Abstract
The Ca2+/calmodulin (CaM)-dependent protein kinase II (CaMKII) and the NMDA-type glutamate receptor are key regulators of synaptic plasticity underlying learning and memory. Direct binding of CaMKII to the NMDA receptor subunit GluN2B (formerly known as NR2B) (i) is induced by Ca2+/CaM but outlasts this initial Ca2+-stimulus, (ii) mediates CaMKII translocation to synapses, and (iii) regulates synaptic strength. CaMKII binds to GluN2B around S1303, the major CaMKII phosphorylation site on GluN2B. We show here that a phospho-mimetic S1303D mutation inhibited CaM-induced CaMKII binding to GluN2B in vitro, presenting a conundrum how binding can occur within cells, where high ATP concentration should promote S1303 phosphorylation. Surprisingly, addition of ATP actually enhanced the binding. Mutational analysis revealed that this positive net effect was caused by four modulatory effects of ATP, two positive (direct nucleotide binding and CaMKII T286 autophosphorylation) and two negative (GluN2B S1303 phosphorylation and CaMKII T305/6 autophosphorylation). Imaging showed positive regulation by nucleotide binding also within transfected HEK cells and neurons. In fact, nucleotide binding was a requirement for efficient CaMKII interaction with GluN2B in cells, while T286 autophosphorylation was not. Kinetic considerations support a model in which positive regulation by nucleotide binding and T286 autophosphorylation occurs faster than negative modulation by GluN2B S1303 and CaMKII T305/6 phosphorylation, allowing efficient CaMKII binding to GluN2B despite the inhibitory effects of the two slower reactions.
Keywords: Calcium Calmodulin-dependent Protein Kinase (CaMK); Glutamate Receptors Ionotropic (AMPA, NMDA); Protein Phosphorylation; Protein Targeting; Protein Translocation; Synapses
Introduction
The Ca2+/Calmodulin (CaM)3-dependent protein kinase II (CaMKII) is a major mediator of Ca2+ signaling in a large variety of cell types; however, it is best known for its functions in brain, where the CaMKII α isoform is expressed in levels up to 1% of total protein (for review, see Refs. 1–5). Glutamate is the major excitatory neurotransmitter in the mammalian brain, and CaMKII mediates long term potentiation (LTP) of excitatory glutamatergic synapse strength (6, 7), a form of synaptic plasticity thought to be crucial for learning and memory (for review see Ref. 1, 8). Specifically, CaMKII is activated by Ca2+ influx through the NMDA-type glutamate receptor, allowing the kinase to potentiate AMPA-type glutamate receptor currents, likely both by increasing the number or receptors at the synapse (9, 10) and by increasing their single channel conductance (11, 12). LTP-inducing NMDA-receptor stimulation also leads to CaMKII translocation to excitatory synapses (13–17), which is thought to be mediated by a Ca2+/CaM-induced direct binding of CaMKII to the NMDA-receptor subunit GluN2B (for review see Refs. 4, 5). Induction of LTP requires CaMKII T286 autophophosphorylation (18, 19), a reaction that occurs between two Ca2+/CaM-stimulated kinase subunits within the 12meric CaMKII holoenzyme (20, 21) and generates “autonomous” activity, i.e. partial kinase activity (∼20%) toward most substrates even after dissociation of Ca2+/CaM (22–25). T286 autophosphorylation also induces CaMKII binding to GluN2B in vitro (26, 27); however, T286 phosphorylation is not required, as Ca2+/CaM stimulation alone is sufficient to induce CaMKII binding to GluN2B (15, 17). This interaction of CaMKII with GluN2B is indeed important in regulation of synaptic strength (28–30). Binding to GluN2B keeps CaMKII in an active conformation, which allows phosphorylation of GluN2B even after the initial Ca2+ stimulus has subsided, and even when T286 is no longer phosphorylated (15, 17, 31, 32). In turn, CaMKII activity is thought to regulate NMDA-receptor currents (33–35). Remarkably, the major CaMKII phosphorylation site on GluN2B, S1303 (36), is located within the major CaMKII binding site on the receptor (15, 27, for review see 4, 5), and S1303 phosphorylation has been shown to interfere with CaMKII binding (27, 37). Conditions that would induce CaMKII binding to GluN2B in cells (CaMKII activation by Ca2+/CaM and/or by T286 autophosphorylation) should also trigger GluN2B S1303 phosphorylation by CaMKII, which in turn should prevent the binding (27). How, then, can CaMKII actually bind to GluN2B within cells, where ATP concentration is high? To address this apparent conundrum, we added ATP to our in vitro binding reactions. Remarkably, ATP actually enhanced Ca2+/CaM-induced binding of CaMKII to GluN2B. Further studies revealed that this positive net effect was the result of four modulatory effects of ATP, two positive (directly by nucleotide binding and indirectly after CaMKII autophosphorylation at T286) and two negative (GluN2B phosphorylation at S1303 and CaMKII autophosphorylation at T305/6). Previous localization studies in cells using corresponding CaMKII or GluN2B mutants indicate biological relevance of several of the regulatory effects of ATP shown here (17, 37, 38). The importance of nucleotide binding for CaMKII translocation to GluN2B both in transfected HEK cells and in primary hippocampal neurons was confirmed by comparing the nucleotide binding-impaired CaMKII mutant K42M with both CaMKII wild type and T286A.
EXPERIMENTAL PROCEDURES
Protein Purification
CaM was purified after bacterial expression, and CaMKIIα was purified from a baculovirus/Sf9 cell expression system (15, 39). GFP-CaMKII wild type and mutants were expressed in HEK 293 cells, and either used in raw extracts (17) (in Fig. 6 and supplemental Fig. S2) or after the same purification method used for unlabeled CaMKII. The GFP protein used for CaMKII fusion was an A207K mutant of EGFP, to eliminate residual GFP dimerization (17, 40), which is of special importance when studying homo-multimeric proteins such as CaMKII. GST-N2B-c, a fusion protein of GST with the cytoplasmic C terminus of GluN2B (amino acids 1,120–1,482), was expressed in bacteria (15) and batch purified using glutathione-Sepharose 4B (GE Healthcare) according to manufacturer's instructions. GST-N2B-c S1303 mutants were generated by mutagenesis using QuikChange (Agilent), and purified in the same manner.
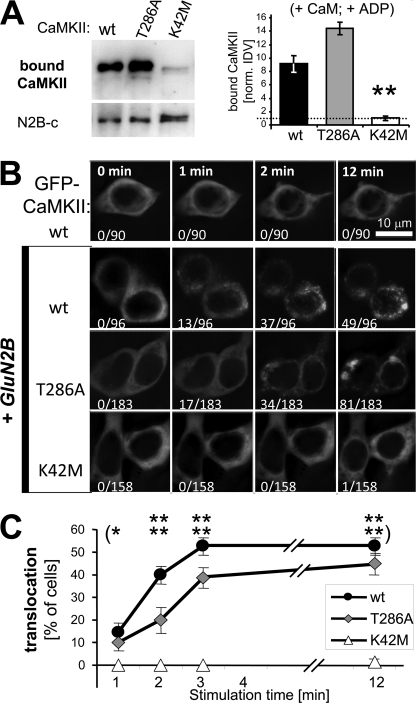
FIGURE 6.
Nucleotide binding is required for stimulus-induced translocation of GFP-CaMKII in heterologous cells co-expressing GluN2B. A, K42M mutation (which impairs nucleotide binding) significantly reduced Ca2+/CaM stimulated binding to GluN2B in vitro (100 μm ADP added), compared with both GFP-CaMKII wild type and T286A, as determined by Western blot analysis of the protein complex (**, p < 0.001; Neuman-Keuls multiple comparison test, after one-way ANOVA). B, GFP-CaMKII localization was monitored in live HEK cells (with or without co-expressed GluN2B) before (0 min) and during an ionomycin induced Ca2+ stimulus (1–12 min). Translocation was seen only after co-expression with GluNRB, and was almost completely abolished by the K42M mutation. Number of cells that show translocation and total number of cells monitored are indicated. C, quantification of the GFP-CaMKII translocation experiment shown in panel B. The mean percentage (± S.E.) of cells showing translocation during each independent stimulation (n = 5; for wild type n = 4) is plotted. The T286A mutant translocated significantly less than wild type after 2 and 3 min of stimulation (p < 0.05) but reached the same level after 12 min. The K42M mutant showed almost no translocation at all, even after 12 min. Statistical difference to T286A (and wild type) translocation is indicated (*, p < 0.05; **, p < 0.001; in Bonferroni post hoc analysis after two-way ANOVA).
Western Blot Analysis
Protein separation, transfer onto PVDF membrane, and immunodetection was done as described (41), using antibodies selective for CaMKIIα (CBα2), for phospho-T305/6 CaMKII (PhosphoSolution), or for GST (Millipore; for detection of the GST-N2B fusion protein). Chemoluminescence detection by Western Lightning (Perkin Elmer) was visualized in a ChemiImager (Alpha Innotech), and the “immuno detection values” (IDV) were quantified using Image J software (after background subtraction). For comparing blots from multiple experiments, the IDVs were normalized, generally to the amount of CaMKII wild type bound in absence of nucleotide.
CaMKII Binding to GluN2B in Vitro
CaMKII/GluN2B binding assays were done as described (15, 17, 41). GST-GluN2B fusion proteins (GST-N2B) were immobilized on anti-GST-antibody-coated microtiter plates (Thermo Scientific), blocked for 30 min with 5% BSA, and then overlaid with 40 nm CaMKII (subunit concentration) in PIPES-buffered saline (pH 7.2) for 15 min at room temperature. After extensive washes in buffer containing 1 mm EGTA, GST-N2B and bound CaMKII was eluted for 12 min in SDS-loading buffer at 95 °C. Addition of Ca2+/CaM (1 mm/1 μm) induces persistent CaMKII binding under these conditions (17). Other conditions tested for binding were: pre-T286-autophosphorylated CaMKII (25) in presence of 1 mm EGTA, and addition of 100 μm (or 2 mm) ATP, ADP, or AMP-PNP. In assays testing the effect of these nucleotides, all parallel conditions additionally contained 10 mm MgCl.
Live Imaging of HEK Cells
HEK cells were grown and transfected with expression vectors for GFP-CaMKII mutants and/or GluN2B (15, 17). For co-expression, the ratio of vector amounts was 10:1 GluN2B to kinase, to assure that all transfected cells identified by GFP-CaMKII fluorescence contain GluN2B. GFP-CaMKII translocation in response to a Ca2+ stimulus induced by 10 μm ionomycin was monitored for 12 min at 32 °C in imaging buffer (0.87× Hanks Balanced Salt Solution, 25 mm Hepes pH 7.4, 2 mm glucose, 2 mm CaCl2, 1 mm MgCl2) by fluorescence microscopy. Images were aquired on a Zeiss Axiovert 200 m equipped with a climate control chamber (41), using SlideBook software (Intelligent Imaging Innovations).
Immunocytochemistry
HEK cells co-expressing GFP-CaMKII and GluN2B were treated as for live imaging, but were fixed and stained for GluN2B as described (15) after 5 min of ionomycin stimulation. Projection images were created from deconvoluted z-stacks and analyzed for mean fluorescence intensity (to compare relative levels of GluN2B and GFP-CaMKII expression between the CaMKII mutants) and for correlation of co-localization (using SlideBook software as described; 42).
Live Imaging of Hippocampal Neurons
Medium density cultures of hippocampal neurons were prepared from newborn rat pups, and transfected on day in vitro (DIV) 12 using Lipofectamine 2000, as described (17, 43). Live imaging was done as for HEK cells on DIV 13, but neurons were stimulated instead by glutamate/glycine (100 μm/10 μm). Z-stacks were acquired prior to glutamate stimulation then again at 4 and 10 min after stimulation. Single plane images were acquired at 30-s intervals for all other time points.
RESULTS
The Phospho-mimetic GluN2B S1303D Mutation Inhibits CaMKII Binding
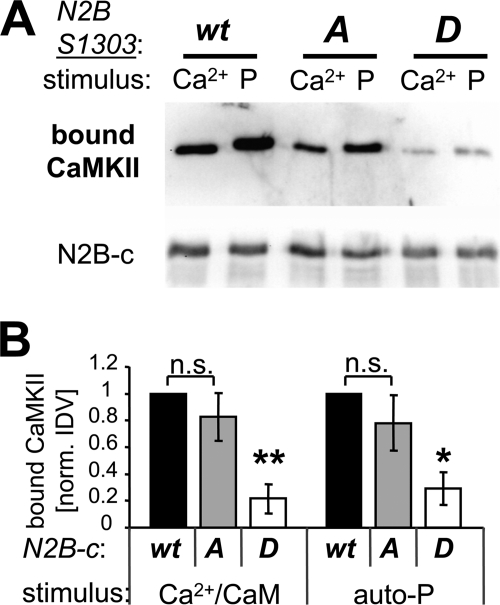
S1303 phosphorylation has been demonstrated to inhibit phospho-T286 CaMKII binding to GluN2B in absence of Ca2+/CaM (27), however, it has recently been suggested that it may not affect Ca2+/CaM-induced CaMKII binding (37). Thus, we compared phospho-T286- and Ca2+/CaM-induced binding of purified CaMKII to purified GST-fusion proteins of the GluN2B cytoplasmic C terminus (GST-N2B-c; amino acids 1,120–1,482) and its mutants S1303A (cannot be phosphorylated) and S1303D (phospho-mimetic) (Fig. 1). For the in vitro binding assays, GST-N2B-c was immobilized on anti-GST-antibody coated microplates and then overlaid with CaMKII for 15 min at room temperature (which induces maximal persistent binding; 17). After extensive washes in presence of EGTA, the complexes were eluted and analyzed by Western blot (Fig. 1A). Quantification of such experiments showed that the phospho-mimetic GluN2B S1303D mutation significantly inhibited CaMKII binding, both when binding was induced by T286-autophosphorylation only (i.e. after subsequent chelation of Ca2+) or by Ca2+/CaM only (i.e. in absence of any ATP) (Fig. 1B). By contrast, mutation of GluN2B S1303 to A did not significantly reduce CaMKII binding (Fig. 1B). Thus, taken together, the phospho-mimetic S1303D mutation of GluN2B reduces any persistent CaMKII binding independent of the inducing stimulus, which is consistent with the proposed model of the CaMKII/GluN2B interaction (15, 17; see also Fig. 9).
FIGURE 1.
The phospho-mimetic GluN2B S1303D mutation impairs CaMKII binding to GluN2B. A, binding of purified CaMKIIα to immobilized GST-N2B-c (a GST-fusion with the cytoplasmic C terminus of GluN2B) was induced by Ca2+/CaM (Ca2+) or by pre-autophosphorylation at T286 (P), both for N2B-c wild type and S1303A, but to a much lesser extent for the phospho-mimetic S1303D mutant. Bound CaMKII (and immobilized GST-N2B-c) was eluted and detected by Western blot analysis. B, quantification of binding experiments as shown in panel A (n = 3). CaMKII bound similarly to N2B-c wild type and S1303A (n.s.) but significantly less to S1303D (**, p < 0.01; *, p < 0.05; in Neuman-Keuls multiple comparison test, after one-way ANOVA), both for CaM-induced and phospho-T286-induced binding (each normalized for binding to N2B wild type; IDV of phospho-T286-induced binding was 1.3–4.0-fold higher in the individual experiment series). Error bars show S.E.
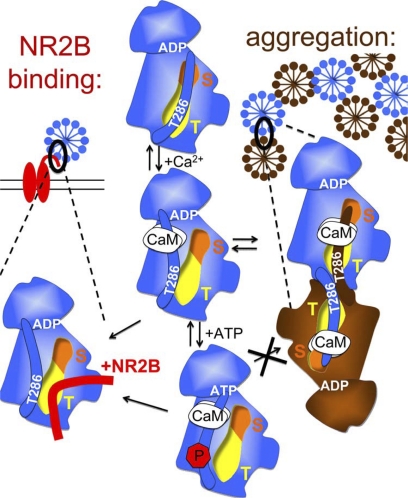
FIGURE 9.
Schematic model: CaMKII binding to GluN2B (left) and aggregation of multiple CaMKII holoenzymes (right) are based on similar molecular interaction mechanisms. Displacement of the auto-inhibitory α-helix (which includes the region around T286) by Ca2+/CaM binding activates the kinase (by allowing substrate access to the S-site; orange) and exposes the T-site (yellow), which then can interact with other binding partners: either the region around S1303 in GluN2B (red) or the T286 region of a kinase subunit on another holoenzyme (brown). Both interactions are enhanced by nucleotide binding to CaMKII. However, while T286 autophosphorylation further enhances binding to GluN2B, it inhibits aggregation of holoenzymes.
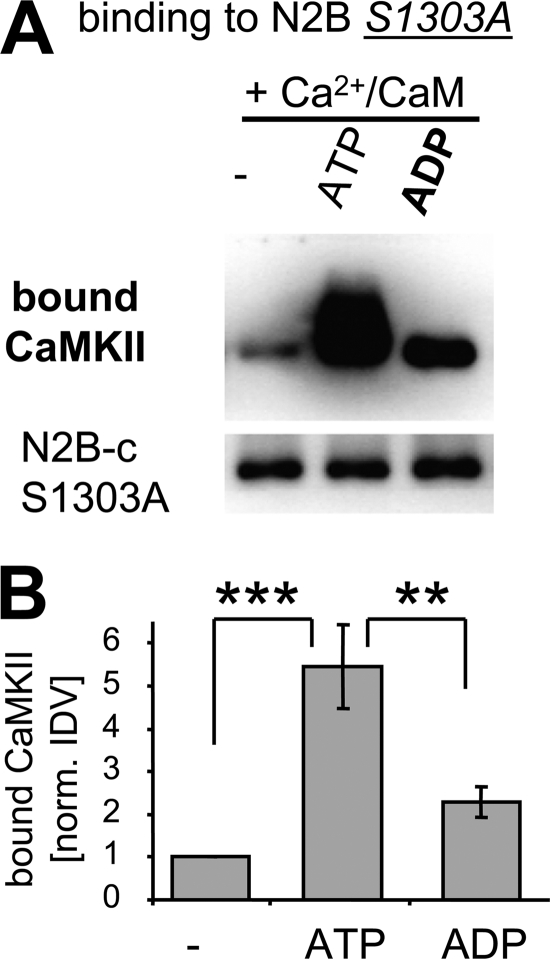
Both ATP and ADP Enhance CaMKII Binding to GluN2B
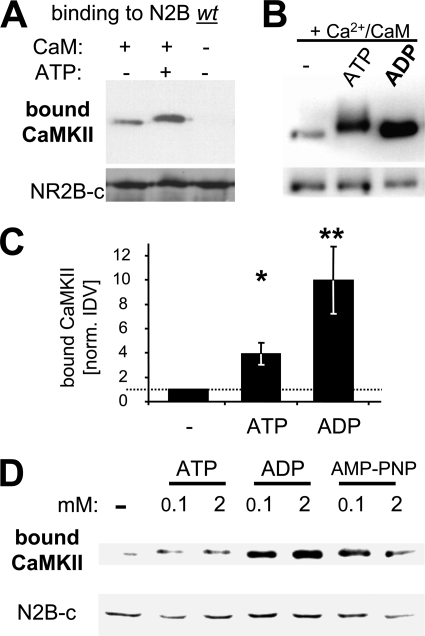
If phosphorylation of S1303, the major CaMKII phosphorylation site on GluN2B, inhibits CaMKII binding to GluN2B, can this binding actually occur in presence of ATP? Surprisingly, addition of ATP (100 μm; >10-fold kDa for CaMKII) actually enhanced Ca2+/CaM-induced CaMKII binding to immobilized GST-N2B-c in vitro (Fig. 2A). A similar effect has been reported previously for ADP and non-hydrolyzable ATP homologues, i.e. for nucleotides that do not support phosphorylation reactions (31). Direct comparison between nucleotides showed that Ca2+/CaM-induced CaMKII binding was even more strongly enhanced by ADP compared with ATP (∼2.5-fold further increase in detected bound CaMKII in presence of ADP compared with ATP, ∼10-fold compared with no nucleotide)(Fig. 2, B and C). The same effect as with ADP was also seen with the non-hydrolyzable ATP analog AMP-PNP (Fig. 2D). The nucleotides reached their maximal effect at a concentration of 100 μm, as increasing their concentrations to 2 mm (which is within the range of the high nucleotide concentration in cells) did not appear to further enhance CaMKII binding to GluN2B (Fig. 2D). By contrast, AMP did not increase CaMKII binding to GluN2B (supplemental Fig. S1), consistent with the >500-fold lower affinity of AMP compared with ADP or ATP described for other kinases (44–46). Thus, taken together, nucleotide binding to CaMKII enhances Ca2+/CaM-induced CaMKII binding to GluN2B, without requirement of a phosphorylation reaction. The results also indicate an additional, negative-regulatory effect of ATP (likely mediated by a phosphorylation reaction), which reduces the enhancement by nucleotide.
FIGURE 2.
ATP and other nucleotides enhance Ca2+/CaM-induced CaMKII binding to GluN2B. A, addition of ATP (100 μm) enhanced Ca2+/CaM-induced CaMKII binding to GluN2B in vitro, as determined by Western blot analysis of the protein complex. B, addition of ADP (100 μm) enhanced Ca2+/CaM-induced CaMKII binding to GluN2B even more than ATP (without the slight band shift caused by autophosphorylation in presence of ATP). C, quantification of experiments as shown in panels A and B (n = 6; for ADP n = 3)). The significantly increased CaMKII binding in presence of ATP (*, p < 0.05) was significantly further increased when ADP was present instead (**, p < 0.01; in Neuman-Keuls multiple comparison test, after one-way ANOVA). Error bars show S.E. D, increasing the amount of nucleotide (ATP, ADP, or AMP-PNP) from 100 μm to 2 mm did not increase CaMKII binding to GluN2B any further, as determined by Western blot analysis.
GluN2B S1303 Phosphorylation Negatively Regulates CaMKII Binding
If GluN2B phosphorylation at S1303 is responsible for the observation that CaMKII binding to GluN2B is enhanced less by ATP than by ADP (see Fig. 2, B and C), mutation of GluN2B S1303 to A should alleviate the differential effect of the two nucleotides. Indeed, the increase of Ca2+/CaM-stimulated CaMKII binding to GluN2B induced by ATP was even more pronounced for the GluN2B S1303A mutant compared with GluN2B wild type (Fig. 3, A and B). In fact, for the GluN2B S1303A mutant, ATP enhanced CaMKII binding even more strongly than ADP did (Fig. 3, A and B), opposite to the observations for GluN2B wild type (see Fig. 2, B and C). Thus, GluN2B S1303 phosphorylation indeed negatively regulates CaMKII binding and is responsible for the lesser enhancement of CaMKII binding by ATP compared with ADP. Additionally, the results indicated that there may be an additional phosphorylation reaction that positively regulates CaMKII binding.
FIGURE 3.
GluN2B S1303A mutation reverses the relative effect of ATP and ADP on enhancing CaMKII binding. A, CaMKII binding to GluN2B S1303A mutant was also further enhance by nucleotide, but more strongly by ATP compared with ADP, as determined by Western blot analysis of the protein complex. B, quantification of experiments as shown in panel A (n = 4). For the GluN2B 1303A mutant, the increased CaMKII binding by nucleotide was significantly greater of ATP (***, p < 0.001) compared with ADP (**, p < 0.01; in Neuman-Keuls multiple comparison test, after one-way ANOVA). Error bars show S.E.
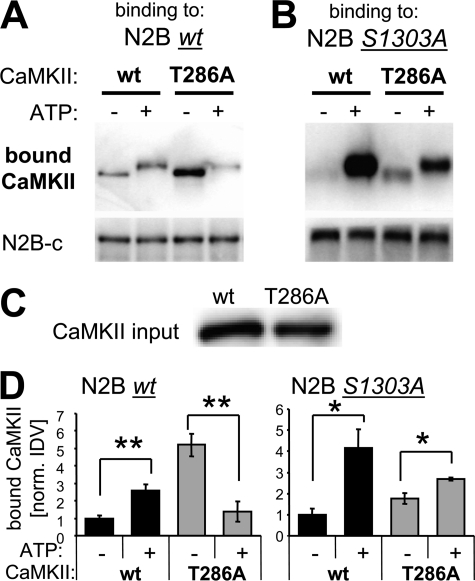
CaMKII T286 Phosphorylation Positively Regulates Binding to GluN2B
A good candidate for the positive-regulatory phosphorylation site in the CaMKII binding to GluN2B (as indicated by the studies using the GluN2B S1303A mutant; see Fig. 3) was T286 on CaMKII, an autophosphorylation site that enhances CaM binding (47) and generates autonomous activity (22–24). Thus, we compared CaMKII wild type with a T286A mutant (Fig. 4A). If T286 is indeed a positive-regulatory site, then ATP should enhance Ca2+/CaM-induced CaMKII binding to GluN2B less for a CaMKII T286A mutant than for CaMKII wild type. Indeed, for the T286A mutant, ATP showed not only less enhancement of the binding, but actually reduced it significantly (Fig. 4B). Thus, lack of positive-regulatory phosphorylation at T286 unmasked a negative-regulatory phosphorylation that becomes dominant for the CaMKII T286A mutant. GluN2B S1303 was already identified as a logical candidate for this negative-regulatory phosphorylation (see above). Indeed, a S1303A mutation prevented the negative effect of ATP on CaMKII T286A binding to GluN2B (Fig. 4C); the S1303A mutation even partially restored the positive effect of ATP also for the CaMKII T286A mutant (Fig. 4C), consistent with the direct positive effect of nucleotide binding indicated by our initial experiments (see Fig. 2). Statistical significance of the effects was demonstrated after quantification (Fig. 4D). Taken together, these results demonstrate that T286 autophosphorylation has a strong positive-regulatory effect on Ca2+/CaM-induced CaMKII binding to GluN2B in presence of ATP, and provide further evidence for positive regulation also by direct nucleotide binding.
FIGURE 4.
ATP enhances CaMKII binding to GluN2B, directly and by inducing T286 autophosphorylation. GFP-CaMKII purified after over-expression in HEK 293 cells was used for these binding experiments. A, increase in CaMKII binding to GluN2B wild type caused by ATP was not only abolished but even reversed for the CaMKII T286A mutant, as determined by Western blot analysis of the protein complex. B, an increase in CaMKII binding to GluN2B S1303 caused by ATP was also seen for the CaMKII T286A mutant, but to a lesser extent than for CaMKII wild type. C, similar amounts of GFP-CaMKII wild type and T286A mutant were used in the binding experiments, as determined by Western blot analysis of the input mix of the binding reaction. D, quantification of experiments as shown in panels A (n = 4) and B (n = 3). Binding to GluN2B wild type was significantly enhanced by ATP for CaMKII wild type, but significantly reduced for CaMKII T286A (**, p < 0.005; t test). By contrast, binding to GluN2B S1303A was enhanced by ATP for both CaMKII wild type and the T286A mutant (*, p < 0.05; t test). Error bars show S.E.
GFP-CaMKII fusion protein purified after overexpression in HEK cells were used for the experiments with CaMKII mutants in Fig. 4. Unpurified GFP-CaMKII from raw HEK cell extracts also showed Ca2+/CaM-induced binding to GluN2B in the same in vitro assay (supplemental Fig. S2). However, addition of exogenous ATP failed to further enhance binding of CaMKII from these extracts (supplemental Fig. S2), likely because of significant nucleotide presence in the extracts (∼40–80 μm in the binding reaction, based on extract dilution and assumption of a ∼4 mm cytosol concentration).
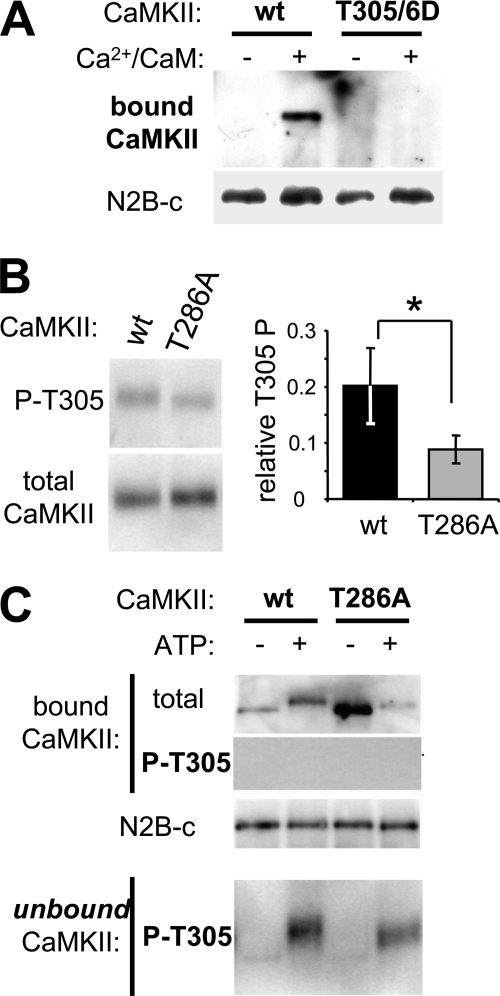
CaMKII T305/306 Autophosphorylation Negatively Regulates Binding to GluN2B
CaMKII autophoshorylation at T305/306 inhibits Ca2+/CaM binding to CaMKII (48–50), which in turn would be expected to interfere with Ca2+/CaM-induced CaMKII binding to GluN2B. Indeed, the T305/306D mutation, which mimics autophosphorylation at these residues, failed to bind to GluN2B in vitro (Fig. 5A), as expected. Thus, the reduced binding of CaMKII T286A to GluN2B after addition of ATP could be explained in part by T305/6 phosphorylation (in addition to the effect caused by GluN2B S1303 phosphorylation demonstrated in Fig. 4), if this mutant is more easily phosphorylated at T305/6 than wild type. However, this was not the case: If any, the T286A mutant autophosphorylates T305/6 to a lesser extend than wild type CaMKII (Fig. 5B). During the GluN2B binding reactions in presence of ATP, both CaMKII wild type and T286A became at least partially autophosphorylated T305/6, as detected in the un-bound supernatant of the binding reaction (Fig. 5C). However, no T305/6 phosphorylation was detected in the bound kinase fraction (Fig. 5C), further indicating that this inhibitory phosphorylation indeed interferes with CaMKII binding to GluN2B.
FIGURE 5.
T305/6 autophosphorylation inhibits CaMKII binding to GluN2B. A, Ca2+/CaM stimulated in vitro GluN2B binding of GFP-CaMKII wild type but not of its T305/6D phospho-mimetic mutant, as determined by Western blot analysis of the protein complex. B, Ca2+/CaM stimulation induced more T305 autophosphorylation in GFP-CaMKII wild type compared with its T286A mutant (*, p < 0.05, t test), as determined by quantification (right; n = 3) of Western blot analysis with phospho-T305 selective antibody (left). C, in a GFP-CaMKII binding reaction to immobilized N2B, CaMKII autophosphorylation at T305 was detected only in the unbound fraction, not in the N2B-bound CaMKII, as detected by Western blot analysis of the protein complex and the unbound supernatant.
CaMKII K42M Mutation Affects Nucleotide-enhanced but Not Basal GluN2B Binding
Presumably, nucleotide-enhancement of CaMKII binding to GluN2B is mediated by nucleotide binding to the nucleotide binding pocket of the kinase. This was directly tested using a CaMKII K42M mutation that impairs nucleotide interaction with this binding pocket. Indeed, in biochemical GluN2B binding assay in presence of ADP (100 μm added), GFP-CaMKII K42M showed much less Ca2+/CaM-induced binding than wild type (Fig. 6A). The difference in binding observed between K42M and wild type (in presence of ADP) was essentially the same as the difference observed between wild type in absence and presence of ADP (Fig. 6A; compared with Fig. 2C). Indeed, in absence of nucleotide, GFP-CaMKII wt and K42M showed the same level of Ca2+/CaM-induced binding to GluN2B (supplemental Fig. S3). Thus, the effect of the K42M mutation on GluN2B binding is mediated by its lack of nucleotide binding and not by other structural impairments.
Nucleotide Dependence of CaMKII Interaction with GluN2B in HEK Cells
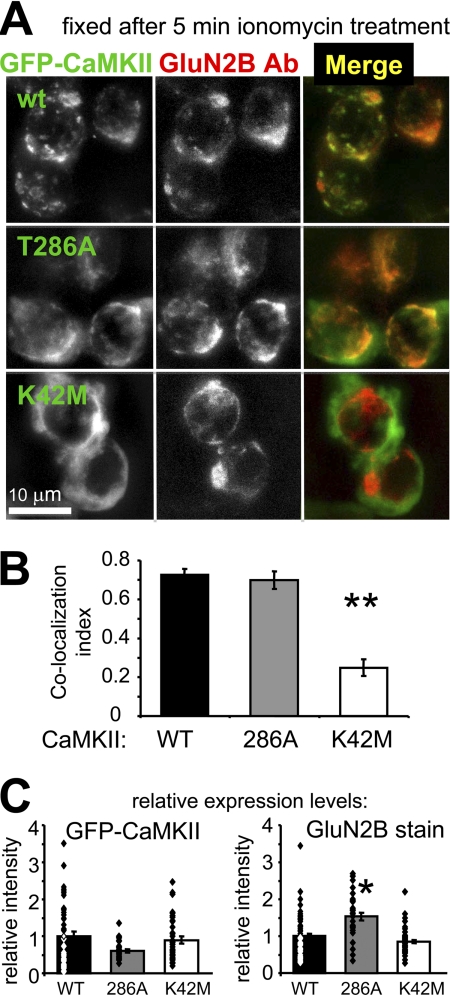
To examine the role of nucleotide binding to CaMKII in cells, we utilized the nucleotide binding-impaired K42M mutant. In transfected HEK cells, the K42M mutation almost completely abolished Ca2+-induced GFP-CaMKII translocation in cells co-expressing GluN2B (Fig. 6B). For these experiments, GFP-CaMKII wild type and mutants were co-expressed with full-length GluN2B. Cells were monitored live by GFP fluorescence microscopy before and during a Ca2+ stimulus induced by ionomycin. Without GluN2B, no CaMKII translocation was observed under the conditions used, even after 12 min of continuous stimulation (Fig. 6B). CaMKII K42M translocated only in 1 out of 158 cells even after 12 min of stimulation, even in presence of GluN2B (compared with 49 of 96 and 81 of 183 for CaMKII wild type and T286A, respectively; Fig. 6B). The time course of translocation was faster for CaMKII wild type compared with T286A, as a two-way ANOVA with Bonferoni post-hoc analysis showed significantly more translocation of wild type after 2 and 3 min of stimulation (Fig. 6C). However, translocation of CaMKII wild type and T286A eventually reached the same level after 12 min of stimulation (Fig. 6C). Significantly stronger translocation for both CaMKII wild type and T286A compared with K42M was immediately obvious, as K42M showed almost no translocation at all (Fig. 6C).
To test if CaMKII translocated indeed to the localization of GluN2B, HEK cells were fixed after 5 min of ionomycin stimulation and immunostained for GluN2B (Fig. 7A). Indeed, co-localization of GFP-CaMKII wild type and T286A, but not K42M, was apparent visually (Fig. 7A) and was confirmed quantitatively by Pearson's correlation of co-localization (Fig. 7B). Both GFP-CaMKII and GluN2B expression levels were undistinguishable between CaMKII wild type and K42M, as quantified based on fluorescence intensity (Fig. 7C), demonstrating that the difference in translocation was not caused by differences in expression levels. The expression levels in T286A-transfected cells also fell within the same range, even though the mean showed statistically significant differences (Fig. 7C).
FIGURE 7.
Expression and co-localization of GFP-CaMKII mutants and GluN2B in heterologous cells after stimulation. A, in HEK cells stimulated with ionomycin (10 μm, 5 min), GFP-CaMKII wild type, and T286A, but not K42M, co-localized with co-expressed GluN2B, detected by immunocytochemistry after fixation. B, quantification of co-localization experiments as shown in panel A (n = 22–23). GFP-CaMKII wild type and T28A showed similar co-localization with GluN2B, while K42M co-localized significantly less (**, p < 0.001 in Neuman-Keuls multiple comparison test, after one-way ANOVA). C, quantification of GFP-CaMKII and GluN2B-immunodetection fluorescence intensity showed that both GFP-CaMKII wild type and T286A expressed to a similar level as the K42M mutant; the level of co-expressed GluN2B was similar for CaMKII wild type and K42M, but slightly higher for T286A (*, p < 0.01; all in Neuman-Keuls multiple comparison test, after one-way ANOVA). Error bars show S.E. in all panels.
Nucleotide Dependence of Synaptic CaMKII Translocation in Hippocampal Neurons
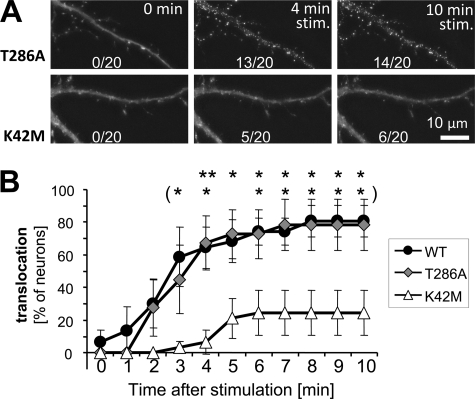
The hippocampus is a brain region important in spatial learning and memory. Glutamate stimulation causes GFP-CaMKII to translocate to post-synaptic sites in hippocampal neurons in dissociated culture (13–15, 17), likely mediated by regulated CaMKII binding to GluN2B (15, 17). To test the effect of nucleotide binding to CaMKII on this translocation, we compared translocation of GFP-CaMKII K42M (nucleotide binding-impaired) to T286A (autophosphorylation incompetent) and wild type in hippocampal neurons in primary culture, during stimulation with glutamate/glycine (100 μm/10 μm) (Fig. 8). As the K42M mutant fails to autophosphorylate, and because T286 phosphorylation enhanced CaMKII binding to GluN2B (see Fig. 4) and increases persistent synaptic translocation (17), comparison to CaMKII T286A in addition to wild type was important. However, no significant difference between translocation of CaMKII wild type and T286A was observed during continuous glutamate stimulation (Fig. 8). This is consistent with previous reports that found rather subtle effects of the T286A mutation, which did not manifest in acute translocation during stimulation (13, 15, 17) but mainly in persistence of localization after removal of the stimulus (17). By contrast, the nucleotide binding deficient K42M mutant showed significantly less stimulation-induced translocation than both GFP-CaMKII wild type and the T286A mutant, even after 5–10 min of continuous stimulation (Fig. 8). Thus, nucleotide binding is required for efficient synaptic translocation of CaMKII in neurons, consistent with its effects on Ca2+/CaM-induced binding of CaMKII to GluN2B in vitro and in heterologous.cells.
FIGURE 8.
Nucleotide binding is required for efficient glutamate-induced synaptic translocation of GFP-CaMKII in hippocampal neurons. A, GFP-CaMKII localization was monitored in live rat hippocampal neurons (DIV 13) before and during a 10 min glutamate/glycine (100 μm/10 μm) stimulus. Translocation to synaptic sites was seen for both wild type and T286A CaMKII within 4 min of stimulation, whereas K42M did not display this pattern of localization even after 10 min of stimulation. Shown are sample projection images created from deconvolved z-stacks of the same neuron at different time points. B, translocation in time lapse images was quantified (for 20 neurons in six independent experiments for each condition). CaMKII wild type and the T286A mutant showed the same translocation, while K42M translocated significantly less than T286A (and wild type) as indicated (*, p < 0.05; **, p < 0.01; in Bonferroni post-hoc analysis after two-way ANOVA).
DISCUSSION
CaMKII binding to the NMDA receptor subunit GluN2B is thought to be important for CaMKII translocation to synapses in response to LTP-inducing stimuli and, in turn, for regulating synaptic plasticity (15, 26, 28; for review see Ref. 4, 5). This study showed that ATP significantly increases Ca2+/CaM-induced CaMKII binding to GluN2B in vitro (despite also inducing negative-regulatory phosphorylation). Nucleotide binding was also required for Ca2+-induced CaMKII translocation to GluN2B co-expressed in heterologous cells, and for efficient glutamate-induced synaptic CaMKII translocation in hippocampal neurons. In vitro, the overall enhancement of CaMKII binding to GluN2B was based on four modulatory effects of ATP, two positive (directly by nucleotide binding and indirectly after CaMKII autophosphorylation at T286) and two negative (GluN2B phosphorylation at S1303 and CaMKII autophosphorylation at T305/6).
Compelling evidence has been collected that indicate direct binding to GluN2B as the mechanism underlying stimulus-induced CaMKII translocation to synapses (for review see Refs. 4, 5). This study clarified the remaining conundrum of how CaMKII could bind to GluN2B under high ATP conditions (as found within cells), which should lead to the GluN2B S1303 phosphorylation (36) shown to prevent CaMKII binding (at least the binding induced by T286 autophosphorylation) (27). Another alternative has recently been suggested, i.e. that phospho-S1303 GluN2B prevents only CaMKII binding induced by T286 autophosphorylation, but not binding induced by Ca2+/CaM (37). However, our results indicated that phospho-S1303 interferes with both phospho-T286- and Ca2+/CaM-induced CaMKII binding, consistent with the binding model (see Fig. 9) and with cellular studies using GluN2B mutated in the S1303 region (27, 28, 37). Another negative-regulatory phosphorylation for CaMKII binding to GluN2B has been suggested previously (51), but was directly demonstrated for the first time in this study: the autophosphorylation at T305/6. This “inhibitory” autophosphorylation prevents Ca2+/CaM binding to CaMKII (48, 50) and thereby also autophosphorylation at T286. Thus, T305/6 phosphorylation was predicted to interfere with CaMKII binding to GluN2B by inhibiting the stimulation required to induce the interaction. Indeed, T305/6D phospho-mimic mutation also impairs synaptic CaMKII translocation in hippocampal neurons (38, 52), while T305/6A mutation enhances synaptic localization (14, 52). Thus, inhibition of GluN2B binding in vitro by T305/6D mutation lends further support for GluN2B binding as underlying mechanism for synaptic CaMKII targeting. Interestingly, T305/6 phosphorylation plays a role in plasticity and flexibility of learning, as T305/6A mutant mice have a lower LTP threshold and impairments in “un-learning” of no longer beneficial tasks (52). Autophosphorylation of T286, which generates autonomous CaMKII activity, is long known to induce CaMKII binding to GluN2B even in absence of Ca2+/CaM-stimulation (15, 26, 27). It has been speculated that it may also further enhance Ca2+/CaM-stimulated binding (for review see Ref. 4), consistent with our previous findings that T286 phosphorylation further increased persistent synaptic targeting after brief stimuli (17). The results of this study show that autophosphorylation at T286 indeed directly enhanced or accelerated Ca2+-stimulated binding to GluN2B, both in vitro and in heterologous cells. Thus, our results provide important new insight about three phosphorylation reactions previously shown or speculated to be involved in regulation of CaMKII binding to GluN2B. All of these findings further support regulated binding of CaMKII to GluN2B as the underlying mechanism for stimulation induced translocation of CaMKII to synapses.
As also found in a previous study (31), ADP enhanced CaMKII binding to GluN2B in vitro, indicating a direct effect of nucleotide binding to CaMKII without requirement for any phosphorylation reaction. Consistent with these findings, CaMKII binding to GluN2B does, vice versa, enhance nucleotide binding to the kinase (53). The present study shows that the positive effect of nucleotide binding on CaMKII/GluN2B interaction is important in order to overcome the negative effect caused by GluN2B S1303 phosphorylation. In fact, efficient CaMKII binding to GluN2B was not only possible in presence of ATP, but even enhanced by it. Importantly, in heterologous cells and in neurons, nucleotide binding was required to enable stimulus-induced CaMKII interaction with GluN2B and efficient synaptic translocation, as both were either almost completely abolished or significantly reduced by the K42M mutation that impairs nucleotide binding to CaMKII. But is the regulation by nucleotide within cells indeed only mediated directly by the binding, or could it involve a phosphorylation reaction? The only known positive regulatory phosphorylation is at T286, and the non-phosphorylatable CaMKII T286A mutant behaved more similarly to wild type than to K42M. In vitro, nucleotide enhanced CaMKII binding to GluN2B even more when phosphorylation was prevented at the same time (for instance by using ADP compared with ATP), due to lack of inhibitory phosphorylation of GluN2B S1303. Thus, direct regulation by the binding of nucleotide is the most plausible explanation also for the effects observed within cells. However, some additional contribution by an unidentified positive regulatory phosphorylation by CaMKII cannot be formally ruled out. In principle, this possibility could be tested by inhibiting CaMKII activity. However, CaMKII inhibitors directly interfere with binding to GluN2B, independently from their effect on kinase activity (i.e. in vitro even in the absence of ATP): KN93 is competitive with activation by Ca2+/CaM, and thus also removes the stimulus required to induce interaction with GluN2B; tatCN21 competes with GluN2B for the same binding region on CaMKII (41, 43). It should be noted that a previous study reported synaptic translocation of another CaMKII mutant with impaired nucleotide binding (13). However, this study used K42R (a more subtle mutation of the ATP binding pocket) and, maybe more importantly, did not quantify the degree of synaptic translocation. While synaptic translocation was significantly reduced in our study by the K42M mutation, it was not completely abolished and some residual translocation was still observed.
Interestingly, direct enhancement by nucleotide binding makes the regulation of CaMKII binding to GluN2B even more similar to the regulation of aggregation of multiple CaMKII holoenzymes into large clusters (the former induced in neurons by physiological glutamate stimuli, the latter by pathological excitotoxic glutamate stimuli). Indeed, both binding reactions were proposed to involve similar molecular interactions with the CaMKII T-site (Fig. 9)(15, 39, 54). In the basal state of CaMKII, the T-site interacts with the region around T286 in the regulatory α-helix (which also blocks access to the adjacent substrate binding S-site; see Fig. 9) (15, 55). After Ca2+/CaM binds to the regulatory helix and displaces it, the T-site becomes accessible for other interactions: with the region around S1303 on GluN2B (which is homologous to the regions around T286 on CaMKII; 15), or with the T286 region on the regulatory helix of a CaMKII subunit in another holoenzyme (thus leading to formation of holoenzyme clusters) (see Fig. 9). Both GluN2B binding and holoenzyme aggregation require stimulation by Ca2+/CaM and involve interaction of the CaMKII T-site with a binding partner that is sensitive to phosphorylation by CaMKII. While aggregation is strictly dependent on presence of nucleotide (39, 56), GluN2B binding was ∼10-fold enhanced by it. The underlying mechanism is unclear, and the kinase domain structure of CaMKII has been solved so far only in absence of nucleotides (55). However, nucleotide can enhance Ca2+/CaM binding independent of T286 autophosphorylation (57), possibly by facilitating displacement of the regulatory helix, which would make the T-site more accessible. Additionally, nucleotide may induce a conformation in or around the T-site that is more favorable for external interaction. In contrast to GluN2B binding, holoenzyme aggregation additionally requires a pH below 6.8 (39, 56), a condition found after the excitotoxic insults that trigger this aggregation within neurons. The most plausible explanation is requirement for protonation of the His-282, which is located at the hinge that connects the regulatory helix with the core kinase domain (55, 58), and may thus enable more dramatic displacement of the regulatory helix. This may not be required for T-site access, but for access to the T286 region, which is necessary for binding between two kinase subunits, but not for GluN2B binding. Thus, both the regulation by pH and the more stringent nucleotide requirement for CaMKII aggregation could be explained by the necessity for more dramatic displacement of the regulatory helix, to allow external interactions of its T286 region.
This study clearly showed that Ca2+/CaM-induced CaMKII binding to GluN2B is possible in presence of ATP, even at physiological mm concentrations. In fact, ATP significantly enhanced this binding, as a positive net result caused by four modulatory effects of ATP, two positive (nucleotide binding; T286 autophosphorylation) and two negative (GluN2B S1303 phosphorylation; T305/6 autophosphorylation). But why was the negative effect of GluN2B S1303 phosphorylation not dominant? Indeed, GluN2B S1303 pre-phosphorylation or phospho-mimetic S1303D mutation dramatically inhibited binding (27) (and this study). Thus, the explanation must be based on the kinetics of the reactions involved. A pre-requisite for any phosphorylation reaction is ATP binding to the kinase, which was shown to already directly enhance binding to GluN2B. The fastest known phosphorylation reaction of CaMKII is autophosphorylation at T286 (with a rate of ∼20 s−1) (59), again promoting binding to GluN2B. This leaves CaMKII bound to ADP, which also directly stimulates binding to GluN2B but without allowing any further phosphorylation until the ADP is exchanged for ATP, with the relatively slow dissociation of ADP (∼20 s−1 compared with ∼500 s−1 for the actual phospho-transfer) generally considered as a rate-limiting step for kinase reactions (44, 60, 61). Only then can CaMKII mediate the inhibitory phosphorylation of GluN2B at S1303. At this point, significant CaMKII binding may have already occurred. Once bound, CaMKII can still phosphorylate GluN2B, but without causing any significant dissociation (17, 27), and it is unclear if this phosphorylation can still happen at S1303 and/or on the same GluN2B subunit. The other inhibitory phosphorylation, at CaMKII T305/6, is inhibited during CaMKII stimulation by Ca2+/CaM binding to an overlapping site on the regulatory helix (48, 50). Thus, fast kinetics of T305/6 autophosphorylation are achieved only by T286 phosphorylated “autonomous” CaMKII and only after dissociation of Ca2+/CaM. These kinetic considerations are also consistent with the observation that aggregation of CaMKII holoenzymes, while dependent on nucleotide binding, is actually inhibited by high physiological concentrations of ATP (39, 56)(which can be significantly reduced under pathological ischemic conditions). In this case, the fast T286 phosphorylation may still enhance T-site access but would at the same time prevent its binding to the now phosphorylated T286 region of another kinase subunit.
GluN2B is not the only binding partner for CaMKII at the synapse (for review see 4, 5). While additional interactions may contribute, they do not appear sufficient to mediate the stimulus induced but persistent synaptic translocation of CaMKII. Similar to T286 autophosphorylation, CaMKII binding to GluN2B also directly generates Ca2+-independent “autonomous” activity (15), which can provide a “molecular memory” of past Ca2+ stimuli (for review see Ref. 62). Recent results have shown that autonomy generated by T286 phosphorylation is required for inducing but not for maintaining LTP at excitatory synapses, and for learning rather than memory (18, 19). In fact, T286 phosphorylation induced by LTP stimuli may only be very short lived (63). However, a recent study indicated that storage of synaptic memory previously proposed to be mediated by T286 phosphorylation can instead be mediated by the NMDA receptor-bound form of CaMKII (30), and it will be interesting to examine the functional contribution to memory in behavioral studies.
Supplementary Material
Acknowledgments
We thank Jacqueline R. Kulbe for technical assistance. We thank Dr. Rebekah Vest for help with the schematic illustration.
The research was supported, in whole or in part, by National Institutes of Health Grants T32GM007635 (to H. O'L., W. H. L., and J. M. R.), P30NS048154 (UCD Center Grant), and R01NS052644 (to K. U. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- CaM
- Ca2+/calmodulin
- NMDA
- N-methyl-d-aspartate
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate
- LTP
- long term potentiation.
REFERENCES
- 1. Lisman J., Schulman H., Cline H. (2002) Nat. Rev. Neurosci. 3, 175–190 [DOI] [PubMed] [Google Scholar]
- 2. Hudmon A., Schulman H. (2002) Annu. Rev. Biochem. 71, 473–510 [DOI] [PubMed] [Google Scholar]
- 3. Sheng M., Kim M. J. (2002) Science 298, 776–780 [DOI] [PubMed] [Google Scholar]
- 4. Colbran R. J. (2004) Biochem. J. 378, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Merrill M. A., Chen Y., Strack S., Hell J. W. (2005) Trends Pharmacol. Sci. 26, 645–653 [DOI] [PubMed] [Google Scholar]
- 6. Malinow R., Schulman H., Tsien R. W. (1989) Science 245, 862–866 [DOI] [PubMed] [Google Scholar]
- 7. Silva A. J., Stevens C. F., Tonegawa S., Wang Y. (1992) Science 257, 201–206 [DOI] [PubMed] [Google Scholar]
- 8. Lee Y. S., Silva A. J. (2009) Nat. Rev. Neurosci. 10, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hayashi Y., Shi S. H., Esteban J. A., Piccini A., Poncer J. C., Malinow R. (2000) Science 287, 2262–2267 [DOI] [PubMed] [Google Scholar]
- 10. Opazo P., Labrecque S., Tigaret C. M., Frouin A., Wiseman P. W., De Koninck P., Choquet D. (2010) Neuron 67, 239–252 [DOI] [PubMed] [Google Scholar]
- 11. Benke T. A., Lüthi A., Isaac J. T., Collingridge G. L. (1998) Nature 393, 793–797 [DOI] [PubMed] [Google Scholar]
- 12. Derkach V., Barria A., Soderling T. R. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3269–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shen K., Meyer T. (1999) Science 284, 162–166 [DOI] [PubMed] [Google Scholar]
- 14. Shen K., Teruel M. N., Connor J. H., Shenolikar S., Meyer T. (2000) Nat. Neurosci. 3, 881–886 [DOI] [PubMed] [Google Scholar]
- 15. Bayer K. U., De Koninck P., Leonard A. S., Hell J. W., Schulman H. (2001) Nature 411, 801–805 [DOI] [PubMed] [Google Scholar]
- 16. Otmakhov N., Tao-Cheng J. H., Carpenter S., Asrican B., Dosemeci A., Reese T. S., Lisman J. (2004) J. Neurosci. 24, 9324–9331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bayer K. U., LeBel E., McDonald G. L., O'Leary H., Schulman H., De Koninck P. (2006) J. Neurosci. 26, 1164–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Giese K. P., Fedorov N. B., Filipkowski R. K., Silva A. J. (1998) Science 279, 870–873 [DOI] [PubMed] [Google Scholar]
- 19. Buard I., Coultrap S. J., Freund R. K., Lee Y. S., Dell'Acqua M. L., Silva A. J., Bayer K. U. (2010) J. Neurosci. 30, 8214–8220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hanson P. I., Meyer T., Stryer L., Schulman H. (1994) Neuron 12, 943–956 [DOI] [PubMed] [Google Scholar]
- 21. Rich R. C., Schulman H. (1998) J. Biol. Chem. 273, 28424–28429 [DOI] [PubMed] [Google Scholar]
- 22. Miller S. G., Kennedy M. B. (1986) Cell 44, 861–870 [DOI] [PubMed] [Google Scholar]
- 23. Lou L. L., Lloyd S. J., Schulman H. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 9497–9501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schworer C. M., Colbran R. J., Soderling T. R. (1986) J. Biol. Chem. 261, 8581–8584 [PubMed] [Google Scholar]
- 25. Coultrap S. J., Buard I., Kulbe J. R., Dell'Acqua M. L., Bayer K. U. (2010) J. Biol. Chem. 285, 17930–17937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Strack S., Colbran R. J. (1998) J. Biol. Chem. 273, 20689–20692 [DOI] [PubMed] [Google Scholar]
- 27. Strack S., McNeill R. B., Colbran R. J. (2000) J. Biol. Chem. 275, 23798–23806 [DOI] [PubMed] [Google Scholar]
- 28. Barria A., Malinow R. (2005) Neuron 48, 289–301 [DOI] [PubMed] [Google Scholar]
- 29. Zhou Y., Takahashi E., Li W., Halt A., Wiltgen B., Ehninger D., Li G. D., Hell J. W., Kennedy M. B., Silva A. J. (2007) J. Neurosci. 27, 13843–13853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sanhueza M., Fernandez-Villalobos G., Stein I. S., Kasumova G., Zhang P., Bayer K. U., Otmakhov N., Hell J. W., Lisman J. (2011) J. Neurosci. 31, 9170–9178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Robison A. J., Bartlett R. K., Bass M. A., Colbran R. J. (2005) J. Biol. Chem. 280, 39316–39323 [DOI] [PubMed] [Google Scholar]
- 32. Pradeep K. K., Cheriyan J., Suma Priya S. D., Rajeevkumar R., Mayadevi M., Praseeda M., Omkumar R. V. (2009) Biochem. J. 419, 123–132 [DOI] [PubMed] [Google Scholar]
- 33. Kitamura Y., Miyazaki A., Yamanaka Y., Nomura Y. (1993) J. Neurochem. 61, 100–109 [DOI] [PubMed] [Google Scholar]
- 34. Kolaj M., Cerne R., Cheng G., Brickey D. A., Randić M. (1994) J. Neurophysiol. 72, 2525–2531 [DOI] [PubMed] [Google Scholar]
- 35. Sessoms-Sikes S., Honse Y., Lovinger D. M., Colbran R. J. (2005) Mol. Cell Neurosci. 29, 139–147 [DOI] [PubMed] [Google Scholar]
- 36. Omkumar R. V., Kiely M. J., Rosenstein A. J., Min K. T., Kennedy M. B. (1996) J. Biol. Chem. 271, 31670–31678 [DOI] [PubMed] [Google Scholar]
- 37. Raveendran R., Devi Suma Priya S., Mayadevi M., Steephan M., Santhoshkumar T. R., Cheriyan J., Sanalkumar R., Pradeep K. K., James J., Omkumar R. V. (2009) J. Neurochem. 110, 92–105 [DOI] [PubMed] [Google Scholar]
- 38. Hudmon A., Lebel E., Roy H., Sik A., Schulman H., Waxham M. N., De Koninck P. (2005) J. Neurosci. 25, 6971–6983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vest R. S., O'Leary H., Bayer K. U. (2009) FEBS Lett. 583, 3577–3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zacharias D. A., Violin J. D., Newton A. C., Tsien R. Y. (2002) Science 296, 913–916 [DOI] [PubMed] [Google Scholar]
- 41. Vest R. S., Davies K. D., O'Leary H., Port J. D., Bayer K. U. (2007) Mol. Biol. Cell 18, 5024–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O'Leary H., Lasda E., Bayer K. U. (2006) Mol. Biol. Cell 17, 4656–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vest R. S., O'Leary H., Coultrap S. J., Kindy M. S., Bayer K. U. (2010) J. Biol. Chem. 285, 20675–20682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cook P. F., Neville M. E., Jr., Vrana K. E., Hartl F. T., Roskoski R., Jr. (1982) Biochemistry 21, 5794–5799 [DOI] [PubMed] [Google Scholar]
- 45. Bhatnagar D., Roskoski R., Jr., Rosendahl M. S., Leonard N. J. (1983) Biochemistry 22, 6310–6317 [DOI] [PubMed] [Google Scholar]
- 46. Aubol B. E., Nolen B., Shaffer J., Ghosh G., Adams J. A. (2003) Biochemistry 42, 12813–12820 [DOI] [PubMed] [Google Scholar]
- 47. Meyer T., Hanson P. I., Stryer L., Schulman H. (1992) Science 256, 1199–1202 [DOI] [PubMed] [Google Scholar]
- 48. Colbran R. J., Soderling T. R. (1990) J. Biol. Chem. 265, 11213–11219 [PubMed] [Google Scholar]
- 49. Lu C. S., Hodge J. J., Mehren J., Sun X. X., Griffith L. C. (2003) Neuron 40, 1185–1197 [DOI] [PubMed] [Google Scholar]
- 50. Hanson P. I., Schulman H. (1992) J. Biol. Chem. 267, 17216–17224 [PubMed] [Google Scholar]
- 51. Leonard A. S., Bayer K. U., Merrill M. A., Lim I. A., Shea M. A., Schulman H., Hell J. W. (2002) J. Biol. Chem. 277, 48441–48448 [DOI] [PubMed] [Google Scholar]
- 52. Elgersma Y., Fedorov N. B., Ikonen S., Choi E. S., Elgersma M., Carvalho O. M., Giese K. P., Silva A. J. (2002) Neuron 36, 493–505 [DOI] [PubMed] [Google Scholar]
- 53. Cheriyan J., Kumar P., Mayadevi M., Surolia A., Omkumar R. V. (2011) PLoS ONE 6, e16495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hudmon A., Kim S. A., Kolb S. J., Stoops J. K., Waxham M. N. (2001) J. Neurochem. 76, 1364–1375 [DOI] [PubMed] [Google Scholar]
- 55. Rosenberg O. S., Deindl S., Sung R. J., Nairn A. C., Kuriyan J. (2005) Cell 123, 849–860 [DOI] [PubMed] [Google Scholar]
- 56. Hudmon A., Aronowski J., Kolb S. J., Waxham M. N. (1996) J. Biol. Chem. 271, 8800–8808 [DOI] [PubMed] [Google Scholar]
- 57. Tzortzopoulos A., Török K. (2004) Biochemistry 43, 6404–6414 [DOI] [PubMed] [Google Scholar]
- 58. Smith M. K., Colbran R. J., Brickey D. A., Soderling T. R. (1992) J. Biol. Chem. 267, 1761–1768 [PubMed] [Google Scholar]
- 59. Bradshaw J. M., Hudmon A., Schulman H. (2002) J. Biol. Chem. 277, 20991–20998 [DOI] [PubMed] [Google Scholar]
- 60. Zhou J., Adams J. A. (1997) Biochemistry 36, 15733–15738 [DOI] [PubMed] [Google Scholar]
- 61. Lew J., Taylor S. S., Adams J. A. (1997) Biochemistry 36, 6717–6724 [DOI] [PubMed] [Google Scholar]
- 62. Lisman J. E., McIntyre C. C. (2001) Curr. Biol. 11, R788–791 [DOI] [PubMed] [Google Scholar]
- 63. Lee S. J., Escobedo-Lozoya Y., Szatmari E. M., Yasuda R. (2009) Nature 458, 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.