Abstract
It is well known that biosynthesis of glycans takes place in organ- and tissue-specific manners and glycan expression is controlled by various factors including glycosyltransferases. The expression mechanism of glycosyltransferases, however, is poorly understood. Here we investigated the expression mechanism of a brain-specific glycosyltransferase, GnT-IX (N-acetylglucosaminyltransferase IX, also designated as GnT-Vb), which synthesizes branched O-mannose glycan. Using an epigenetic approach, we revealed that the genomic region around the transcriptional start site of the GnT-IX gene was highly associated with active chromatin histone marks in a neural cell-specific manner, indicating that brain-specific GnT-IX expression is under control of an epigenetic “histone code.” By EMSA and ChIP analyses we identified two regulatory proteins, NeuroD1 and CTCF that bind to and activate the GnT-IX promoter. We also revealed that GnT-IX expression was suppressed in CTCF- and NeuroD1-depleted cells, indicating that a NeuroD1- and CTCF-dependent epigenetic mechanism governs brain-specific GnT-IX expression. Several other neural glycosyltransferase genes are also found to be regulated by epigenetic histone modifications. This is the first report demonstrating a molecular mechanism at the chromatin level underlying tissue-specific glycan expression.
Keywords: Gene Regulation, Glycoconjugate, Glycoprotein, Glycoprotein Biosynthesis, Glycosylation, Glycosyltransferase, N-acetylglucosaminyltransferase
Introduction
Glycosylation is a major post-translational modification reaction of proteins and plays various roles in mammals during early development, neural plasticity, inflammation, and disease progression (1, 2). Glycan is synthesized in a stepwise manner by ER- or Golgi-resident glycosyltransferases (3), and organ- and tissue-specific glycan biosynthesis is achieved by glycosyltransferases, which are also themselves expressed in a tissue-specific manner. For example, polysialic acid and human natural killer-1 (HNK-1) epitope are synthesized in the nervous system and involved in synaptic plasticity (4, 5), and specific N-glycan branching on pancreatic beta-cells is required for insulin secretion (6). During the last few decades, most glycosyltransferase genes have been cloned (7), and some of their promoters have been characterized by conventional reporter assay methods. These studies revealed that most glycosyltransferase genes have TATA-less, CpG-associated promoters; however, very limited studies have been reported on the underlying molecular mechanism by which glycans are expressed (8). Moreover, although epigenetic regulation mechanisms, such as DNA methylation, histone modification, chromatin remodeling, and non-coding RNA, are fundamental for controlling gene expression (9), there have been few studies addressing the epigenetic mechanisms of tissue-specific glycosyltransferase expression.
Previously, we cloned a brain-specific glycosyltransferase, GnT-IX (MGAT5B), as a homolog of GnT-V (10). GnT-V, which is a well-known β1,6-branching enzyme toward the α1–6 mannose arm of N-glycan, is ubiquitously expressed and is involved in growth factor receptor signaling (11). GnT-V is up-regulated by the oncogenic transcription factor, ETS1, and is associated with cancer progression (12, 13). In contrast, GnT-IX exhibits β1,6-branching activity toward O-mannose type glycan and is exclusively expressed in the brain (14). O-Mannosyl glycan is a rare modification in mammals, and this glycan attached to α-dystroglycan has a pivotal role for skeletal muscle integrity through binding with extracellular proteins such as laminin and agrin (15), while branched type O-mannosyl glycan was suggested to be involved in neural cell adhesion and migration through β-catenin signaling (16). Furthermore, overexpression of GnT-IX in PC12 cells enhanced nerve growth factor (NGF)-mediated neurite outgrowth probably by modulating integrin-extracellular matrix interaction (17). These observations indicate an intrinsic and specific role of GnT-IX in the nervous system.
In this study, we show that neural GnT-IX expression is under the control of neural cell-specific histone modifications (18). The histone code hypothesis predicts that the post-translational modifications of histones, alone or in combination, function to direct specific and distinct DNA-templated programs and that the post-translationally modified histones serve as extremely selective binding platforms for specific regulatory proteins that drive distinct nuclear processes (19). Meanwhile, we identified two nuclear proteins, CTCF and NeuroD1, which bound to and activated the GnT-IX promoter in neural cells. Moreover, we demonstrated that expression of other neural glycosyltransferases is likely to be regulated by a similar epigenetic mechanism.
EXPERIMENTAL PROCEDURES
Materials
The following antibodies were used in this study: CTCF (ab70303), H3K27me3 (ab6002), N-Myc (ab16898), actin (Sigma, AC-40), H3K9ac (07-352), and H3K4me3 (07-473) and H3K9me2 (07-521) from Millipore, NeuroD1 (Cell Signaling Technology, D35G2), ZEB1 (Novus Biologicals, NBP1–05987). Trichostatin A (TSA)2 was prepared as described previously (20). 5-Aza-2′-deoxycytidine (5-Aza) was from Sigma. Human fetal brain cDNA was from Clontech.
Cloning and Plasmid Construction
Genomic DNA from C57BL/6 mouse tail biopsies was extracted and purified according to a standard protocol using phenol/chroloform extraction after proteinase K digestion. For P1 and P2 constructs, DNA fragments were amplified using genomic DNA as a template and then ligated into pCR4Blunt-TOPO (Invitrogen). Using these plasmids as a template, serial deletion constructs were generated. Resulting DNA fragments, which had been amplified and digested with restriction enzymes were ligated into promoter-less, enhancer-less pGV-B2 (Toyo Ink). Point mutations were generated by QuickChange II XL Site-directed Mutagenesis Kit (Stratagene) using pGV/−313 + 606 as a template. All the following cDNAs were subcloned into pcDNA6/myc-His A (Invitrogen). Mouse ZEB1 cDNA (purchased from Openbiosystems) was PCR amplified and then subcloned using EcoRI and EcoRV. Human N-Myc cDNA was amplified by PCR from fetal brain cDNA and ligated into EcoRI-EcoRV sites of the vector. Human CTCF cDNA (cloned from MCF7 cells cDNA), mouse NeuroD1, NeuroD2, Math2, Neurogenin1, Olig1 and CaMKIIα cDNAs (cloned from 10-week-old mouse brain cDNA) were ligated into pCR4Blunt-TOPO (Invitrogen), then digested and ligated using EcoRI. The NeuroD1 point mutation (S336A) was generated using QuickChange II XL Site-directed Mutagenesis kit. Primers are listed in supplemental Fig. S8.
Luciferase Assay
Cells, plated on 6-cm culture dishes, were transfected with 1 μg of each pGV construct and 0.1 μg of pRL-TK (Toyo Ink) using FUGENE6 transfection reagent (Roche). 24 h after transfection, the cells were washed twice with PBS and then lysed with 500 μl of lysis buffer (PicaGene Dual SeaPansy Luminescence Kit, Toyo Ink). Luciferase assays were performed according to the manufacturer's instructions with a luminometer, TR717 (Applied Biosystems). The luminescence value of firefly luciferase was divided by that of Renilla luciferase (pRL-TK), and the resultant value is shown in figures as “luciferase activity.” As a negative control, cells were transfected with pGV-B2 empty vector.
For CTCF or NeuroD1 overexpression, 1 μg of expression plasmid was co-transfected with the luciferase vectors. For CTCF knockdown, cells at 30% confluency were transfected with 80 pmol of siRNA using 8 μl of Lipofectamine 2000 (Invitrogen). After 4 h, the medium was replaced with fresh medium. 24 h post-transfection, the luciferase plasmids were transfected using FUGENE6 as described above.
Nuclear Protein Extraction
Cells (semi-confluent on 10-cm dishes) were washed twice with PBS and collected. The cell pellet was then resuspended in 300 μl of hypotonic buffer A (10 mm HEPES pH 7.9, 10 mm KCl, 0.1 mm EGTA, 0.1 mm EDTA, 1 mm DTT, and protease inhibitors) and incubated on ice for 15 min. After adding 18.75 μl of 10% Nonidet P-40, the sample was vortexed for 10 s and then centrifuged at 500 × g for 3 min. The pellet was then washed with 300 μl of buffer A and centrifuged. After repeating the wash and centrifugation, the resultant pellet was resuspended in 200 μl of high salt buffer B (20 mm HEPES pH 7.9, 400 mm NaCl, 1 mm EGTA, 1 mm EDTA, 1 mm DTT, and protease inhibitors) and then incubated on ice for 30 min. After centrifugation at 13,000 × g for 10 min, the supernatant was used as a nuclear extract. Protein concentration was measured using a BCA Protein Assay (Thermo Scientific).
EMSA
Oligonucleotides were biotinylated using a Biotin 3′-end DNA labeling kit (Pierce) according to the manufacturer's protocol. The oligonucleotides used were GCGAGACTGCGGCCACCAGATGGCGCTCAGGCGCCGAGG and its complementary sequence. The labeled complementary oligonucleotides were mixed in buffer containing 10 mm Tris-HCl, pH 8.0, 1 mm EDTA and 50 mm NaCl and incubated at 95 °C for 2 min. Then, the solution was gradually cooled to facilitate annealing. The same oligonucleotides, which were not labeled, were annealed in the same way and used as a competitor. EMSA was performed using a LightShift Chemiluminescent EMSA kit (Pierce) according to the provider's protocol with modifications. Binding mixture was incubated for 20 min at room temperature in 20 μl of buffer containing 1× binding buffer (supplied in the kit), 2 mm MgCl2, 0.1 mm ZnSO4, 7.5% glycerol, 10 ng/μl poly (dAdT) (Sigma), 1 μg of bovine serum albumin, 4.5 μg of nuclear extract and 20 fmol of DNA probe. When competitor DNA was used, 4 pmol (200 molar excess) of DNA was added to the mixture. For antibody addition, antibody was pre-incubated with the mixture without the probe for 30 min at room temperature. The mixture was electrophoresed in 4% polyacrylamide gels containing 0.5× TBE buffer, and then transferred to a positively charged nylon membrane (Roche) using a semi-dry transfer blotter. Staining and detection procedures were carried out according to the kit protocol.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP was performed using a SimpleChIP Enzymatic Chromatin IP kit (Cell Signaling Technology). Approximately, 2 × 107 cells were used as starting material. 1% of the chromatin was reserved as input sample before immunoprecipitation. For detection, real-time PCR was performed.
For tissue ChIP, 10-week-old C57BL/6 female mice were first deeply anesthetized and then sacrificed. Organs were collected and immediately minced on ice. Tissue pieces (200 mg) were incubated in 1% formaldehyde/PBS for 10 min, and then glycine was added (125 mm at final concentration) followed by incubation for 5 min. After washing twice with PBS, the tissues were homogenized in 4 ml of buffer A (supplied in the kit) with a Dounce homogenizer. Subsequent procedures were carried out as for cultured cells. All the animal experiments were approved by the Animal Experiment Committee of RIKEN.
Cell Culture
3T3-L1 and Neuro2A cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin.
Computational Search for Transcription Factor Consensus Sites
Putative binding proteins to a given DNA sequence were searched for using JASPAR CORE Vertebrata database and MatInspector.
Isolation of Total RNA, Reverse Transcription and RT-PCR
Total RNA from cultured cells was extracted using an RNeasy Mini kit (Qiagen). Total RNA from adult C57BL/6 mice was extracted using TRIzol (Invitrogen). 1 μg of total RNA was reverse transcribed using the SuperScript III First-Strand Synthesis System (Invitrogen) with random hexamers. Primers used for RT-PCR are listed in supplemental Fig. S8.
Western Blotting
Proteins were separated by 4–20% gradient SDS-PAGE with the buffer system of Laemmli and then transferred to PVDF or nitrocellulose membranes. After blocking with 5% nonfat dried milk in TBS containing 0.05% Tween 20, the membranes were incubated with primary antibodies, followed by HRP-conjugated secondary antibodies. Proteins were detected with SuperSignal West Dura Extended Duration Substrate (Thermo Scientific) using a Luminoimage Analyzer LAS-1000 (Fujifilm).
Overexpression and siRNA Transfection
For overexpression, cells, plated on 10-cm dishes at 80% confluency, were transfected with 4–8 μg of each expression vector using 10–20 μl of Lipofectamine 2000. After 4 h, the medium was replaced with fresh medium. 24 h post-transfection, the cells were used for other experiments.
For knockdown, cells at 30% confluency on 10-cm dishes were transfected with 200 pmol of control siRNA (AllStars negative control siRNA, Qiagen) or mouse CTCF siRNA (5′-GGUGCAAUUGAGAACAUUATT-3′ and 5′-UAAUGUUCUCAAUUGCACCTG-3′) or mouse NeuroD1 siRNA (5′-CGAAUUUCGUGUAGCUGUATT-3′ and 5′-UACAGCUACACGAAAUUCGTG-3′) using 20 μl of Lipofectamine 2000. After 4 h, the medium was replaced with fresh medium. 48 h post-transfection, cells were used for other experiments.
Quantitative Real-time PCR
For GnT-IX and GAPDH primers and probes, we used Assays-on-Demand gene expression products, and cDNAs were added to TaqMan Universal PCR Master Mix (Applied Biosystems). The probe for GnT-IX was labeled with FAM at its 5′-end and with quencher MGB at its 3′-end. The probe for GAPDH was labeled with VIC at its 5′-end and with quencher TAMRA at its 3′-end. The cDNAs were amplified by one cycle at 50 °C for 2 min, one cycle at 95 °C for 10 min, and 40 cycles at 95 °C for 15 s and 60 °C for 1 min in a total volume of 20 μl using an ABI PRISM 7900HT sequence detection system (Applied Biosystems). The levels of GnT-IX mRNA were measured in duplicate and normalized to the corresponding GAPDH levels. For 3T3-L1 cells, the level of GnT-IX mRNA was indicated as a percentage relative to the level in Neuro2A cells, because endogenous GnT-IX mRNA was below the detectable limit in non-treated 3T3-L1 cells.
ChIP products were quantified using the primers and probes in supplemental Fig. S8. Genomic DNA was amplified by one cycle at 50 °C for 2 min, one cycle at 95 °C for 10 min, and 40 cycles at 95 °C for 15 s and 62 °C for 1 min in a total volume of 20 μl. The amount of precipitated DNA was measured in duplicate and indicated as a relative percentage to the level of the input chromatin of each sample.
TSA and 5-Aza Treatment
These compounds were dissolved in DMSO. TSA and 5-Aza were added to the culture medium at final concentrations of 0.1 μm and 2.5 μm, respectively. In the control sample, the same amount of DMSO was added as vehicle. After 24 h, cells were used for subsequent experiments.
For CTCF knockdown experiments, cells at 30% confluency were transfected with CTCF siRNA as described above. 24 h post-transfection, TSA was added to the culture medium, and cells were incubated for an additional 24 h before further analysis.
Bisulfite Sequencing
Genomic DNA is extracted from mouse tissues using PureLink Genomic DNA Mini kit (Invitrogen). Unmethylated cytosines were converted to uracils using MethylCode Bisulfite Conversion kit (Invitrogen). The conversion reaction was carried out at 64 °C for 150 min using 100 ng of genomic DNA as a starting material. Then, a part of CpG island in GnT-IX promoter was amplified using MightyAmp DNA Polymerase (TAKARA) by one cycle at 98 °C for 2 min, and 42 cycles at 98 °C for 10 s, 50 °C for 20 s and 68 °C for 15 s, and then cloned and sequenced (8 clones for each tissue). Primers are ATGTTTTAGGAAGTTGTATGGTTTAG and CATTATTCCCTCTAAAAAAAC.
To validate efficiency of the CT conversion, we used methylated plasmid DNA (pGV/−313 + 606). The plasmid was methylated by GCGC-specific methyltransferase HhaI (New England Biolabs), and then purified by phenol/chloroform extraction.
RESULTS
Neural Cell-specific Chromatin Activation around the GnT-IX TSS Region
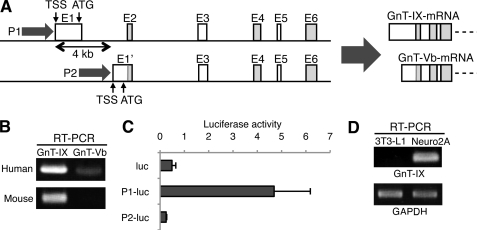
Although human GnT-IX gene (MGAT5B) generates two mRNA sequences (10, 21), which are generated by two alternative promoters (P1 for GnT-IX mRNA and P2 for GnT-Vb mRNA, Fig. 1A), our RT-PCR analysis demonstrated that P1-driven GnT-IX mRNA is the major form in normal brain (Fig. 1B). Consistently, P1 exhibited markedly higher promoter activity in neuroblastoma cells compared with P2 in luciferase assay (Fig. 1C). Based on these results, we focused on P1 and the GnT-IX mRNA isoform in this study. 5′-RACE revealed that GnT-IX mRNA are generated by several transcriptional start sites (TSS) (supplemental Fig. S1), and one of major TSSs are designated as +1. We detected endogenous GnT-IX mRNA in mouse neuroblastoma cells (Neuro2A), while 3T3-L1 cells showed no expression (Fig. 1D). Therefore, we used these cells to investigate how GnT-IX mRNA transcription is specifically regulated in the brain.
FIGURE 1.
Alternative promoters and a dominant mRNA isoform of the GnT-IX gene. A, schematic diagram and partial gene structure of the GnT-IX gene (MGAT5B) are shown (left). Two transcriptional start sites (TSS) are 4 kb apart. In human, two mRNA isoforms, GnT-IX and GnT-Vb, are driven by promoter 1 and 2 (P1 and P2), respectively (right). P1 has a CpG-island but no TATA box. B, RT-PCR was performed with specific primers for GnT-IX or GnT-Vb mRNA using reverse-transcribed total RNA of fetal human brain or 10-week-old C57BL/6 mouse brain. C, activities of mouse P1 and 2 in Neuro2A cells were measured by luciferase assay. One kb upstream from the initiation ATG (for P1) or from the start site of exon 2 (for P2) was used in this experiment. Control experiments were performed using a promoter-less, enhancer-less luciferase plasmid, pGV-B2 (indicated as luc). Relative luciferase activities against internal control Renilla luciferase are shown. Data are presented as the mean ± S.E. (n = 3). D, GnT-IX mRNA was detected by RT-PCR in Neuro2A cells but not in 3T3-L1 cells. GAPDH is shown as a reference gene.
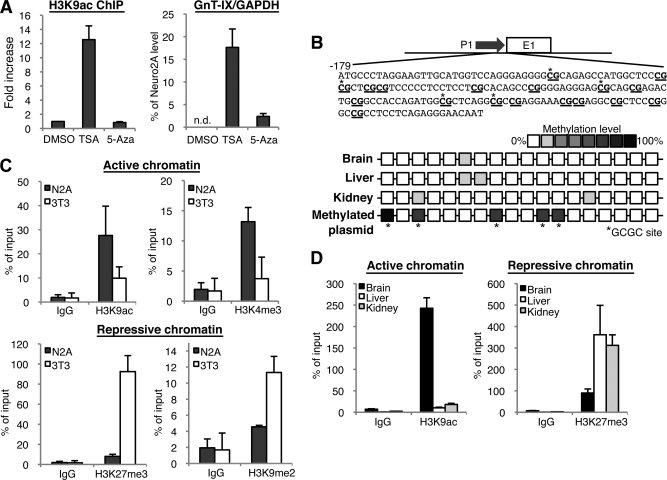
We first expected GnT-IX expression to be controlled by neural specific chromatin structure, because epigenetic events play key roles for specific gene expression. To see if drugs that modify chromatin would affect GnT-IX expression, we first treated GnT-IX-negative cells (3T3-L1) with a HDAC inhibitor, TSA (20), to forcibly remodel chromatin structure. We confirmed by ChIP analysis that TSA treatment caused a marked increase of acetylation on histone 3 lysine 9 (H3K9ac), which corresponds to a typical active chromatin mark, around the GnT-IX TSS (Fig. 2A, left). Concomitantly, TSA treatment strongly induced GnT-IX transcription in 3T3-L1 cells (Fig. 2A, right). We also treated the cells with a DNA methyltransferase inhibitor, 5-Aza (22), but 5-Aza treatment produced only slight GnT-IX induction. Moreover, bisulfite sequencing of genomic DNA from mouse tissues revealed that CpG sites in GnT-IX promoter are barely methylated in all tissues tested including brain (Fig. 2B). These results indicate that chromatin activation associated with histone modification but not with DNA hypo-methylation seems to be pivotal for GnT-IX transcription. From these results, we speculated that the histone tail modifications around the GnT-IX TSS region would be different between neural and non-neural cells. We, therefore, examined histone code-based chromatin activation state around the GnT-IX core promoter and the TSS region by ChIP analysis in Neuro2A and 3T3-L1 cells. In Neuro2A cells the GnT-IX TSS region was highly co-precipitated with antibodies against active chromatin marks, such as H3K9ac and trimethylated H3 lysine 4 (H3K4me3) (Fig. 2C, top), whereas in 3T3-L1 cells the same genomic region was highly associated with repressive chromatin marks, such as trimethylated H3 lysine 27 (H3K27me3) and dimethylated H3 lysine 9 (H3K9me2) (Fig. 2C, bottom). Moreover, by tissue-ChIP analysis, we found that the same genomic region was highly associated with an active chromatin mark in mouse brain and with a repressive chromatin mark in other tissues such as liver and kidney (Fig. 2D). These results strongly suggest that the chromatin of the GnT-IX core promoter and TSS region is open and in a highly active state, specifically in neural cells, which correlates well with the predominant neural expression of GnT-IX.
FIGURE 2.
Neural-specific chromatin activation in the GnT-IX core promoter. A, 3T3-L1 cells were treated with DMSO (vehicle), 0.1 μm TSA or 2.5 μm 5-Aza for 24 h, and then ChIP using anti-H3K9ac antibody (left) or real-time PCR (right) was performed. For detection of ChIP, precipitated GnT-IX core promoter region including the TSS (about 200 bp) was amplified and quantified by real-time PCR. In the left graph, relative amounts of precipitated DNA to that of DMSO-treated sample were shown as the mean ± S.E. (n = 3). In the right graph, the amounts of GnT-IX mRNA relative to GAPDH were quantified by real-time PCR. Expression levels are shown as the mean ± S.E. of the ratio to that in Neuro2A cells (n = 3). In DMSO-treated 3T3-L1 cells, endogenous expression of GnT-IX mRNA was not detected. B, methylation levels of GnT-IX promoter from mouse tissues were analyzed by bisulfite sequencing. Schematic representation of GnT-IX promoter (−179 to −1) was shown in which CpG sites are underlined. Eight clones were selected and sequenced from each tissue. Methylation levels are shown as color density. The same analysis was performed for a control GnT-IX promoter construct, which had been methylated by HhaI (bottom). In most clones HhaI-recognition GCGC sites were methylated. C, ChIP experiments were performed with anti-H3K9ac, anti-H3K4me3 (upper), or H3K27me3, H3K9me2 (lower). The same region of the GnT-IX promoter was amplified as A. Amount of precipitated DNA was quantified by real-time PCR. Relative percentage against 1% input sample is shown. Data are presented as the mean ± S.E. (n = 3). D, ChIP experiments similar to those in C were performed using chromatin from 10-week-old C57BL/6 mouse tissues (brain, liver, and kidney). Data are presented as the mean ± S.E. (n = 3).
Identification of CTCF as a Regulatory Cis-element Binding Protein
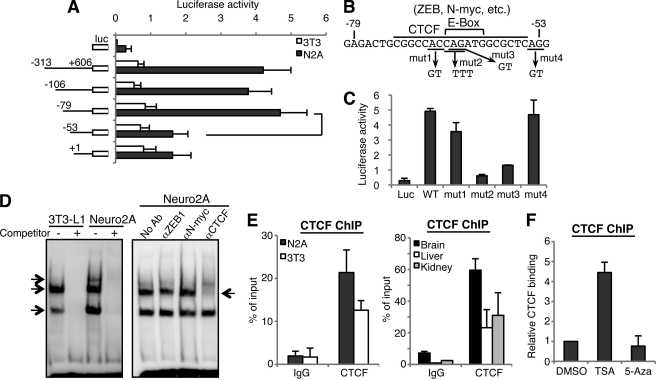
We next attempted the identification of transcription factors that bind to and drive the GnT-IX promoter. We cloned P1 as a 4.7 kb fragment extending upstream from the initiation ATG. We then generated serial deletion constructs and performed luciferase assays using 3T3-L1 and Neuro2A cells (Fig. 3A). The promoter activities of all the constructs tested were higher in Neuro2A cells. Although no obvious enhancer elements were found in the region upstream from −313 (Fig. S2), we found that deletion of −79 to −53 dramatically reduced promoter activity in Neuro2A cells (Fig. 3A), indicating that this element close to the TSS is important for GnT-IX expression. Bioinformatic analysis showed that this element has several consensus binding sites for DNA binding proteins (Fig. 3B). We then introduced point mutations within this element as shown in Fig. 3B and again performed luciferase assays. When we mutated the central CAG sequence (mut2), which is critical for binding of several transcription factors, including CTCF (23) and E-box binding proteins (N-myc, ZEB1 etc.), the promoter activity was drastically reduced in Neuro2A cells (Fig. 3C). A similar mutation (mut3), which is considered to disrupt the CTCF-site and E-box also significantly reduced the promoter activity. While another mutation (mut1 or 4), that is considered to have less or no impact on the binding of these transcription factors, had a little or no effect on the GnT-IX promoter activity. This suggests that these factors may bind to the promoter to regulate GnT-IX expression.
FIGURE 3.
Identification of CTCF as a binding protein to GnT-IX promoter. A, activity of mouse P1 was analyzed by luciferase assay in 3T3-L1 and Neuro2A cells. Relative luciferase activities against internal control Renilla luciferase are shown as the mean ± S.E. (n = 4). Promoter regions analyzed are shown to the left of the graph. B, sequence of the identified cis-element, putative binding sites for transcriptional factors, and the sites for mutagenesis (mut1, mut2, mut3, and mut4) are shown. C, part of the mouse P1 construct (−313 to +606) and its mutants were transfected into Neuro2A cells and luciferase activities were measured. Relative luciferase activities against internal control Renilla luciferase are shown as the mean ± S.E. (n = 3). D, EMSA was performed using nuclear extracts from 3T3-L1 or Neuro2A cells and biotinylated oligonucleotide probe (from −82 to −44 in P1) (left). Competitor is a non-labeled oligonucleotide with the same sequence as the probe. Arrows indicate specific binding proteins. A similar EMSA experiment was performed using Neuro2A nuclear extract with anti-ZEB1, N-Myc, or CTCF antibody (right). An arrow indicates signal reduction of the middle band with anti-CTCF antibody. E, ChIP experiment was performed with anti-CTCF antibody using cultured cells (left) or mouse tissues (right). Real-time PCR was performed to quantify the amount of precipitated GnT-IX core promoter region. Relative percentage against 1% input sample was shown as the mean ± S.E. (n = 3). F, 3T3-L1 cells were treated with DMSO (vehicle), 0.1 μm TSA or 2.5 μm 5-Aza for 24 h, and then ChIP was performed with anti-CTCF antibody. The amounts of precipitated DNA relative to that of DMSO-treated sample are shown as the mean ± S.E. (n = 3).
Next, to identify proteins that bind to this sequence, EMSA were performed using an oligonucleotide that contained the identified element. Several nuclear proteins bound to this oligonucleotide, and this binding was completely inhibited by non-labeled competitor (Fig. 3D, left panel). To identify the proteins, we used several antibodies against candidate transcription factors (Fig. 3D, right panel). Addition of an anti-CTCF antibody reduced the signal intensity of the middle band. Moreover, overexpression of CTCF in Neuro2A cells clearly increased the signal intensity of this band (supplemental Fig. S3A). Based on these results, we concluded that CTCF was identified as a binding protein in vitro. We next analyzed the binding of CTCF to the GnT-IX promoter region in vivo using the ChIP assay. As shown in Fig. 3E, left graph, CTCF binding was observed in both Neuro2A and 3T3-L1 cells. Intriguingly, binding intensity appeared to be higher in Neuro2A than in 3T3-L1 cells, although the in vitro binding of CTCF in the EMSA experiment was comparable in these cell lines (Fig. 3D, left panel). Similarly, we demonstrated by tissue-ChIP that CTCF was enriched at the GnT-IX promoter in mouse brain (Fig. 3E, right graph). These observations suggest that CTCF may be preferentially recruited to the GnT-IX promoter region in neural cells. Considering the close proximity of the CTCF site to TSS, our ChIP analysis of histone modifications indicates that chromatin around the CTCF site is also activated in neural cells. We, therefore, assumed that CTCF binding to the GnT-IX promoter would be dependent on chromatin activation status. To test this hypothesis, we examined the CTCF binding after forced chromatin activation by TSA in 3T3-L1 cells. As expected, CTCF was highly recruited to the GnT-IX promoter after forced chromatin activation (Fig. 3F), indicating that CTCF binding to the GnT-IX promoter depends on chromatin activation state.
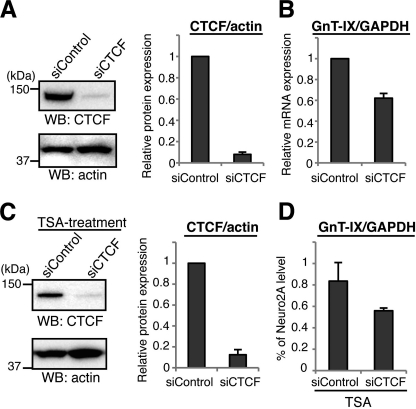
CTCF has recently been reported to have multiple functions, including transcriptional regulation and insulation (24). To examine whether CTCF regulates GnT-IX transcription, CTCF was overexpressed or knocked down by siRNA in Neuro2A cells and GnT-IX promoter activity was measured (supplemental Fig. S3, B and C). Overexpression of CTCF enhanced promoter activity, while siRNA treatment significantly reduced promoter activity, indicating that CTCF positively regulates GnT-IX transcription. To assess whether CTCF actually regulates endogenous GnT-IX expression in vivo, CTCF was knocked down by siRNA in Neuro2A cells and then GnT-IX mRNA levels were quantified by real-time PCR. As shown in Fig. 4, A and B, when CTCF protein was suppressed to be ∼8% of the control level, the level of GnT-IX mRNA was reduced to ∼60% of control, indicating that CTCF positively regulates GnT-IX transcription in vivo. To further demonstrate the importance of CTCF for GnT-IX transcription, 3T3-L1 cells were treated with TSA after CTCF knockdown (Fig. 4C). In CTCF-depleted cells, up-regulation of GnT-IX mRNA by TSA was suppressed compared with TSA-treated control cells (Fig. 4D). These results suggest that transcription of GnT-IX is mediated by the binding of CTCF.
FIGURE 4.
Knockdown of CTCF down-regulates GnT-IX transcription. A, CTCF was knocked down in Neuro2A cells by siRNA transfection. Cells were lysed and then subjected to Western blotting with anti-CTCF or anti-actin antibody (left). The bars represent the relative immunoreactivities of CTCF to actin, shown as the mean ± S.E. (n = 3) (right). B, under CTCF-depleted conditions, the levels of GnT-IX and GAPDH mRNA were quantified by real-time PCR. The expression levels of GnT-IX were normalized to the corresponding GAPDH levels, and the relative level to that of control sample is shown as the mean ± S.E. (n = 3). C, 3T3-L1 cells were transfected with control- or CTCF-specific siRNA before TSA treatment. 24 h after TSA addition, levels of CTCF and actin as a control were examined by Western blotting (left). The bars represent the relative immunoreactivities of CTCF to actin shown as the mean ± S.E. (n = 3) (right). D, levels of GnT-IX mRNA after TSA treatment relative to those of GAPDH were quantified by real-time PCR. Levels of GnT-IX are shown as a percentage relative to that of Neuro2A cells under normal conditions. Data are presented as the mean ± S.E. (n = 3).
Identification of a Second Regulatory Factor, NeuroD1
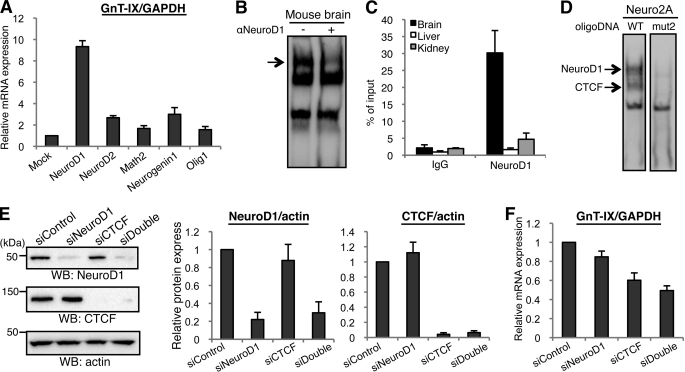
We showed that depletion of CTCF by siRNA had a definite but limited effect on GnT-IX expression. Also, CTCF is almost ubiquitously expressed (25), and a lower level of CTCF bound to GnT-IX promoter in 3T3-L1 cells. These data indicated the existence of an additional mechanism of brain-specific GnT-IX transcription. Indeed, EMSA analysis using nuclear extracts from 3T3-L1 and Neuro2A cells revealed that one of two unidentified binding proteins is likely to be neural specific (Fig. 3D, left panel, upper band), suggesting that a neural-specific protein recognizes this element. Consequently, we focused on an E-box sequence (CANNTG) which overlaps with the binding site of CTCF (Fig. 3B). Even though the E-box can be recognized by various kinds of transcription factors, including the ZEB family and basic helix-loop-helix (bHLH) type factors, we focused on several brain-specific bHLH factors, such as NeuroD, Neurogenin, and the Olig families (26). We overexpressed these neural factors (NeuroD1, NeuroD2, Math2, Neurogenin1, and Olig1) in Neuro2A cells to examine their effect on GnT-IX expression. Overexpression of all the factors tested induced GnT-IX transcription. In particular, NeuroD1 drastically up-regulated GnT-IX expression (Fig. 5A), strongly suggesting that NeuroD1 is an important transactivator of the GnT-IX gene in the brain. We confirmed that the promoter activity of GnT-IX is increased by NeuroD1 overexpression (supplemental Fig. S4A). To examine whether NeuroD1 binds to the GnT-IX promoter, EMSA was performed using nuclear extract from mouse brain. As shown in Fig. 5B, anti-NeuroD1 antibody reduced the signal intensity of the upper band (indicated by an arrow), and overexpression of NeuroD1 increased the signal intensity of this band (supplemental Fig. S4B). Because overexpression of NeuroD1 did not reduce the signal intensity of the middle band, corresponding to the CTCF-DNA complex, binding of these two identified factors to this sequence seems to be independent. Moreover, NeuroD1 binding does not require any divalent cation, which is different from Zn2+-dependent CTCF binding. We also performed EMSA analysis using mutated oligonucleotides in the presence of EDTA to see the specific binding of NeuroD1 (supplemental Fig. S4C). The binding was impaired when the E-box was mutated, indicating that NeuroD1 bound to this E-box. Also, ChIP analysis demonstrated in vivo binding of NeuroD1 to the GnT-IX promoter, specifically in the brain (Fig. 5C). We confirmed that the critical mutation in Fig. 3B (mut2), which drastically reduced the promoter activity, almost completely lost the binding of CTCF and NeuroD1 in EMSA (Fig. 5D), supporting the importance of CTCF and NeuroD1 for GnT-IX transcription.
FIGURE 5.
Identification of NeuroD1 as a positive regulator of GnT-IX transcription. A, series of neural bHLH transcription factors (NeuroD1, NeuroD2, Math2, Neurogenin1, or Olig1) were overexpressed in Neuro2A cells and then GnT-IX mRNA was quantified by real-time PCR relative to GAPDH mRNA. Relative expression levels to that of the mock transfected cells are shown as the mean ± S.E. (n = 3). B, EMSA experiments were performed using nuclear extracts from mouse brain and biotinylated oligonucleotide probe (from −82 to −44 in P1) in the presence or absence of anti-NeuroD1 antibody. An arrow indicates reduced signal intensity in the right lane. C, ChIP was performed with anti-NeuroD1 antibody using various mouse tissues. Real-time PCR was performed to quantify the amount of precipitated GnT-IX core promoter region. Relative percentage against 1% input sample is shown as the mean ± S.E. (n = 3). D, nuclear extracts of Neuro2A cells that had been transfected with NeuroD1 expression plasmid were prepared. EMSA was performed using two kinds of biotinylated oligonucleotide probe (from −82 to −44 in P1). In mut2 oligonucleotide, the E-box sequence and CTCF recognition site were completely disrupted. Arrows indicate the loss of NeuroD1 and CTCF binding to the mutated oligonucleotide. E, Neuro2A cells were transfected with NeuroD1 siRNA or CTCF siRNA, or co-transfected with NeuroD1 and CTCF siRNAs. Cells were lysed and then subjected to Western blotting with anti-NeuroD1, anti-CTCF or anti-actin antibody. The bars represent the immunoreactivities of NeuroD1 or CTCF relative to actin, shown as the mean ± S.E. (n = 3) (middle and right). F, under NeuroD1-, CTCF- or double-depleted conditions, relative levels of GnT-IX mRNA to those of GAPDH were quantified. Data are presented as the mean ± S.E. (n = 3).
To investigate whether NeuroD1 actually regulates GnT-IX transcription, NeuroD1 expression in Neuro2A cells was knocked down by siRNA, either singly or together with CTCF. siRNA against NeuroD1 effectively suppressed NeuroD1 expression (Fig. 5E) and moderately down-regulated GnT-IX expression (Fig. 5F). GnT-IX transcription was suppressed more effectively by double knockdown of NeuroD1 and CTCF (to 49% of control), indicating that NeuroD1, as well as CTCF, positively regulates GnT-IX expression in vivo. However, GnT-IX mRNA is still expressed at certain level after double knockdown of CTCF and NeuroD1, suggesting that another important regulator may exist in vivo.
Brain-specific Chromatin Activation of Other Neural Glycosyltransferase Genes
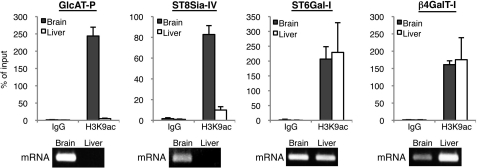
So far, we showed the involvement of CTCF and NeuroD1 in the activation of GnT-IX expression. However, our findings in which CTCF binding is highly dependent on chromatin activation and overexpression of NeuroD1 did not upregulate GnT-IX expression in 3T3-L1 cells (supplemental Fig. S5) suggest that neural specific chromatin regulation is a critical event for GnT-IX transcription. To demonstrate our hypothesis that neural glycosyltransferase genes are generally regulated by a histone code, we focused on two neural glycosyltransferases, GlcAT-P and ST8Sia-IV (PST), which biosynthesize neural-specific glycans, HNK-1 and polysialic acid, respectively (4, 5). For comparison, we also chose two ubiquitous glycosyltransferases, ST6Gal-I and β4GalT-I, which were reported to have constitutive promoters (27, 28). We confirmed by RT-PCR that GlcAT-P and ST8Sia-IV were expressed in mouse brain but not in liver, while ST6Gal-I and β4GalT-I were expressed both in mouse brain and liver (Fig. 6, bottom). We then examined the chromatin activation state around the TSS by ChIP assay (Fig. 6, top) (27–30). The TSS regions of neural GlcAT-P and ST8Sia-IV were associated with an active chromatin mark in brain but not in liver. In contrast, the TSS regions of the ubiquitous ST6Gal-I and β4GalT-I were associated with an active chromatin mark in both brain and liver. These results suggest that tissue-specific chromatin activation is a general mechanism for tissue-specific glycan expression.
FIGURE 6.
Chromatin activation analysis of other neural glycosyltransferases. Genomic regions around the TSSs of four glycosyltransferase genes were subjected to ChIP analysis with anti-H3K9ac (active chromatin mark) antibody using 20-week-old mouse brain or liver (top). Real-time PCR was performed to quantify the amount of precipitated DNA. Relative percentage against 1% input sample is shown. Data are presented as the mean ± S.E. (n = 3). mRNA level of each glycosyltransferase in adult mouse brain or liver was analyzed by RT-PCR (bottom).
DISCUSSION
In this study, we showed that brain-specific GnT-IX expression is regulated by a neural specific epigenetic mechanism. Our ChIP analysis indicates that the GnT-IX core promoter region encompassing the CTCF and NeuroD1 binding sites consists of active chromatin in neural cells, while the same region consists of inactive chromatin in non-neural cells. We also found that core promoters of other neural glycosyltransferase genes are within active chromatin in the brain (Fig. 6). We here propose a new model in which epigenetic chromatin activation determines tissue-specific glycan expression.
CTCF has various roles, such as transcriptional repression (31), activation (32), insulation (33), and control of imprinted genes (34). In particular, recent genome-wide studies revealed its central role as a boundary molecule for insulation (35). CTCF often locates to the boundary between two distinct regions (33, 36) to block the abnormal spread of inactive chromatin (37). In our case, however, knockdown of CTCF did not cause an obvious change in the chromatin state around the GnT-IX TSS in Neuro2A cells (supplemental Fig. S6). Meanwhile, overexpression of CTCF enhanced promoter activity in a reporter assay (supplemental Fig. S3B), suggesting that it is more likely that binding of CTCF to the GnT-IX promoter directly up-regulates GnT-IX transcription. Interestingly, CTCF directly binds the largest subunit of RNA polymerase II (pol-II) and only a single CTCF binding site activates reporter transcription in cells (38). A similar mechanism might exist for the GnT-IX promoter.
We identified NeuroD1 as another regulatory molecule of GnT-IX, and its transactivation potential toward GnT-IX is exerted only in neural cells (Fig. 5A and supplemental Fig. S5). To our surprise, we found that overexpression of NeuroD1 cannot induce GnT-IX expression in 3T3-L1 cells, even in the presence of TSA (supplemental Fig. S5), suggesting that histone acetylation is not sufficient for NeuroD1 to upregulate GnT-IX gene in non-neural cells. NeuroD1 is functionally phosphorylated by a neuronal kinase, CaMKII (39). However, overexpression of mutant NeuroD1 in which phosphorylation site was disrupted (serine 336 to alanine) also enhanced GnT-IX expression in Neuro2A cells. Furthermore, overexpression of CaMKIIα did not influence on GnT-IX up-regulation by NeuroD1 in Neuro2A cells (supplemental Fig. S7). These data suggest that another neural factor other than phosphorylation is required for NeuroD1-dependent activation of the GnT-IX promoter.
In our experiments, NeuroD1 induced the highest level of GnT-IX transcription among the neural bHLH factors tested (Fig. 5A), but single knockdown of NeuroD1 in Neuro2A cells slightly suppressed GnT-IX expression (Fig. 5F), suggesting that other neural bHLH factors can contribute to brain specific expression of GnT-IX. NeuroD1 mainly functions in terminally differentiating post-mitotic neurons (40) and impaired neuronal development is observed in NeuroD1-deficient mice (41, 42). While we detected high GnT-IX expression in neurons in the mouse brain, we could also detect moderate expression in glial cells,3 indicating that other neural bHLH factors, such as Olig family, may also be involved in GnT-IX expression in the brain.
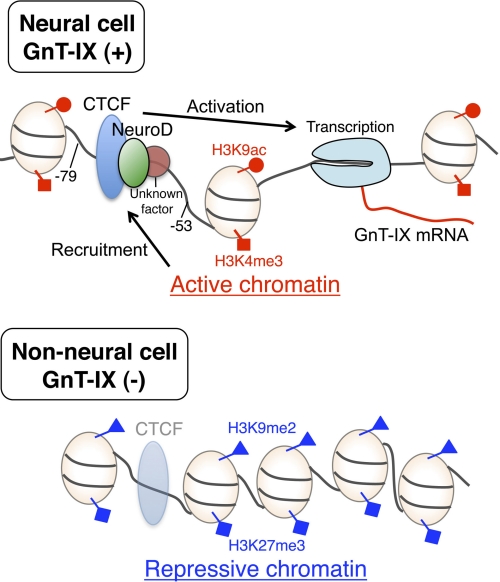
Our present study revealed a gene-regulation mechanism for brain-specific GnT-IX expression, which involves brain-specific chromatin activation and binding proteins (CTCF and NeuroD1). Combining all our results in this study, a model is depicted in Fig. 7. One of important issues would be to clarify how chromatin around glycosyltransferase promoters is activated in specific tissues.
FIGURE 7.
Schematic model of the mechanism of brain-specific GnT-IX expression. In GnT-IX positive cells (upper), histones around the core promoter region are modified with active marks (H3K9Ac and H3K4me3), thus CTCF and NeuroD (and an additional factor) are recruited to activate GnT-IX mRNA transcription. While in GnT-IX-negative cells (lower), chromatins are repressive (marked by H3K9me2 and H3K27me3) to which the transcription factors weakly or do not bind, and GnT-IX mRNA is not transcribed.
Supplementary Material
Acknowledgments
We thank Drs. Yasushi Kawaguchi (The Institute of Medical Science, The University of Tokyo), Tatsuya Hirano (Chromosome Dynamics Laboratory, RIKEN), and Shigeaki Kato (Institute of Molecular and Cellular Biosciences, The University of Tokyo) for valuable discussions and technical advice. We also thank Masaki Kato (Systems Glycobiology Research Group, RIKEN) for bioinformatic analysis.
This work was supported in part by grants from the Systems Glycobiology Research project (to N. T.), a Grant for the Strategic programs for R&D of RIKEN ASI, Global COE Program of Osaka University, and Grants-in-Aid for Scientific Research (A) (to N. T.) and for Young Scientists (B) (to Y. K.) from the Japan Society for the Promotion of Science (JSPS).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S8.
N. Taniguchi, unpublished results.
- TSA
- Trichostatin A
- 5-Aza
- 5-aza-2′-deoxycytidine
- EMSA
- electromobility shift assay
- β4GalT-I
- β-1,4-galactosyltransferase 1
- ChIP
- chromatin immunoprecipitation
- GlcAT-P
- glucuronyltransfearse-P
- GnT-IX
- N-acetylglucosaminyltransferase-IX
- HDAC
- histone deacetylase
- HNK-1
- human natural killer-1
- ST6Gal-I
- β-galactoside α-2,6-sialyltransferase 1
- ST8Sia-IV
- α-2,8-sialyltransferase-IV
- TSS
- transcriptional start site.
REFERENCES
- 1. Ohtsubo K., Marth J. D. (2006) Cell 126, 855–867 [DOI] [PubMed] [Google Scholar]
- 2. Taniguchi N., Miyoshi E., Gu J., Honke K., Matsumoto A. (2006) Curr. Opin. Struct. Biol. 16, 561–566 [DOI] [PubMed] [Google Scholar]
- 3. Tu L., Banfield D. K. (2010) Cell Mol. Life Sci. 67, 29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eckhardt M., Bukalo O., Chazal G., Wang L., Goridis C., Schachner M., Gerardy-Schahn R., Cremer H., Dityatev A. (2000) J. Neurosci. 20, 5234–5244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yamamoto S., Oka S., Inoue M., Shimuta M., Manabe T., Takahashi H., Miyamoto M., Asano M., Sakagami J., Sudo K., Iwakura Y., Ono K., Kawasaki T. (2002) J. Biol. Chem. 277, 27227–27231 [DOI] [PubMed] [Google Scholar]
- 6. Ohtsubo K., Takamatsu S., Minowa M. T., Yoshida A., Takeuchi M., Marth J. D. (2005) Cell 123, 1307–1321 [DOI] [PubMed] [Google Scholar]
- 7. Taniguchi N., Honke K., Fukuda M. (2002) Handbook of Glycosyltransferases and Related Genes, Springer, Tokyo [Google Scholar]
- 8. Chen G. Y., Osada H., Santamaria-Babi L. F., Kannagi R. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16894–16899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Goldberg A. D., Allis C. D., Bernstein E. (2007) Cell 128, 635–638 [DOI] [PubMed] [Google Scholar]
- 10. Inamori K., Endo T., Ide Y., Fujii S., Gu J., Honke K., Taniguchi N. (2003) J. Biol. Chem. 278, 43102–43109 [DOI] [PubMed] [Google Scholar]
- 11. Partridge E. A., Le Roy C., Di Guglielmo G. M., Pawling J., Cheung P., Granovsky M., Nabi I. R., Wrana J. L., Dennis J. W. (2004) Science 306, 120–124 [DOI] [PubMed] [Google Scholar]
- 12. Ko J. H., Miyoshi E., Noda K., Ekuni A., Kang R., Ikeda Y., Taniguchi N. (1999) J. Biol. Chem. 274, 22941–22948 [DOI] [PubMed] [Google Scholar]
- 13. Granovsky M., Fata J., Pawling J., Muller W. J., Khokha R., Dennis J. W. (2000) Nat. Med. 6, 306–312 [DOI] [PubMed] [Google Scholar]
- 14. Inamori K., Endo T., Gu J., Matsuo I., Ito Y., Fujii S., Iwasaki H., Narimatsu H., Miyoshi E., Honke K., Taniguchi N. (2004) J. Biol. Chem. 279, 2337–2340 [DOI] [PubMed] [Google Scholar]
- 15. Barresi R., Michele D. E., Kanagawa M., Harper H. A., Dovico S. A., Satz J. S., Moore S. A., Zhang W., Schachter H., Dumanski J. P., Cohn R. D., Nishino I., Campbell K. P. (2004) Nat. Med. 10, 696–703 [DOI] [PubMed] [Google Scholar]
- 16. Abbott K. L., Matthews R. T., Pierce M. (2008) J. Biol. Chem. 283, 33026–33035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee I., Guo H. B., Kamar M., Abbott K., Troupe K., Lee J. K., Alvarez-Manilla G., Pierce M. (2006) J. Neurochem. 97, 947–956 [DOI] [PubMed] [Google Scholar]
- 18. Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 19. Bannister A. J., Kouzarides T. (2011) Cell Res. 21, 381–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yoshida M., Kijima M., Akita M., Beppu T. (1990) J. Biol. Chem. 265, 17174–17179 [PubMed] [Google Scholar]
- 21. Kaneko M., Alvarez-Manilla G., Kamar M., Lee I., Lee J. K., Troupe K., Zhang W., Osawa M., Pierce M. (2003) FEBS Lett. 554, 515–519 [DOI] [PubMed] [Google Scholar]
- 22. Jones P. A., Taylor S. M. (1980) Cell 20, 85–93 [DOI] [PubMed] [Google Scholar]
- 23. Schmidt D., Schwalie P. C., Ross-Innes C. S., Hurtado A., Brown G. D., Carroll J. S., Flicek P., Odom D. T. (2010) Genome Res. 20, 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Phillips J. E., Corces V. G. (2009) Cell 137, 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Loukinov D. I., Pugacheva E., Vatolin S., Pack S. D., Moon H., Chernukhin I., Mannan P., Larsson E., Kanduri C., Vostrov A. A., Cui H., Niemitz E. L., Rasko J. E., Docquier F. M., Kistler M., Breen J. J., Zhuang Z., Quitschke W. W., Renkawitz R., Klenova E. M., Feinberg A. P., Ohlsson R., Morse H. C., 3rd, Lobanenkov V. V. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6806–6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bertrand N., Castro D. S., Guillemot F. (2002) Nat. Rev. Neurosci. 3, 517–530 [DOI] [PubMed] [Google Scholar]
- 27. Wuensch S. A., Huang R. Y., Ewing J., Liang X., Lau J. T. (2000) Glycobiology 10, 67–75 [DOI] [PubMed] [Google Scholar]
- 28. Rajput B., Shaper N. L., Shaper J. H. (1996) J. Biol. Chem. 271, 5131–5142 [DOI] [PubMed] [Google Scholar]
- 29. Yamamoto S., Oka S., Saito-Ohara F., Inazawa J., Kawasaki T. (2002) J. Biochem. 131, 337–347 [DOI] [PubMed] [Google Scholar]
- 30. Takashima S., Yoshida Y., Kanematsu T., Kojima N., Tsuji S. (1998) J. Biol. Chem. 273, 7675–7683 [DOI] [PubMed] [Google Scholar]
- 31. Filippova G. N., Fagerlie S., Klenova E. M., Myers C., Dehner Y., Goodwin G., Neiman P. E., Collins S. J., Lobanenkov V. V. (1996) Mol. Cell. Biol. 16, 2802–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vostrov A. A., Quitschke W. W. (1997) J. Biol. Chem. 272, 33353–33359 [DOI] [PubMed] [Google Scholar]
- 33. Bell A. C., West A. G., Felsenfeld G. (1999) Cell 98, 387–396 [DOI] [PubMed] [Google Scholar]
- 34. Bell A. C., Felsenfeld G. (2000) Nature 405, 482–485 [DOI] [PubMed] [Google Scholar]
- 35. Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 36. Hark A. T., Schoenherr C. J., Katz D. J., Ingram R. S., Levorse J. M., Tilghman S. M. (2000) Nature 405, 486–489 [DOI] [PubMed] [Google Scholar]
- 37. Witcher M., Emerson B. M. (2009) Mol. Cell 34, 271–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chernukhin I., Shamsuddin S., Kang S. Y., Bergström R., Kwon Y. W., Yu W., Whitehead J., Mukhopadhyay R., Docquier F., Farrar D., Morrison I., Vigneron M., Wu S. Y., Chiang C. M., Loukinov D., Lobanenkov V., Ohlsson R., Klenova E. (2007) Mol. Cell. Biol. 27, 1631–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gaudillière B., Konishi Y., de la Iglesia N., Yao G., Bonni A. (2004) Neuron 41, 229–241 [DOI] [PubMed] [Google Scholar]
- 40. Cho J. H., Tsai M. J. (2004) Mol. Neurobiol. 30, 35–47 [DOI] [PubMed] [Google Scholar]
- 41. Liu M., Pleasure S. J., Collins A. E., Noebels J. L., Naya F. J., Tsai M. J., Lowenstein D. H. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 865–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Miyata T., Maeda T., Lee J. E. (1999) Genes Dev. 13, 1647–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.