Abstract
Human natural killer-1 (HNK-1) carbohydrate is highly expressed in the nervous system and is involved in synaptic plasticity and dendritic spine maturation. This unique carbohydrate, consisting of a sulfated trisaccharide (HSO3-3GlcAβ1–3Galβ1–4GlcNAc-), is biosynthesized by the successive actions of β-1,4-galactosyltransferase (β4GalT), glucuronyltransferase (GlcAT-P and GlcAT-S), and sulfotransferase (HNK-1ST). A previous study showed that mice lacking β4GalT-II, one of seven β4GalTs, exhibited a dramatic loss of HNK-1 expression in the brain, although β4GalT-I-deficient mice did not. Here, we investigated the underlying molecular mechanism of the regulation of HNK-1 expression. First, focusing on a major HNK-1 carrier, neural cell adhesion molecule, we found that reduced expression of an N-linked HNK-1 carbohydrate caused by a deficiency of β4GalT-II is not likely due to a general loss of the β1,4-galactose residue as an acceptor for GlcAT-P. Instead, we demonstrated by co-immunoprecipitation and endoplasmic reticulum-retention analyses using Neuro2a (N2a) cells that β4GalT-II physically and specifically associates with GlcAT-P. In addition, we revealed by pulldown assay that Golgi luminal domains of β4GalT-II and GlcAT-P are sufficient for the complex to form. With an in vitro assay system, we produced the evidence that the kinetic efficiency kcat/Km of GlcAT-P in the presence of β4GalT-II was increased about 2.5-fold compared with that in the absence of β4GalT-II. Finally, we showed that co-expression of β4GalT-II and GlcAT-P increased HNK-1 expression on various glycoproteins in N2a cells, including neural cell adhesion molecule. These results indicate that the specific enzyme complex of β4GalT-II with GlcAT-P plays an important role in the biosynthesis of HNK-1 carbohydrate.
Keywords: Carbohydrate Biosynthesis; Endoplasmic Reticulum (ER); Enzymes; Glycoprotein; Glycosylation; Golgi; HNK-1; beta-1,4-Galactosyltransferase-II; Glucuronyltransferase; Polysialic Acid
Introduction
Glycosylation is a major post-translational modification, especially for cell surface and extracellular proteins, and plays important roles in cellular functions such as adhesion, endocytosis, and receptor signaling (1, 2). In general, glycan is biosynthesized in a stepwise manner by ER3- or Golgi-resident glycosyltransferases. As most of these glycosyltransferases have been cloned, there is an overall understanding of the pathway of production. However, the expression of a given glycosyltransferase does not always reflect that of its product, because it has become clear that activities of glycosyltransferases are regulated by oligomerization or proteolytic cleavage (3, 4). In addition, some glycosyltransferases form heterocomplexes that alter their activities, substrate specificity, or distribution in the ER or Golgi apparatus (5, 6). Thus, to understand the functions and biosynthesis of glycans, it is necessary to clarify how the activity of individual glycosyltransferases is regulated in cells.
Human natural killer-1 (HNK-1) carbohydrate is highly expressed on several cell adhesion molecules in the nervous system, including the neural cell adhesion molecule (NCAM) (7, 8). HNK-1 carbohydrate has a unique structure, including a sulfated trisaccharide (HSO3-3GlcAβ1–3Galβ1–4GlcNAc-) (9, 10), and is biosynthesized by the successive actions of β-1,4-galactosyltransferase (β4GalT), a glucuronyltransferase (GlcAT-P or GlcAT-S), and a sulfotransferase (HNK-1ST) (11–13). We previously generated mice lacking the gene for GlcAT-P, a major glucuronyltransferase in the nervous system (14). The GlcAT-P-deficient mice showed an almost complete loss of HNK-1 expression in the brain and exhibited reduced long term potentiation at hippocampal CA1 synapses along with impaired maturation of dendritic spines, indicating an important role for this glycan in synaptic plasticity (14, 15). In terms of the expression, we have been uncovering well controlled mechanisms of HNK-1 biosynthesis. For instance, GlcAT-P (S) and HNK-1ST form a functional complex, and the distribution of GlcAT-P in the Golgi is regulated by a small GTPase (16). However, it is still unclear how the inner N-acetyllactosamine structure (Galβ1–4GlcNAc) for HNK-1 carbohydrate is synthesized and how HNK-1 is selectively attached to limited kinds of carrier proteins.
Although the β4GalT family is known to have seven members (β4GalT-I to VII) in vivo (17, 18), the biological role of each is not fully understood. Some of them are able to synthesize N-acetyllactosamine on glycoproteins. which is potentially further modified by GlcAT-P. We recently reported that β4GalT-II-deficient mice showed similar phenotypes to GlcAT-P-deficient mice with a reduction in HNK-1 expression and impaired spatial learning and memory (19), although HNK-1 expression was unaltered in the β4GalT-I-deficient mouse brain (20) despite the similarity in primary structure and acceptor specificity of these two enzymes (21, 22). These results indicated that β4GalT-II has specific roles in the biosynthesis of HNK-1 carbohydrate. However, the precise molecular mechanism has not been explored.
Polysialic acid (PSA), a well known neural specific glycan, plays crucial roles in the development of the nervous system, attenuating cellular interactions because of its large and highly negative charge (23, 24). PSA has features in common with HNK-1 in that it is mainly expressed on NCAM and has an N-acetyllactosamine residue (Galβ1–4GlcNAc-) in its backbone structure synthesized by β4GalT (25). Surprisingly, however, β4GalT-II-deficient mice as well as β4GalT-I-deficient mice expressed PSA at the same level as wild-type mice (19, 20), indicating the biosynthesis of HNK-1 and PSA to be more complicated mechanisms than was previously thought.
In this study, we demonstrated that β4GalT-II but not β4GalT-I physically interacts with GlcAT-P but not with a polysialyltransferase (PST), which explains the loss of or remaining expression of HNK-1 or PSA in these knock-out mice. Furthermore, overexpression of β4GalT-II enhanced HNK-1 biosynthesis by GlcAT-P compared with β4GalT-I, suggesting that β4GalT-II is specifically required for production of HNK-1.
EXPERIMENTAL PROCEDURES
Materials
The monoclonal antibody (mAb) M6749 against HNK-1 carbohydrate was a gift from Dr. H. Tanaka (Kumamoto University). HNK-1 mAb was purchased from the American Type Culture Collection. The rat anti-mouse NCAM mAb (clone H28) was kindly provided by Dr. K. Ono (Kyoto Prefectural University). RCA120 was purchased from Seikagaku Corp., Tokyo, Japan. The mouse anti-FLAG M2 mAb and rabbit anti-FLAG polyclonal antibody (pAb) were obtained from Sigma. The mouse anti-Myc mAb and rabbit anti-Myc pAb were from Millipore and Abcam, respectively. The rabbit anti-GlcAT-P pAb (GP2) was generated as described previously (16). The mouse anti-GM130 mAb was purchased from BD Biosciences. HRP-conjugated anti-mouse IgG, anti-mouse IgM, anti-rabbit IgG, and anti-rat IgG were obtained from Invitrogen. Protein G-Sepharose TM4 Fast Flow and IgG-Sepharose TM6 Fast Flow were from GE Healthcare. The expression vector pcDNA3.1/myc-His B was from Invitrogen, and p3×FLAG-CMV-10 and p3×FLAG-CMV-14 were from Sigma. The plasmid pEF-BOS was kindly provided by Dr. S. Nagata (Kyoto University).
Mice
β4GalT-I-deficient mice and β4GalT-II-deficient mice were generated as described previously (19, 26). β4GalT-II-deficient mice backcrossed to C57BL/6 mice for more than 10 generations were used for the experiments, whereas β4GalT-I-deficient mice on a mixed background of 129/Sv and C57BL/6 were used because the mice with the C57BL/6 background were lethal.4 The animal experiments were conducted according to the Fundamental Guidelines for Proper Conduct of Animal Experiments and Related Activities in Academic Research Institutions under the jurisdiction of the Ministry of Education, Culture, Sports, Science and Technology of Japan and approved by the Committees on Animal Experimentation of Kanazawa University and Kyoto University.
Preparation of Brain Homogenate and Membrane Fraction
Whole brains from 4-week-old mice were homogenized with a Polytron homogenizer in 9 volumes of 50 mm Tris-HCl, pH 7.4, containing 150 mm NaCl, 1 mm EDTA, and protease inhibitors (Nacalai Tesque, Kyoto, Japan). The homogenate was centrifuged at 1,000 × g for 10 min at 4 °C to remove the nuclei and then centrifuged again at 105,000 × g for 1 h at 4 °C. The resulting pellet was used as the membrane fraction.
Peptide N-Glycosidase F Digestion
The membrane fraction of mouse brain was dissolved and denatured with phosphate-buffered saline (PBS) containing 0.5% SDS, 1% 2-mercaptoethanol, and 20 mm EDTA. To reduce the concentration of SDS, the sample was diluted with 4 volumes of PBS containing 0.5% Triton X-100. Two units of peptide N-glycosidase F (Roche Applied Science) were added, and the solution was incubated for 16 h at 37 °C.
Expression Plasmids
The subcloning of rat GlcAT-P cDNA into pEF-BOS was performed as described previously (11). The expression plasmid for GlcAT-P-AAA (pEF-BOS/GlcAT-P-AAA) was constructed as reported (16). The mouse β4GalT-I and β4GalT-II coding sequences were amplified by PCR using the primers listed below to create HindIII and EcoRV (skipping stop codon) sites and then cloned into pcDNA3.1/myc-His B. The mouse PST coding sequence was cloned into p3×FLAG-CMV-14 as described previously (16). The expression plasmid for PST-AAA-FLAG (p3×FLAG-CMV-14/PST-AAA) was constructed as follows. p3×FLAG-CMV-14/PST-AER-FLAG was constructed using QuikChange Lightning site-directed mutagenesis kits (Stratagene) according to the manufacturer's directions using the primers listed below, with p3×FLAG-CMV-14/PST-FLAG as a template. Then, p3×FLAG-CMV-14/PST-AAA-FLAG was constructed as mentioned above using the primers listed below, with p3×FLAG-CMV-14/PST-AER-FLAG as a template. cDNAs encoding the Golgi luminal domains of mouse β4GalT-I and -II (from Ser-43 and Asp-33 to the C terminus) were amplified by PCR using the primers listed below and cloned into pEF-BOS-protein A (27), which contains the insulin signal sequence and IgG-binding domain of protein A. For the constructions of prot.A-GalT-Icat and -IIcat, cDNAs encoding the catalytic domains of mouse β4GalT-I and -II (from Leu-128 and Ile-91 to the C terminus) (28) were amplified by PCR using the primers listed below and cloned into pEF-BOS-protein A. pEF-BOS-protein A was digested with SmaI. Then the blunt end fragments of β4GalTs were cloned. GlcAT-P-sol was amplified by PCR using the primers listed below with pEF-BOS/GlcAT-P as a template to create EcoRV and NotI sites, and cloned into pEF-1/V5-His A into which the signal sequence of insulin had been inserted between EcoRI and EcoRV sites.
The primers used for β4GalT-I-myc were as follows: GGGAAGCTTGCGGGCCGTCCTCTCAGCCG and TCTCGGTGTCCCGATGTCCACTGTG; β4GalT-II-myc, GGGAAGCTTCTTGCGGGATGAGCAGACTG and GCCTTGAGTGAGCCACGAGA; PST-AER-FLAG, ATGCGCTCAATTGCAGAACGGTGGACCATCTGCACTATAAGTC and GACTTATAGTGCAGATGGTCCACCGTTCTGCAATTGAGCGCAT; PST-AAA-FLAG, ATGCGCTCAATTGCAGCAGCGTGGACCATCTGCACTATAAGTC and GACTTATAGTGCAGATGGTCCACGCTGCTGCAATTGAGCGCAT; prot.A-GalT-I (Ser-43), CTGGCCGCGATCTGAGCCG and CTATCTCGGTGTCCCGATGTCCACT; prot.A-GalT-II (Asp-33), ACGTCTATGCCCAGCACCTGG and TCAGCCTTGAGTGAGCCACGACATG; prot.A-GalT-Icat (Leu-128), TGCCAGCTTGCCCTGAGGAG and CTATCTCGGTGTCCCGATGTCCACT; prot.A-GalT-IIcat (Ile-91), TCATTCCGCCCTGTCCTGAC and TCAGCCTTGAGTGAGCCACGACATG; and GlcAT-P-sol, CCCCGCCATGAGGCACCACC and TGAGCGGCCGCTCAGATCTCCACCGAGGGGT.
Cell Culture and Transfection
N2a cells were cultured in minimum Eagle's medium supplemented with 10% fetal bovine serum at 37 °C until 50–70% confluent. For transfection, cells were plated on 60-mm tissue culture dishes, grown overnight, and then transfected with various expression constructs using FuGENE 6 (Roche Applied Science) according to the manufacturer's directions. Briefly, a 2.5 volume of FuGENE 6 and 1 μg of each DNA were incubated with 100 μl of minimum Eagle's medium for 15 min at room temperature, and then the mixture was added to the tissue culture dishes.
Cell Lysis and Immunoprecipitation of Transiently Expressed Proteins
Cells were collected 24 h after transfection and lysed with a buffer consisting of 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, and a protease inhibitor mixture (Nacalai Tesque). After centrifugation, the clarified lysate was incubated with the anti-FLAG rabbit pAb or anti-Myc rabbit pAb for 0.5 h. The mixture was then incubated with protein G-Sepharose TM4 Fast Flow for 2 h with gentle shaking. The beads were precipitated by centrifugation (500 × g for 1 min) and washed three times with an excess volume of wash buffer consisting of 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 0.1% Tween 20. Proteins bound to the Sepharose beads were eluted by boiling in Laemmli sample buffer.
Immunostaining of N2a Cells
At 24 h post-transfection, cells were washed with PBS, fixed with ice-cold methanol, and incubated with primary antibodies followed by Alexa Fluor-conjugated secondary antibodies (Invitrogen). Cells were visualized with a Fluoview laser confocal microscope system (Olympus).
SDS-PAGE and Western Blot and Lectin Blot Analyses
Proteins were separated by SDS-PAGE with the buffer system of Laemmli and transferred to nitrocellulose membranes. For Western blotting, after being blocked with 5% skim milk in PBS containing 0.05% Tween 20, the membrane was incubated with primary antibodies followed by HRP-conjugated secondary antibodies. For lectin blotting, after being blocked with PBS containing 0.05% Tween 20 (T-PBS), the membrane was incubated with sialidase (Roche Applied Science) according to the manufacturer's instructions, and then incubated with HRP-conjugated lectin in T-PBS. Protein bands were detected with ECL (Pierce) using a LAS3000 Luminoimage analyzer (Fujifilm).
Pulldown Assays
N2a cells were transiently transfected with prot.A-GalT-I (Ser-43), -Icat (Leu-128), -II (Asp-33), or -IIcat (Ile-91) and GlcAT-P-sol. After 6 h of incubation, the culture medium was replaced with serum-free Opti-MEM I (Invitrogen) and incubation continued for another 2 days. Normal human IgG-conjugated Sepharose beads were added to the culture medium containing secreted proteins. The beads were precipitated by centrifugation (500 × g for 1 min) and then washed three times with an excess volume of wash buffer consisting of 20 mm Tris-HCl, pH 7.4, 150 mm NaCl, and 0.1% Tween 20. Proteins bound to the Sepharose beads were eluted by boiling in Laemmli sample buffer.
Preparation of Protein A-fused GalTs
COS-1 cells plated on 175-cm2 tissue culture flasks were transfected with prot.A-GalT-I (Ser-43) or prot.A-GalT-II (Asp-33) using FuGENE 6 transfection reagent. After 5 h of incubation, the culture medium was replaced with serum-free ASF104 medium (Ajinomoto), followed by incubation for another 3 days. Then, the culture medium containing secreted proteins was applied to IgG-Sepharose TM6 fast flow column (GE Healthcare). Unbound proteins were washed out with more than 10 column volumes of PBS. Bound proteins were eluted with 100 mm glycine-HCl, pH 2.5, and then the eluate was immediately neutralized with 3 m Tris-HCl, pH 8.0. The purified prot.A-GalTs were used for the following kinetic analysis of GlcAT-P.
Measurement of Glucuronyltransferase Activity
The FLAG-tagged GlcAT-P catalytic domain (FLAG-P) used was expressed in Escherichia coli and purified as described previously (29). The enzymatic activity of GlcAT-P toward glycoprotein acceptors was measured according to the procedure described previously (10) with slight modification. In brief, prior to the assay, FLAG-P (25 ng) was incubated in the absence or presence of prot.A-GalT-I (Ser-43) or prot.A-GalT-II (Asp-33) (100 ng each) at room temperature for 15 min. The preincubated enzyme solution was added to a reaction mixture with a final volume of 25 μl consisting of 200 mm MES, pH 6.5, 0.2% Nonidet P-40, 20 mm MnCl2, 0.1–20 μg ASOR, 100 μm UDP-[14C]GlcA (100,000 dpm), and 0.5 mm ATP. After incubation at 37 °C for 2 h, the assay mixture was spotted onto a Whatman No. 1 filter paper. The filter paper was washed with a 10% (w/v) trichloroacetic acid solution three times, followed by with ethanol/ether (2:1, v/v), and then with ether. The filter paper was air-dried, and then the radioactivity of [14C]GlcA-ASOR was counted with a liquid scintillation counter.
RESULTS
Unaltered Expression of N-Acetyllactosamine on a Major HNK-1 Carrier in β4GalT-II-deficient Mice
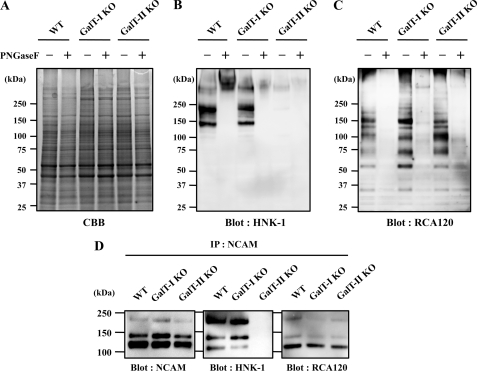
Recently, we reported that HNK-1 expression is substantially reduced in β4GalT-II-deficient mice, although an earlier study revealed that β4GalT-I-deficient mice expressed normal levels of HNK-1 (19, 20). To directly compare the expression of HNK-1 in these mutant mice, brain membrane fractions were prepared from wild-type and β4GalT-I, II-deficient mice (Fig. 1A) and Western-blotted with the HNK-1 mAb (Fig. 1B). Consistent with previous reports, HNK-1 expression was dramatically decreased in β4GalT-II-deficient mice but was normal in β4GalT-I-deficient mice. Most HNK-1 epitopes in the wild-type mice disappeared after treatment with peptide N-glycosidase F (Fig. 1B, 2nd lane), indicating that HNK-1 carbohydrates are mainly expressed on N-glycan in the membrane fraction. This suggests that β4GalT-II synthesizes most HNK-1 carbohydrates on N-glycans in the brain. To explore the molecular mechanism involved, we first confirmed that the GlcAT-P transcript was expressed normally in these knock-out mice brains (supplemental Fig. 1). Next, to examine whether the N-acetyllactosamine residue remains in these knock-out mice, a lectin blot analysis was performed using RCA120, which predominantly recognizes the βGal residue of N-acetyllactosamine. Because the nonreducing end of N-acetyllactosamine is often capped by sialic acid, the membrane was treated with sialidase prior to RCA120 blotting. As a result, β4GalT-I and -II-deficient mice showed similar reactivity for RCA120 to wild-type mice, most of which disappeared after peptide N-glycosidase F treatment, indicating that the N-acetyllactosamine on N-glycans was normally expressed in these knock-out mice and that several β4GalTs contribute to the biosynthesis of N-linked N-acetyllactosamine in the brain. This excludes the possibility that the reduction in HNK-1 is due to a general loss of N-acetyllactosamine in the β4GalT-II-deficient brain.
FIGURE 1.
Expression of HNK-1 carbohydrate and N-acetyllactosamine in mouse brains. Membrane fractions of 4-week-old C57BL/6 (WT), β4GalT-I-deficient (GalT-I-KO), and β4GalT-II-deficient (GalT-II-KO) mouse brain were subjected to SDS-PAGE followed by Coomassie Brilliant Blue (CBB) staining (A), Western blotting with HNK-1 mAb (B), or lectin blotting with RCA120 (C). D, membrane fractions of 4-week-old mouse brains were immunoprecipitated (IP) with anti-NCAM antibody, subjected to SDS-PAGE, and blotted with anti-NCAM mAb, HNK-1 mAb, and RCA120. PNGase F, peptide:N-glycosidase F.
Another possibility is that β4GalT-II is the only β4GalT in HNK-1-expressing cell or a specific β4GalT for HNK-1 carrier proteins. To test this, we focused on NCAM. Because NCAM is a major carrier of HNK-1 carbohydrates in the nervous system, it must be expressed in HNK-1-positive cells. NCAM was immunoprecipitated from the brain membrane fraction and blotted with anti-NCAM mAb, HNK-1 mAb, and RCA120. As shown in Fig. 1D, three major splicing isoforms of NCAMs (NCAM-120, NCAM-140, and NCAM-180) are detected, and no difference in expression levels of each NCAM isoform (Blot: NCAM) or in reactivity with Ricinus communis agglutinin (Blot: R. communis agglutinin was observed among three genotypes. In contrast, HNK-1 is predominantly expressed on NCAM-180 and NCAM-140 rather than on NCAM120 in wild-type mice brain, which is consistent with a previous report (30). The HNK-1 on NCAM disappeared in β4GalT-II-deficient mice (Blot: HNK-1). These results indicate that other β4GalTs synthesize N-acetyllactosamine even onto NCAM in β4GalT-II-deficient mice and also suggest that β4GalT-II is not the only enzyme in HNK-1-expressing cells and not responsible for the general production of N-acetyllactosamine on HNK-1 carrier molecules. Therefore, it is possible that GlcAT-P transfers glucuronic acid (GlcA) only to the N-acetyllactosamine structure that β4GalT-II synthesizes, prompting us to speculate that GlcAT-P specifically associates with β4GalT-II.
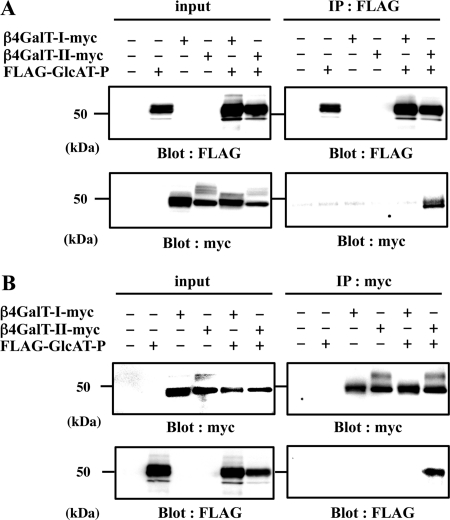
Co-immunoprecipitation of GlcAT-P and β4GalT-II
To investigate the interaction between β4GalT-II and GlcAT-P, we performed co-immunoprecipitation experiments. Myc-tagged β4GalT-I or -II (β4GalT-I-myc or β4GalT-II-myc) and FLAG-tagged GlcAT-P (FLAG-GlcAT-P) were transiently expressed in N2a cells, and cell lysates were incubated with anti-FLAG pAb to precipitate FLAG-GlcAT-P (Fig. 2A, IP: FLAG). Subsequent Western blotting was conducted with anti-Myc mAb to detect β4GalT-I and -II-myc. As shown in Fig. 2A, right lower panel, β4GalT-II-myc was co-immunoprecipitated by FLAG-GlcAT-P, whereas β4GalT-I-myc was not. To determine whether FLAG-GlcAT-P could be conversely co-precipitated by β4GalT-II-myc, we conducted immunoprecipitation using anti-Myc pAb. As shown in Fig. 2B, right lower panel, FLAG-GlcAT-P was co-immunoprecipitated by β4GalT-II-myc, but not by β4GalT-I-myc. These results revealed that GlcAT-P specifically interacted with β4GalT-II in cells.
FIGURE 2.
Co-immunoprecipitation of GlcAT-P and β4GalT-II in N2a cells. Lysate of N2a cells transiently expressing FLAG-GlcAT-P, β4GalT-I-myc, or β4GalT-II-myc was immunoprecipitated (IP) with anti-FLAG pAb (A, IP: FLAG) or anti-Myc pAb (B, IP: myc), subjected to SDS-PAGE, and Western-blotted (Blot) with anti-FLAG mAb or anti-Myc mAb. To examine the level of each protein, the cell lysates were directly subjected to SDS-PAGE and Western blotting with anti-FLAG mAb or anti-Myc mAb (input).
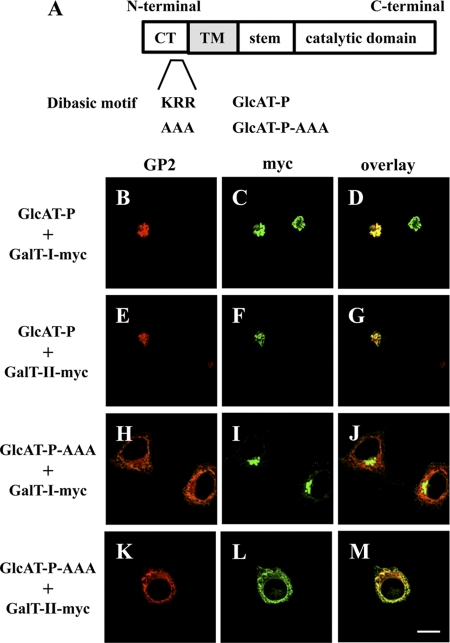
Effect of ER-retained GlcAT-P on β4GalT-II Localization
Next, we performed an ER-retention assay (6, 31) to visualize the interaction. Normally, GlcAT-P and β4GalT-I and -II mainly occur in Golgi, as was confirmed by co-localization with a Golgi marker, GM130, in N2a cells (supplemental Fig. 2). GlcAT-P appeared to co-localize with β4GalT-I as well as β4GalT-II under normal conditions (Fig. 3, B–G). In the ER-retention assay, we employed an ER-retained version of GlcAT-P in which a dibasic motif (K/R)(X)(K/R) was mutated (Fig. 3A). Some glycosyltransferases have this motif in their cytoplasmic tail, which is required for transport from the ER to the Golgi apparatus (32). Actually, GlcAT-P lacking this motif (GlcAT-P-AAA) was retained in the ER (Fig. 3, H and K, and also see supplemental Fig. 3, A and D) (16). To investigate the effect of GlcAT-P-AAA on the distribution of β4GalT-I and -II, N2a cells expressing GlcAT-P-AAA and β4GalT-I or -II-myc were immunostained. As a result, the location of β4GalT-II-myc changed to the ER from the Golgi (Fig. 3L and supplemental Fig. 3E), which overlapped with GlcAT-P-AAA (Fig. 3M and supplemental Fig. 3F), whereas β4GalT-I-myc was still localized to the Golgi regardless of the expression of GlcAT-P-AAA (Fig. 3, I and J, and also see supplemental Fig. 3, B and C). These results reinforce the notion of selective interaction between GlcAT-P and β4GalT-II in cells.
FIGURE 3.
ER retention assays using GlcAT-P-AAA. A, schematic diagrams of GlcAT-P and GlcAT-P-AAA. CT, cytoplasmic tail, TM, transmembrane domain. B–M, N2a cells were transiently co-transfected with GlcAT-P or GlcAT-P-AAA and β4GalT-I-myc or β4GalT-II-myc. GlcAT-P and GlcAT-P-AAA were detected with GP2 pAb (B, E, H, and K) and Alexa 546-conjugated secondary antibodies. β4GalT-myc (I and II) was detected with anti-Myc mAb (C, F, I, and L) and Alexa 488-conjugated secondary antibodies. D, G, J, and M, overlaid images. Bar, 10 μm.
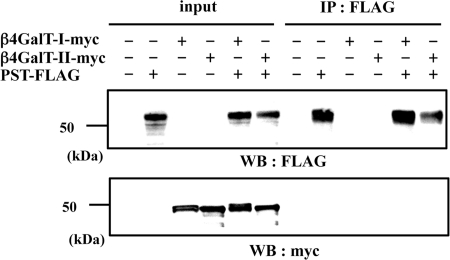
β4GalT-II and β4GalT-I Do Not Associate with PST
To examine the specificity of the binding between β4GalT-II and GlcAT-P, we used a PST instead of GlcAT-P. PST is one of the α2,8-polysialyltransferases involved in the biosynthesis of PSA (33, 34), and PSA is attached to monosialylated N-acetyllactosamine on NCAM (35). Our previous study revealed that PSA was expressed in the brain of β4GalT-II-deficient mice at the same level as in wild-type mice (19). This is also the case in β4GalT-I-deficient mice (20). Therefore, we expected that β4GalT-I and -II would not associate with PST. β4GalT-I- or -II-myc and PST-FLAG were expressed in N2a cells, immunoprecipitated with anti-FLAG pAb, and subjected to Western blotting with anti-Myc mAb. As shown in Fig. 4, lower panel, β4GalT-I and -II were not co-precipitated with PST, suggesting that these enzymes do not interact in cells.
FIGURE 4.
Immunoprecipitation using PST-FLAG and β4GalT-myc in N2a cells. Lysate of N2a cells transiently expressing PST-FLAG, β4GalT-I-myc, or β4GalT-II-myc were immunoprecipitated (IP) with anti-FLAG pAb (IP: FLAG), subjected to SDS-PAGE, and Western-blotted with anti-FLAG mAb or anti-Myc mAb. To examine the level of each protein, the cell lysates were directly subjected to SDS-PAGE and then Western blotting (WB) with anti-FLAG mAb or anti-Myc mAb (input).
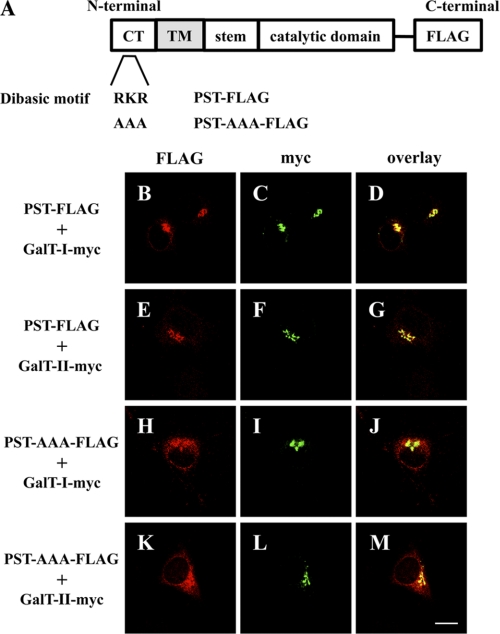
To support the immunoprecipitation data, an ER retention assay was performed using PST-AAA-FLAG (Fig. 5A), a mutant of the dibasic motif (K/R)(X)(K/R) in PST. PST is also localized to the Golgi apparatus, as confirmed by the co-localization of PST-FLAG with β4GalT-I, and -II-myc in the Golgi (Fig. 5, B–G). As expected, the altered distribution of PST-AAA-FLAG was observed as shown in Fig. 5, H and K, and also in supplemental Fig. 3, G and J. The distribution of PST-AAA is slightly different from GlcAT-P-AAA, i.e. PST-AAA was detected not only in ER but in Golgi, suggesting that the effect of the mutation of dibasic motif on ER distribution varies by an individual glycosyltransferase as described previously (32). However, the altered distribution of PST-AAA did not perturb the Golgi-based localization of β4GalT-I and -II-myc (Fig. 5, I, J, L, and M, and also see supplemental Fig. 3, H, I, K, and L). These results revealed that β4GalT-I and -II do not associate with PST, supporting our idea that β4GalT-II and GlcAT-P form a specific complex.
FIGURE 5.
ER retention assays using PST-AAA-FLAG. A, schematic diagrams of PST-FLAG and PST-AAA-FLAG. B–M, N2a cells were transiently co-transfected with PST-FLAG or PST-AAA-FLAG and β4GalT-I-myc or β4GalT-II-myc. PST-FLAG or PST-AAA-FLAG was detected with anti-FLAG pAb (B, E, H, and K), and Alexa 546-conjugated secondary antibodies. β4GalT-myc (I and II) was detected with anti-Myc mAb (C, F, I, and L) and Alexa 488-conjugated secondary antibodies. D, G, J, and M, overlaid images. Bar, 10 μm. CT, cytoplasmic tail, TM, transmembrane domain.
Interaction between GlcAT-P and β4GalT-II through Their Luminal Domains
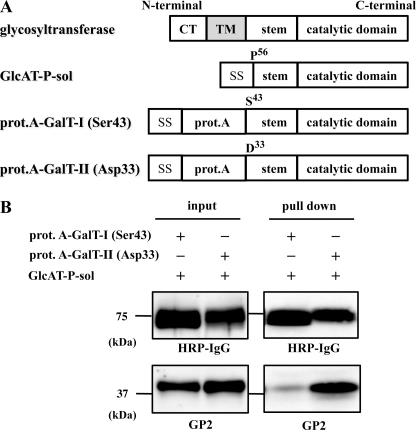
Glycosyltransferases, such as GlcAT-P and β4GalT, are type II transmembrane proteins with a short cytoplasmic tail, a hydrophobic single-pass transmembrane domain, a stem region, and a catalytic domain (Fig. 6A) (36). We previously reported that GlcAT-P and HNK-1ST formed an enzyme complex through their catalytic domains in the Golgi lumen (37). Thus, we examined whether GlcAT-P and β4GalT-II also form a complex via their Golgi luminal domains. We constructed an expression vector encoding the luminal domain of GlcAT-P downstream of the insulin signal sequence (GlcAT-P-sol). Similarly, expression vectors for luminal domains of β4GalT-I and -II fused with the signal sequence and protein A (prot.A-GalT-I (Ser-43) and prot.A-GalT-II (Asp-33), respectively) were constructed (Fig. 6A). The culture medium of N2a cells expressing GlcAT-P-sol and prot.A-GalT-I (Ser-43) or -II (Asp-33) was incubated with IgG-Sepharose beads to pull down the prot.A-tagged enzymes. Subsequent Western blotting was conducted with HRP-conjugated IgG and anti-GlcAT-P pAb (GP2) to detect prot.A-fused enzymes and GlcAT-P, respectively. As shown in Fig. 6B, GlcAT-P-sol was highly co-precipitated with prot.A-GalT-II compared with prot.A-GalT-I, indicating that Golgi luminal domains are enough to form the enzyme complex. To narrow down the binding region of Golgi luminal domains of β4GalTs, we also constructed expression vectors for catalytic domains of β4GalT-I and -II fused with the signal sequence and protein A (prot.A-GalT-Icat (Leu-129) and prot.A-GalT-IIcat (Ile-91), respectively) and performed pulldown assay as described above. However, neither prot.A-GalT-Icat (Leu-129) nor prot.A-GalT-IIcat (Ile-91) interacts with GlcAT-P-sol (data not shown), suggesting that the deleted stem region are necessary for the specific binding of GlcAT-P and β4GalT-II. Therefore, prot.A-GalT-I (Ser-43) and prot.A-GalT-II (Asp-33) were used for the following kinetic analysis of GlcAT-P.
FIGURE 6.
Pulldown assays using soluble forms of GlcAT-P-sol, prot.A-GalT-I, and prot.A-GalT-II. A, schematic diagrams of GlcAT-P-sol, prot.A-GalT-I (Ser-43), and prot.A-GalT-II (Asp-33). SS, signal sequence. B, culture medium of N2a cells transiently expressing GlcAT-P-sol and prot.A-GalT-I (Ser-43) or prot.A-GalT-II (Asp-33) was incubated with IgG-Sepharose TM6 Fast Flow (pulldown), subjected to SDS-PAGE, and Western-blotted with HRP-conjugated normal rabbit IgG or GP2 pAb. To examine the level of each protein, the culture medium was directly subjected to SDS-PAGE and then Western-blotted with HRP-conjugated normal rabbit IgG or GP2 pAb (input). CT, cytoplasmic tail, TM, transmembrane domain.
Kinetic Analysis of GlcAT-P
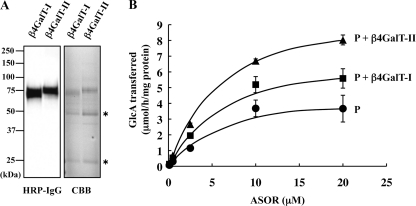
Next, to investigate whether or not a specific association of β4GalT-II affects GlcAT-P activity, we examined the dependence of the rate of the glucuronyltransferase reaction on the concentration of ASOR as a glycoprotein acceptor in the presence or absence of β4GalT-I or β4GalT-II. For this kinetic analysis, we used FLAG-tagged human GlcAT-P (from Thr-58 to the C terminus, FLAG-P) expressed in E. coli. The method for expression and purification of FLAG-P had been already established (29). Human GlcAT-P is highly homologous to mouse GlcAT-P, especially 99.3% of amino acids are identical in the region we used (Thr-58 to C terminus). prot.A-GalT-I (Ser-43) and prot.A-GalT-II (Asp-33) were purified from culture media of COS-1 cells that had been transfected with their expression vectors as shown in Fig. 7A. The rate of glucuronyltransferase reaction of FLAG-P in the absence of prot.A-GalTs was compared with that in the presence of prot.A-GalT-I (Ser-43) or prot.A-GalT-II (Asp-33) using different concentrations of ASOR. As shown in Fig. 7B, the rate of FLAG-P reaction was increased depending on the concentration of ASOR and enhanced in the presence of GalT-I or -II. The data were analyzed by means of Lineweaver-Burk plotting, and the kinetic parameters were then determined (Table 1). Apparent Vmax value was increased about 2-fold, and the apparent Km value was slightly decreased by β4GalT-II. Thus, β4GalT-II resulted in an approximate 2.5-fold increase in the catalytic efficiency kcat/Km of FLAG-P. To our surprise, β4GalT-I, which does not associate with GlcAT-P, also enhanced the catalytic efficiency kcat/Km about 1.7-fold. However, the catalytic efficiency in the presence of β4GalT-II is still higher than that of β4GalT-I. These results suggest that the enzyme complex of GlcAT-P and β4GalT-II is indeed capable of facilitating the expression of HNK-1.
FIGURE 7.
Effect of the co-presence of β4GalT-II on the in vitro glucuronyltransferase reaction of GlcAT-P. A, prot.A-GalT-I (Ser-43) (β4GalT-I) or -II (Asp-33) (β4GalT-II) was expressed in COS-1 cells and purified from the culture media. The purified enzymes were subjected to Western blotting with HRP-conjugated rabbit IgG and Coomassie Brilliant Blue (CBB) staining. Asterisks indicate heavy and light chains of immunoglobulin derived from human IgG Sepharose. B, rate of glucuronyltransferase reaction of FLAG-P was measured in the absence of β4GalTs (P), or in the presence of prot.A-GalT-I (Ser-43) (P + β4GalT-I) or prot.A-GalT-II (Asp-33) (P + β4GalT-II) using different concentrations of ASOR as substrates. All experiments were employed in triplicate, and error bars indicate S.E.
TABLE 1.
Kinetic parameters of GlcAT-P
| Km | Vmax | kcat | kcat/Km | Relative kcat/Km | |
|---|---|---|---|---|---|
| μm | μmol/h/mg | s−1 | μm−1s−1 | -fold | |
| GlcAT-P | 7.46 | 4.90 | 0.0545 | 7.30 | 1 |
| GlcAT-P + β4GalT-I | 6.62 | 7.53 | 0.0837 | 12.6 | 1.73 |
| GlcAT-P + β4GalT-II | 6.09 | 9.86 | 0.110 | 18.0 | 2.47 |
Enhanced HNK-1 Biosynthesis by Enzyme Complex in N2a Cells
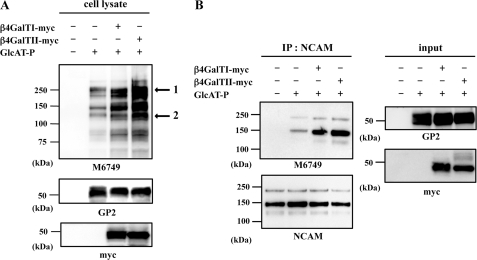
To determine the effect of the interaction on HNK-1 carbohydrate production in cells, a Western blot analysis was performed with the M6749 mAb. This mAb recognizes nonsulfated HNK-1 carbohydrate (GlcAβ1–3Galβ1–4GlcNAc) (38), and N2a cells do not express endogenous GlcAT-P or HNK-1 carbohydrate (Fig. 8A, 1st lane). By employing this antibody, we can examine the level of nonsulfated HNK-1 carbohydrate, which was produced by GlcAT-P (Fig. 8A, 2nd lane). GlcAT-P and β4GalT-I or -II-myc were co-expressed in N2a cells, and then cell lysates were subjected to SDS-PAGE and Western-blotted with the M6749 mAb, GP2 pAb, and anti-Myc mAb. As shown in Fig. 8A, M6749 immunoreactivity was increased when co-expressed with β4GalT-I and -II-myc despite the comparable expression of GlcAT-P (compare 2nd lane with 3rd and 4th lanes), probably because of the increased amount of N-acetyllactosamine, which can be utilized as an acceptor by GlcAT-P. Intriguingly, however, M6749 immunoreactivity was increased more when GlcAT-P was co-expressed with β4GalT-II-myc than with β4GalT-I-myc (Fig. 8A, compare 3rd with 4th lane). To confirm these results, we quantitated two bands (see Fig. 8A, band 1 and 2) by means of densitometric analysis using image analysis software ImageGauge (FujiFilm). As shown in Table 2, co-expression of GlcAT-P and β4GalT-II-myc caused about 3-fold (see Fig. 8A, bands 1 and 2) increases in M6749 immunoreactivity compared with that of GlcAT-P, and a 2- (band 1) or 1.4-fold (band 2) increase compared with that of GlcAT-P and β4GalT-I-myc. These suggest that β4GalT-II is better able to promote HNK-1 biosynthesis by associating with GlcAT-P. We carried out a similar analysis with immunoprecipitated NCAM, which is endogenously expressed in N2a cells. As shown in Fig. 8B, the highest expression of the M6749 epitope was observed when GlcAT-P was co-expressed with β4GalT-II-myc. These results suggest that the complex of GlcAT-P and β4GalT-II contributes to the enhancement of HNK-1 synthesis.
FIGURE 8.
Effect of the enzyme complex on HNK-1 expression in N2a cells. A, lysates of N2a cells transiently expressing GlcAT-P, β4GalT-I-myc, or β4GalT-II-myc were subjected to SDS-PAGE and then Western blotted with M6749 mAb, GP2 pAb, or anti-Myc mAb. Arrows indicate bands used for quantification by densitometric analyses. B, lysates of N2a cells transiently expressing GlcAT-P and β4GalT-I-myc or β4GalT-II-myc were immunoprecipitated (IP) with anti-NCAM mAb and subjected to SDS-PAGE and then Western-blotted with M6749 mAb or anti-NCAM mAb.
TABLE 2.
Quantitative analysis of the effect of the enzyme complex
| Mock | GlcAT-P | GlcAT-P + β4GalT-I-myc | GlcAT-P + β4GalT-II-myc | |
|---|---|---|---|---|
| Band 1 | ||||
| Intensity (a.u.)a | 0 | 0.48 | 0.77 | 1.53 |
| -Fold | 1.00 | 1.60 | 3.20 | |
| Band 2 | ||||
| Intensity (a.u.) | 0 | 0.21 | 0.43 | 0.60 |
| -Fold | 1.00 | 2.03 | 2.86 | |
a The immunoreactivity with M6749 mAb was calculated and normalized to GlcAT-P expression and is shown as intensity in arbitrary units (a.u.).
DISCUSSION
The β4GalT family has seven members, but the biological roles have not been fully understood. We recently reported that β4GalT-II-deficient mice showed a marked reduction in levels of HNK-1 carbohydrate and impaired spatial memory, and the phenotypes are similar to those of GlcAT-P-deficient mice (19). Moreover, β4GalT-II is highly expressed in the brain although β4GalT-I is ubiquitous (39). These findings suggest intrinsic roles in vivo, although β4GalT-I and -II show the highest sequence similarity among β4GalTs. Analyses using these gene-deficient mice revealed that β4GalT-II is crucial to the production of HNK-1, but the underlying mechanism was not elucidated. In this study, we produced evidence that β4GalT-II physically associates with GlcAT-P (Figs. 2 and 3) probably through the luminal stem domain of β4GalT-II. Considering the similar domain organizations and molecular sizes of GlcAT-P and β4GalT-II, the stem domain of GlcAT-P may also be involved in this interaction. Because we had revealed that GlcAT-P bound to HNK-1ST through the catalytic region (37), β4GalT-II seems not to compete with HNK-1ST for the binding to GlcAT-P; rather, these three enzymes may simultaneously form a large functional complex to synthesize HNK-1 efficiently in living cells. Actually, we revealed that β4GalT-II is capable of increasing the catalytic efficiency kcat/Km of the GlcAT-P catalytic reaction using in vitro enzyme assay system (Fig. 7). Also in N2a cells, the co-expression of β4GalT-II and GlcAT-P enhanced HNK-1 biosynthesis (Fig. 8). These results suggest that HNK-1 expression is regulated by the interaction of these two enzymes.
We found that both β4GalT-I- and -II-deficient mice showed normal levels of N-acetyllactosamine on N-glycans, indicating that several β4GalTs actually contribute to galactosylation in the brain. The specific loss of HNK-1 in β4GalT-II-deficient mice can be explained in several ways. The present analysis suggests that β4GalT-II is not the only enzyme for N-acetyllactosamine synthesis in HNK-1-expressing cells because N-acetyllactosamine is likely expressed at normal levels in β4GalT-II knock-out mice even on NCAM. Our experiment showed that β4GalT-II does not specialize in the galactosylation of HNK-1-expressing molecules such as NCAM by recognizing their polypeptide backbones, because the galactosylation of NCAM is unlikely to be impaired in β4GalT-II knock-out mice. β4GalT-II might act on a specific N-glycosylation site or specific branch of N-glycan that is preferentially modified further by GlcAT-P. We demonstrated here that simply the presence of N-acetyllactosamine or GlcAT-P is not sufficient for HNK-1 biosynthesis and that both N-acetyllactosamines synthesized by β4GalT-II and GlcAT-P are required. Although β4GalT-II is involved in HNK-1 biosynthesis, this enzyme is not a chaperone-like molecule for GlcAT-P, because a recombinant GlcAT-P from E. coli was fully active without β4GalT-II in vitro (29). Rather, we speculate that β4GalT-II would be in a specialized complex with GlcAT-P in a specific Golgi compartment to cooperatively catalyze their transfer reactions for HNK-1 synthesis on specific glycoproteins.
To our surprise, β4GalT-I also had the potential to activate HNK-1 biosynthesis (Fig. 8), even though its effect was weaker than β4GalT-II. This result could be explained by the overproduction of N-acetyllactosamine residues by β4GalT-I overexpression even without enzyme-enzyme interaction. Meanwhile, our in vitro analysis also showed that β4GalT-I weakly activated GlcAT-P catalytic activity (Fig. 7). Although we could not detect the interaction between GlcAT-P and β4GalT-I in cell-based co-immunoprecipitation assays, weaker binding was observed in pulldown assays (Fig. 6B). These results suggest that β4GalT-I could compensate for the loss of HNK-1 biosynthesis in β4GalT-II-deficient cells if it could be overexpressed. Because HNK-1 expression almost disappeared in the β4GalT-II-deficient brain, it suggests that the levels of other β4GalTs in HNK-1-expressing cells may be under the levels that are required for compensation for the loss of β4GalT-II.
We previously reported that β4GalT-I- and -II-deficient mice showed the same level of PSA expression as wild-type mice, indicating that the biosynthesis of PSA does not depend on these enzymes (19, 20). Also, we found that PST does not associate with β4GalT-I or -II (Fig. 4). Regarding the biosynthesis of PSA, another β4GalT may be associated with polysialyltransferase to specifically synthesize the inner structure of PSA. Alternatively, polysialyltransferase may not distinguish between the inner N-acetyllactosamines synthesized by several β4GalTs. Other β4GalTs could compensate for the loss of deficiency of a given β4GalT. Moreover, acidic amino acid residues in the fibronectin type III domain adjacent to the fifth Ig domain of NCAM, which has N-glycosylation sites attaching to PSA, were reported to be required for polysialylation (40). PSA biosynthesis would be mainly dependent on the polypeptide backbone of NCAM rather than the enzyme responsible for production of the inner N-acetyllactosamine. It is indicated that the biosynthesis of PSA is considerably different from that of HNK-1, despite some common features between these two neural glycans expressed on NCAM.
In recent years, many groups have reported that enzyme complexes of glycosyltransferases have enhanced catalytic activities. Seko and Yamashita (5) showed that a complex of β3GnT2 and β3GnT8 produced more poly-N-acetyllactosamine. Our previous study demonstrated that GlcAT-P interacts with HNK-1ST, and the interaction enhances the enzymatic activity of HNK-1ST (37). Also, it has been reported that the activities of some glycosyltransferases are regulated through interaction with nonglycosyltransferases. For example, GnT-I activity was inhibited by interaction with GnT-I-IP (GnT-I inhibitory protein), resulting in the expression of high mannose-type N-glycans during spermatogenesis (41). T-synthase (C1β3Gal-T) forms a complex with core 1 β3GalT-specific molecular chaperone, which functions as a specific chaperone. Core 1 β3GalT-specific molecular chaperone is essential for the T-synthase activity and synthesizing T-antigen (42). Actually, core 1 β3GalT-specific molecular chaperone is a factor responsible for Tn syndrome, a genetic disease involving a deficiency in T-antigen but no defect in T-synthase (43). In this study, we found that HNK-1 expression is not regulated simply by GlcAT-P but also by the interaction of the enzymes. Supporting this finding, the distribution of GlcAT-P mRNA did not completely match the pattern of HNK-1 expression in the brain. For example, HNK-1 antibody staining showed parasagittal stripes in the molecular layer in the cerebellum (44) because of HNK-1 carbohydrate expression on a limited population of Purkinje cells, whereas GlcAT-P mRNA is likely expressed in most Purkinje cells (45). It would be of interest to compare the distribution of β4GalT-II mRNA and HNK-1 carbohydrate in the brain. Taken together, to understand the precise mechanisms of expression and proper functions of glycans, it is important to recognize that many glycosyltransferases form complexes in vivo.
HNK-1 carbohydrate has an important role in synaptic plasticity and spine morphogenesis in the nervous system (14, 15). These functions are based on its specific expression on certain carriers such as NCAM and GluR2, which is a subunit of the α-amino-3-hydroxy-5-methylisoxazole propionate-type glutamate receptor. It should be noted that another subunit, GluR1, which forms a complex with GluR2 to make a functional α-amino-3-hydroxy-5-methylisoxazole propionate receptor in vivo, is not modified with HNK-1 (46) even if several complex-type N-glycans are expressed on GluR1. This indicates that HNK-1 carbohydrate greatly contributes to the specific function of GluR2, but the underlying mechanism of this selective modification is poorly understood. It could be involved in the selective modification to form the complex of β4GalT-II and GlcAT-P. As well as HNK-1 carbohydrate, many glycans are expressed on specific target proteins, thereby regulating their functions. Thus, it is of great interest to study the mechanisms by which the functions of individual glycosyltransferases are regulated to produce glycoconjugates.
Supplementary Material
This work was supported in part by Grant-in-aid for Scientific Research (B) 21370053 (to S. O.) from the Ministry of Education, Culture, Sports, Science and Technology and in part by the Ministry of Health, Labor, and Welfare of Japan Health and Labour Sciences Research Grant on Comprehensive Research on Disability Health and Welfare, H21-012.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
M. Asano and N. Hashimoto, unpublished results.
- ER
- endoplasmic reticulum
- β4GalT
- β1, 4-galactosyltransferase
- GlcAT
- glucuronyltransferase
- HNK-1
- human natural killer-1
- PSA
- polysialic acid
- PST
- polysialyltransferase
- NCAM
- neural cell adhesion molecule
- ASOR
- asialo-orosomucoid.
REFERENCES
- 1. Ohtsubo K., Marth J. D. (2006) Cell 126, 855–867 [DOI] [PubMed] [Google Scholar]
- 2. Lowe J. B., Marth J. D. (2003) Annu. Rev. Biochem. 72, 643–691 [DOI] [PubMed] [Google Scholar]
- 3. Sugimoto I., Futakawa S., Oka R., Ogawa K., Marth J. D., Miyoshi E., Taniguchi N., Hashimoto Y., Kitazume S. (2007) J. Biol. Chem. 282, 34896–34903 [DOI] [PubMed] [Google Scholar]
- 4. Seko A. (2006) Trends Glycosci. Glycotechnol. 18, 209–230 [Google Scholar]
- 5. Seko A., Yamashita K. (2008) J. Biol. Chem. 283, 33094–33100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee P. L., Kohler J. J., Pfeffer S. R. (2009) Glycobiology 19, 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schwarting G. A., Jungalwala F. B., Chou D. K., Boyer A. M., Yamamoto M. (1987) Dev. Biol. 120, 65–76 [DOI] [PubMed] [Google Scholar]
- 8. Yoshihara Y., Oka S., Watanabe Y., Mori K. (1991) J. Cell Biol. 115, 731–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Voshol H., van Zuylen C. W., Orberger G., Vliegenthart J. F., Schachner M. (1996) J. Biol. Chem. 271, 22957–22960 [DOI] [PubMed] [Google Scholar]
- 10. Oka S., Terayama K., Kawashima C., Kawasaki T. (1992) J. Biol. Chem. 267, 22711–22714 [PubMed] [Google Scholar]
- 11. Terayama K., Oka S., Seiki T., Miki Y., Nakamura A., Kozutsumi Y., Takio K., Kawasaki T. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6093–6098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seiki T., Oka S., Terayama K., Imiya K., Kawasaki T. (1999) Biochem. Biophys. Res. Commun. 255, 182–187 [DOI] [PubMed] [Google Scholar]
- 13. Bakker H., Friedmann I., Oka S., Kawasaki T., Nifant'ev N., Schachner M., Mantei N. (1997) J. Biol. Chem. 272, 29942–29946 [DOI] [PubMed] [Google Scholar]
- 14. Yamamoto S., Oka S., Inoue M., Shimuta M., Manabe T., Takahashi H., Miyamoto M., Asano M., Sakagami J., Sudo K., Iwakura Y., Ono K., Kawasaki T. (2002) J. Biol. Chem. 277, 27227–27231 [DOI] [PubMed] [Google Scholar]
- 15. Morita I., Kakuda S., Takeuchi Y., Kawasaki T., Oka S. (2009) Neuroscience 164, 1685–1694 [DOI] [PubMed] [Google Scholar]
- 16. Kizuka Y., Tonoyama Y., Oka S. (2009) J. Biol. Chem. 284, 9247–9256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lo N. W., Shaper J. H., Pevsner J., Shaper N. L. (1998) Glycobiology 8, 517–526 [DOI] [PubMed] [Google Scholar]
- 18. Hennet T. (2002) Cell. Mol. Life Sci. 59, 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yoshihara T., Sugihara K., Kizuka Y., Oka S., Asano M. (2009) J. Biol. Chem. 284, 12550–12561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kido M., Asano M., Iwakura Y., Ichinose M., Miki K., Furukawa K. (1998) Biochem. Biophys. Res. Commun. 245, 860–864 [DOI] [PubMed] [Google Scholar]
- 21. Almeida R., Amado M., David L., Levery S. B., Holmes E. H., Merkx G., van Kessel A. G., Rygaard E., Hassan H., Bennett E., Clausen H. (1997) J. Biol. Chem. 272, 31979–31991 [DOI] [PubMed] [Google Scholar]
- 22. Guo S., Sato T., Shirane K., Furukawa K. (2001) Glycobiology 11, 813–820 [DOI] [PubMed] [Google Scholar]
- 23. Rutishauser U. (2008) Nat. Rev. Neurosci. 9, 26–35 [DOI] [PubMed] [Google Scholar]
- 24. Mühlenhoff M., Eckhardt M., Gerardy-Schahn R. (1998) Curr. Opin. Struct. Biol. 8, 558–564 [DOI] [PubMed] [Google Scholar]
- 25. Kudo M., Kitajima K., Inoue S., Shiokawa K., Morris H. R., Dell A., Inoue Y. (1996) J. Biol. Chem. 271, 32667–32677 [DOI] [PubMed] [Google Scholar]
- 26. Asano M., Furukawa K., Kido M., Matsumoto S., Umesaki Y., Kochibe N., Iwakura Y. (1997) EMBO J. 16, 1850–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kakuda S., Sato Y., Tonoyama Y., Oka S., Kawasaki T. (2005) Glycobiology 15, 203–210 [DOI] [PubMed] [Google Scholar]
- 28. Qasba P. K., Ramakrishnan B., Boeggeman E. (2008) Curr. Drug Targets 9, 292–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kakuda S., Oka S., Kawasaki T. (2004) Protein Expr. Purif. 35, 111–119 [DOI] [PubMed] [Google Scholar]
- 30. Kruse J., Mailhammer R., Wernecke H., Faissner A., Sommer I., Goridis C., Schachner M. (1984) Nature 311, 153–155 [DOI] [PubMed] [Google Scholar]
- 31. Nilsson T., Hoe M. H., Slusarewicz P., Rabouille C., Watson R., Hunte F., Watzele G., Berger E. G., Warren G. (1994) EMBO J. 13, 562–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Giraudo C. G., Maccioni H. J. (2003) Mol. Biol. Cell 14, 3753–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eckhardt M., Mühlenhoff M., Bethe A., Koopman J., Frosch M., Gerardy-Schahn R. (1995) Nature 373, 715–718 [DOI] [PubMed] [Google Scholar]
- 34. Kojima N., Yoshida Y., Tsuji S. (1995) FEBS Lett. 373, 119–122 [DOI] [PubMed] [Google Scholar]
- 35. Nakayama J., Fukuda M. (1996) J. Biol. Chem. 271, 1829–1832 [DOI] [PubMed] [Google Scholar]
- 36. Breton C., Mucha J., Jeanneau C. (2001) Biochimie 83, 713–718 [DOI] [PubMed] [Google Scholar]
- 37. Kizuka Y., Matsui T., Takematsu H., Kozutsumi Y., Kawasaki T., Oka S. (2006) J. Biol. Chem. 281, 13644–13651 [DOI] [PubMed] [Google Scholar]
- 38. Tagawa H., Kizuka Y., Ikeda T., Itoh S., Kawasaki N., Kurihara H., Onozato M. L., Tojo A., Sakai T., Kawasaki T., Oka S. (2005) J. Biol. Chem. 280, 23876–23883 [DOI] [PubMed] [Google Scholar]
- 39. Nakamura N., Yamakawa N., Sato T., Tojo H., Tachi C., Furukawa K. (2001) J. Neurochem. 76, 29–38 [DOI] [PubMed] [Google Scholar]
- 40. Mendiratta S. S., Sekulic N., Lavie A., Colley K. J. (2005) J. Biol. Chem. 280, 32340–32348 [DOI] [PubMed] [Google Scholar]
- 41. Huang H. H., Stanley P. (2010) J. Cell Biol. 190, 893–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ju T., Cummings R. D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16613–16618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ju T., Cummings R. D. (2005) Nature 437, 1252. [DOI] [PubMed] [Google Scholar]
- 44. Marzban H., Sillitoe R. V., Hoy M., Chung S. H., Rafuse V. F., Hawkes R. (2004) J. Neurocytol. 33, 117–130 [DOI] [PubMed] [Google Scholar]
- 45. Inoue M., Kato K., Matsuhashi H., Kizuka Y., Kawasaki T., Oka S. (2007) Brain Res. 1179, 1–15 [DOI] [PubMed] [Google Scholar]
- 46. Morita I., Kakuda S., Takeuchi Y., Itoh S., Kawasaki N., Kizuka Y., Kawasaki T., Oka S. (2009) J. Biol. Chem. 284, 30209–30217 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.