Abstract
A zinc finger protein, ZPR9, has been identified as a physiological substrate of murine protein serine/threonine kinase 38 (MPK38), which is involved in various cellular responses, including the cell cycle, apoptosis, embryonic development, and oncogenesis. Here, ZPR9 was found to physically interact with apoptosis signal-regulating kinase 1 (ASK1) through a disulfide linkage involving Cys1351 and Cys1360 of ASK1 and Cys305 and Cys308 of ZPR9. ASK1 directly phosphorylated ZPR9 at Ser314 and Thr318, suggesting that ZPR9 can act as an ASK1 substrate. Ectopic expression of wild-type ZPR9, but not an S314A/T318A mutant, stimulated ASK1 kinase activity and positively regulated ASK1-mediated signaling to both JNK and p38 kinases by destabilizing complex formation between ASK1 and its negative regulators, Trx and 14-3-3, or by increasing complex formation between ASK1 and its substrate MKK3. ZPR9 functionally stimulated ASK1-induced AP-1 transcriptional activity as well as H2O2-mediated apoptosis in a phosphorylation-dependent manner. ASK1-mediated phosphorylation of ZPR9 at Ser314 and Thr318 was also responsible for ZPR9-induced apoptosis. Moreover, ZPR9 inhibited PDK1-mediated signaling through ASK1 activation. These results suggest that ZPR9 functions as a novel positive regulator of ASK1.
Keywords: Apoptosis, Phosphatidylinositol-dependent kinase-1 (PDK1), Protein Kinases, Protein Phosphorylation, Protein-Protein Interactions, Zinc Finger
Introduction
Apoptosis signal-regulating kinase 1 (ASK1),2 a member of the mitogen-activated protein kinase kinase kinase (MAPKKK) family, is activated by various cellular stresses, including reactive oxygen species, tumor necrosis factor α (TNFα), lipopolysaccharide, Fas ligand, endoplasmic reticulum stress, calcium influx, and cancer chemotherapeutic agents (1–6). ASK1 activates both c-Jun NH2-terminal kinase (JNK) and p38 signaling cascades via the phosphorylation and activation of mitogen-activated protein kinase kinases (MAPKKs) such as MKK3, -4, -6, and -7 (1–3). ASK1 activity is tightly regulated within cells and positively or negatively regulated through protein-protein interactions by a variety of intracellular proteins (1, 6, 7–18). Many proteins, including thioredoxin (Trx), glutaredoxin, GSTμ, heat shock protein 72, 14-3-3, Akt, p21Cip1, protein serine/threonine phosphatase 5, and serine-threonine kinase receptor-associated protein (STRAP), were found to play an inhibitory role in ASK1 activity through physical interaction. Meanwhile, other ASK1-interacting proteins, including tumor necrosis factor receptor-associated factor, Daxx, JNK/stress-activated protein kinase-associated protein 1/JNK-interacting protein 3, calcineurin, and murine protein serine/threonine kinase 38 (MPK38), function as positive regulators of ASK1 function. It is possible that others are yet to be discovered, and if so, these will aid in understanding how ASK1 is regulated in cells.
Zinc finger protein ZPR9 was originally identified as a physiological substrate of MPK38, also named maternal embryonic leucine zipper kinase (Melk). MPK38 is responsible for various cellular functions, including early T cell activation, embryonic development, apoptosis, and oncogenesis (19–21). ZPR9 is a zinc finger protein containing three zinc finger motifs. It is mainly localized in the cytoplasm, but its interacting partner MPK38 redirects the subcellular localization of ZPR9 from the cytoplasm to the nucleus through direct interaction (19). ZPR9 also interacts with B-myb and stimulates B-myb transcriptional activity (22). ZPR9 has been shown to enhance retinoic acid- or TNF-α-induced apoptosis, suggesting ZPR9 as a potential pro-apoptotic protein (22). However, the mechanisms underlying the pro-apoptotic properties of ZPR9 are unknown at present.
In this study we found that ZPR9 plays an important role in the regulation of the ASK1-mediated signaling pathway, in which it acts as a positive regulator of ASK1 by modulating complex formation between ASK1 and its negative regulators, Trx and 14-3-3, or its substrate MKK3. We also provide evidence that ASK1-mediated ZPR9 phosphorylation at Ser314 and Thr318 is critical for stimulating ASK1 activity, leading to the enhancement of JNK-mediated transactivation and H2O2-induced apoptosis.
MATERIALS AND METHODS
Cell Culture, Antibodies, and Plasmids
HEK293, NIH 3T3, HaCaT, SK-N-BE(2)C, and 293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum as previously described (23, 24). Anti-FLAG (M2), anti-His, anti-hemagglutinin (HA), anti-ASK1, anti-ZPR9, anti-Trx, anti-14-3-3, anti-GST, anti-MKK3, anti-ATF2, anti-poly(ADP-ribose)polymerase (PARP), anti-caspase-3, anti-phospho-MKK3/6(Ser189/207), anti-p38, anti-phospho-p38(Thr180/Tyr182), anti-phospho-ATF2(Thr71), anti-phospho-ASK1(Thr845), anti-phospho-ASK1(Ser83), anti-phospho-ASK1 (Ser967), anti-phospho-Ser/Thr, and anti-β-actin antibodies have been previously described (10, 18). Anti-phospho-Akt(Ser473) antibody was from Cell Signaling Technology (Danvers, MA). Wild-type ASK1, ASK1-N, ASK1-K, ASK1-C, activator protein 1 (AP-1)-Luc reporter, c-Fos, pSuper vector, glutathione S-transferase (GST)-tagged MKK6(K82A), p38, and ATF2 have been previously described (10, 25).
Stable Cell Lines
SK-N-BE(2)C cells stably expressing pcDNA3-His-ZPR9 plasmid (ZPR9(OE)) were screened in the presence of 850 μg/ml G418 until control parental SK-N-BE(2)C cells completely died. To prepare HEK293 cells stably expressing ZPR9-specific shRNA (ZPR9(KD)), double-stranded oligonucleotides (forward, 5′-GATCCCCGCGACGTACACCTGCATAATTCAAGAGATTATGCAGGTGTACGTCGCTTTTTGGAAA-3′; reverse, 5′-AGCTTTTCCAAAAAGCGACGTACACCTGCATAATCTCTTGAATTATGCAGGTGTACGTCGCGGG-3′; the ZPR9 sequence is underlined) were cloned into the pSuper vector. ZPR9(KD) cells were screened in the presence of 1.5 μg/ml puromycin until all control parental HEK293 cells died, as described previously (10). The inducible ZPR9 shRNA HEK293 cell line was generated using the following oligonucleotides, as described previously (26): forward, 5′-TCGAGGGCGACGTACACCTGCATAATTCAAGAGATTATGCAGGTGTACGTCGCCTTTTTTA-3′; reverse, 5′-AGCTTAAAAAAGGCGACGTACACCTGCATAATCTCTTGAATTATGCAGGTGTACGTCGCCC-3′; the ZPR9 sequence is underlined. These double-stranded oligonucleotides were cloned into the XhoI/HindIII sites of pSingle-tTS-shRNA vectors (Clontech). HEK293 cells were transfected with pSingle-tTS-shRNA harboring ZPR9-specific shRNA (ZPR9 shRNA), pSingle-tTS-shRNA containing scrambled shRNA (Sc shRNA), or pSingle-tTS-shRNA empty vector using WelFect-ExTM Plus (WelGENE, Daegu, Korea). Inducible ZPR9 shRNA stable clones were screened in the presence of 450 μg/ml G418 for ∼14 days. To confirm the knockdown of endogenous ZPR9, stable clones were isolated, treated with 1 μg/ml doxycycline for 72 h (18), and subjected to immunoblot analysis with an anti-ZPR9 antibody.
ZPR9 Substitution Mutants
ZPR9 mutants were generated by PCR as described previously (10). In brief, wild-type ZPR9 was used as the template for amplification with either the ZPR9 forward (5′-GCGAATTCATGGCGACGTACACCTGCATA-3′) or reverse (5′-GCCTCGAGTCAGAATCTCACTTGGACCCG-3′) primer, in conjunction with one of the following mutant primers containing alterations in the nucleotide sequence of ZPR9: for C221S, forward (5′-GATGAAGAATTGGAATCTGAGGATACTGAAGCA-3′) and reverse (5′-TGCTTCAGTATCCTCAGATTCCAATTCTTCAT-3′; for C254S, forward (5′-ATCCCTATCACGGACTCCTTATTTTGTTCCCAT-3′) and reverse (5′-ATGGGAACAAAATAAGGAGTCCGTGATAGGGAT-3′); for C257S, forward (5′-ACGGACTGCTTATTTTCTTCCCATCATTCCAGC-3′) and reverse (5′-GCTGGAATGATGGGAAGAAAATAAGCAGTCCGT-3′); for C305S, forward (5′-GGTGTTGGCAAGATTTCCTTGTGGTGCAACGAG-3′) and reverse (5′-CTCGTTGCACCACAAGGAAATCTTGCCAACACC-3′); for C308S, forward (5′-AAGATTTGCTTGTGGTCCAACGAGAAAGGGAAG-3′) and reverse (5′-CTTCCCTTTCTCGTTGGACCACAAGCAAATCTT-3′); for C331S, forward (5′-AATGACAAAAGCCACTCTAAGCTCCTCACAGAT-3′) and reverse (5′-ATCTGTGAAGAGCTTAGAGTGGCTTTTGTCATT-3′); for S314A, forward (5′-GAGAAAGGGAAGGCCTTCTACTCC-3′) and reverse (5′-GGAGTAGAAGGCCTTCCCTTTCTC-3′); for T318A, forward (5′-CTTCTACTCCGCAGAAGCTGTACA-3′) and reverse (5′-TGTACAGCTTCTGCGGAGTAGAAG-3′). The reaction parameters were 5 min at 94 °C for 1 cycle; 30 s at 94 °C, 2 min at 42–55 °C, and 2 min at 72 °C for 20 cycles; 5 min at 72 °C for 1 cycle. The amplified PCR mixtures were subjected to a second round of PCR amplification in the absence of primers using the following parameters: 5 min at 94 °C for 1 cycle; 30 s at 94 °C, 2 min at 50 °C, and 2 min at 72 °C for 7 cycles. A third round of PCR amplification was then performed using ZPR9 forward and reverse primers and the following parameters: 5 min at 94 °C for 1 cycle; 30 s at 95 °C, 2 min at 55 °C, and 2 min at 72 °C for 18 cycles. To generate the C305S/C308S double mutant, pGEX4T-1-C305S or pEBG-C305S was used as the template, and the C308S forward and reverse primers were used for PCR amplification. The ASK1-C substitution mutants (C1351S, C1360S, and C1351S/C1360S) were described previously (18).
In Vivo and in Vitro Interaction
In vivo binding assays were performed as previously described (23, 24). To assess the in vitro interaction of ASK1 with ZPR9, native polyacrylamide gel electrophoresis (PAGE) was carried out as previously described using autophosphorylated ASK1 or in vitro translated 35S-labeled ZPR9 (18).
In Vitro Kinase Assays for ASK1, MKK3, p38, and JNK
Assays were performed in the presence of each kinase buffer containing ∼5 μg of recombinant GST-tagged substrates as described previously (10, 23, 24).
Small Interfering RNA (siRNA) Experiments
siRNA analysis was performed as described previously (27). The ZPR9-specific siRNA #1 (5′-GCGACGUACACCUGCAUAA-3′) targeting a coding region (amino acids 2–7) and #2 (5′-GCACCGUUUGCAGUAAGAA-3′) targeting a coding region (amino acids 69–75) on human ZPR9 (GenBankTM accession number AY046059) were used for transfection experiments in HEK293 cells. The ASK1-specific siRNA and scrambled siRNA were described previously (10, 23).
Luciferase Reporter Assay
The AP-1-Luc (18) was transfected together with the indicated expression vectors into 293T cells. Luciferase activity was measured by a luciferase assay kit (Promega) according to the manufacturer's instructions.
Apoptosis Assay
Apoptosis analysis was performed in HEK293 cells transfected with the indicated expression vectors using green fluorescent protein (GFP) and terminal deoxynucleotide transferase-mediated dUTP nick end labeling (TUNEL) staining for visualization as described previously (10).
Statistical Analysis
Statistical analysis was carried out using Student's t test; p < 0.05 was considered to be significant. Values represent the means ± S.E. from at least three independent experiments.
RESULTS
ZPR9 Interacts with ASK1 in Cells
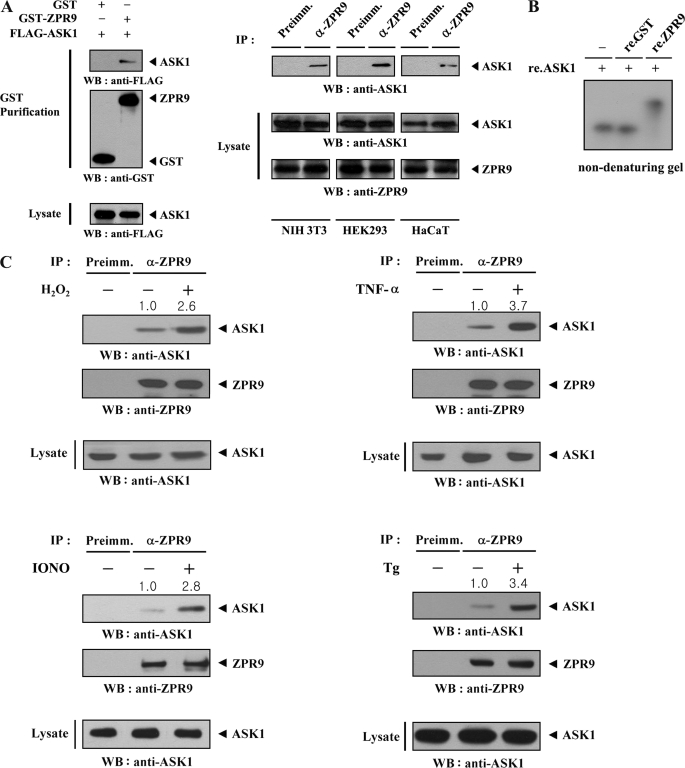
MPK38 has been shown to stimulate signaling downstream of ASK1 through direct interaction and phosphorylation (10). In addition, a previous study identified ZPR9 as a direct substrate of MPK38 (19). Based on the findings from these studies, we hypothesized that ZPR9 might affect the function of ASK1. To determine whether ZPR9 physically interacts with ASK1 in cells, cotransfection experiments were performed using GST-ZPR9 and FLAG-ASK1. ASK1 was only detected in the GST precipitate when coexpressed with GST-ZPR9 and not with control GST alone (Fig. 1A, left). The endogenous interaction between ASK1 and ZPR9 was also detected in NIH 3T3, HEK293, and HaCaT cells through coimmunoprecipitation experiments (Fig. 1A, right), supporting the functional association between these two proteins in vivo. Further non-denaturing PAGE experiments were conducted to demonstrate the direct interaction between ASK1 and ZPR9. A shift in the mobility of autophosphorylated ASK1 was detectable in the presence of recombinant ZPR9 but not in the presence of recombinant GST alone (Fig. 1B). This result indicates that ASK1 and ZPR9 interact directly with each other in vitro. Using coimmunoprecipitation experiments, it was then assessed whether ASK1 signals can affect ASK1-ZPR9 complex formation in cells. Exposure of HEK293 cells to ASK1 stimuli, including H2O2, TNF-α, thapsigargin (for endoplasmic reticulum stress), and ionomycin (for calcium overload) resulted in a remarkable increase in the endogenous interaction between ASK1 and ZPR9 compared with the untreated control (Fig. 1C). These data suggest that ZPR9 is functionally linked with ASK1 signaling.
FIGURE 1.
Association of ZPR9 with ASK1 in vivo and in vitro. A, shown is the physical association of ZPR9 with ASK1. GST alone and GST-ZPR9 were cotransfected with FLAG-ASK1 into HEK293 cells. GST fusion proteins were purified on glutathione-Sepharose beads (GST Purification), and the amount of complex formation was determined by anti-FLAG antibody immunoblot (left). Cell lysates from NIH 3T3, HEK293, or HaCaT cells were subjected to immunoprecipitation using either rabbit preimmune serum (Preimm.) or anti-ZPR9 antibody (α-ZPR9) followed by immunoblot analysis using an anti-ASK1 antibody to determine the degree of complex formation between endogenous ZPR9 and ASK1 (right). B, native PAGE analysis of the ASK1-ZPR9 complex is shown. Purified recombinant (re.) ASK1 was autophosphorylated as described previously (18). Autophosphorylated ASK1 (2–3 μg) was incubated with unlabeled recombinant GST as a control or ZPR9 as indicated (5 μg of each) at room temperature for 1 h. C, modulation of ASK1-ZPR9 interaction by ASK1 signals is shown. HEK293 cell lysates were treated with or without the following stimuli: H2O2 (2 mm, 30 min), TNF-α (500 ng/ml, 30 min), ionomycin (IONO; 1 μm, 24 h), or thapsigargin (Tg; 20 μm, 30 min) and then immunoprecipitated with either rabbit preimmune serum (Preimm.) or anti-ZPR9 antibody (α-ZPR9) followed by immunoblotting with an anti-ASK1 antibody to determine the endogenous association between ASK1 and ZPR9.
ASK1-ZPR9 Interaction Is Mediated by Cysteine Residues
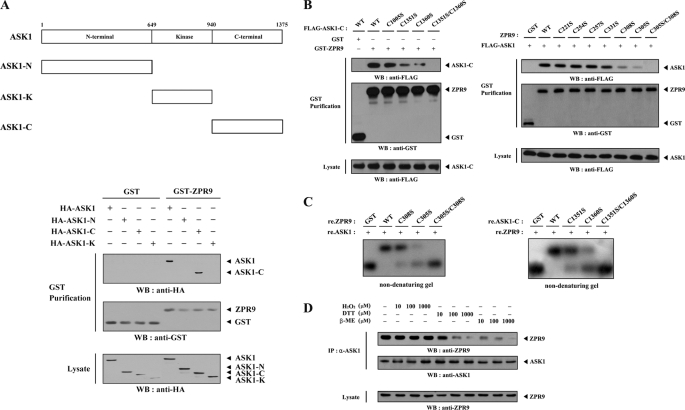
To identify the interaction domain of ASK1 involved in ZPR9 binding, in vivo binding assays were performed using three ASK1 deletion mutants (ASK1-N, comprising the amino-terminal domain (amino acids 1–648); ASK1-K, harboring the kinase domain (amino acids 649–940); ASK1-C, comprising the carboxyl-terminal regulatory domain (amino acids 941–1375) as described previously (10). ASK1-C was found to be responsible for ZPR9 binding, whereas ASK1-N and ASK1-K were unable to bind with ZPR9 (Fig. 2A), suggesting that the carboxyl-terminal domain of ASK1 is required for ZPR9 binding. The previous observation (18) that complex formation between ASK1 and its negative regulator STRAP requires cysteine residues present in each of the two proteins prompted us to investigate whether cysteine residues of ASK1 and ZPR9 participate in ASK1-ZPR9 complex formation. To this end, four cysteine-to-serine amino acid substitution mutants of ASK1-C (C1005S, C1351S, C1360S, and C1351S/C1360S) were used for in vivo binding assays to identify the potential cysteine residues of ASK1 that are responsible for ZPR9 binding. ZPR9 strongly interacted with ASK1-C and its mutant C1005S but weakly with two other mutants of ASK1-C, C1351S and C1360S. Moreover, this interaction was not detectable in the presence of the C1351S/C1360S double mutant (Fig. 2B, left). These results suggest that the Cys1351 and Cys1360 residues of ASK1 are involved in ZPR9 binding. Next, to search the potential cysteine residues of ZPR9 required for ASK1 binding, in vivo binding assays were also performed using HEK293 cells transfected with seven cysteine-to-serine amino acid substitution mutants of ZPR9 (C221S, C254S, C257S, C305S, C308S, C331S, and C305S/C308S). ASK1 interacted strongly with four ZPR9 mutants (C221S, C254S, C257S, and C331S) and weakly with two additional ZPR9 mutants (C305S and C308S). However, no ASK1 was detected when the C305S/C308S double mutant was coexpressed (Fig. 2B, right), indicating the involvement of Cys305 and Cys308 of ZPR9 in ASK1 binding. A non-denaturing PAGE analysis was then performed to confirm that the Cys1351 and Cys1360 residues of ASK1 and Cys305 and Cys308 residues of ZPR9 play key roles in complex formation. A shift in the mobility of autophosphorylated ASK1 was not detectable in the presence of the C305S/C308S ZPR9 double mutant (Fig. 2C, left). In addition, the C1351S/C1360S double mutant of ASK1-C had no effect on the mobility of 35S-labeled ZPR9 (Fig. 2C, right). These data led us to examine the importance of redox status in the interaction between ASK1 and ZPR9. The presence of the reductants dithiothreitol (DTT) and β-mercaptoethanol (β-ME) decreased complex formation between ASK1 and ZPR9 in a dose-dependent manner, whereas the oxidant H2O2 had no effect (Fig. 2D). These results suggest that the redox-dependent interaction of ASK1 with ZPR9 occurs in cells. Together, these data clearly indicate that ASK1-ZPR9 complex formation is mediated by cysteine residues present in each of these two proteins, Cys1351 and Cys1360 of ASK1 and Cys305 and Cys308 of ZPR9.
FIGURE 2.
Mapping of the ASK1 and ZPR9 interaction domains. A, shown is the schematic structure of wild-type ASK1 and three different ASK1 deletion mutants. Numbers indicate the amino acid residues corresponding to the domain boundaries. HEK293 cells transfected with the indicated expression vectors were lysed, precipitated using glutathione-Sepharose beads (GST purification), and then immunoblotted with anti-HA antibody to determine the level of ASK1-ZPR9 complex formation. B, shown is the effect of cysteine residues on ASK1-ZPR9 complex formation. HEK293 cells, transfected with the expression vectors indicated, were lysed, and the GST precipitates were analyzed for complex formation between ASK1 and ZPR9 by immunoblot analysis using anti-FLAG antibody. C, shown is native PAGE analysis of the ASK1-ZPR9 complex. Autophosphorylated ASK1 (left) or in vitro translated 35S-labeled ZPR9 (right) was incubated with unlabeled recombinant (re.) GST as a control or wild-type and mutant forms of ZPR9 or ASK1-C as indicated at room temperature for 1 h as described previously (18). D, shown is redox-dependence of the endogenous ASK1-ZPR9 interaction. HEK293 cell lysates were treated with the indicated concentration of H2O2, DTT, and β-mercaptoethanol (β-ME) on ice for 0.5–1 h and subjected to immunoprecipitation using anti-ASK1 antibody (IP). Immune complexes were analyzed for the presence of ZPR9 by immunoblot analysis using anti-ZPR9 antibody (top panel). The amount of immunoprecipitated ASK1 and the expression level of ZPR9 in total cell lysates were determined by immunoblot analysis using anti-ASK1 and anti-ZPR9 antibodies, respectively.
ASK1 Phosphorylates ZPR9 at Ser314 and Thr318
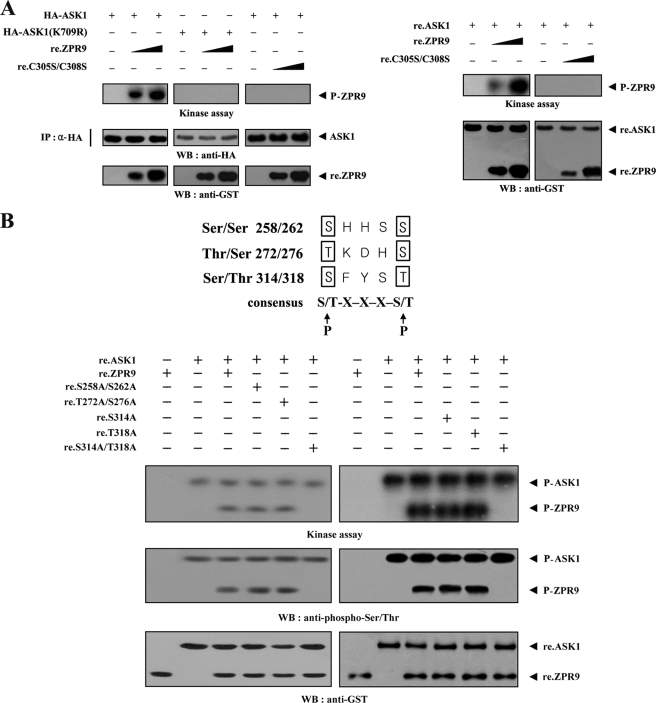
Given that ASK1 physically interacts with ZPR9 (see Fig. 1), it is possible that ASK1 phosphorylates ZPR9 through direct interaction. To assess the extent of ASK1-mediated ZPR9 phosphorylation, immunoprecipitated wild-type and kinase-dead ASK1 was incubated with [γ-32P]ATP to allow phosphorylation of the recombinant wild-type and mutant ZPR9 proteins as part of the ASK1 kinase assay. Wild-type ASK1 induced ZPR9 phosphorylation in a dose-dependent manner (Fig. 3A, left), suggesting that ZPR9 may be a potential substrate of ASK1. However, ZPR9 phosphorylation was not detected in the presence of kinase dead ASK1. To determine whether ASK1 directly phosphorylates ZPR9, we carried out in vitro kinase assays using recombinant ASK1 and ZPR9 proteins. ZPR9 phosphorylation was clearly detected in the presence of ASK1, whereas the C305S/C308S double mutant, which was unable to bind with ASK1 (see Fig. 2B), was not phosphorylated by ASK1 (Fig. 3A, right). These results indicate that ASK1 is directly phosphorylating ZPR9 through physical interaction. To identify potential ASK1 phosphorylation sites on ZPR9, in vitro kinase assays were performed using three ZPR9 amino acid substitution mutants (S258A/S262A, T272A/S276A, and S314A/T318A) that were screened by searching consensus ASK1 phosphorylation sites ((S/T)XXX(S/T)) in ZPR9. As shown in Fig. 3B, the S314A/T318A double mutant completely abolished ASK1-mediated phosphorylation, whereas ASK1-mediated phosphorylation was detected in the presence of the other double mutants (S258A/S262A and T272A/S276A) as well as the two single mutants (S314A and T318A). These findings suggest that Ser314 and Thr318 of ZPR9 represent potential phosphorylation sites for ASK1.
FIGURE 3.
Phosphorylation of ZPR9 at Ser314 and Thr318 by ASK1. A, ZPR9 phosphorylation by ASK1 is shown. HA-immunoprecipitated wild-type and kinase-dead (K709R) ASK1 (left) or recombinant ASK1 (right) was assayed for its kinase activity in the presence of kinase buffer (10) containing increasing amounts of recombinant wild-type (re.ZPR9) and mutant (re.C305S/C308S) ZPR9 as substrates. HA-immunoprecipitated ASK1(K709R) was used for the kinase assay as a control. B, shown is identification of ASK1 phosphorylation sites on ZPR9. Recombinant ASK1 was analyzed for its kinase activity by an in vitro kinase assay using recombinant wild-type ZPR9 and one of its substitution mutants (S258A/S262A, T272A/S276A, S314A, T318A, or S314A/T318A) as substrates. The phosphorylation of ZPR9 was also confirmed by immunoblotting with an anti-phospho-Ser/Thr antibody (second panel).
ZPR9 Positively Regulates ASK1 Signaling to p38
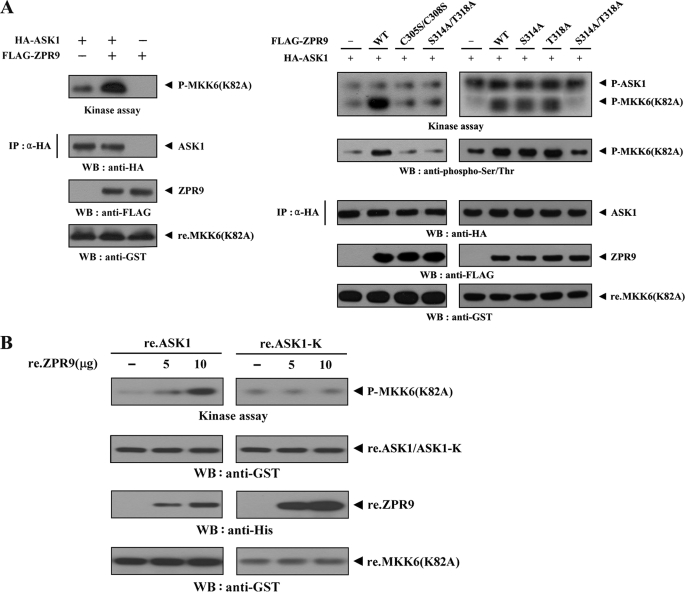
To investigate the effect of ZPR9 on ASK1 activity, ASK1 kinase activity was first examined in HEK293 cells transfected with ASK1 alone or cotransfected with ZPR9 using an in vitro kinase assay. ASK1 kinase activity was markedly increased when ASK1 was coexpressed with wild-type ZPR9 (Fig. 4A, left). Next, to determine whether direct interaction and phosphorylation between ASK1 and ZPR9 are essential for the positive regulation of ASK1 kinase activity, in vitro kinase assays were performed using two ZPR9 double mutants, C305S/C308S and S314A/T318A, which are unable to bind with ASK1 (see Fig. 2 and supplemental Fig. S1) as well as two ZPR9 single mutants, S314A and T318A. The results showed that the two defective mutants (C305S/C308S and S314A/T318A) of ZPR9 in ASK1 binding had no effect on ASK1 kinase activity, whereas the two single mutants, S314A and T318A, were able to increase the ASK1 kinase activity (Fig. 4A, right). This result suggests that ZPR9 functions as a positive regulator of ASK1 through direct interaction and phosphorylation. The direct role of ZPR9 in ASK1 kinase activity was also confirmed using an in vitro experiment with recombinant ASK1 and ZPR9 proteins. As shown in Fig. 4B, recombinant ZPR9 increased ASK1 kinase activity in a dose-dependent manner. However, ASK1 kinase activity was not affected by recombinant ZPR9 in the presence of ASK1-K, which is defective in ZPR9 binding (see Fig. 2A), supporting again the importance of direct interaction between ASK1 and ZPR9 for the regulation of ASK1 activity.
FIGURE 4.
Enhancement of ASK1 kinase activity by ZPR9. A, ZPR9 enhances ASK1 kinase activity through physical interaction. HEK293 cells were transiently transfected with HA-ASK1 in the presence or absence of wild-type (WT) and mutant forms (C305S/C308S, S314A, T318A, and S314A/T318A) of FLAG-tagged ZPR9. Cell lysates were then subjected to immunoprecipitation with anti-HA antibody (IP), and the resulting immunoprecipitates were subjected to an in vitro kinase assay using recombinant MKK6(K82A) as a substrate to determine ASK1 kinase activity. P-MKK6(K82A) and P-ASK1 indicate phosphorylated MKK6(K82A) and autophosphorylated ASK1, respectively. The presence of equivalent amounts of substrate was verified by immunoblotting with anti-GST antibody (bottom panels). B, direct stimulation of ASK1 kinase activity by recombinant ZPR9 is shown. In 30 μl of 20 mm Tris-HCl buffer (pH 7.5), 4 μg of recombinant wild-type ASK1 (re.ASK1) or ASK1-K (re.ASK1-K) proteins were incubated at room temperature for 1 h with the indicated amount of recombinant ZPR9 (re.ZPR9) and then analyzed for ASK1 kinase activity by an in vitro kinase assay using recombinant MKK6(K82A) as a substrate.
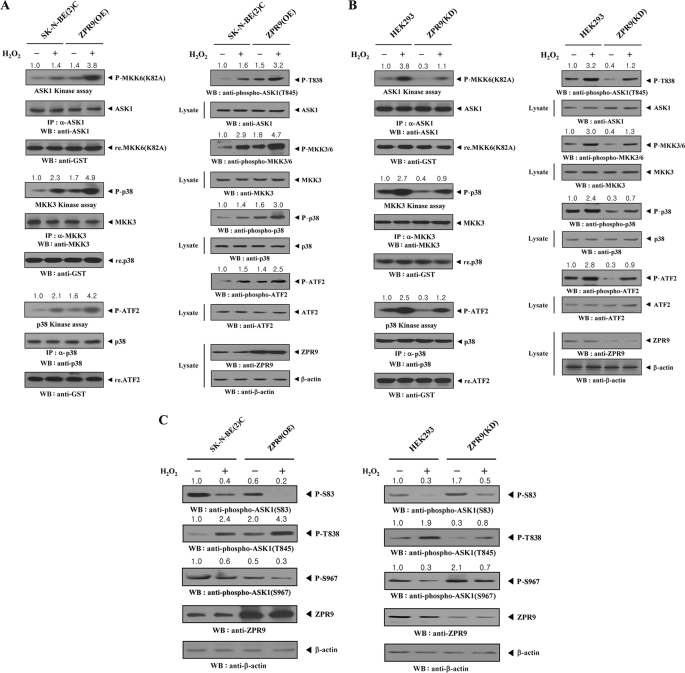
Based on these findings, it was next examined whether ZPR9 affects signaling downstream of ASK1. Endogenous ASK1, MKK3, and p38 kinase activities were determined by in vitro kinase assays using parental SK-N-BE(2)C cells and SK-N-BE(2)C cells stably expressing pCMV-ZPR9 (ZPR9(OE)). Overexpression of ZPR9 markedly increased the endogenous kinase activities of ASK1, MKK3, and p38 compared with parental SK-N-BE(2)C cells (Fig. 5A, left). These results were also confirmed in immunoblot analysis using anti-phospho-specific antibodies for ASK1 Thr845 (corresponding to Thr838 in human), MKK3/6, p38, and ATF2. ZPR9 overexpression induced higher phosphorylation levels of ASK1, MKK3/6, p38, and ATF2, which accompany ASK1-mediated signaling (Fig. 5A, right). Consistent with this, knockdown of endogenous ZPR9 showed an opposite trend in the modulation of endogenous ASK1, MKK3, and p38 kinase activities and phosphorylations (Fig. 5B), indicating that ZPR9 functions as a positive regulator of ASK1.
FIGURE 5.
Stimulation of ASK1 downstream signaling by ZPR9. A and B, parental SK-N-BE(2)C and HEK293 cells, SK-N-BE(2)C cells stably overexpressing ZPR9 (ZPR9(OE)), or HEK293 cells stably expressing ZPR9-specific shRNA (ZPR9(KD)) were incubated with or without 2 mm H2O2 for 30 min. The endogenous activities of ASK1, MKK3, or p38 were determined by in vitro kinase assays and immunoblot analysis using the indicated immunoprecipitates (left) and total cell lysates (right). The presence of equal amounts of each substrate in these assays was confirmed by immunoblotting with an anti-GST antibody. Overexpression or knockdown level of endogenous ZPR9 was determined by anti-ZPR9 immunoblotting. The relative level of kinase activity was quantitated by densitometric analyses, and the -fold increase relative to untreated samples in parental SK-N-BE(2)C or HEK293 cells was calculated. P-, phosphorylated. C, the effect of ZPR9 on ASK1 phosphorylation is shown. Parental SK-N-BE(2)C and HEK293 cells, ZPR9(OE) cells, or ZPR9(KD) cells were incubated with or without 2 mm H2O2, as described above. ASK1 phosphorylation was determined by immunoblot analysis with the indicated phospho-specific antibodies using total cell lysates.
It was also determined whether the stimulatory effect of ZPR9 on ASK1-p38/JNK signaling is dependent on ASK1 phosphorylations involved in ASK1 activation (at Thr838) and inactivation (at Ser83 and Ser967) using immunoblot analysis in ZPR9(OE) and ZPR9(KD) cells. Overexpression of ZPR9 considerably decreased ASK1 phosphorylation at Ser83 and Ser967 compared with control parental SK-N-BE(2)C cells. In contrast, ASK1 phosphorylation at Thr838 was increased by ZPR9 (Fig. 5C, left). Consistently, knockdown of endogenous ZPR9 clearly showed an opposite trend in the modulation of Ser83, Thr838, and Ser967 phosphorylation (Fig. 5C, right). These results indicate that ZPR9-mediated stimulation of ASK1 activity apparently embodies the concordant modulation of ASK1 phosphorylation at Ser83, Thr838, and Ser967.
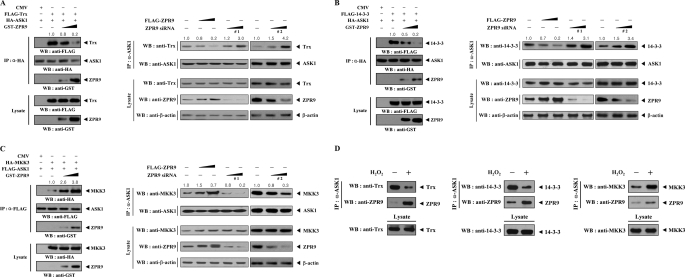
ZPR9 Modulates Complex Formation between ASK1 and Its Regulators (Trx and 14-3-3) or Substrate (MKK3)
Given that Trx and 14-3-3 physically associate with ASK1 and inhibit ASK1 activity (3, 14), in vivo binding assays were performed using HEK293 cells transfected with FLAG-Trx (or FLAG-14-3-3), HA-ASK1, and increasing amounts of ZPR9 to investigate whether ZPR9 affects complex formation between ASK1 and Trx or 14-3-3. Complex formation was markedly decreased in a dose-dependent manner in the presence of ZPR9 (Fig. 6, A and B, left). These results suggest that the modulation of the association between ASK1 and its negative regulators, Trx and 14-3-3, is possibly involved in ZPR9-mediated regulation of ASK1 activity. Consistent with this result, knockdown of endogenous ZPR9 increased complex formation between ASK1 and its negative regulators, Trx and 14-3-3 (Fig. 6, A and B, right). Using the same approach, the effect of ZPR9 on MKK3 binding to ASK1 was also examined, as MKK3 is a direct substrate of ASK1. The results showed that ZPR9 increased ASK1-MKK3 complex formation in a dose-dependent manner (Fig. 6C), resulting in the stimulation of ASK1 activity. A similar trend was also observed at endogenous levels in the analysis of the complex formation between ASK1 and its regulators (Trx, 14-3-3, MKK3, and ZPR9) (Fig. 6D). Together, these data indicate that ZPR9 positively regulates ASK1 activity by decreasing the association between ASK1 and its negative regulators, Trx and 14-3-3, and increasing complex formation between ASK1 and its substrate MKK3.
FIGURE 6.
Modulation of the physical association between ASK1 and its regulators (Trx and 14-3-3) or substrate (MKK3) by ZPR9. A and B, shown is the effect of ZPR9 on the association between ASK1 and its negative regulators, Trx and 14-3-3. HEK293 cells were transfected with the indicated combinations of expression vectors as well as ZPR9-specific siRNAs (#1 and #2). Cell lysates were then subjected to immunoprecipitation with anti-HA antibody or anti-ASK1 antibody, and the resulting immunoprecipitates were analyzed by immunoblot analysis with the indicated antibodies to determine the association between ASK1 and Trx (A) or 14-3-3 (B). C, the effect of ZPR9 on the association between ASK1 and its substrate MKK3 is shown. Complex formation between ASK1 and MKK3 was determined by immunoblotting with the indicated antibodies. The relative level of complex formation was quantitated by densitometric analysis and the -fold increase was calculated relative to controls expressing ASK1 and its interactors (Trx, 14-3-3, or MKK3) in the absence of ZPR9 or untransfected samples in parental HEK293 cells. D, endogenous association of ASK1 with its regulators (Trx, 14-3-3, MKK3, and ZPR9) in cells. Cell lysates from HEK293 cells treated with or without H2O2 were subjected to immunoprecipitation using anti-ASK1 antibody followed by immunoblot analyses with the indicated antibodies to determine the complex formation between endogenous ASK1 and its regulators (Trx, 14-3-3, MKK3, and ZPR9).
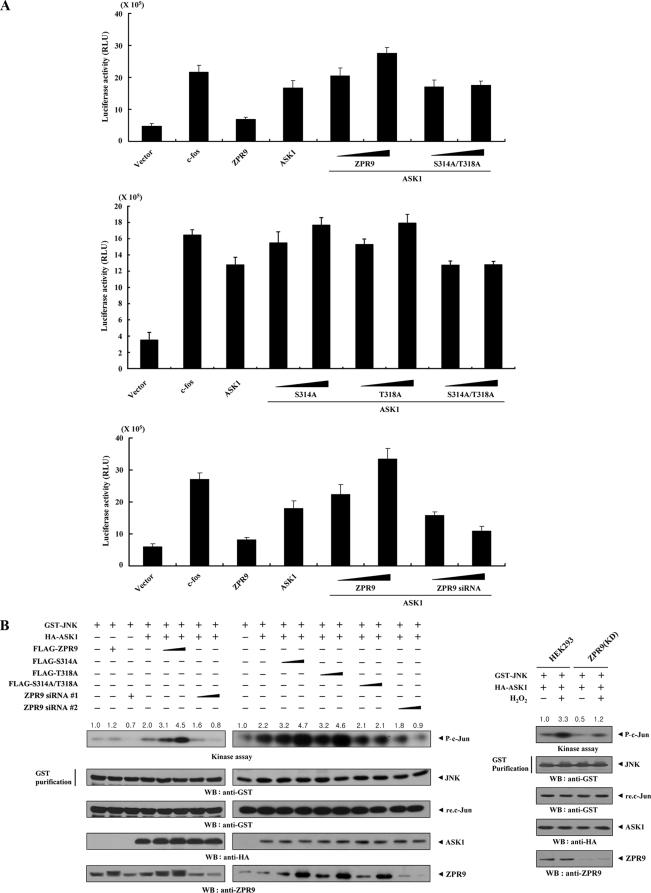
ZPR9 Enhances ASK1-induced AP-1 Transactivation in a Phosphorylation-dependent Manner
Because ZPR9 stimulates ASK1 activity through physical interaction (see Figs. 4 and 5), it is possible that ZPR9 can potentiate ASK1-induced AP-1 transcriptional activity. To examine this, reporter assays were performed using an AP-1 luciferase reporter gene. ZPR9 increased ASK1-induced AP-1 transcriptional activity in a dose-dependent manner, whereas the S314A/T318A mutant of ZPR9, which is defective in ASK1-mediated phosphorylation (see Fig. 3), had no effect (Fig. 7A, top and middle). This suggested that ZPR9 phosphorylation at Ser314 and Thr318 by ASK1 plays an important role in the ZPR9-mediated enhancement of ASK1-induced AP-1 transcriptional activity. Consistently, knockdown of endogenous ZPR9 decreased AP-1 transcriptional activity in a dose-dependent manner (Fig. 7A, bottom). Next, to investigate whether the ZPR9-mediated enhancement of AP-1 transcriptional activity is accompanied by ASK1-mediated JNK activation, in vitro kinase assays were also performed with c-Jun as a substrate to assess JNK activity. ZPR9 increased ASK1-mediated JNK activation in a dose-dependent manner, whereas the S314A/T318A mutant had no effect (Fig. 7B, left). This result was confirmed in a ZPR9 knockdown system induced by ZPR9-specific siRNAs. JNK activity was decreased in a dose-dependent manner by ZPR9 knockdown (Fig. 7B, left). A similar trend was also observed in HEK293 cells stably expressing an shRNA targeting ZPR9 (ZPR9(KD)) (Fig. 7B, right). Together, these data suggest a critical role for ASK1-mediated phosphorylation of ZPR9 at Ser314 and Thr318 in the ZPR9-mediated enhancement of ASK1-induced AP-1 transactivation.
FIGURE 7.
Stimulation of JNK-mediated transcription by ZPR9. A, enhancement of ASK1-mediated AP-1 transcriptional activity by ZPR9 is shown. In the presence or absence of c-Fos (0.6 μg), 293T cells were transfected with 0.2 μg of AP-1 luciferase plasmid and increasing amounts of wild-type and mutant forms (S314A, T318A, and S314A/T318A) of ZPR9 (0.2 and 0.6 μg) or ZPR9-specific siRNA #1 (50 and 200 nm) as indicated. RLU, relative light units. B, shown is stimulation of JNK kinase activity by ZPR9. HEK293 cells were cotransfected using GST-JNK (2 μg) and HA-ASK1 (3 μg) in the presence or absence of increasing amounts of wild-type and mutant forms (S314A, T318A, and S314A/T318A) of ZPR9 (4 and 8 μg) or ZPR9-specific siRNAs (each 100 and 200 nm). An in vitro kinase assay for JNK activity was performed as described previously (10). ZPR9(KD) cells transfected with GST-JNK and HA-ASK1 were also precipitated using glutathione-Sepharose beads (GST purification), and the precipitates were subjected to an in vitro kinase assay using recombinant c-Jun as a substrate to determine JNK kinase activity (right). The relative level of JNK kinase activity was quantitated by densitometric analysis, and the -fold increase was calculated relative to controls expressing JNK alone or untreated HEK293 cells expressing JNK and ASK1.
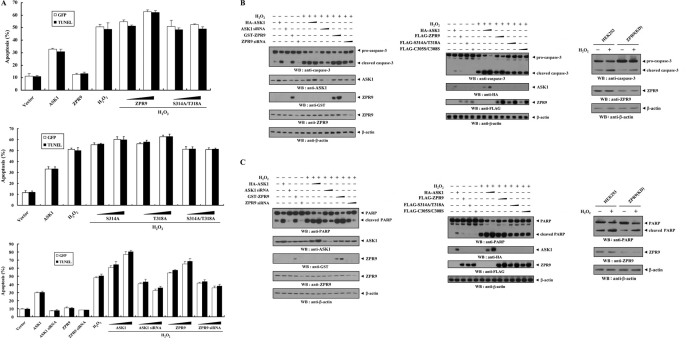
ZPR9 Stimulates H2O2-induced Apoptosis in a Phosphorylation-dependent Manner
To investigate whether ZPR9 can regulate cell death induced by an ASK1 stimulator, H2O2, the effect of ZPR9 on H2O2-induced cell death was examined using apoptosis assays as determined by GFP system and TUNEL staining (10, 18) in HEK293 cells. The results showed that wild-type ZPR9 increased H2O2-induced cell death in a dose-dependent manner (Fig. 8A, top, fourth lane versus fifth and sixth lanes). A dose-dependent increase in H2O2-induced cell death was also detected in the presence of the two ZPR9 single mutants, S314A and T318A (Fig. 8A, middle). However, no difference in H2O2-induced apoptosis was observed with the S314A/T318A double mutant compared with the control, which was treated with H2O2 in the absence of ZPR9 (Fig. 8A, top and middle). These results suggest that ASK1-mediated phosphorylation of ZPR9 at Ser314 and Thr318 is essential for the regulation of H2O2-induced apoptosis. The stimulatory role of ZPR9 in H2O2-induced cell apoptosis was confirmed using a ZRP9 knockdown system. Transfection of ZPR9-specific siRNA resulted in a dose-dependent decrease in H2O2-mediated apoptosis (Fig. 8A, bottom, sixth lane versus thirteenth and fourteenth lanes), suggesting that ZPR9 functions as a positive regulator of ASK1 that is involved in H2O2-induced apoptosis. Next, to investigate the ability of ZPR9 to induce caspase-3 activation and PARP cleavage, immunoblot analysis was also performed with anti-caspase-3 and anti-PARP antibodies, respectively, using HEK293 cells transfected with wild-type and mutant forms (S314A/T318A and C305S/C308S) of ZPR9 or ZPR9-specific siRNA as well as ZPR9(KD) cells stably expressing an shRNA targeting ZPR9. The results showed that ZPR9-mediated stimulation of H2O2-induced apoptosis involves in vivo caspase-3 activation, leading to proteolytic cleavage of PARP (Fig. 8, B and C).
FIGURE 8.
Stimulation of H2O2-induced apoptosis by ZPR9. A, HEK293 cells were transiently transfected with the indicated combinations of expression vectors encoding wild-type and mutant forms (S314A, T318A, and S314A/T318A) of ZPR9 (1 and 3 μg), wild-type ASK1 (1 and 3 μg), and ASK1- or ZPR9-specific siRNAs (100 and 200 nm) in the presence or absence of H2O2 (1 mm, 9 h). Apoptotic cell death was determined using the GFP expression system (GFP) or TUNEL (24). B, caspase-3 activation by ZPR9. HEK293 cells were transiently transfected with increasing amounts of wild-type and mutant (S314A/T318A and C305S/C308S) ZPR9 (0.8 and 1.6 μg), wild-type ASK1 (0.8 and 1.6 μg), and ASK1- or ZPR9-specific siRNAs (100 and 200 nm) in the presence or absence of H2O2. Cell lysates were then analyzed by immunoblot analysis with an anti-caspase-3 antibody to determine the degradation of caspase-3 (top panels). HEK293 cells stably expressing ZPR9-specific shRNA (ZPR9(KD)) were incubated with 2 mm H2O2 for 30 min. Cells were lysed and then subjected to immunoblot analysis using the indicated antibodies (right). C, stimulation of PARP degradation by ZPR9 is shown. HEK293 cells transfected with the indicated expression vectors were lysed and subjected to immunoblot analysis using the indicated antibodies. PARP degradation was also measured by immunoblotting with anti-PARP antibody in ZPR9(KD) cells treated with or without H2O2 (right).
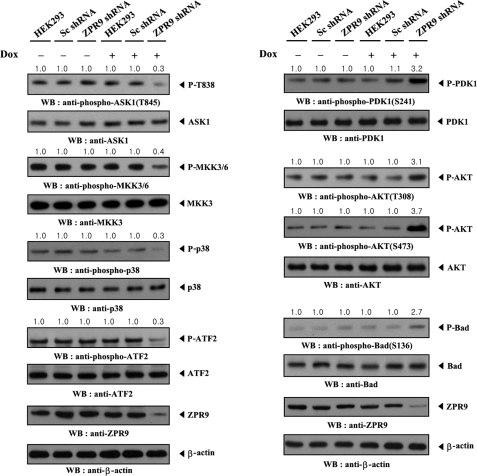
ZPR9 Potentiates ASK1-mediated Suppression of PDK1 Signaling
Given that ASK1 inhibits PDK1 activity through direct interaction and phosphorylation (24), it was next assessed whether PDK1-mediated signaling suppressed by ASK1 could be regulated by ZPR9 using an inducible ZPR9 knockdown system. As expected, the knockdown of endogenous ZPR9 markedly decreased signaling downstream of ASK1 as determined by immunoblot analysis using anti-phospho-specific antibodies for ASK1 Thr845, MKK3/6, p38, and ATF2 (Fig. 9, left). However, the opposite trend in PDK1-mediated signaling was observed upon knockdown of endogenous ZPR9 (Fig. 9, right). These results clearly indicate that, physiologically, ZPR9 acts as a positive regulator of ASK1 and subsequently leads to the inhibition of PDK1-mediated signaling.
FIGURE 9.
ZPR9 potentiates ASK1-mediated suppression of PDK1 signaling. HEK293 cells harboring stably integrated pSingle-tTS-shRNA vector containing scrambled shRNA (Sc shRNA) or ZPR9-specific shRNA (ZPR9 shRNA) were cultured in the presence or absence of 1 μg/ml doxycycline (Dox) for 72 h to determine the activities of ASK1, MKK3/6, p38, PDK1, or Akt as determined by immunoblot analysis. Inducible silencing of endogenous ZPR9 expression by doxycycline was assessed by immunoblotting using anti-ZPR9 antibody. β-Actin was used as a loading control.
DISCUSSION
In this study it was found that ZPR9, a physiological substrate of MPK38 serine/threonine kinase (19), physically interacted with ASK1 and stimulated ASK1 function. Furthermore, ZPR9 phosphorylation at Ser314 and Thr318 induced by ASK1 was critical for the regulation of ASK1-mediated signaling.
The finding that ZPR9 associated with ASK1 prompted us to investigate how ZPR9 regulates ASK1 activity. To this end, it was determined whether the direct interaction and phosphorylation between ASK1 and ZPR9 plays an important role in ZPR9-mediated regulation of ASK1 activity using in vitro kinase assays (Figs. 4 and 5), luciferase reporter assays (Fig. 7), and apoptosis analyses (Fig. 8). The results showed that the S314A/T318A double mutant of ZPR9, which is defective in ASK1-mediated phosphorylation (see Fig. 3), had no effect on ASK1-mediated signaling to both JNK and p38 kinases, whereas a marked stimulation of ASK1 function was detected in the presence of wild-type ZPR9. These data strongly indicate that ZPR9-mediated stimulation of ASK1 signaling is dependent on ZPR9 phosphorylation at Ser314 and Thr318 by ASK1. In addition, it is possible that ASK1-mediated phosphorylation of ZPR9 at Ser314 and Thr318 may have an effect on the physical association between ASK1 and ZPR9. To test this, an in vivo binding assay was performed with expression vectors for GST-ASK1 and FLAG-tagged wild-type and mutant forms (S314A/T318A and C305S/C308S) of ZPR9. The results showed that the S314A/T318A double mutant, like the C305S/C308S double mutant (see Fig. 2B), failed to form a complex with ASK1 (supplemental Fig. S1), further supporting that it has an important role for the direct phosphorylation of ZPR9 at Ser314 and Thr318 in the positive regulation of ASK1 signaling.
We previously showed that STRAP, a TGF-β receptor-interacting protein, physically interacts with ASK1 through cysteine residues, Cys152 and Cys270 of STRAP and Cys1351 and Cys1360 of ASK1, and inhibits ASK1 function (18). Meanwhile, our present results show that Cys305 and Cys308 of ZPR9 and Cys1351 and Cys1360 of ASK1 are responsible for complex formation (Fig. 2), indicating that both ZPR9 and STRAP utilize the same cysteine residues (Cys1351 and Cys1360) of ASK1 for their binding to ASK1. Based on this, it was investigated whether the direct interaction between ASK1 and ZPR9, a putative positive regulator of ASK1, can influence complex formation between ASK1 and its negative regulator, STRAP, using in vivo binding assays. As expected, a marked decrease in the association between ASK1 and STRAP was observed in the presence of wild-type ZPR9 compared with the control expressing ASK1 and STRAP in the absence of ZPR9. Meanwhile, the ZPR9 double mutants (C305S/C308S and S314A/T318A) that lack the ability to bind with ASK1 had no effect on the ASK1-STRAP association (supplemental Fig. S2). These results suggest that, in addition to the modulation of known regulators of ASK1, such as Trx and 14-3-3 (Fig. 6), ZPR9 stimulates ASK1-mediated signaling by destabilizing complex formation between ASK1 and STRAP, recently identified as a negative regulator of ASK1 (18), probably through competition with STRAP.
We previously found that ZPR9 was involved in TNF-α- or retinoic acid-induced apoptosis (22). However, the mechanisms by which ZPR9 induces apoptotic cell death remain unknown. To evaluate one possible explanation for this, the role of ASK1-mediated ZPR9 phosphorylation in the regulation of apoptosis induced by TNF-α or retinoic acid was investigated. The effect of wild-type ZPR9 on apoptosis in response to TNF-α or retinoic acid was compared with that in the presence of the S314A/T318A double mutant of ZPR9. The results showed that the S314A/T318A double mutant had no effect on apoptosis induced by the apoptotic stimuli TNF-α and retinoic acid when compared with the control in the absence of ZPR9 (supplemental Fig. S3). A similar trend was also observed in the analysis of apoptosis induced by other apoptotic stimuli tested, including H2O2, thapsigargin, ionomycin, and TGF-β (data not shown). These data suggest that ZPR9 phosphorylation at Ser314 and Thr318 may play an important role in the regulation of ZPR9-mediated apoptosis induced by various apoptotic stimuli. However, our present results do not rule out the possibility that ZPR9 phosphorylation at other sites may contribute to the regulation of ZPR9-mediated apoptosis. This is because ZPR9 was also phosphorylated by MPK38 serine/threonine kinase (data not shown), which was previously shown to participate in ASK1- and Smad-mediated signaling (10, 23). In this context, the further identification and characterization of potential phosphorylation sites of ZPR9 for other intracellular kinases may aid in the deciphering of the exact mechanism through which ZPR9 induces apoptotic cell death in response to apoptotic stimuli. In addition, the broad expression of ZPR9 in various human tissues and cell lines (19) together with its role in apoptosis may explain in part the phenotype of ZPR9 knock-out mice, which show embryonal lethality (data not shown). We imagine that the ZPR9-mediated inhibition of PDK1 signaling likely plays a role that is important in the ZPR9 null phenotype rather than the ZPR9-mediated MKK3/6 activation. Additionally, we speculate that ZPR9 directly and indirectly contributes to the inhibition of PDK1 signaling by regulating Akt phosphorylation at Thr308 and Ser473 through interference with PDK1 (Fig. 9) and mTORC2 (mammalian target of rapamycin complex 2; data not shown), respectively.
In summary, our studies indicate that ZPR9, previously identified as a MPK38-interacting protein (19), induces the activation of ASK1-mediated signaling to both JNK and p38 kinases and that the ASK1-mediated phosphorylation of ZPR9 at Ser314 and Thr318 is responsible for ASK1 activation. This potential role of ZPR9 as a novel stimulator of ASK1-mediated signaling may help to clarify the regulatory mechanism(s) of ASK1-mediated signaling in cells. Moreover, the present findings regarding ZPR9 phosphorylation-dependent regulation of apoptosis in response to various apoptotic stimuli tested in this study will increase our understanding of the putative mechanism(s) through which ZPR9 induces apoptotic cell death. However, to clarify this issue, it will be necessary to further investigate other possible functional links between ASK1 and other ZPR9-mediated signaling pathways as ASK1 signaling can associate with MPK38 signaling pathways in which ZPR9 acts as a physiological substrate.
Supplementary Material
This work was supported by National Research Foundation of Korea Grant R0A-2007-000-20006-0 and in part by Chungbuk National University Grant 2010.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- ASK1
- apoptosis signal-regulating kinase 1
- MPK38
- murine protein serine/threonine kinase 38
- PARP
- poly(ADP-ribose)polymerase
- ZPR9
- zinc finger-like protein 9
- AP-1
- activator protein 1
- Trx
- thioredoxin
- STRAP
- serine-threonine kinase receptor-associated protein
- PKB/Akt
- protein kinase B
- WB
- Western blot
- IP
- immunoprecipitate.
REFERENCES
- 1. Chang H. Y., Nishitoh H., Yang X., Ichijo H., Baltimore D. (1998) Science 281, 1860–1863 [DOI] [PubMed] [Google Scholar]
- 2. Ichijo H., Nishida E., Irie K., ten Dijke P., Saitoh M., Moriguchi T., Takagi M., Matsumoto K., Miyazono K., Gotoh Y. (1997) Science 275, 90–94 [DOI] [PubMed] [Google Scholar]
- 3. Saitoh M., Nishitoh H., Fujii M., Takeda K., Tobiume K., Sawada Y., Kawabata M., Miyazono K., Ichijo H. (1998) EMBO J. 17, 2596–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen Z., Seimiya H., Naito M., Mashima T., Kizaki A., Dan S., Imaizumi M., Ichijo H., Miyazono K., Tsuruo T. (1999) Oncogene 18, 173–180 [DOI] [PubMed] [Google Scholar]
- 5. Nishitoh H., Matsuzawa A., Tobiume K., Saegusa K., Takeda K., Inoue K., Hori S., Kakizuka A., Ichijo H. (2002) Genes Dev. 16, 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Liu Q., Wilkins B. J., Lee Y. J., Ichijo H., Molkentin J. D. (2006) Mol. Cell. Biol. 26, 3785–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gotoh Y., Cooper J. A. (1998) J. Biol. Chem. 273, 17477–17482 [DOI] [PubMed] [Google Scholar]
- 8. Liu H., Nishitoh H., Ichijo H., Kyriakis J. M. (2000) Mol. Cell. Biol. 20, 2198–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Matsuura H., Nishitoh H., Takeda K., Matsuzawa A., Amagasa T., Ito M., Yoshioka K., Ichijo H. (2002) J. Biol. Chem. 277, 40703–40709 [DOI] [PubMed] [Google Scholar]
- 10. Jung H., Seong H. A., Ha H. (2008) J. Biol. Chem. 283, 34541–34553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Song J. J., Rhee J. G., Suntharalingam M., Walsh S. A., Spitz D. R., Lee Y. J. (2002) J. Biol. Chem. 277, 46566–46575 [DOI] [PubMed] [Google Scholar]
- 12. Cho S. G., Lee Y. H., Park H. S., Ryoo K., Kang K. W., Park J., Eom S. J., Kim M. J., Chang T. S., Choi S. Y., Shim J., Kim Y., Dong M. S., Lee M. J., Kim S. G., Ichijo H., Choi E. J. (2001) J. Biol. Chem. 276, 12749–12755 [DOI] [PubMed] [Google Scholar]
- 13. Park H. S., Cho S. G., Kim C. K., Hwang H. S., Noh K. T., Kim M. S., Huh S. H., Kim M. J., Ryoo K., Kim E. K., Kang W. J., Lee J. S., Seo J. S., Ko Y. G., Kim S., Choi E. J. (2002) Mol. Cell. Biol. 22, 7721–7730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang L., Chen J., Fu H. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8511–8515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim A. H., Khursigara G., Sun X., Franke T. F., Chao M. V. (2001) Mol. Cell. Biol. 21, 893–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhan J., Easton J. B., Huang S., Mishra A., Xiao L., Lacy E. R., Kriwacki R. W., Houghton P. J. (2007) Mol. Cell. Biol. 27, 3530–3541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Morita K., Saitoh M., Tobiume K., Matsuura H., Enomoto S., Nishitoh H., Ichijo H. (2001) EMBO J. 20, 6028–6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jung H., Seong H. A., Manoharan R., Ha H. (2010) J. Biol. Chem. 285, 54–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Seong H. A., Gil M., Kim K. T., Kim S. J., Ha H. (2002) Biochem. J. 361, 597–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gil M., Yang Y., Lee Y., Choi I., Ha H. (1997) Gene 195, 295–301 [DOI] [PubMed] [Google Scholar]
- 21. Heyer B. S., Warsowe J., Solter D., Knowles B. B., Ackerman S. L. (1997) Mol. Reprod. Dev. 47, 148–156 [DOI] [PubMed] [Google Scholar]
- 22. Seong H. A., Kim K. T., Ha H. (2003) J. Biol. Chem. 278, 9655–9662 [DOI] [PubMed] [Google Scholar]
- 23. Seong H. A., Jung H., Ha H. (2010) J. Biol. Chem. 285, 30959–30970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seong H. A., Jung H., Ichijo H., Ha H. (2010) J. Biol. Chem. 285, 2397–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seong H. A., Jung H., Ha H. (2007) J. Biol. Chem. 282, 12075–12096 [DOI] [PubMed] [Google Scholar]
- 26. Jung H., Seong H. A., Ha H. (2008) J. Biol. Chem. 283, 20383–20396 [DOI] [PubMed] [Google Scholar]
- 27. Seong H. A., Jung H., Choi H. S., Kim K. T., Ha H. (2005) J. Biol. Chem. 280, 42897–42908 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.