR sequences of miR-183, miR-96, and miR-182 into the pRNAT-CMV3.1 vector (supplemental Fig. S4A). For miR targeting assays, cells were cultured in 24-well plates and transfected with 500 ng of control pRNAT-CMV3.1 vector or plasmids overexpressing miRs-183, -96, or -182 along with 160 ng of a reporter construct, either pGL3P-4×Neurod4-3′-UTR, pGL3P-4×Arrdc3-3′-UTR, pGL3P-4×Casp2-3′-UTR, or empty pGL3P. Forty ng of pRL-CMV-Renilla plasmid was also transfected as an internal control. Expression levels of miR-183, miR-96, and miR-182 were measured by EGFP fluorescence. The luciferase reporter assays (Promega, Madison, WI) were performed 48 h after transfection, and luciferase activity was determined with an LmaxTM microplate luminometer (Molecular Devices, Sunnyvale, CA). Relative luciferase activities were calculated as ratios of firefly to Renilla luciferase activities.
Light-induced Retinal Damage
Two- to 4-month-old mice were dark-adapted for 1 week before use for light-induced retinal damage experiments. After pupillary dilation by 1% tropicamide, mice were exposed to white fluorescent light (10,000 lux) (150-watt spiral lamp, Commercial Electric, Cleveland, OH) for either 10 or 30 min in a white bucket (Papersmith, San Marcos, TX) and then kept under normal cyclic light conditions. No more than two mice were kept in a bucket at the same time. OCT examination was performed 2 weeks after light exposure.
Effect of Caspase 2 Inhibitor on Light-induced Retinal Damage
Two- to 4-month-old mice were dark-adapted for 1 week before use for the Casp2 inhibitor experiment. After pupillary dilation with 1% tropicamide and exposure to 10,000 lux white light for 30 min, mice were anesthetized by intraperitoneal injection of a mixture (20 μl/g body weight) containing ketamine (6 mg/ml) and xylazine (0.44 mg/ml) in 10 mm sodium phosphate, pH 7.2, with 100 mm NaCl. One μl of Z-VDVAD-FMK (R&D Systems, Minneapolis, MN; 1.6 mm in 2% DMSO) was injected intravitreally into the right eyes with a NanoFil 100-μl syringe (World Precision Instruments, Sarasota, FL) and a 33-gauge needle (World Precision Instruments). Left eyes were injected in parallel with 2% DMSO as the control. After injection, a drop of antiseptic solution (Daiichi Sankyo Co., Ltd., Tokyo, Japan) was applied to the cornea. OCT examination and immunocytochemistry analysis were performed 2 weeks later.
Statistical Analyses
Results are presented as means ± S.D., and the number of experiments is indicated. ImageJ 1.42q (National Institutes of Health) was used for quantification. Statistical significance was assessed by using the two-tailed Student's t test.
RESULTS
miR-183/96/182 Cluster Is Specifically Expressed in Photoreceptor Cells
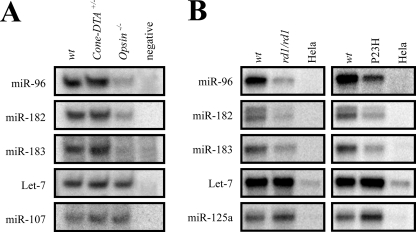
To determine whether miR-183/96/182 cluster miRs are expressed in rods and/or cones, we used two knock-out mouse models of photoreceptor degeneration. Cones are absent in Cone-DTA+/− mice, but the density and morphology of rods are normal (27). In contrast, neither cones nor rods are present in 4-month-old Opsin−/− mice (28). Total RNAs from the retina of 4-month-old WT, Cone-DTA+/−, and Opsin−/− mice were extracted and subjected to splinted ligation to measure miR abundance with the ubiquitous miR, let-7, used as an internal control. None of the three miRs showed any changes in Cone-DTA+/− mice, but all were significantly reduced in Opsin−/− mice (Fig. 1A). miR-107, known to be expressed in human and rat retina (8), exhibited little change in the splinted ligation assay. The reduced expression of miR-96, -182, and -183 in Opsin−/− mice strongly suggests that these miRs are expressed in photoreceptors. But as cones account for fewer than 5% of total photoreceptors, it is unclear whether these miRs are expressed in cones as well.
FIGURE 1.
miR-183/96/182 clusters are specifically expressed in photoreceptor cells. A, quantification of selected miRs by splinted ligation in 4-month-old wild type (wt), Cone-DTA+/−, and Opsin−/− mouse retinas. RNase-free water was used for a negative control and Let-7 as an internal control. miR-183, miR-96, and miR-182 were all significantly decreased in Opsin−/− mice but unchanged in Cone-DTA+/− mice. Compared with WT, miR-107 was unchanged in Cone-DTA+/− and Rho−/− mice. B, quantification of selected miRs by splinted ligation in the retina of 4-month-old WT mice, rd1/rd1 mutant mice (29), and P23H Opsin knock-in homozygous mice (P23H) (30). HeLa cells, derived from a human cervical cancer cell line, were used as a negative control because these cells do not express the miR-183/96/182 cluster. Compared with WT retina, miR-183, miR-96, and miR-182 were decreased in rd1/rd1 and P23H mutant retinas but Let-7 stayed the same, and miR-125a even increased slightly in mutant mice.
To confirm expression of the miR-183/96/182 cluster in photoreceptor cells, we examined its expression in RNA isolated from the retina of 4-month-old mutant mice with either retinal degeneration (rd1/rd1 mice) or the P23H Opsin mutation, both of which lack photoreceptors but retain the remaining cellular constituents of neural retina (29, 30). Splinted ligation results showed that, compared with wild type retina, miRs-183, -96, and -182 were decreased 2.4-, 2.0-, and 2.1-fold, respectively, in rd1/rd1 mutant retina, and 2.3-, 2.0-, and 2.5-fold, respectively, in P23H Opsin mutant retina (Fig. 1B). miR-125a (which is expressed in retina, brain, and other organs (12)) and the ubiquitous miR let-7 were used as controls for this assay. Expression of let-7 was unchanged in both rd1/rd1 mutant and P23H Opsin mutant retinas, and miR-125a showed a modest increase in expression (1.4- and 1.6-fold in rd1/rd1 and P23H Opsin mutant retinas). Thus, miR-183, miR-96, and miR-182 were all specifically decreased in retinas of mutant mice that exhibited different types of photoreceptor degeneration, suggesting that miR-183/96/182 clusters are highly expressed in photoreceptor cells.
Creation of miR-183/96/182 Cluster Sponge Transgenic Mice
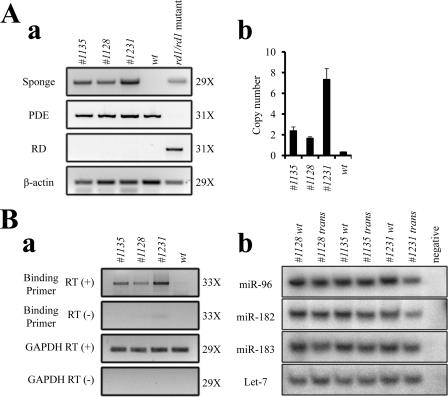
miR-183/96/182 miRs share significant sequence similarity in their seed regions (supplemental Fig. S2A, panel b). Indeed, there is extensive overlap in the predicted targets of these three miRs. This functional redundancy complicates interpreting the results of disrupting each member of this cluster. To overcome this problem and better understand the roles of the miR-183/96/182 cluster in vivo, we generated miR-183/96/182 cluster sponge transgenic mouse lines. The miR-183/96/182 cluster sponge element was composed of 10 copies each of sequences complementary to miR-183, miR-96, and miR-182 with mismatches at positions 9–12 to enhance stability. This sequence was inserted into the 3′-UTR of EGFP in a pNOVA expression vector regulated by an Opsin promoter and ending with an SV40 polyadenylation signal (supplemental Fig. S2A). Several transgenic mouse lines were confirmed by PCR analyses of genomic DNAs. Because the target sequence was injected into fertilized eggs of BL6/SJL hybrid mice and there is a retinal degeneration allele Pde6brd1 harbored in SJL background mice, we also eliminated Pde6brd1 mutant mice after genotyping PCR analyses. Three transgenic mouse lines were established and analyzed (Fig. 2A, panel a). Real time PCR analyses of F1 genomic DNA showed that line SP1231 harbored the highest copy number of the transgene, followed by SP1135 and SP1128. The transgene numbers were 7.0 ± 1.0, 2.1 ± 0.4, and 1.3 ± 0.1 copies per haploid genome, respectively (calculated by real time PCR, Fig. 2A, panel b, and supplemental Fig. S6A).
FIGURE 2.
Creation of miR-183/96/182 cluster sponge transgenic mice. A, PCR genotyping of the miR-183/96/182 cluster sponge in transgenic mice. Panel a, three transgenic lines are shown with different levels of transgene detected by PCR. Rd_For/Pde_Rev and Pde_For/Pde_Rev primers were used to confirm the absence of the Pde6brd1 mutant allele. PCR of β-actin was used as a control for genomic DNA preparations. Panel b, shown are absolute transgene copy numbers in heterozygotes calculated by real time PCR in different mouse lines. These were 2.1 ± 0.4 in SP1135, 1.3 ± 0.1 in SP1128, and 7.0 ± 1.0 in SP1231, respectively. Error bars represent means ± S.E. B, validation of sponge element expression and function. Panel a, mRNA expression levels of sponge elements in retina were measured by semiquantitative RT-PCR. RT-minus (RT(−)) and Gapdh (Gapdh RT (+)) reactions were used as negative and positive controls. Panel b, splinted ligation shows that mature miR-183, miR-96, and miR-182 were only slightly down-regulated in all three transgenic mouse lines. Amplification cycle numbers for A, panel a, and B, panel a, are indicated on the right.
Although miR sponge elements are designed to disrupt miR activity by sequestering mature miRs, introduction of a miR sponge can alter miR levels in some cases (31). To determine whether sponge components were transcribed, and the endogenous miR-183/96/182 cluster levels changed, total RNAs from retinas of transgenic and WT mice were extracted and analyzed by semi-quantitative RT-PCR or splinted ligation. Semi-quantitative RT-PCR demonstrated that line SP1231 expressed the highest level of the sponge element in the retina, followed by lines SP1135 and SP1128 (Fig. 2B, panel a). The splinted ligation assay showed only a marginal decrease of miR-183, miR-96, and miR-182 in all three sponge transgenic mouse lines (Fig. 2B, panel b, and supplemental Fig. S6C). This result, consistent with that of a previous study (13), indicates that our sponge element disrupted the activity of the miRs primarily by sequestration rather than destabilization. To verify the spatial expression pattern of sponge elements, we measured sponge element expression levels in different tissues of sponge transgenic mice by RT-PCR. This revealed that the sponge element mRNA was detectable only in retina and not in brain, heart, liver, or intestine (supplemental Fig. S6, B and C). We also tested whether endogenous Opsin expression was affected by the presence of multiple Opsin promoter elements in these transgenic mice. Immunoblotting results showed that Opsin expression levels were similar in WT and all three transgenic mouse lines (supplemental Fig. S6D).
Bright Light Induces Severe Retinal Degeneration in miR-183/96/182 Cluster Sponge Transgenic Mice
Because levels of the miR-183/96/182 cluster decreased dramatically in mutant mouse models of retinal degeneration (Fig. 1) (12), we tested whether inhibition of the miR-183/96/182 cluster in sponge transgenic mice would result in retinal degeneration. To our surprise, no morphological or functional changes in retinas from either 2- or 6-month-old transgenic mice were identified by histological or ERG analysis (data not shown).
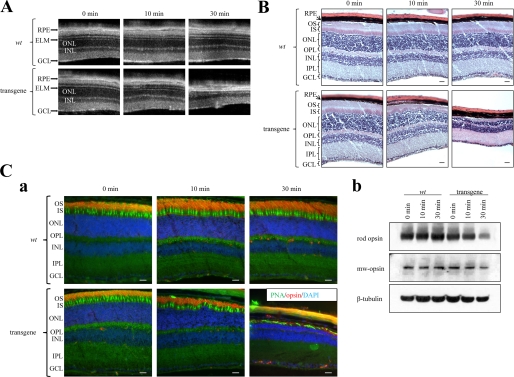
Considering that the miR-183/96/182 cluster was previously shown to be up-regulated in response to light (13), we hypothesized that these miRs might play a role in protecting the retina from acute light-induced retinopathy. Both WT and sponge transgenic mice were exposed to 10,000 lux white light for 10 or 30 min. Then OCT and H&E staining of retinas was performed 2 weeks later. In the superior retina, sponge transgenic mice exhibited a 30 ± 3% loss of photoreceptors after 10 min of light exposure and an 80 ± 5% loss after 30 min of exposure, whereas WT control mice exhibited no significant retinal degeneration after either exposure (Fig. 3, A and B). To distinguish retinal rods from cones, we performed immunocytochemistry with 1D4 antibody (rods) and PNA (cones). After the 10-min light exposure, rod cell death was noted, and both the outer segment and outer nuclear layer were thinner, but cone damage was less pronounced. After a 30-min exposure, rods had almost disappeared and few cones remained. With support from rods lacking, the remaining cones were highly disordered (Fig. 3C, panel a). This phenotype was confirmed by immunoblotting. In transgenic mouse retina, the Opsin level decreased by 30 and 60% after 10 and 30 min of light exposure. But the Mw-opsin level decreased only slightly after 30 min of light exposure. Both Opsin and Mw-opsin levels remained unchanged in wild type mouse retina (Fig. 3C, panel b, and supplemental Table S3). It should be noted that the EGFP expression did not cause retinal degeneration because EGFP expression in transgenic mice was not reliably detected by immunocytochemistry. Repetitive miR-183/96/182 complementary sequences in the 3′-UTR of EGFP likely blocked the EGFP expression.
FIGURE 3.
Bright light induces severe degeneration in the superior retina of 4-month-old miR-183/96/182 sponge cluster transgenic mice. A, B-scanned averaged SD-OCT images of superior mouse retina were taken at 0, 10, and 30 min after 10,000 Lux light exposure. A significant decrease in ONL thickness occurred in transgenic retinas after 10 and 30 min of light exposure. B, representative retinal histology after H&E staining is shown for both WT and transgenic mice after indicated periods of bright light exposure. Transgenic mice exhibited photoreceptor cell death after 10 and 30 min of light exposure, whereas WT control mice displayed no significant retinal degeneration. C, panel a, immunohistochemistry of retinas stained with PNA for cones and anti-Opsin (1D4) antibody for rods; DAPI was used as a counterstain to detect cell nuclei. In transgenic mice, rod cell death occurred after 10 min of light exposure as noted by thinner outer segment and outer nuclear layers. However, cone damage was less pronounced. After 30 min of light exposure, rods nearly disappeared and only a few cones remained. Rods and cones of WT mice were unaffected by the 30-min light exposure. Abbreviations used are as follows: GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; ELM, external limiting membrane; IS, inner segment; OS, outer segment; RPE, retinal pigmented epithelium. Scale bars, 10 μm. Panel b, immunoblotting studies show that the Opsin level decreased in transgenic mouse retina after 10 min of light exposure, and both Opsin and Mw-opsin levels decreased after 30 min light exposure. In contrast, Opsin and Mw-opsin levels in WT mouse retina remained unchanged after 10 or 30 min of light exposure.
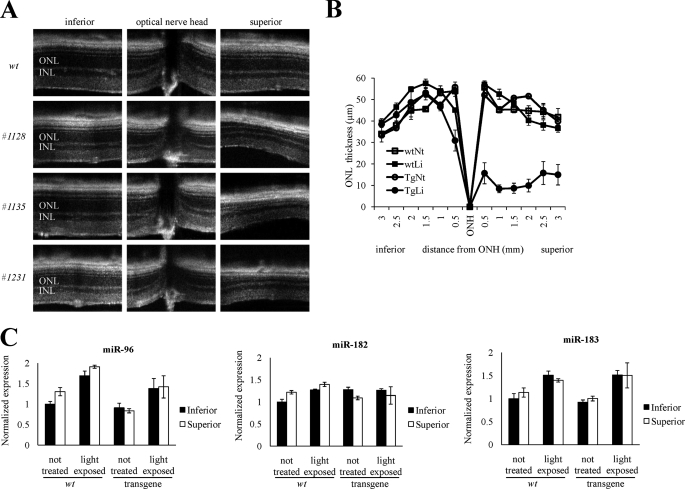
The vulnerability of transgenic mouse retina to intense light differed by region. Thus, ONL thickness in the inferior retina was nearly identical in unexposed and light-stressed mice, but in the superior retina it decreased from about 50 to 20 μm after light exposure in transgenic mice (Fig. 4, A and B). To check whether the regional selectivity of light-induced retinal degeneration was related to the spatial expression pattern of miR-183/96/182 cluster, we examined the expression level of miR-183, miR-96, and miR-182 in both inferior and superior retinas under different conditions by laser-capture microscopy and LNA-based real time PCR. LNA-based real time PCR indicated no difference between the superior and inferior retina in the expression of these three miRs, although miR-96 and miR-183 but not miR-182 increased after 30 min of light exposure in both WT and transgenic mice (Fig. 4C). The number of transgene copies did not affect this result as this phenotype was reproducible in all three transgenic mouse lines (Fig. 4A). Thus sponge transgenic mice displayed severe degeneration of the superior retina after 30 min of intense light exposure.
FIGURE 4.
Retinal damage is more severe in the superior retina than in other regions. A, B-scanned averaged SD-OCT images of retinas from three 4-month-old transgenic mice reveal that the superior area was more damaged than the optic disc and inferior area. B, thickness of the outer nuclear retinal layer measured 2 weeks after light exposure is shown. ONL thicknesses were almost identical in the inferior retina of untreated and light-exposed mice, but they were reduced from about 50 to 20 μm in the superior retina of transgenic mice after bright light exposure. Abbreviations used are as follows: INL, inner nuclear layer; ONL, outer nuclear layer; ONH, optic nerve head. Li, light; Nt, not exposed to light. Error bars represent then means ± S.D. (n = 4). C, expression levels of the miR-183/96/182 cluster were the same in mouse superior and inferior retina; locked nucleic acid-based real time PCR showed that miR-96 and miR-183 but not miR-182 increased after light exposure in both WT and transgenic mice. None of these three miRs were differentially expressed in the superior and inferior retina under these conditions. Error bars represent means ± S.E. (n = 3).
Identification and Validation of Potential Target Genes of miR-183/96/182 Cluster miRs
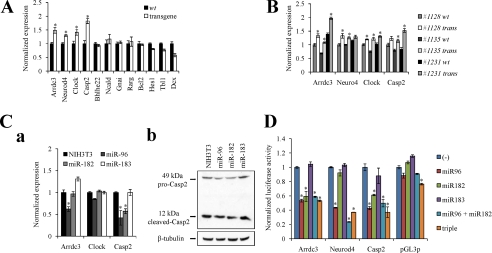
To better understand the biological roles of miR-183/96/182 cluster miRs in mouse retina, we compiled a list putative target genes of the miR-183/96/182 cluster by using three computational target prediction algorithms: PicTar, TargetScan 5.1, and MicroCosm. We also identified genes expressed in WT retina by massive parallel RNA sequencing (32). Of all putative targets, we focused only on those expressed in the retina with 3′-UTRs conserved in vertebrates. Several targets with functions involved in nervous system development, photo-transduction, visual cycle, circadian rhythm, or apoptosis are listed in Table 1. These targets were validated by real time PCR with Gapdh used as an internal control. Four genes out of 11 candidates, namely Arrdc3, Neurod4, Clock, and Casp2 (Fig. 5A), showed significantly increased expression in transgenic mice. Arrdc3 is an arrestin domain-containing protein, which can recruit the Nedd4 E3 ubiquitin ligase (33) important for nervous system development. Neurod4 is involved in the differentiation of horizontal, amacrine, and bipolar cells (34); repression of its expression in photoreceptors could adversely affect photoreceptor cell fate. Clock, the core component of the circadian clock, forms a heterodimer with Bmal1 and binds to circadian E-box promoter elements that enhance the transcription of Period 1 and 2 and cryptochrome 1 and 2 (35). Casp2 mediates apoptosis induced by many cellular stresses, including reactive oxygen species, endoplasmic reticulum stress, and DNA damage (24). We further confirmed these target genes by repeating the real time PCR experiment with all three transgenic mouse lines, using their littermates as controls. All four genes were up-regulated in these cell lines, although some increases were not statistically significant (Fig. 5B).
TABLE 1.
Predicted miR183 cluster downstream gene candidates
List of putative conserved miR-183/96/182 targets in retina. The 1st column represents the official symbol of the candidate target; the 2nd column lists the accession number; the 3rd column indicates which members of the miR-183/96/182 cluster are predicted to target each mRNA; the 4th column shows the mRNA level of the candidate target in retina as measured by deep sequencing; and the 5th column indicates the known function of the candidate target.
| Name | mRNA | miR | Expression level in retina | Function |
|---|---|---|---|---|
| Arrdc3 | NM_001042591 | miR-96, miR-182 | 12.8 | Nervous system development |
| Neurod4 | NM_007501 | miR-96 | 30.71 | |
| Hes1 | NM_008235 | miR-96, miR-182 | 11.3 | |
| Dcx | NM_001110222 | miR-96, miR-182, miR-183 | 0.35 | |
| Ncald | NM_001170866 | miR-96, miR-182 | 29.3 | Phototransduction and visual cycle |
| Rarg | NM_001042727 | miR-182 | 2.48 | |
| Gnai3 | NM_010306 | miR-96, miR-182 | 9 | |
| Clock | NM_007715 | miR-96, miR-182 | 15.62 | Circadian rhythm |
| Bhlhe22 | NM_021560 | miR-182 | 11.28 | |
| Casp2 | NM_007610 | miR-96, miR-182 | 7.17 | Apoptosis |
| Bcl2 | NM_009741 | miR-96, miR-182 | 3.41 |
FIGURE 5.
Identification of candidate targets of miR-183/96/182 cluster miRs. Predicted miR-183/96/182 targets are listed in Table 1 where their NCBI reference sequences, putative binding miRs, expression levels in retina, and known functions are also shown. A, comparison of candidate target mRNA levels between 4-month-old WT and transgenic mice by real time PCR. Four genes, namely Arrdc3, Neurod4, Clock, and Casp2, out of 11 candidates showed significant increases in transgenic mice. B, confirmation of these target genes by real time PCR. mRNA levels were compared between transgenic mice and their WT littermates. Arrdc3, Neurod4, Clock, and Casp2 mRNAs were all up-regulated in retinas from all three transgenic mouse lines. Real time PCR results were all normalized to Gapdh. Error bars represent means ± S.E. (n = 3). *, p < 0.01. C, target gene changes in stable cell lines. Panel a, mRNA levels of indicated genes in three stable cell lines were detected by real time PCR; Gapdh was used as an internal control. mRNA levels of Arrdc3 in miR-96 stable cells were 62% of those in NIH3T3 cells but were essentially unchanged in miR-182 stable cells and even increased 1.3-fold in miR-183 stable cells. The Clock mRNA levels remained the same in all cell lines. mRNA levels of Casp2 in miR-96 and miR-182 stable cells were 42 and 57% those in NIH3T3 cells but were unchanged in miR-183 stable cells. Error bars represent the means ± S.E. (n = 3), *, p < 0.01. Panel b, protein levels of both pro-Casp2 and cleaved Casp2 in three stable cell lines were documented by immunoblotting with an anti-Casp2 antibody. Both pro-Casp2 and cleaved Casp2 were decreased in miR-96 and miR-182 stable cells but not in miR-183 stable cells. β-Tubulin was used as the internal control. D, confirmation of miR-183/96/182 cluster targets by luciferase assay. miR-96 repressed luciferase activity by 47% in Arrdc3, 57% in Neurod4, and 57% in Casp2 compared with the parental pGL3P vector; miR-182 repressed luciferase activity by 41% in Arrdc3 and 39% in Casp2 compared with the empty vector; and miR-183 made no difference in all target genes. As a negative control, luciferase activity of the empty pGL3P vector was not affected by any miR-183/96/182 cluster miRs. All data were normalized to those obtained with the pGL3P vector alone. Error bars represent means ± S.E. (n = 3), *, p < 0.01.
Having demonstrated that disruption of the miR183/96/182 cluster leads to up-regulation of these candidate target genes, we next wanted to confirm that these genes can be directly targeted by these miRs. Thus, we established stable cell lines overexpressing miR-183, miR-96, and miR-182, respectively. Partial pri-miR sequences packed in pMXs-IRES-EGFP retrovirus (supplemental Fig. S4A) were used to infect NIH3T3 cells, and the top 5% of cells exhibiting the greatest EGFP fluorescence intensity were sorted and saved as overexpressing stable cell lines. First, we confirmed miR expression levels in these three cell lines for both pri-miRs (supplemental Fig. S4B, panel a) and mature miRs (supplemental Fig. S4B, panel b) by RT-PCR and splinted ligation. Next, we measured the effect of miR overexpression on the expression of putative miR-183/96/182 target genes. mRNA levels of Arrdc3 were 38% lower in miR-96 stable cells than NIH3T3 cells, but they showed little change in miR-182 stable cells and increased 1.3-fold in miR-183 stable cells. The mRNA level of Clock remained the same in all cell lines. mRNA levels of Casp2 dropped to 42 and 57% in miR-96 and miR-182 stable cells but exhibited no change in miR-183 stable cells (Fig. 5C, panel a). Neurod4 was barely detectable in NIH3T3 cells, so we could not evaluate its reaction to the miR-183/96/182 cluster in overexpressing cell lines. Immunoblotting analyses were also performed with extracts of all cell lines with β-tubulin used as an internal control. Casp2 antibody, which recognizes both the 49-kDa pro-Casp2 and 12-kDa cleaved Casp2, revealed that both pro-Casp2 and cleaved Casp2 were decreased in miR-96 and miR-182 stable cells but not in miR-183 stable cells (Fig. 5C, panel b, and supplemental Table S4).
To further evaluate the role of the miR-183/96/182 cluster in regulating its target genes, we generated luciferase reporters with four copies of the predicted target regions from the 3′-UTR of each target gene and co-transfected the reporters with miR-183, miR-96, and miR-182 overexpression plasmids. These luciferase assay results showed that miR-96 repressed luciferase activity by 47% in the reporter derived from the Arrdc3-targeted region, 57% for Neurod4, and 57% for Casp2 as compared with the empty vector. miR-182 repressed luciferase activity by 41% for Arrdc3 and 39% for Casp2 compared with the empty vector; and miR-183 had no effect on any target genes. As a negative control, luciferase activity of cells containing the empty pGL3P vector was not affected by any miR-183/96/182 cluster miR. No obvious additive effects were detected when miR-96 and miR-182 or all three miRs were transfected together (Fig. 5D). These reporter assays provide clear evidence that miR-96 specifically targets the 3′-UTR of Arrdc3, Neurod4, and Casp2; miR-182 specifically targets the 3′-UTR of Arrdc3 and Casp2; and miR183 does not affect the 3′-UTR of any of the tested target genes. Taken together, the above data indicate that Arrdc3, Neurod4, and Casp2 are targets of miR-183/96/182 cluster miRs.
Repression of Casp2 Expression Is One Reason That miR-183/96/182 Cluster miRs Protect the Retina from Light-induced Degeneration
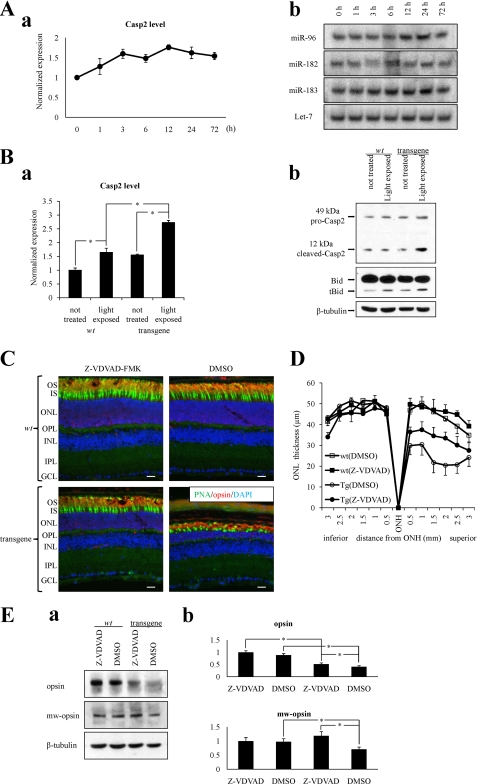
Bright light-induced retinal degeneration is thought to be Casp-dependent (36, 37). Because miR-183/96/182 sponge transgenic mice showed severe acute light-induced retinal degeneration and Casp2 was one of the targets of miR-183/96/182, we hypothesized that repression of Casp2 expression was one reason why the miR-183/96/182 cluster protected against light-induced retinal degeneration. To test this hypothesis, we first measured Casp2 mRNA levels as well as cluster miR levels in WT mice after a 30-min 10,000 lux white light exposure. Real time PCR results revealed an increase of Casp2 after light exposure (Fig. 6A, panel a) similar to that reported in Sprague-Dawley rats (36). Casp2 mRNA peaked at 12 h after light induction, and the fold change was about 1.8 ± 0.1. Splinted ligation results showed that miR-183 and miR-96 but not miR-182 increased significantly 12–24 h after light exposure (Fig. 6A, panel b, and supplemental Table S5). These experiments were repeated twice, and densitometric analysis indicated that miR-96 and miR-183 expression in animals 24 h after light exposure were 2.5 and 2.1-times higher than in dark-adapted mice, whereas miR-182 expression remained unchanged.
FIGURE 6.
miR-183/96/182 cluster miRs repress Casp2 during light exposure. A, both miR-183/96/182 cluster miRs and Casp2 expression levels increased in 4-month-old wild type mice after 10,000 Lux white bright light exposure. Panel a, real time PCR shows Casp2 was modestly up-regulated 12–24 h after light exposure. Panel b, splinted ligation data show that miR-96 and miR-183 but not miR-182 increased 12–24 h after light exposure. B, repression of Casp2 is not observed in sponge transgenic mice. Panel a, Casp2 mRNA levels increased more after light exposure in transgenic mice than in WT mice as demonstrated by real time PCR. Panel b, immunoblotting analysis confirms the greater increase of Casp2 protein levels and the enhanced cleavage of Bid protein in light-exposed transgenic mice. C, immunohistochemistry of retinas stained with PNA for cones and anti-Opsin (1D4) antibody for rods; DAPI was used as a counterstain to detect cell nuclei. DMSO-treated 4-month-old sponge transgenic mice showed severe retinal degeneration after light exposure, whereas Z-VDVAD-FMK-treated 4-month-old sponge transgenic mice showed only moderate retinal damage. WT mouse retinas appeared normal in both conditions. Scale bars: 10 μm. D, thicknesses of the outer nuclear retinal layer measured 2 weeks after light exposure are shown. Injection of Z-VDVAD-FMK had a partial protective effect against light-induced structural damage to the retina of transgenic mice. As in transgenic mice, ONL thicknesses in retinas of Z-VDVAD-FMK-treated transgenic mice were about 30 μm in the superior area, whereas in the same area, ONL thicknesses of DMSO-treated transgenic mice were only about 20 μm. Retinas from WT mice were normal, with ONL thicknesses of about 50 μm. Error bars represent means ± S.E. (n = 4). Abbreviations used are as follows: GCL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer; ELM, external limiting membrane; IS, inner segment; OS, outer segment; RPE, retinal pigmented epithelium; ONH, optic nerve head. E, panel a, immunoblotting data show that without Z-VDVAD-FMK treatment, the Opsin level in 4-month-old sponge transgenic mouse retina was only 40% that in WT littermate retina after light exposure, and the Mw-opsin level in the transgenic mouse retina was only 70% that in WT retina. But with Z-VDVAD-FMK treatment, the Opsin level in 4-month-old sponge transgenic mouse retina was 50% that in the retina of WT littermates after light exposure, and the Mw-opsin level in transgenic retina was even slightly increased. Statistical analyses are shown in panel b. Error bars represent means ± S.E. (n = 3), *, p < 0.05.
Data in WT mice suggested that miR-183/96/182 cluster miRs and Casp2 are both regulated similarly after bright light exposure. So we measured Casp2 expression in sponge transgenic mice after light exposure. Both WT and transgenic mice were exposed or unexposed to light. Then retinas were collected, and total RNA and protein were isolated to determine Casp2 levels. Both real time PCR and immunoblotting results showed that without protection against degeneration by miR-183/96/182 cluster miRs, Casp2 levels increased much more in response to bright light in transgenic mice than in WT mice (Fig. 6B, panels a and b, and supplemental Table S6). As predicted, cleavage of Bid, a substrate of Casp2 (24), also was enhanced in transgenic mice after bright light exposure (Fig. 6B, panel b, and supplemental Table S6).
To check whether inhibition of Casp2 could protect the retina against light-induced damage in transgenic mice, the Casp2 inhibitor Z-VDVAD-FMK was injected intravitreally into the right eyes of both WT and transgenic mice before light exposure; 2% DMSO in PBS was injected into the left eyes of the same mouse as a control. Immunocytochemistry results showed that, compared with the pseudo treatment in left eyes, Z-VDVAD-FMK partially relieved light-induced retinopathy in the right eyes of transgenic mice (Fig. 6C). In transgenic mice, the ONL thickness of Z-VDVAD-FMK-treated retina was about 30 μm in the superior area but only about 20 μm in the comparable area of DMSO-treated retina. In WT mouse retina unaffected by either the light exposure or the treatment, the ONL thickness was about 50 μm (Fig. 6D). Immunoblots also demonstrated that Z-VDVAD-FMK treatment can slightly attenuate the decrease of both rod Opsin and Mw-opsin after light exposure in transgenic mice. Statistical analyses showed that without Z-VDVAD-FMK treatment, the Opsin level in transgenic mouse retina was only 40% that of WT retina after light exposure, whereas the Mw-opsin level was 70% that of WT retina. With Z-VDVAD-FMK treatment, however, the Opsin level in transgenic mouse retina was 50% that of WT retina and the Mw-opsin level even marginally increased relative to that in WT retina after light exposure (Fig. 6E).
DISCUSSION
This research reveals an important role for the miR-183/96/182 cluster in protecting against bright light-induced retinal degeneration in vivo and identifies a connection between this cluster and an apoptotic pathway. The findings highlight yet another protective mechanism against light-induced photoreceptor degeneration and also demonstrate a clear role of miRs in a postmitotic tissue in vivo. The observed phenotype is unambiguous and manifests when miR-183/96/182 cluster miRs are simply sequestered rather than destroyed.
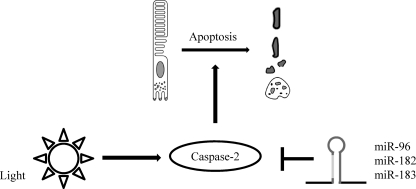
Exposure to excess light induces photoreceptor apoptosis, which has been used as a model for previous studies of retinal degeneration. However, the underlying pathogenic mechanisms remain obscure. Here, we provide evidence for involvement of the miR-183/96/182 cluster in this process. Repression of Casp2 expression appears to be one mechanism by which this cluster protects mice against retinal degeneration after bright light exposure (Fig. 7).
FIGURE 7.
Model of miR-183/96/182 cluster-mediated regulation of Casp2 protein and function in retinal degeneration. miR-183/96/182 cluster miRs repress Casp2 expression after bright light exposure, thereby protecting photoreceptors from apoptosis.
Involvement of caspases in retinal light-induced damage is still under active investigation. Chahory et al. (38) did not observe Casp3 activation in the retina of Fisher rats after bright light exposure, and a similar result was obtained in BALB/c mice by Donovan et al. (39). But other studies reported activation of both Casp3 and Casp2 after light exposure in Sprague-Dawley rats (36) and implicated caspase proteases in retinal degeneration by showing that inhibition of Casp1 or Casp3 reduced photoreceptor cell death (37). Differences in light intensity and duration may explain these different findings. Previous work also demonstrated two distinct pathways involved in light-induced retinal degeneration (40). Bright light-induced apoptosis does not depend on transducin and is accompanied by induction of Ap-1, whereas low light-induced apoptosis is transducin-dependent. Studies by the Chahory and Donovan groups (38, 39) that did not observe Casp3 activation involved a relatively low dose of light (2 days of exposure to 1,000 lux white light or 2 h of exposure to 5,000 lux white light, respectively). But those who observed Casp3 activation applied a higher intensity and duration of light exposure (12–24 h exposure to 2,700 lux white light by the Tomita group (36); 24 h of exposure to 3,400 lux white light by the Perche group (37)). We also tested Casp2 expression under higher light intensity exposure. Immunoblotting data demonstrated that Casp2 was only up-regulated with 5,000 and 10,000 lux light intensity exposure (supplemental Fig. S7 and supplemental Table S7). So we used a 30-min exposure to 10,000 lux white light, a high dose that probably accounts for the observed caspase activation.
Although high level expression of the miR-183/96/182 cluster in retina was identified several years ago, the in vivo function and downstream targets of these miRs remain obscure. Adcy6 was confirmed to be one target of the miR-183/96/182 cluster by Xu et al. (6) who suggested that this cluster may be involved in circadian rhythm. SLC1A1, a voltage-dependent glutamate transporter, was validated as another target by Krol et al. (13), who suggested this cluster functions in glutamate scavenging. Our research now indicates an important role of the miR-183/96/182 cluster in preventing bright light-induced retinal degeneration in vivo and defines Casp2 as a new target that mediates this effect. In fact, phylogenic studies of the 3′-UTR of Casp2 in different mammals defined highly conserved seed regions for miR-183/96/182 (supplemental Fig. S1B). A previous in vivo study of miR-182 knock-out mice showed no phenotypic changes, perhaps because of the functional redundancy of the cluster miR components. Indeed, we found that miR-96 and miR-183 but not miR-182 were up-regulated after bright light exposure suggesting that miR-96 and miR-183 are the cluster components primarily involved in retinal protection. Additional phylogenetic support for this interpretation is that miR-96 and miR-183 but not miR-182 are present in amphioxus (41). miR-183, -96, and -182 are suggested to be transcribed in the same pri-miR, and all of them were up-regulated in light-adapted retinas (13). But their different expression levels and responses after bright light exposure suggest that their expression, subsequent processing, and/or stability are regulated in different ways.
Another intriguing finding of our study was the regional selectivity of light-induced retinal degeneration. The superior retina was far more damaged than the inferior retina. However, we did not detect an expression difference of the miR-183/96/182 cluster between superior and inferior retina in any conditions. The same regional difference of vulnerability to intense light was noted previously. In genetically altered mice with defects in the visual cycle, such as Abca4−/−Rdh8−/− and Rdh8−/−Rdh12−/− double knockouts (42, 43) and an arrestin single knock-out (44), retinal degeneration was localized to the inferior region. But in BALB/cJ mice (45) and albino rats (46), retinal damage was more severe in the superior retina than the inferior retina. Differential gene expression in mouse superior and inferior retina has previously been examined by microarray analysis (47), with 27 genes showing an expression difference of >1.5-fold. Among those genes, miR-183/96/182 cluster target sites were predicted in just two, the Efnb2 and Gprc5b, 3′-UTRs. Efnb2 is a member of the ephrin family of proteins implicated in development, especially in the nervous system and in erythropoiesis. Gprc5b is a G-protein-coupled receptor with an unknown specific function (48). However, the latter protein was suggested to mediate the cellular effects of retinoic acid on the G-protein signal transduction cascade. How these genes are regulated by the miR-183/96/182 cluster and what their roles are in retinal degeneration require future study.
Supplementary Material
Acknowledgments
We thank Dr. Leslie T. Webster, Jr., Dr. Thomas Sundermeier, Dr. Michael E. Maguire, and members of the Palczewski laboratory for critical comments about the manuscript. We also thank Dr. Yang Yu, Patricia Maroney, and Timothy Nilsen from Center for RNA Molecular Biology, Case Western Reserve University (Cleveland, OH) for their guidance during initiation of this study.
This work was supported, in whole or in part, by National Institutes of Health Grants EY008061 and K08EY019880.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Tables S1–S7.
- miR
- microRNA
- Casp
- caspase
- EGFP
- enhanced green fluorescent protein
- ERG
- electroretinography
- OCT
- optical coherence tomography
- ONL
- outer nuclear layer
- PNA
- peanut agglutinin lectin
- Z
- benzyloxycarbonyl
- FMK
- fluoromethyl ketone.
REFERENCES
- 1. Bartel D. P. (2009) Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bartel D. P. (2004) Cell 116, 281–297 [DOI] [PubMed] [Google Scholar]
- 3. Tanzer A., Stadler P. F. (2004) J. Mol. Biol. 339, 327–335 [DOI] [PubMed] [Google Scholar]
- 4. Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. (2009) Genome Res. 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. (2002) Curr. Biol. 12, 735–739 [DOI] [PubMed] [Google Scholar]
- 6. Xu S., Witmer P. D., Lumayag S., Kovacs B., Valle D. (2007) J. Biol. Chem. 282, 25053–25066 [DOI] [PubMed] [Google Scholar]
- 7. Rodieck R. W. (1998) The First Steps in Seeing, pp. 38–55, Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 8. Arora A., McKay G. J., Simpson D. A. (2007) Invest. Ophthalmol. Vis. Sci. 48, 3962–3967 [DOI] [PubMed] [Google Scholar]
- 9. Karali M., Peluso I., Marigo V., Banfi S. (2007) Invest. Ophthalmol. Vis. Sci. 48, 509–515 [DOI] [PubMed] [Google Scholar]
- 10. Damiani D., Alexander J. J., O'Rourke J. R., McManus M., Jadhav A. P., Cepko C. L., Hauswirth W. W., Harfe B. D., Strettoi E. (2008) J. Neurosci. 28, 4878–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Georgi S. A., Reh T. A. (2010) J. Neurosci. 30, 4048–4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Loscher C. J., Hokamp K., Kenna P. F., Ivens A. C., Humphries P., Palfi A., Farrar G. J. (2007) Genome Biol. 8, R248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krol J., Busskamp V., Markiewicz I., Stadler M. B., Ribi S., Richter J., Duebel J., Bicker S., Fehling H. J., Schübeler D., Oertner T. G., Schratt G., Bibel M., Roska B., Filipowicz W. (2010) Cell 141, 618–631 [DOI] [PubMed] [Google Scholar]
- 14. Jin Z. B., Hirokawa G., Gui L., Takahashi R., Osakada F., Hiura Y., Takahashi M., Yasuhara O., Iwai N. (2009) Mol. Vis. 15, 523–533 [PMC free article] [PubMed] [Google Scholar]
- 15. Mencía A., Modamio-Høybjør S., Redshaw N., Morín M., Mayo-Merino F., Olavarrieta L., Aguirre L. A., del Castillo I., Steel K. P., Dalmay T., Moreno F., Moreno-Pelayo M. A. (2009) Nat. Genet. 41, 609–613 [DOI] [PubMed] [Google Scholar]
- 16. Jomary C., Cullen J., Jones S. E. (2006) Invest. Ophthalmol. Vis. Sci. 47, 1620–1629 [DOI] [PubMed] [Google Scholar]
- 17. Katai N., Kikuchi T., Shibuki H., Kuroiwa S., Arai J., Kurokawa T., Yoshimura N. (1999) Invest. Ophthalmol. Vis. Sci. 40, 1802–1807 [PubMed] [Google Scholar]
- 18. Jomary C., Neal M. J., Jones S. E. (2001) Mol. Cell. Neurosci. 18, 335–346 [DOI] [PubMed] [Google Scholar]
- 19. Deleted in proof.
- 20. Hahn P., Lindsten T., Lyubarsky A., Ying G. S., Pugh E. N., Jr., Thompson C. B., Dunaief J. L. (2004) Cell Death Differ. 11, 1192–1197 [DOI] [PubMed] [Google Scholar]
- 21. Yoshizawa K., Kiuchi K., Nambu H., Yang J., Senzaki H., Kiyozuka Y., Shikata N., Tsubura A. (2002) Graefes Arch. Clin. Exp. Ophthalmol. 240, 214–219 [DOI] [PubMed] [Google Scholar]
- 22. Bode C., Wolfrum U. (2003) Mol. Vis. 9, 144–150 [PubMed] [Google Scholar]
- 23. Liu C., Li Y., Peng M., Laties A. M., Wen R. (1999) J. Neurosci. 19, 4778–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kumar S. (2009) Nat. Rev. Cancer 9, 897–903 [DOI] [PubMed] [Google Scholar]
- 25. Ebert M. S., Neilson J. R., Sharp P. A. (2007) Nat. Methods 4, 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maroney P. A., Chamnongpol S., Souret F., Nilsen T. W. (2008) Nat. Protoc. 3, 279–287 [DOI] [PubMed] [Google Scholar]
- 27. Soucy E., Wang Y., Nirenberg S., Nathans J., Meister M. (1998) Neuron 21, 481–493 [DOI] [PubMed] [Google Scholar]
- 28. Jaissle G. B., May C. A., Reinhard J., Kohler K., Fauser S., Lütjen-Drecoll E., Zrenner E., Seeliger M. W. (2001) Invest. Ophthalmol. Vis. Sci. 42, 506–513 [PubMed] [Google Scholar]
- 29. Pittler S. J., Baehr W. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 8322–8326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sakami S., Maeda T., Bereta G., Okano K., Golczak M., Sumaroka A., Roman A. J., Cideciyan A. V., Jacobson S. G., Palczewski K. (2011) J. Biol. Chem. 286, 10551–10567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ebert M. S., Sharp P. A. (2010) RNA 16, 2043–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mustafi D., Kevany B. M., Genoud C., Okano K., Cideciyan A. V., Sumaroka A., Roman A. J., Jacobson S. G., Engel A., Adams M. D., Palczewski K. (2011) FASEB J., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nabhan J. F., Pan H., Lu Q. (2010) EMBO Rep. 11, 605–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ohsawa R., Kageyama R. (2008) Brain Res. 1192, 90–98 [DOI] [PubMed] [Google Scholar]
- 35. Tosini G., Pozdeyev N., Sakamoto K., Iuvone P. M. (2008) BioEssays 30, 624–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tomita H., Kotake Y., Anderson R. E. (2005) Invest. Ophthalmol. Vis. Sci. 46, 427–434 [DOI] [PubMed] [Google Scholar]
- 37. Perche O., Doly M., Ranchon-Cole I. (2007) Invest. Ophthalmol. Vis. Sci. 48, 2753–2759 [DOI] [PubMed] [Google Scholar]
- 38. Chahory S., Keller N., Martin E., Omri B., Crisanti P., Torriglia A. (2010) Neurochem. Int. 57, 278–287 [DOI] [PubMed] [Google Scholar]
- 39. Donovan M., Carmody R. J., Cotter T. G. (2001) J. Biol. Chem. 276, 23000–23008 [DOI] [PubMed] [Google Scholar]
- 40. Hao W., Wenzel A., Obin M. S., Chen C. K., Brill E., Krasnoperova N. V., Eversole-Cire P., Kleyner Y., Taylor A., Simon M. I., Grimm C., Remé C. E., Lem J. (2002) Nat. Genet. 32, 254–260 [DOI] [PubMed] [Google Scholar]
- 41. Luo Y., Zhang S. (2009) Gene 428, 41–46 [DOI] [PubMed] [Google Scholar]
- 42. Maeda A., Maeda T., Golczak M., Palczewski K. (2008) J. Biol. Chem. 283, 26684–26693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maeda A., Maeda T., Sun W., Zhang H., Baehr W., Palczewski K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19565–19570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen J., Simon M. I., Matthes M. T., Yasumura D., LaVail M. M. (1999) Invest. Ophthalmol. Vis. Sci. 40, 2978–2982 [PubMed] [Google Scholar]
- 45. Zhang C., Lei B., Lam T. T., Yang F., Sinha D., Tso M. O. (2004) Invest. Ophthalmol. Vis. Sci. 45, 2753–2759 [DOI] [PubMed] [Google Scholar]
- 46. Tanito M., Kaidzu S., Ohira A., Anderson R. E. (2008) Exp. Eye Res. 87, 292–295 [DOI] [PubMed] [Google Scholar]
- 47. Zhu Y., Natoli R., Valter K., Stone J. (2010) Mol. Vis. 16, 740–755 [PMC free article] [PubMed] [Google Scholar]
- 48. Robbins M. J., Michalovich D., Hill J., Calver A. R., Medhurst A. D., Gloger I., Sims M., Middlemiss D. N., Pangalos M. N. (2000) Genomics 67, 8–18 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.