Abstract
Human T-cell leukemia virus type 1 (HTLV-1) has two late domain (LD) motifs, PPPY and PTAP, which are important for viral budding. Mutations in the PPPY motif are more deleterious for viral release than changes in the PTAP motif. Several reports have shown that the interaction of PPPY with the WW domains of a Nedd4 (neuronal precursor cell-expressed developmentally down-regulated-4) family ubiquitin ligase (UL) is a critical event in virus release. We tested nine members of the Nedd4 family ULs and found that ITCH is the main contributor to HTLV-1 budding. ITCH overexpression strongly inhibited release and infectivity of wild-type (wt) HTLV-1, but rescued the release of infectious virions with certain mutations in the PPPY motif. Electron microscopy showed either fewer or misshapen virus particles when wt HTLV-1 was produced in the presence of overexpressed ITCH, whereas mutants with changes in the PPPY motif yielded normal looking particles at wt level. The other ULs had significantly weaker or no effects on HTLV-1 release and infectivity except for SMURF-1, which caused enhanced release of wt and all PPPY− mutant particles. These particles were poorly infectious and showed abnormal morphology by electron microscopy. Budding and infectivity defects due to overexpression of ITCH and SMURF-1 were correlated with higher than normal ubiquitination of Gag. Only silencing of ITCH, but not of WWP1, WWP2, and Nedd4, resulted in a reduction of HTLV-1 budding from 293T cells. The binding efficiencies between the HTLV-1 LD and WW domains of different ULs as measured by mammalian two-hybrid interaction did not correlate with the strength of their effect on HTLV-1 budding.
Keywords: Gene Silencing, Protein Motifs, Retrovirus, Ubiquitin Ligase, Virus Assembly, HTLV-1, Late Domain
Introduction
Retroviruses, like several other enveloped viruses, use the host multivesicular body (MVB)3 machinery to bud from infected cells (recently reviewed in Refs. 1–6). The MVB complex consists of a network of class E vacuolar protein sorting proteins, which form four distinct heteromeric endosomal sorting complexes required for transport known as ESCRT-0, -I, -II, and -III (7, 8); these four complexes are required for the formation and release of the vesicles of the MVB. Their major function in the cell is to transport cargo proteins, such as activated cell surface receptors, from the early endosomal membrane to be released into the lysosome in small vesicles for degradation (9). The proteins are targeted to the endosomal degradation pathway by modification with mono- to tetraubiquitin chains.
Retroviruses use this pathway as it allows for the formation of plasma membrane vesicles with the right topology, i.e. budding away from the cytoplasm. They recruit components of the MVB pathway through highly conserved motifs in the Gag polyprotein. These motifs have been called late domain (LD) motifs because they control retroviral budding, a late step in the viral life cycle (10, 11). Three motifs have been identified in different retroviruses; they are PPXY, PT/SAP, and YPXL/LXXLF. The PTAP motif recruits ESCRT-I by interacting with Tsg101 (tumor susceptibility gene 101) (12–14), whereas YPXL/LXXLF binds AIP/Alix, which interacts with ESCRT-II (15, 16). The PPXY motif has been shown to interact with the WW domains of the Nedd4 (neuronal precursor cell-expressed developmentally down-regulated-4) family ubiquitin ligases (UL) (17–21). Nedd4 family ULs have three functional domains, an N-terminal C2 domain for membrane binding, WW domains for protein/protein interaction targeting mainly proline-rich sequences, and C-terminal HECT (homologous to the E6-AP carboxyl terminus) domains containing the active site. There are nine Nedd4 family ULs in humans (Nedd4, Nedd4L, NEDL1 (BUL1), NEDL2 (BUL2), WWP1, WWP2, ITCH, SMURF-1, and SMURF-2) (22–24). Nedd4 family ULs add monoubiquitin to cargo protein to flag it for recognition by ESCRT-0 (24), the first complex in the MVB pathway, and they also ubiquitinate components of ESCRT-I and ESCRT-II to stabilize their interactions (25–27). In addition, Nedd4 family ULs participate in other cellular pathways through the addition of mono-, oligo-, and polyubiquitin chains to target proteins (28). It is clear that all retroviruses, including those with a PPXY motif as their primary LD, use the ESCRT-III/vacuolar protein sorting 4 complex for the final fission step as dominant-negative mutants of vacuolar protein sorting 4 potently inhibit virus release (29–31). However, it is not known how the initial interaction of the PPXY motif with a UL and this last step are integrated and which other MVB components are involved.
Most retroviruses have more than one LD, for instance, HIV-1 has a PTAP and a LXXLF motif located in p6, the C-terminal peptide of Gag. Mutations in PTAP cause a more pronounced decrease in HIV-1 release. However, overexpression of AIP, which interacts with the LXXLF motif in cooperation with Nedd4-1, can rescue the budding of a PTAP-deficient HIV-1 mutant (16, 32, 33). In addition, HIV-1 Gag is able to recruit Nedd4 ULs in various other ways involving sequences outside of p6 (32, 34, 35). These findings indicate that HIV has several ways to recruit the ESCRT system to ensure its release, albeit at different efficiencies.
HTLV-1 has both a PPPY and a PTAP motif located at the C-terminal end of the matrix (MA) region in Gag. We and several other groups have shown that the PPPY motif is dominant; mutations in this motif cause loss of budding that cannot be rescued by the PTAP motif (36–40). Several other viruses, including Mason-Pfizer monkey virus (17, 41), murine leukemia virus (MLV) (42, 43), rhabdovirus (21), and Ebola virus (20, 29) have both PPXY and PT/SAP motifs and, for most of these viruses, the PPPY motif is also more important for efficient budding. PPPY-deficient mutants of these viruses show the typical late domain mutant phenotype with immature particles tethered to the cell or to each other by thin stalks of membrane. In contrast, the phenotype of HTLV-1 PPPY− mutants is significantly different: almost no particles are formed and Gag protein accumulates under the plasma membrane of the infected cell showing only minor contraction (37–40). Several ULs have been reported to be involved in the budding process (36–39). HTLV-1 Gag is mono- and diubiquitinated on Lys-74, and HTLV-1 K74R mutants release 5-fold fewer particles than wild-type (44). Although this reduction shows that ubiquitination at Lys-74 is important for efficient budding, it also indicates that it is not as essential as the interaction with the UL per se.
Martin-Serrano et al. (42) tested the nine Nedd4 family ULs and showed that WWP1 is the most efficient in promoting release of wt and mutant MLV. Budding was dependent on WWP1 even when the PPPY motif was in a somewhat different context than is normal for MLV-Gag. Furthermore, the WW domains of WWP1 had the highest binding efficiency, as measured in a yeast two-hybrid assay, for a number of viral PPPY motifs. Although both WWP1 and Nedd4 have been shown to interact with the PPPY motif of HTLV-1 (36–38), there has been no systematic comparative analysis to quantitate the effects of the nine Nedd4 ULs.
We wanted to know whether one of the nine ULs is more important for HTLV-1 budding to facilitate the identification of proteins that act further downstream from it. In this study we compared the nine members of the Nedd4 family in their ability to affect the release of wild-type and LD mutant HTLV-1. We found that overexpression of ITCH and SMURF-1 reduced the release of wt HTLV-1, whereas the other family members had only marginal effects. ITCH was also the only UL that was able to rescue the infectivity of PPPY mutant virus, although SMURF-1 was able to rescue budding. However, the SMURF-1-rescued mutant particles were deformed and non-infectious. Cell lines in which ITCH expression was silenced showed a budding defect; silencing of the other Nedd4 ULs had no effect.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfections, Virus Preparation, and Infection
HEK293T (293T) cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, penicillin, and streptomycin. Transfections were done essentially as previously described (38). 5 × 106 293T cells in 10-cm dishes or 3 × 105 cells/well in 12-well plates were transfected with a total of 5 or 0.2 μg of viral reporter plasmids, respectively, at a ratio of 1 packaging to 1.5 reporter to 0.1 envelope plasmid plus varied amounts of expression plasmids for FLAG-tagged ubiquitin ligase and/or empty vector DNA. FuGENE-6 (Roche Applied Science) was used at a ratio of 2.5 μl of reagent to 1 μg of DNA as the transfection reagent.
Plasmids
The HTLV-1 packaging plasmid pCMVHT-1MΔenv expresses all of the HTLV-1 gene products except Env and is identical to pCMVHT-1Δenv (45), except that it carries the pol gene of the HTVL-1 virus isolated from MT2 cells (46). The plasmid pCMV-VSV-G has been described (47) and the pCRU5inGlucβ construct is detailed under “Results.” Late domain mutants PPPG and AAAP affecting the PPPY and PTAP motifs, respectively, were described previously (38). Mutations APPY, PAPY, PPPD, and PPPF were generated by overlapping PCR and transferred into the appropriate expression plasmids on restriction fragments, which were sequenced in their entirety. Plasmid DNAs were isolated using Qiagen (Hilden, Germany) columns and protocols. All Nedd4 family UL expressing plasmids were gifts from Dr. Wes Sundquist (University of Utah) (34).
HTLV-1 Infectivity Assay
293T cells were transfected with four plasmids; a FLAG-tagged Nedd4 family UL expressing plasmid, transfer vector pCRU5inGluc, packaging plasmid pCMVHT-1MΔenv, and pCMV-VSV-G. The infectivity of virus-like particles was assessed 48–60 h post-transfection by measuring the luciferase activity in the supernatant using the Gaussia luciferase reagent from Targeting Systems (El Cajon, CA). The transfer vector encodes luciferase, which can be assayed in the supernatant as a measure of the amount of infectious virus produced in the experiment. Infectivity was expressed relative to the infectivity measured in the absence of exogenous Nedd4 UL expression, which was set at 100%.
Immunoblot Analysis
293T cells were seeded into 12-well plates and transfected the following day using the FuGENE 6 transfection reagent according to the manufacturer's protocols. For immunoblots the cell supernatants containing virus-like particles (VLP) were collected 48–60 h after transfection, filtered through 0.45-μm pore filters (Millipore, Bedford, MA), and pelleted for 2 h at 22,000 × g in a microcentrifuge at 4 °C. Cells were washed with phosphate-buffered saline (PBS) and lysed at RT in radioimmunoprecipitation assay buffer (RIPA) (100 mm NaCl, 10 mm Tris, pH 7.4, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100) with protease inhibitor mixture (Sigma) for 15 min and then kept on ice. Cell lysates were cleared at 20,000 × g for 15 min. Denaturing protein gel electrophoresis was carried out on 4 to 12% BisTris Bio-Rad gels in MOPS buffer according to the manufacturer's directions (Bio-Rad). The proteins in the gels were transferred to Immobilon P membranes (Millipore, Bedford, MA) and blocked with 3% dried milk in Tris-buffered saline. Blots were probed in the same solution containing one of the following antibodies: anti-HTLV-1 MA (p19) mouse monoclonal antibody (Zeptometrix, Buffalo, NY), affinity-purified rabbit anti-HTLV-1 MA peptide (SRPAPPPPSSPTHDPPDSDP) antisera, mouse anti-FLAG antibody (Sigma), rabbit anti-ITCH antibody (Sigma), or mouse anti-ubiquitin antibody (Santa Cruz Biotechnologies, Inc.). Washing and exposure to the appropriate horseradish peroxidase-linked secondary antibody (Cell Signaling) were done in Tris-buffered saline with 0.05% Tween. Blots were developed with Supersignal Dura or Femto reagent (ThermoScientific) and visualized on an AlphaInnotech Imager.
Ubiquitination Assay
For detection of ubiquitinated Gag proteins, 293T cells were transiently co-transfected with plasmid encoding 3 tandem copies of HA-tagged ubiquitin.4 Ubiquitinated proteins were detected with anti-HA antibodies (Covance, Princeton, NJ).
Transmission Electron Microscopy (TEM)
Detailed procedures for standard thin-section TEM were previously described (38), with the following modifications. 293T cells were transfected with pCMVHT-1MΔenv and the indicated UL expression plasmids in 10-cm dishes. Supernatants were harvested 48 h after transfection, filtered, and the VLP were pelleted through a 20% glycerol in PBS cushion for 1.5 h at 150,000 × g. The pellets were fixed in 2% glutaraldehyde in cacodylate buffer (0.1 m, pH 7.4) overnight at 4 °C followed by 1% osmium fixation for 1 h at room temperature. VLP pellets were dehydrated in graded ethanol (e.g. 35, 50, 70, 95, and 100%) and propylene oxide (100%). Pellets were infiltrated overnight in a 1:1 mixture of propylene oxide and epoxy resin, embedded in pure resin, and cured at 55 °C for 48 h. The cured block was thin sectioned, mounted on naked 200-mesh copper grids, and stained with uranyl acetate and lead citrate. Images were obtained with an electron microscope equipped with a digital camera system.
Generation of Stable Nedd4 Family ULs Knockdown Cell Lines
To generate stable 293T cell clones with silenced Nedd4 family ULs we used pGIPZ lentiviral shRNAmir vectors from Open Biosystems using the protocols provided by the manufacturer. We generated stable lines and optimal knockdown was achieved for WWP1 with constructs V2LHS_5907 and V2LHS_13521, for Nedd4 with V2LHS_72553 and V2LHS_72555, for Nedd4L with V2LHS_80459 and V2LHS_80461, and for ITCH with V2LHS_202105 and V2LHS_202846. The efficiency of silencing of Nedd4 family ULs was determined by measuring the expression of these ULs by using qualitative RT-PCR and immunoblot analysis.
Mammalian Two-hybrid Assay
The pM and pVP16 mammalian two-hybrid vectors from the MatchmakerTM system (Clontech) were used to generate fusion protein constructs for use in two-hybrid assays. As reporter we used the plasmid pGL5-Luc, which expresses the firefly luciferase gene under the control of five copies of Gal4 consensus binding sites. The nucleotide sequences encoding each of the WW domains from Nedd4, WWP1, ITCH, and SMURF-1 were amplified by PCR using the following primers: ITCH, 5′-CGGGATCCGCCCTGTAACTCAAGCTCCCTTG and 5′-CTTCTGCAGGGCTATCTGAGGTCCATTGTC; SMURF1; 5′-GCGGATCCGCCAGACGCCCCAGAACGACCAC and 5′-CTGCAGTGTGGTGTAACCTTGGGTCTGTAAACTG; Nedd4; 5′-GCGGATCCATTTGCAGCAACAACAAGAACC, and 5′-CGATGCATGGCACTGCTGGTCCAGTTATTGC. The fragments were cloned into pCR2.1-TOPO-TA (Invitrogen), sequenced, and transferred into the activation domain-expressing plasmid pVP16. The proline-rich domain of SMAD (amino acid residues 133 to 266) was amplified on pOTB-SMAD (OpenBiosystems clone ID 4109682) with oligos 5′-GAGTAGAATTCCCTGTACTTCCTCCTGTG and 5′-CACCAGTGTTTTGGATCCTCATAAGCAAC and cloned into pVP16. The plasmid encoding the DNA binding partner (pM) expressed amino acids 2 to 153 of HTLV-1 Gag containing wt or mutant late domains. Inserts were generated by PCR using primers containing appropriate restriction sites. Transfections for mammalian Matchmaker experiments were done in duplicate in 24-well plates seeded with 293T cells. Luciferase assays were performed by using a kit (Promega, Madison, WI) according to the manufacturer's protocol.
RESULTS
Overexpression of Wild-type ITCH and SMURF-1 Inhibits HTLV-1 Infectivity
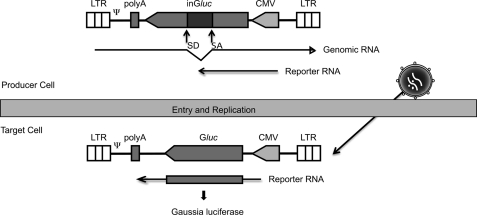
As a first approach to determine which of the different ULs interacts with HTLV-1 Gag, we analyzed the consequences of overexpressing each of the wt ULs on HTLV-1 budding and infectivity. We used a new variant of the recently described HTLV-1 transfer vector depicted in Fig. 1 (48) to measure the infectivity. Both the old and new vectors are based on the principle of the replication-dependent reporter vectors developed for measuring retrotransposition of endogenous retroviruses and long interspersed element (LINE)-1 (49–51). The new vector, pCRU5-inGlucβ, expresses the Gaussia luciferase (Gluc), which is secreted into the supernatant of the culture, instead of the cytoplasmic firefly luciferase. The Gluc transcription unit is inserted in antisense orientation relative to transcription of the viral genomic RNA and is interrupted by the second intron of the rabbit β-globin gene. The intron is placed in sense orientation relative to the viral RNA, but in antisense orientation relative to the Gluc cassette. Thus, no functional luciferase is expressed in the producer cells, but only in the target cell after a retroviral life cycle has been completed. This allows for the measurement of luciferase activity as an indicator of viral infectivity directly in a small aliquot of the supernatant of the transfected culture. The remainder of the supernatant and cells can be used for further analyses.
FIGURE 1.
Replication-dependent HTLV-1 reporter vector. Schematic showing the replication-dependent HTLV-1 reporter vector pCRU5inGlucβ, which contains an intron-interrupted Gluc expression cassette transcribed in opposite orientation relative to the retroviral genomic RNA. The splice donor and acceptor signals in Gluc are in sense orientation in the retroviral transcript. Thus transcription, splicing, reverse transcription, and integration are required to generate an uninterrupted Gluc gene.
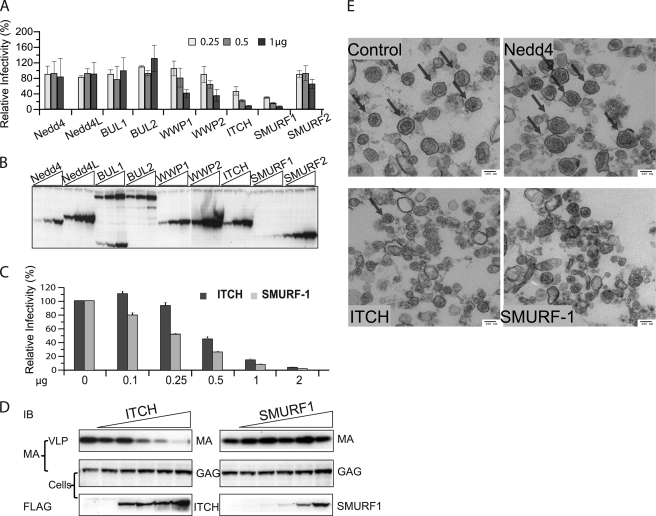
HEK293T cells were cotransfected with pCRU5inGlucβ, packaging vector pCMVHT-1MΔenv, pCMV-VSV-G, and 0.25, 0.5, and 1 μg of plasmid encoding FLAG-tagged Nedd4 family UL proteins (pCI-FlagUL). The experiment showed that overexpression of most of the ULs had little effect on the infectivity (2-fold or less for the highest amount of the expression construct). The exceptions were ITCH and SMURF-1; overexpression of these ULs reduced the infectivity 2-fold, when the lowest amount of UL expression construct was added and 20-fold for the highest (Fig. 2A). All of the ULs were expressed at comparable levels except for SMURF-1, which was consistently expressed at lower levels (Fig. 2B). Near identical results were obtained in single cycle infection experiments when we used a conventional reporter construct (45) containing an un-interrupted luciferase gene in the forward orientation. In this case we transfected 293T cells and used the supernatants to infect HeLa cells (data not shown).
FIGURE 2.
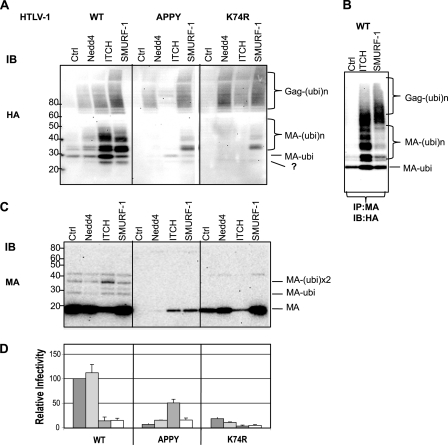
Overexpression of ITCH and SMURF-1 reduces the infectivity and release of HTLV-1 wt. A, relative infectivity of HTLV-1 VLPs in the presence of different ULs. 3 × 105 293T cells were transfected with 0.2 μg of pCMVHT-1MΔenv, 0.3 μg of pCRU5inGlucβ, and 0.03 μg of pCMV-VSV-G and three different concentrations (0.25, 0.5, and 1 μg) of plasmids encoding FLAG-tagged Nedd4 family ULs. The infectivity of the VLPs was determined by measuring the Gluc activity in the supernatant 48 h after transfection. The infectivity of HTLV-1 wt VLPs without exogenous UL was set as 100%. The data represent the mean ± S.D. of three independent experiments. B, representative anti-FLAG immunoblot of cell extracts used in the experiment in A showing the expression levels of the different ULs. C, the infectivity of wt HTLV-1 is reduced in the presence of increasing amounts of ITCH and SMURF-1. 293T cells were transfected with the retroviral vectors constructs as in A and 0, 0.1, 0.25, 0.05, 1, or 2 μg of pCI-FLAG ITCH or pCI-FLAG SMURF-1. The infectivity of the VLPs was determined by measuring Gluc activity in the supernatants 48 h after transfection. The data represent the mean ± S.D. of three independent experiments. D, the overexpression of ITCH, but not SMURF-1, inhibits release of HTLV-1 wt. Immunoblot analysis of pelleted virus particle and cell lysates of transfected cultures generated in D using the indicated antibodies. E, ITCH and SMURF-1 impact the quantity and quality of HTLV-1 wt virus particles. Representative TEM images of HTLV-1 wt VLP pellets produced in by co-transfection of 5 × 106 293T cells with 5 μg of pCMVHT-1MΔenv and 10 μg of pCI-FLAG-Nedd4, -ITCH, or -SMURF-1. Arrows indicate particles with HTLV-1 wt morphology.
We repeated the infection experiment with an expanded range of concentrations for ITCH and SMURF-1 expression constructs (Fig. 2C) and again observed concentration-dependent inhibition. Immunoblot analysis using anti-MA antibody showed that the amount of VLPs released into the supernatant decreased concomitantly with the reduction of infectivity in the presence of increasing amounts of ITCH, whereas the concentration of p55 Gag in cell extracts increased (Fig. 2D) indicating that the block was in virus release rather than in Gag synthesis. Surprisingly, the same quantity of VLPs was released from cultures expressing no or increasing amounts of SMURF-1 (Fig. 2D) despite the considerable reduction in infectivity (Fig. 2C). Again, SMURF-1 was expressed less efficiently than ITCH. Attempts at increasing the level of SMURF-1 expression were not successful as higher amounts of transfected plasmid eventually caused cytotoxicity (not observed under the experimental conditions shown here).
We used TEM to analyze the morphology of VLP pelleted from supernatants of cultures co-transfected with pCMVHT-1MΔenv with and without the different ULs. The micrographs showed that similar numbers of normal particles were present in all samples except those obtained from cultures overexpressing ITCH or SMURF-1. Fig. 2E, upper right panel, shows the particles produced in the presence of Nedd4 as an example. In contrast, very few particles could be identified in the samples obtained from ITCH overexpressing cells (Fig. 2E, lower left panel) and the particles produced by SMURF-1 overexpressing cells were aberrant in size and morphology (Fig. 2E, lower right panel).
ITCH Overexpression Rescues the Budding and Infectivity Defects of Some PPPY Mutants
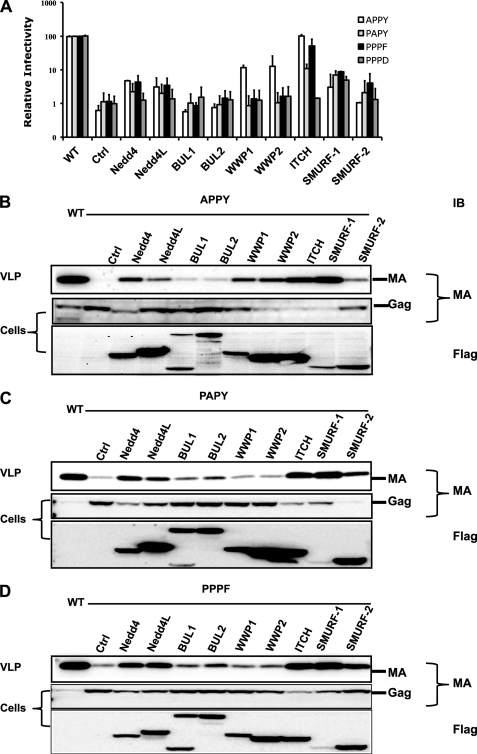
Previous reports demonstrated that in several instances ULs were not only able to influence the release of wt virus but also to rescue the release of some LD mutant viruses (32–34). To test the ability of the ULs to promote release of HTLV-1 with mutations in the PPPY motif, we generated four mutants in the pCMVHT-1MΔenv packaging construct replacing the PPPY motif with APPY, PAPY, PPPF, or PPPD (Fig. 3A). The infectivity of the first three mutants was severely reduced to about 1% of wt, whereas the PPPD mutant did not generate any infectious particles when co-transfected with reporter and VSV-G envelope plasmids; the PPPD mutant behaved in all other respects like the previously described PPPG mutant (38). When 0.5 μg of UL encoding plasmid was added to the transfection mixtures, ITCH caused a significant rescue of the APPY and PPPF mutants with infectivity approaching wt levels, whereas the infectivity of PAPY and PPPD mutants was 12 and 1.5% of wt, respectively. SMURF-1 was able to reconstitute about 5–8% of wt infectivity to all the PPPY mutants. Nedd4 restored some infectivity to all of the mutants except PPPD, whereas WWP1 and WWP2 were able to rescue the infective of the APPY construct to some extent (15% of wt).
FIGURE 3.
Overexpression of Nedd4 family ULs rescues the infectivity of HTLV-1 late domain mutants. A, the relative infectivity of HTLV-1 APPY, PAPY, PPPF, and PPPD mutant in 293T cells in the presence of different Nedd4-like ULs. 0.2 μg of pCMVHT-1MΔenv carrying the indicated mutations, 0.3 μg of pCRU5inGlucβ, and 0.03 μg of pCMV-VSV-G and 0.5 μg of the indicated pCI-Flag-UL construct were transfected into 293T cells. Gluc was measured in the supernatant 60 h after transfection. The data represent the mean ± S.D. of three independent experiments. B–D, ITCH and SMURF-1 rescue virus release most efficiently. Immunoblots of cell and pelleted VLP lysates of HTLV-1-APPY, -PAPY, and -PPPF constructs, respectively, co-transfected in 293T cells with FLAG-tagged Nedd4 family ULs as described above. Viral proteins and ULs were detected using the indicated antibodies. MA, monoclonal antibody.
VLP release in the cultures was analyzed with anti-MA antibodies using immunoblots of pelleted supernatants and cell lysates (Fig. 3, B–D). Although relative budding seemed to be more enhanced than infectivity, overall in most cases, the amount of particles in the supernatant correlated with infectivity. The exceptions were the SMURF-1 samples; here the pelleted supernatants for all mutants contained as much MA as supernatants harvested from cultures expressing wt pCMVHT-1MΔenv without additional UL expression plasmids. As seen before, the level of SMURF-1 expression in all cases was relatively low, whereas those of the other ULs were similar to each other. In summary, HTLV-1 mutants APPY and PPPF can be completely rescued by overexpression of ITCH, whereas high-level overexpression of Nedd4, WWP1, and WWP2 resulted in an intermediate level of release of the APPY mutant but had a marginal effect for the PPPF mutant. All of the mutants bud at wt levels in the presence of SMURF-1, but the maximal infectivity was only 8% of wt.
ITCH and SMURF-1 Interact Differently with HTLV-1 Gag
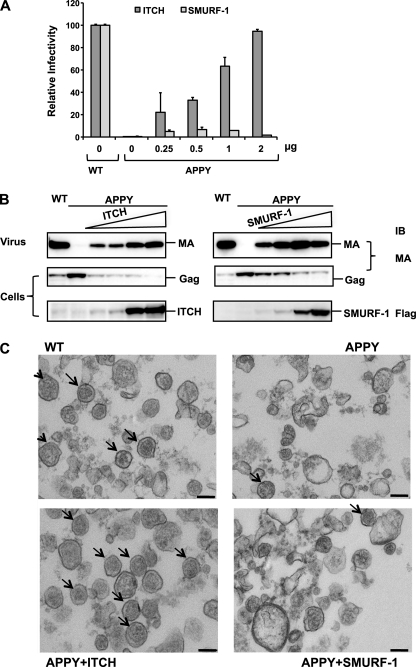
The previous experiments suggested that the effect of overexpressing the ULs on HTLV-1 release depends on the affinity of the UL for Gag, and that ITCH had the largest effect both in its ability to inhibit wt budding and to rescue the budding of LD-mutant HTLV-1. In contrast, SMURF-1 was able to promote high levels of budding of the LD-mutant VLPs, but was much less potent in terms of rescuing infectivity. To ensure that these differences were not the consequence of different expression levels, we compared the infectivity and release of HTLV-1 APPY VLPs in the presence of increasing amounts of SMURF-1 and ITCH using 0.25, 0.5, 1, and 2 μg of UL expression plasmid. The infectivity of the VLPs in the presence of ITCH in this series of experiments increased in parallel with the amount of the ITCH expression plasmid added to the transfection, reaching 100% of wt HTLV-1 at 2 μg (Fig. 4A). Low levels of SMURF-1 overexpression caused a slight increase in infectivity reaching about 8% at 0.5 μg of pCI-FlagSMURF-1 added, but higher levels led to a decrease in infectivity. VLP release was determined by immunoblot analysis and showed that budding of HTLV-1 APPY increased proportional to the amount of ITCH present and reached levels comparable with HTLV-1 wt (Fig. 4B). SMURF-1 was even more effective at rescuing HTLV-1 APPY budding. The amount of HTLV-1 APPY particles released from cells transfected with 0.5 μg of SMURF-1 expression plasmid was similar to the amount of wt HTLV-1 released from control cells (Fig. 4B).
FIGURE 4.
Increasing concentrations of ITCH, but not of SMURF-1, rescue the infectivity of HTLV-1 APPY. A, transfections and determination of infectivity were carried out as described in the legend to Fig. 2 using the indicated amounts of pCI-Flag-ITCH or pCI-Flag-SMURF-1. The infectivity of HTLV-1 wt was set to 100%. The results are the average of at least three experiments and the S.D. are shown. B, overexpression of ITCH and SMURF-1 rescues the release of HTLV-1 APPY. Immunoblots of cell and VLP lysates from the above transfection were analyzed with the indicated antibodies. C, ITCH overexpression rescues both the release and morphology of the APPY virions, but SMURF-1 overexpression does not. Representative TEM images of HTLV-1 wt VLP pellets produced by transfection of 5 × 106 293T cells with 5 μg of the indicated HTLV-1 construct and 10 μg of pCI-Flag-ITCH, pCI-Flag-SMURF-1, or empty vector.
TEM analysis showed that the morphology of the VLPs in the virus pellets from cultures transfected with the pCMVHT-1Δenv APPY and 1 μg of pCI-FLAG-ITCH was indistinguishable from those obtained with pCMVHT-1Δenv alone in terms of quantity and quality of particles (Fig. 4C). In contrast, only few particles with normal morphology were found in samples of HTLV-1 APPY produced in the presence of SMURF-1. As expected pCMVHT-1Δenv APPY alone produced almost no VLPs.
Silencing of ITCH Reduces HTLV-1 Particle Release from 293T
The results from the overexpression experiments indicated that ITCH had the most pronounced effects on HTLV-1 budding, but that Nedd4, Nedd4L, WWP1, and WWP2 could also affect HTLV-1 release. However, because the endogenous expression levels for the various ULs differ based on qualitative RT-PCR (data not shown) and the levels of the overexpressed ULs were not identical, it is difficult to compare the relative effects. We reasoned that if a specific UL was more important for the release of HTLV-1, silencing it should have a more pronounced effect. However, if their function in HTLV-1 release were redundant, the other ULs would compensate for the silencing of any one UL. Based on their activity in the previous experiments, we concentrated on Nedd4, WWP1, WWP2, and ITCH.
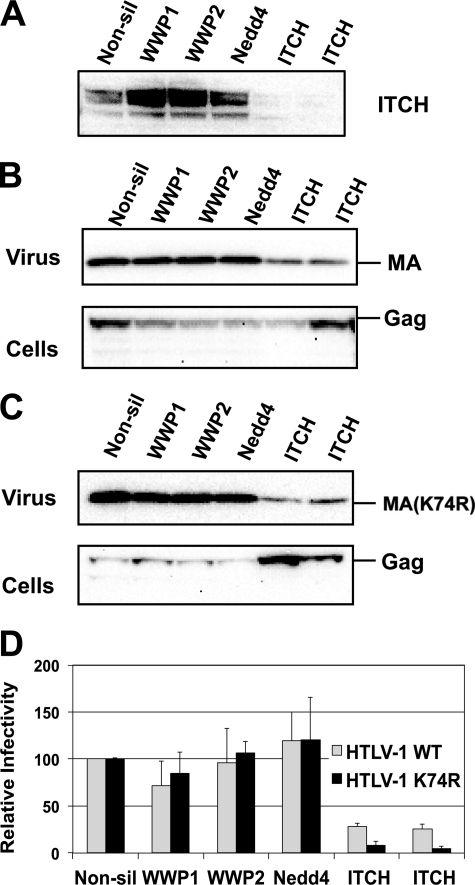
We used lentiviral vectors expressing shRNAs to infect HEK293T cells and generated stable lines, in which Nedd4, WWP1, WWP2 (data not shown), and ITCH (Fig. 5A) were specifically down-regulated at least 80% based on mRNA (data not shown) and protein levels. Virus release and single cycle infectivity were only reduced in cells, in which ITCH expression was knocked down. Fig. 5B shows that 293T/ITCHshRNA cell lines only released 20% of infectious virus compared with 293T derived lines expressing shRNAs for Nedd4, WWP1, WWP2, or control shRNAs. This result was confirmed in assays that measured VLP release by immunoblot. Again we saw that VLP release was reduced to about 20% of that seen in control cells or cells expressing shRNAs silencing other Nedd4 family ULs.
FIGURE 5.
Release of HTLV-1 wt is inhibited in ITCH-silenced 293T cells. 3 × 105 293T derived cells stably silenced for the indicated UL were transfected with 0.2 μg of pCMVHT-1MΔenv or pCMVHT-1MΔenv K74R, 0.3 μg of pCRU5inGlucβ, 0.03 μg of pCMV-VSV-G. A, anti-ITCH immunoblot of cell extracts of the different lines showing the silencing of ITCH expression. Immunoblots of cell and pelleted VLP lysates show the reduced budding of wt HTLV-1 (B) and HTLV-1 K74R (C) in ITCH-silenced cells. D, relative infectivity of wt and K74R in stable cell lines silenced for the indicated UL. The infectivity in the cell line expressing a non-silencing shRNA was set at 100%. MA, monoclonal antibody.
The effect was even more pronounced when we used pCMVHT-1ΔenvK74R in the experiments. We have previously shown that Lys-74 is the major substrate for ubiquitination in HTLV-1 Gag (44). The K74R mutation does not impact budding as severely as a mutation in the PPPY motif, but causes a 4–5-fold drop in virus release and infectivity and renders the virus hypersensitive to dominant-negative forms of Nedd4 family ULs. In ITCH-silenced cells, infectivity of the K74R mutant was reduced to less than 1% of the infectivity seen with the same construct in control cells or in cells with silenced expression of the other ULs in the assay (Fig. 5D). This was reflected in the immunoblot analysis, which showed a more pronounced decrease in the release of K74R-Gag from cells silenced for ITCH than from control cells or cells in which the expression of the other ULs was silenced (Fig. 5C). These results indicate that the other ULs cannot compensate for the missing ITCH, even though some of them are expressed more highly in 293T cells. The expression pattern of the ULs in 293T cells was similar to those of CD4+ PBLs, Jurkat cells, and chronically HTLV-1-infected cell line MS9 (data not shown).
The Role of ITCH and SMURF-1 Ubiquitin Ligase Activity in Budding
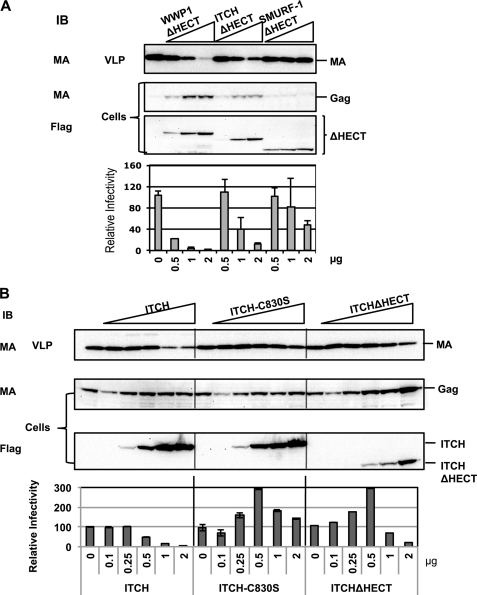
We have previously shown that overexpression of WWP1 with the C890S mutation in the active center of the HECT domain only had a minimal inhibitory effect on release and infectivity of HTLV-1 (44), whereas a WWP1 mutant with a deletion of the HECT domain had a dominant-negative phenotype. We compared the effect of overexpressing WWP1ΔHECT, ITCH-ΔHECT, and SMURF-1ΔHECT (Fig. 6A). To our surprise, ITCHΔHECT was less inhibitory than WWP1ΔHECT suggesting that the affinity between the Gag and WWP1ΔHECT is higher than between Gag and ITCHΔHECT. SMURF-1ΔHECT was expressed at similar levels as ITCHΔHECT and WWP1ΔHECT, but had little effect on budding and infectivity of wt HTLV-1 resulting in a 2-fold reduction of infectivity at the highest input level. This suggests that the endogenous SMURF-1 is not important for HTLV-1 budding and that SMURF-1 does not directly compete for the same binding sites that the other ULs occupy.
FIGURE 6.
The role of ubiquitin ligase activity in HTLV-1 release. 3 × 105 293T cells were transfected with 0.2 μg of pCMVHT-1MΔenv, 0.3 μg of pCRU5inGlucβ, 0.03 μg of pCMV-VSV-G, and the amounts of plasmids encoding FLAG-tagged UL constructs indicated in the infectivity plot. The infectivity of VLP was assessed by measuring Gluc activity in the supernatant 48 h after transfection. The infectivity of HTLV-1 wt VLPs was set as 100%. Cell and pelleted VLP lysates were analyzed by immunoblot (IB) with the indicated antibodies. A, ΔHECT versions of WWP1 and ITCH inhibit HTLV-1 release and infectivity, whereas SMURF-1ΔHECT does not. B, enzymatically inactive ITCH-C830S does not inhibit HTLV-1 release or infectivity. MA, monoclonal antibody.
We next generated a mutation (C830S) in the active center of ITCH to address the question whether the ubiquitin ligase activity was needed for the negative impact of overexpression of ITCH on HTLV-1 wt budding. The C830S mutant was expressed at levels comparable with wt ITCH, but did not result in loss of infectivity or budding of wt HTLV-1 (Fig. 6B) at levels where both wt ITCH and ITCHΔHECT reduced infectivity by at least 20-fold. Interestingly, expression of relatively low levels of either the ITCH-C830S or ITCHΔHECT enhanced infectivity (Fig. 6, A and B). It was surprising that the C830S mutant did not have a dominant-negative effect similar to the ΔHECT construct. However, disruption of the active center was evident, when we observed no rescue of HTLV-1-APPY with ITCH-C830S (data not shown) by indicating that the ubiquitin ligase activity is necessary for this ITCH function.
The Major Ubiquitination Site Lys-74 Is Needed for Full Activity
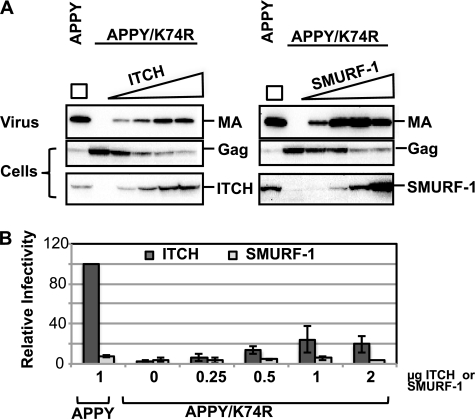
Several recent reports have suggested that the addition of ubiquitin to the Gag polyprotein creates a site that aids in the recruitment of the ESCRT machinery, indicating that the ubiquitin adduct functions as an alternate LD (52, 53). We tested whether overexpression of ULs could rescue the K74R defect or whether budding was further compromised. We found that, as with wt HTLV-1, ITCH and SMURF-1 negatively affected infectivity of K74R virus, whereas overexpression of the other ULs had little effect; ITCH reduced both release and infectivity, whereas SMURF-1 only impacted infectivity (data not shown). The K74R-APPY double mutant has a severe budding defect and, like HTLV-1 APPY, it was only marginally rescued by overexpression of most ULs. The exceptions were ITCH and SMURF-1, which could reconstitute some budding (Fig. 7A) and infectivity (Fig. 7B); however, the highest level that was achieved by ITCH overexpression was comparable with the budding of the K74R single mutant indicating that Lys-74 is necessary for efficient release. SMURF-1 overexpression resulted in complete rescue of budding (Fig. 7A), but the particles had only about 5% of the infectivity of wt particles without exogenous UL expression (Fig. 7B). The highest level of infectivity (5%) was obtained at intermediate levels of SMURF-1 expression and particles were even less infectious when more SMURF-1 was present in the cells.
FIGURE 7.
Effects of ITCH and SMURF-1 on the infectivity and release of HTLV-1 APPY/K74R. 3 × 105 293T cells were transfected 0.2 μg of pCMVHT-1-MΔenv-APPY or APPY/K74R, 0.3 μg of pCRU5inGlucβ, 0.03 μg of pCMV-VSV-G and the amounts of plasmids encoding FLAG-tagged-ITCH or -SMURF-1 constructs indicated in the infectivity plot in B. Representative results from one of three experiments are shown. A, overexpression of ITCH and SMURF-1 rescues the release of HTLV-1 APPY/K74R. Immunoblots of cell and pelleted VLP lysates were reacted with the indicated antisera. B, Lys-74 is necessary for complete rescue of HTLV-1. The infectivity of VLPs was determined by measuring Gluc activity in the supernatant 48 h after transfection. The infectivity of HTLV-1 APPY achieved in a co-transfection with 1 μg of ITCH was set as 100%. MA, monoclonal antibody.
Interaction of Wt and Mutant LD with WW Domains of Different ULs by Mammalian Two-hybrid Assay
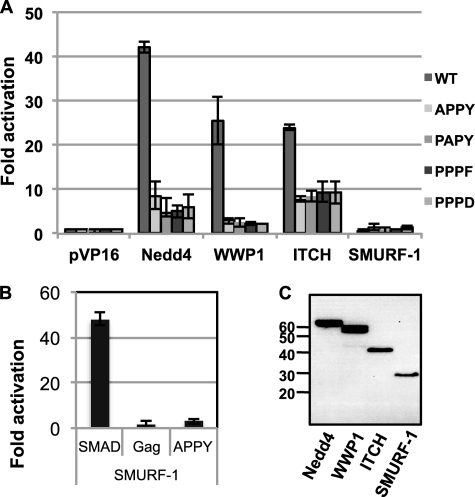
To understand the basis of the differences in the ability of the various ULs to affect HTLV-1 release, we used the mammalian two-hybrid system to measure the binding affinity between the WW domain regions of Nedd4, WWP1, ITCH, and SMURF-1, and the wt, APPY-, PAPY-, PPPF- and PPPD-mutant LD motifs. The WW domains were cloned into pVP16 and expressed as chimeras with the activation domain of herpesvirus VP16 (AD-WW). Residues 4–153 of the wt or APPY-, PAPY-, PPPF-, and PPPD-mutant Gags were fused to the DNA-binding domain of yeast Gal4-transcriptional activator (DB-MA) encoded by plasmid pM. Interaction of the two chimeric proteins results in transcription of the luciferase gene under control of five copies of the Gal4 binding site. Values were corrected for the differences in the expression levels of the various WW constructs (Fig. 8C), which were quantified by image analysis. Co-expression of the Nedd4-AD-WW and wt DB-MA showed the highest level of luciferase activity, 43-fold (Fig. 8A). Co-expression of the WWP1- or ITCH-WW-AD with wt MA-DB resulted in 25-fold activation of luciferase.
FIGURE 8.
Interaction of HTLV-1 wt and late domain mutants with WW domains of Nedd4, WWP1, ITCH, and SMURF-1. A, pM-p19 wt and the related late domain mutant plasmids express the Gal4 DNA-binding domain fused to residues 5–154 of Gag. The pVP16-WW plasmids express fusions between the WW domains of the indicated UL and the VP16 activation domain. The reporter plasmid pGL5-Luc expresses the luciferase gene under control of five copies of the Gal4 consensus binding site. Average of three experiments with standard deviations is shown. Activation is expressed as the fold-induction over that seen with the pM-p19 and the parental pVP16 plasmid, values were corrected for the expression level of the WW chimeras as determined by image analysis as shown in C. B, VP16-SMURF-1WW can bind Gal4DB-SMAD1PR. The SMAD PR region encodes residues 133–266, which have previously been shown to interact with the WW domain of SMURF-1. C, anti-VP16 immunoblot showing expression of the VP16-WW domains of the indicated ULs.
The DB-MA chimeras with mutations in the PPPY motif were significantly less able to recruit any of the WW domains resulting in 3–8-fold activation. However, the level of activation was dependent on the WW domain and not on the mutation in Gag. The AD-WWP1-WW/mutant DB-MA interactions always were the weakest, resulting in 3-fold activation, whereas other combinations caused activation levels between 5- and 8-fold. Even the PPPD DB-MA construct showed this level of activation, despite the fact that the PPPD mutant did not show any infectivity or enhancement of release in the presence of any of the overexpressed ULs except SMURF-1. We could not detect any interaction between the SMURF-1 WW domain and any of the LD motifs by mammalian two-hybrid analysis. To show that the AD-SMURF-1 WW construct was expressed in a functional form, we assayed its ability to induce luciferase expression, when the proline-rich domain containing a PPXY motif of SMAD-1 was used in the DB construct (54, 55). With this partner, the SMURF-1 WW domain caused 48-fold activation of luciferase expression. This result suggests that different domains in SMURF-1 and/or Gag are involved in the interaction or that an intermediary protein, which is not accessible in the context of the mammalian two-hybrid assay, links the two proteins. To test the first possibility we repeated the assay using chimeras with full-length Gag or SMURF-1, but these constructs were not functional in the assay. In summary, the differences in the affinities between the WW domains of the various ULs and the PPPY region of HTLV-1 Gag, as measured in the mammalian two-hybrid assay, do not correlate with the different biological effects of the individual ULs on virus release, suggesting that additional factors are involved.
The Role of Gag-Ubiquitination in Virus Release
The role of Gag ubiquitination in retroviral budding is being debated. Free ubiquitin has been reported to be present in the particles of all retroviral species and there is evidence in most viruses that a fraction of the Gag precursors is monoubiquitinated (44, 56–58). The fact that a mutant primate foamy virus Gag without any lysines can still bud with wt efficiency (52) suggests that ubiquitination is not essential for virus release. However, other studies found that removing all of the lysines in the HIV Gag caused significant budding defects (59). Furthermore, work by Joshi et al. (53) showed that an equine infectious anemia virus Gag-ubiquitin chimera could bud in the absence of any other LD; however, the resulting particles were deformed. Similar results for primate foamy virus ubiquitin chimeras were recently reported by Zhadina and Bieniasz (60). These findings indicate that the ubiquitin modification of Gag may be another level of redundancy in addition to the two or three LDs already present in most retroviral Gags to recruit components of the ESCRT machinery in different cell types and under different growth conditions.
Next we asked whether the main role of the UL binding to the PPPY motif is to ubiquitinate Gag and/or to recruit other ESCRT components. We compared the ubiquitination patterns of wt and LD-mutant MA and Gag from VLPs produced in cells overexpressing different ULs. Normally, wt HTLV-1 Gag is mono- and diubquitinated almost exclusively on Lys-74, as there is no mono- or diubiquitinated Gag proteins in viral particles of the K74R mutant (44) (shown here in Fig. 9C). We expressed pCMVHT-1MΔenv in the presence of HA-tagged ubiquitin and ULs, and analyzed the MA modifications in the VLPs by anti-HA immunoblot. As can be seen in Fig. 9C, in wt particles the major bands represent the mono- and diubiquitinated forms of MA. In cells that overexpressed Nedd4, the overall ubiquitination of MA increased slightly, however, budding efficiency was not affected (Fig. 9C) and the level of infectivity is similar to wt (Fig. 9D). High levels of ITCH caused a reduction in the release and infectivity of VLPs (Fig. 9, C and D) and the amount of ubiquitin modification was higher than in the control (Fig. 9A). In addition, a band migrating at about 25 kDa just below the MA-Ubi band was visible in the anti-HA blot. Overexpression of SMURF-1 did not affect the release of VLP (Fig. 9C), but severely compromised the infectivity of the particles (Fig. 9D). It was also associated with an increase in overall ubiquitination and the appearance of the 25-kDa band (Fig. 9A). The same analyses were performed for the APPY and K74R HTLV-1 mutants. Very few VLPs were formed by the APPY construct alone and no ubiquitinated protein was detected on the immunoblot (Fig. 9, A and C). As before, APPY-VLPs released in the presence of overexpressed ITCH were infectious and their ubiquitination pattern was similar to that of wt VLP alone. In contrast, overexpression of Nedd4 caused higher levels of polyubiquitination and less monoubiquitination and did result in particle release. SMURF-1 overexpression, which permitted Gag budding, resulted in high levels of ubiquitination in the particles and led to the appearance of a prominent band at 25 kDa. As before, very little infectivity was associated with the released particles. As expected, immunoblots of K74R mutant VLP alone showed no ubiquitinated bands (Fig. 9A). In the presence of all three ULs ubiquitinated proteins can be seen in blots suggesting that polyubiquitination can occur at alternative sites. Again the presence of the 25-kDa band was visible in all of the VLPs produced in the presence of the overexpressed ULs.
FIGURE 9.
ITCH-mediated ubiquitination of VLPs of wt HTLV-1, HTLV-1-K74R, and HTLV-1-APPY. 3 × 105 293T cells were co-transfected with 0.2 μg of pCMVHT-1MΔenv, 0.3 μg of pCRU5inGlucβ, 0.03 μg of pCMV-VSV-G, 0.5 μg of pCI-Flag-Nedd4, -ITCH, or -SMURF-1 or pCI-FLAG (empty vector control) and 0.5 μg of pCMV-3xHA-Ubi. Pelleted VLP lysates were analyzed by immunoblots with anti-HA (A) or anti-MA (C) antibodies. B, viral pellets were lysed in the presence of 2% SDS and 0.1 m DTT, diluted with PBS, and precipitated with anti-MA antibodies. The pellets were analyzed by anti-HA immunoblot. D, the infectivity of VLPs from the transfections was determined as described in the legend to Fig. 1. The infectivity of HTLV-1 wt was set as 100%. The results shown are representative of three experiments. MA, monoclonal antibody.
To prove that the bands that were seen in the immunoblots were MA and/or full-length Gag conjugated to ubiquitin, we isolated VLPs from the supernatants of cultures transfected with viral expression constructs, plasmids encoding ubiquitinated HA, and different ULs or the empty vectors. The proteins in the VLP pellets were denatured and reduced in a small volume of 2% SDS, 0.5 m DTT, diluted, and precipitated with anti-MA. After fractionation by PAGE, anti-HA blots revealed that the high molecular bands correspond to polyubiquitinated forms of MA and Gag; most of the bands in the immunoprecipitates corresponded to bands seen in direct immunoblots (Fig. 9B). As expected, the 25-kDa band was not precipitated. The size of this protein is consistent with a ubiquitinated form of NC and we tried to identify it using rabbit anti-NC antibodies. However, the results were inconclusive (data not shown).
These findings suggest that monoubiquitination of Gag, converted in viral particles to monoubiquitinated MA, is associated with efficient budding of infectious virions. In contrast, polyubiquitinated forms of Gag or the ubiquitinated moiety migrating at about 25 kDa are associated with problems in particle assembly and with VLP that have reduced infectivity.
DISCUSSION
It has been well established over the past 10 years that the budding of all retroviruses depends on the MVB machinery. The entry points into, and the progression through, the pathway for viruses that have PTAP and YPXL late motifs are well understood and described in several recent reviews (5, 6, 61). The connection between the PPPY LD and the ESCRT complexes is less clear, except for the fact that the PPPY motifs in different viruses interact with WW domains of Nedd4 family ubiquitin ligases. Several ULs have been shown to function in release of different viruses; however, only one systematic comparison study has been done and identified WWP1 as most active, when the PPPY motifs plus immediate flanking regions of different origins were tested on the MLV Gag backbone (42). The goal of this project was to determine whether WWP1 or a different Nedd4 family ubiquitin ligase is the primary enzyme interacting with the PPPY late domain motif in HTLV-1 Gag to promote viral morphogenesis and release, or whether all or several of them could function interchangeably. We analyzed the effects of overexpression and silencing of individual ULs on the release of HTLV-1 with wt and mutant LD motifs. Overall, ITCH showed the highest activity in affecting HTLV-1 budding in all assays, whereas most of the other ULs had either no effect (BUL1, BUL2, and SMURF-2) or significantly less effect (Nedd4, Nedd4L, WWP1, and WWP2). SMURF-1 impacted HTLV-1 in a different manner than the other ULs (see below). Most indicative of the importance of ITCH for HTLV-1 budding was the finding that the knockdown of ITCH expression significantly reduced HTLV-1 release, whereas silencing of other Nedd4 family ULs had no consequences. This result demonstrates that ITCH activity could not be replaced by other ULs, although some of them are expressed at higher levels in 293T cells than ITCH.
ITCH also was best able to reconstitute budding of viruses with mutations in PPPY, although not all mutants could regain function. HTLV-1 constructs, in which the PPPY motif had been replaced with APPY or PPPF, produced wt levels of infectious particles in the presence of overexpressed ITCH. In contrast, other mutants with different substitutions in the PPPY motif were rescued less efficiently (e.g. PAPY) or not at all (e.g. PPPD and PPPG). These results agree well with data presented by Martin-Serrano et al. (42) and it seems likely that the higher amount of ITCH was able to compensate for the lower affinity of the APPY and PPPF mutant for the WW domain(s), resulting in sufficient recruitment of the UL; this was also apparent in the reconstitution of Gag ubiquitination to wt levels resulting in normal patterns of mono- and diubiquitination. In contrast, substitution of the aromatic residue in the late domain seemed to abolish all interaction with ITCH. Taken together these data suggest that the ITCH/HTLV-1 Gag interaction via the WW-PPPY (or similar) interface is necessary and cannot be replaced through alternative binding sites. This indicates that ITCH functions differently in HTLV-1 and MLV release. A study by Jadwin et al. (62) showed that ITCH could rescue a MLV mutant with a PPPY to AAAY replacement by binding outside of the L domain.
When wt Gag was produced in the presence of ITCH, already relatively modest overexpression inhibited virus release and infectivity. The effects were dependent on the enzymatic activity of ITCH and correlated with increased ubiquitination of Gag, especially apparent in higher di- and polyubiquitination. Interestingly, budding of Gag with a mutation in the ubiquitination target site (K74R) was not rescued but further inhibited by ITCH overexpression. The ubiquitination patterns of wt and K74R Gag were similar, except that the band corresponding to monoubiquitinated MA was lacking in the K74R sample, suggesting that the additional ubiquitinations were inhibitory rather than enhancing to virus release. One interpretation of these results is that the main purpose of the recruitment of ITCH is the ubiquitination of Gag on Lys-74. However, the fact that the enzymatically inactive ITCH-C830S did not act as a dominant-negative mutant, but rather enhanced infectivity of wt HTLV-1, suggests that the ubiquitination itself is not necessary and that Lys-74 is more important as a binding site than as a substrate site. This result is surprising, especially in light of several recent papers suggesting that a ubiquitin moiety on Gag could act as an additional LD (34, 53, 60, 63). However, several lines of evidence support our statement that ubiquitination is not necessary for budding: 1) we had observed a similar effect before with WWP1ΔHECT and WWP1-C890S, where only the HECT deleted mutant acted as a dominant-negative mutant for HTLV-1 release (44); 2) Martin-Serrano et al. (42) showed that the active site mutant was 50-fold less inhibitory for MLV release than the HECT deletion construct; 3) Weiss et al. (63) showed that recruitment of the HECT domain to the site of particle assembly was more important for HIVΔPTAP release than ubiquitination itself; 4) we did not observe rescue of HTLV-1 APPY budding when ITCH C830S was overexpressed proving that the construct was defective in ubiquitin ligase activity.
Conversely, whereas the ubiquitin ligase activity of the HECT domain may not be essential, the presence of the HECT domain is necessary, as the ΔHECT versions of ITCH and WWP1 had strong dominant-negative effects. The finding that the ΔHECT version of WWP1 was more inhibitory than ITCHΔHECT suggests that the specificity of ITCH for HTLV-1 is mainly dependent on the ITCH HECT domain and, at least in part, on the interaction between the ITCH-HECT and the Lys-74 region in HTLV-1 Gag. This may explain why K74R mutants are more highly sensitive to the dominant-negative effect of ΔHECT mutants. If the K74R mutant cannot bind the HECT domain, binding to the endogenous ITCH is dependent only on the PPPY/WW-domain interaction. As the endogenous ITCH would also only have one contact site, it could be more efficiently competed by the overexpressed ΔHECT protein. Similarly, the depletion of ITCH in the knockdown cell lines would have a stronger effect on the K74R mutant than on wt Gag due to the loss of the cooperative binding between the partners. Likewise, the interaction between the Gag-APPY protein and ITCH-C830S would be weakened in both binding sites preventing rescue even with overexpressed ITCH-C830S.
The consequences of SMURF-1 overexpression on the production of VLPs encoded by wt and LD-mutant HTLV-1 further lend support to this argument. The quantity of wt VLP budding was not significantly affected, but the particles were abnormal in shape and non-infectious concomitantly with abnormal and high level ubiquitination of Gag. Several lines of evidence point to the conclusion that the SMURF-1/Gag interaction is basically different from that between ITCH and Gag. 1) We did not observe binding between the SMURF-1 WW domains and HTLV-1 MA in the mammalian two-hybrid system. 2) The SMURF-1ΔHECT construct did not act as a dominant-negative mutant also suggesting that SMURF-1 binding sites are different from those of the other ULs. 3) SMURF-1 was efficient in rescuing budding of all LD mutants, but particles were misshapen and had low infectivity. The ubiquitination patterns in the presence of SMURF-1 always showed a high degree of polyubiquitination suggesting that the aberrant ubiquitin adducts are able to recruit the ESCRT components necessary for vesicle release, but interfere with correct virus morphogenesis. Reports by several groups have previously shown that Nedd4, Nedd4L, and ITCH interact with the Gags of different viruses outside the immediate LD regions and rescue budding and even some infectivity in the absence of any of the canonical viral LD or UL WW domains (17, 32, 34, 52, 63, 64). The effect of SMURF-1 on budding of HTLV-1 with PPPY defects is comparable with the effect of ubiquitin appended to Gag in the form of a Gag-ubiquitin chimera. As ubiquitin is used as a tag by components in the ESCRT pathway to recognize each other, one would expect that the added ubiquitin can recruit ESCRT components resulting in the assembly of budding complexes. Although most of the resulting particles are misshapen, a few will be infectious. If the level of ubiquitination is too high budding becomes too disregulated to allow for the formation of any infectious particles.
In summary, our data demonstrate that ITCH is the UL most responsible for HTLV-1 particle formation. Surprisingly, the ubiquitin ligase activity is not necessary for this function, suggesting that ITCH may serve as an assembly platform for the recruitment of ESCRT components necessary for particle formation. We are currently further defining which domains in ITCH are involved, to allow us to identify the interacting partners.
Acknowledgments
We thank Dr. W. Sundquist for the generous gift of the pCI-FLAG UL plasmids. We are grateful to Drs. Stephen Hughes and Thomas Fanning for discussions and help with the manuscript.
This work was supported by the Intramural Research Program of the National Institutes of Health, NCI, Center for Cancer Research.
This paper is dedicated to the memory of Dr. David Derse.
G. Heidecker, unpublished data.
- MVB
- multivesicular body
- LD
- late domain
- UL
- ubiquitin ligase
- Gluc
- Gaussia luciferase
- TEM
- transmission electron microscopy
- MLV
- murine leukemia virus
- VSV
- vesicular stomatitis virus
- VLP
- virus-like particles
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Morita E., Sundquist W. I. (2004) Annu. Rev. Cell Dev. Biol. 20, 395–425 [DOI] [PubMed] [Google Scholar]
- 2. Pornillos O., Garrus J. E., Sundquist W. I. (2002) Trends Cell Biol. 12, 569–579 [DOI] [PubMed] [Google Scholar]
- 3. Demirov D. G., Freed E. O. (2004) Virus Res. 106, 87–102 [DOI] [PubMed] [Google Scholar]
- 4. Bieniasz P. D. (2006) Virology 344, 55–63 [DOI] [PubMed] [Google Scholar]
- 5. Adamson C. S., Freed E. O. (2007) Adv. Pharmacol. 55, 347–387 [DOI] [PubMed] [Google Scholar]
- 6. Usami Y., Popov S., Popova E., Inoue M., Weissenhorn W. G., Göttlinger H. (2009) Biochem. Soc. Trans. 37, 181–184 [DOI] [PubMed] [Google Scholar]
- 7. Babst M., Katzmann D. J., Snyder W. B., Wendland B., Emr S. D. (2002) Dev. Cell 3, 283–289 [DOI] [PubMed] [Google Scholar]
- 8. Raiborg C., Stenmark H. (2009) Nature 458, 445–452 [DOI] [PubMed] [Google Scholar]
- 9. Marmor M. D., Yarden Y. (2004) Oncogene 23, 2057–2070 [DOI] [PubMed] [Google Scholar]
- 10. Freed E. O. (2002) J. Virol. 76, 4679–4687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martin-Serrano J. (2007) Traffic 8, 1297–1303 [DOI] [PubMed] [Google Scholar]
- 12. VerPlank L., Bouamr F., LaGrassa T. J., Agresta B., Kikonyogo A., Leis J., Carter C. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7724–7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martin-Serrano J., Zang T., Bieniasz P. D. (2001) Nat. Med. 7, 1313–1319 [DOI] [PubMed] [Google Scholar]
- 14. Demirov D. G., Orenstein J. M., Freed E. O. (2002) J. Virol. 76, 105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin-Serrano J., Yarovoy A., Perez-Caballero D., Bieniasz P. D., Yaravoy A. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12414–12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Strack B., Calistri A., Craig S., Popova E., Göttlinger H. G. (2003) Cell 114, 689–699 [DOI] [PubMed] [Google Scholar]
- 17. Yasuda J., Hunter E., Nakao M., Shida H. (2002) EMBO Rep. 3, 636–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kikonyogo A., Bouamr F., Vana M. L., Xiang Y., Aiyar A., Carter C., Leis J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11199–11204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Strack B., Calistri A., Accola M. A., Palu G., Gottlinger H. G. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13063–13068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harty R. N., Brown M. E., Wang G., Huibregtse J., Hayes F. P. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13871–13876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harty R. N., Paragas J., Sudol M., Palese P. (1999) J. Virol. 73, 2921–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen K., Bachtiar I., Piszczek G., Bouamr F., Carter C., Tjandra N. (2008) Biochemistry 47, 1928–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harvey K. F., Kumar S. (1999) Trends Cell Biol. 9, 166–169 [DOI] [PubMed] [Google Scholar]
- 24. Rotin D., Kumar S. (2009) Nat. Rev. Mol. Cell Biol. 10, 398–409 [DOI] [PubMed] [Google Scholar]
- 25. Hoeller D., Crosetto N., Blagoev B., Raiborg C., Tikkanen R., Wagner S., Kowanetz K., Breitling R., Mann M., Stenmark H., Dikic I. (2006) Nat. Cell Biol. 8, 163–169 [DOI] [PubMed] [Google Scholar]
- 26. Patnaik A., Chau V., Wills J. W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13069–13074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hicke L. (2001) Nat. Rev. Mol. Cell Biol. 2, 195–201 [DOI] [PubMed] [Google Scholar]
- 28. Woelk T., Sigismund S., Penengo L., Polo S. (2007) Cell Div. 2, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Licata J. M., Simpson-Holley M., Wright N. T., Han Z., Paragas J., Harty R. N. (2003) J. Virol. 77, 1812–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Urata S., Yokosawa H., Yasuda J. (2007) Virol. J. 4, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garrus J. E., von Schwedler U. K., Pornillos O. W., Morham S. G., Zavitz K. H., Wang H. E., Wettstein D. A., Stray K. M., Côté M., Rich R. L., Myszka D. G., Sundquist W. I. (2001) Cell 107, 55–65 [DOI] [PubMed] [Google Scholar]
- 32. Usami Y., Popov S., Popova E., Göttlinger H. G. (2008) J. Virol. 82, 4898–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sette P., Jadwin J. A., Dussupt V., Bello N. F., Bouamr F. (2010) J. Virol. 84, 8181–8192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chung H. Y., Morita E., von Schwedler U., Müller B., Kräusslich H. G., Sundquist W. I. (2008) J. Virol. 82, 4884–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dussupt V., Javid M. P., Abou-Jaoudé G., Jadwin J. A., de La Cruz J., Nagashima K., Bouamr F. (2009) PLoS Pathog. 5, e1000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blot V., Perugi F., Gay B., Prévost M. C., Briant L., Tangy F., Abriel H., Staub O., Dokhélar M. C., Pique C. (2004) J. Cell Sci. 117, 2357–2367 [DOI] [PubMed] [Google Scholar]
- 37. Bouamr F., Melillo J. A., Wang M. Q., Nagashima K., de Los Santos M., Rein A., Goff S. P. (2003) J. Virol. 77, 11882–11895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heidecker G., Lloyd P. A., Fox K., Nagashima K., Derse D. (2004) J. Virol. 78, 6636–6648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang H., Machesky N. J., Mansky L. M. (2004) J. Virol. 78, 1503–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Le Blanc I., Prévost M. C., Dokhélar M. C., Rosenberg A. R. (2002) J. Virol. 76, 10024–10029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gottwein E., Bodem J., Müller B., Schmechel A., Zentgraf H., Kräusslich H. G. (2003) J. Virol. 77, 9474–9485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Martin-Serrano J., Eastman S. W., Chung W., Bieniasz P. D. (2005) J. Cell Biol. 168, 89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yuan B., Campbell S., Bacharach E., Rein A., Goff S. P. (2000) J. Virol. 74, 7250–7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Heidecker G., Lloyd P. A., Soheilian F., Nagashima K., Derse D. (2007) J. Virol. 81, 9769–9777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Derse D., Hill S. A., Lloyd P. A., Chung H. k., Morse B. A. (2001) J. Virol. 75, 8461–8468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mitchell M. S., Bodine E. T., Hill S., Princler G., Lloyd P., Mitsuya H., Matsuoka M., Derse D. (2007) J. Virol. 81, 4422–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F. H., Verma I. M., Trono D. (1996) Science 272, 263–267 [DOI] [PubMed] [Google Scholar]
- 48. Mazurov D., Ilinskaya A., Heidecker G., Lloyd P., Derse D. (2010) PLoS Pathog. 6, e1000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Heidmann O., Heidmann T. (1991) Cell 64, 159–170 [DOI] [PubMed] [Google Scholar]
- 50. Moran J. V., Holmes S. E., Naas T. P., DeBerardinis R. J., Boeke J. D., Kazazian H. H., Jr. (1996) Cell 87, 917–927 [DOI] [PubMed] [Google Scholar]
- 51. Curcio M. J., Garfinkel D. J. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 936–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhadina M., McClure M. O., Johnson M. C., Bieniasz P. D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20031–20036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Joshi A., Munshi U., Ablan S. D., Nagashima K., Freed E. O. (2008) Traffic 9, 1972–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhu H., Kavsak P., Abdollah S., Wrana J. L., Thomsen G. H. (1999) Nature 400, 687–693 [DOI] [PubMed] [Google Scholar]
- 55. Ebisawa T., Fukuchi M., Murakami G., Chiba T., Tanaka K., Imamura T., Miyazono K. (2001) J. Biol. Chem. 276, 12477–12480 [DOI] [PubMed] [Google Scholar]
- 56. Ott D. E., Coren L. V., Copeland T. D., Kane B. P., Johnson D. G., Sowder R. C., 2nd, Yoshinaka Y., Oroszlan S., Arthur L. O., Henderson L. E. (1998) J. Virol. 72, 2962–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Putterman D., Pepinsky R. B., Vogt V. M. (1990) Virology 176, 633–637 [DOI] [PubMed] [Google Scholar]
- 58. Ott D. E., Coren L. V., Sowder R. C., 2nd, Adams J., Nagashima K., Schubert U. (2002) J. Virol. 76, 3038–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gottwein E., Jäger S., Habermann A., Kräusslich H. G. (2006) J. Virol. 80, 6267–6275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhadina M., Bieniasz P. D. (2010) PLoS Pathog. 6, e1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Carlton J. G., Martin-Serrano J. (2009) Biochem. Soc. Trans. 37, 195–199 [DOI] [PubMed] [Google Scholar]
- 62. Jadwin J. A., Rudd V., Sette P., Challa S., Bouamr F. (2010) J. Virol. 84, 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Weiss E. R., Popova E., Yamanaka H., Kim H. C., Huibregtse J. M., Gottlinger H. (2010) PLoS Pathog. 6, e1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Calistri A., Del Vecchio C., Salata C., Celestino M., Celegato M., Göttlinger H., Palù G., Parolin C. (2009) J. Cell. Physiol. 218, 175–182 [DOI] [PMC free article] [PubMed] [Google Scholar]