Abstract
The oligosaccharyltransferase complex catalyzes the transfer of oligosaccharide from a dolichol pyrophosphate donor en bloc onto a free asparagine residue of a newly synthesized nascent chain during the translocation in the endoplasmic reticulum lumen. The role of the less known oligosaccharyltransferase (OST) subunits, DC2 and KCP2, recently identified still remains to be determined. Here, we have studied DC2 and KCP2, and we have established that DC2 and KCP2 are substrate-specific, affecting amyloid precursor protein (APP), indicating that they are not core components required for N-glycosylation and OST activity per se. We show for the first time that DC2 and KCP2 depletion affects APP processing, leading to an accumulation of C-terminal fragments, both C99 and C83, and a reduction in full-length mature APP. This reduction in mature APP levels was not due to a block in secretion because the levels of sAPPα secreted into the media were unaffected. We discover that DC2 and KCP2 depletion affects only the γ-secretase complex, resulting in a reduction of the PS1 active fragment blocking Aβ production. Conversely, we show that the overexpression of DC2 and KCP2 causes an increase in the active γ-secretase complex, particularly the N-terminal fragment of PS1 that is generated by endoproteolysis, leading to a stimulation of Aβ production upon overexpression of DC2 and KCP2. Our findings reveal that components of the OST complex for the first time can interact with the γ-secretase and affect the APP processing pathway.
Keywords: Alzheimer Disease, Amyloid, Glycoprotein Biosynthesis, Glycosylation, Presenilin, Secretases
Introduction
Alzheimer disease (AD)2 is the primary cause of adult onset dementia, with a dramatic increase in the incidence of AD apparent in our aging population. AD is pathologically characterized by the accumulation of tangles and senile plaques. Senile plaques are composed of the amyloid-β (Aβ) peptides, Aβ40 and Aβ42 (1). The early onset familial form of AD is linked to three genes, amyloid precursor protein (APP) and presenilin (PS1 and PS2) (2, 3), strongly suggesting that the production of Aβ is a key factor in the pathogenesis of AD. Aβ is generated by proteolysis of APP, driven by the secretases found in the cell. Prior to proteolysis, APP undergoes a number of post-translational modifications, including N-glycosylation in the endoplasmic reticulum (ER) and O-glycosylation in the Golgi apparatus. In order to generate Aβ40 and Aβ42, APP is first cleaved by β-secretase and then by γ-secretase. For the cleavage of APP, β-secretase competes with α-secretase, which produces non-amyloidogenic peptides (4). γ-Secretase is an aspartyl protease complex composed of four core components, including presenilins (PS1/PS2), presenilin enhancer 2 (PEN2), nicastrin, and anterior pharynx-defective 1 (APH1) (2). Presenilin is the catalytic core of the γ-secretase complex consisting of nine transmembrane domains (5) and is cleaved by an unknown protease called “presenilinase” or self-cleavage stimulated by PEN2 binding of the cytosolic loop between transmembrane domains 6 and 7, releasing N- and C-terminal PS1 fragments, that contributes to γ-secretase activity (6, 7). Therefore, the identification of novel cellular factors that suppress the generation of Aβ could provide important drug targets for the treatment of AD.
N-Glycosylation is the most common type of protein modification that occurs at the ER in eukaryotic cells. This process is facilitated by the oligosaccharyltransferase (OST), an enzyme complex that catalyzes the attachment of a high mannose oligosaccharide en bloc onto suitable asparagine residues of newly synthesized polypeptide chains during their translocation into the ER lumen (8). What is intriguing is that most eukaryotes possess rather elaborate OST complexes that contain several additional subunits of poorly defined function. For the mammalian OST complex, we have proposed that the role of one such subunit, ribophorin I, is to selectively facilitate the N-glycosylation of certain precursors and more recently that ribophorin I acts as a substrate-specific enhancer (9–11). Hence, there still remains the opportunity to investigate the role of less well defined subunits of the OST.
The role of the less known OST subunits, DC2 and KCP2 (keratinocyte-associated protein 2), recently identified still remains to be determined (12). Here we show for the first time that DC2 and KCP2 are not core components globally required for N-glycosylation and OST activity. However, DC2 and KCP2 could be substrate-specific because we discover that APP N-glycosylation is disrupted in their absence. We show that both DC2 and KCP2 depletion leads to a reduction of the endoproteolytic cleavage of PS1 involved in the γ-secretase cleavage pathway of APP. Conversely, induction of DC2 and KCP2 expression results in a dosage increase of PS1 N-terminal fragments. Consequently, there is a stimulation of Aβ production, which could be due to an elevated level of γ-secretase activity. Hence, these results demonstrate for the first time that alteration of the OST subunits, DC2 and KCP2, specifically alters the γ-cleavage of APP and suggest that these subunits act specifically as modulators of γ-secretase activity.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Cell culture reagents were from Invitrogen or LONZA, T7 RNA polymerase and rabbit reticulocyte lysate were from Promega, SP6 RNA polymerase was from New England Biolabs, and EasyTag l-[35S]methionine was from PerkinElmer Life Sciences. All other chemicals were from Sigma or BDH/Merck. All sera are rabbit polyclonal unless otherwise stated. Antisera specific for the C terminus of nicastrin (Sigma) or BACE1 (Merck), mouse monoclonal p58 (Sigma), mouse monoclonal Hsc70 (Novus), mouse monoclonal β-actin (Sigma), and PEN-2 (Abcam) were purchased. Antisera recognizing ribophorin I, ribophorin II, STT3A DadI, and OST48 were described previously (10). Antisera specific for DC2, KCP2, Sec61β, TRAM, and APP made to order by Eurogentec were gifts from Professor Stephen High (University of Manchester), and Notch 1 (H-131) and Notch 1 intracellular domain fragment (C-20), available from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), were gifts from Dr. Fabrice Lalloué (Université de Limoges). cDNA constructs for Notch 1 were a kind gift from Dr. Martin Baron (University of Manchester), and PS1 from Prof. Konrad Beyreuther (ZMBH, University of Heidelberg).
Molecular Cloning and DNA Manipulation
The expressed sequence tag clones of human DC2 (IMAGE: 10008408; NM_021227) and human KCP2 (IMAGE: 100011011; NM_173832) were obtained from Geneservice and sequenced to confirm the correct full-length coding sequence. Both DC2 and KCP2 cDNA were cloned into the vector, pDNA5FRT/V5/TA, with a C-terminal V5 tag (Invitrogen) for transient expression studies and subcloned into BamHI/NotI sites of the tetracycline-responsive expression vector pDNA5/FRT/TO (Invitrogen). Site-directed mutagenesis was used to add a stop codon to generate untagged versions of DC2 and KCP2. Subcloned and mutagenized constructs were confirmed by DNA sequencing.
Cell Culture and Transfection
HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 2 mm glutamine, and 1% non-essential amino acids and grown at 37 °C with 5% CO2. SH-SY5Y cells were frown in RPMI 640 medium supplemented with 10% fetal calf serum and 5 mm glutamine. Invitrogen Tet-On HEK293 cells were grown in DMEM containing 100 μg/ml zeocin (Invitrogen). For both transient and stable transfection, cells were transfected by using Lipofectamine 2000 (Invitrogen) and analyzed after 18–24 h in the case of the transients. Stable HEK293T-REx cell lines expressing DC2, KCP2, and ribophorin I under the control of a tetracycline-inducible promoter were selected after 48 h post-transfection in the presence of 100 μg/ml hygromycin. Clonal lines were selected based on level of inducible expression as judged by Western blotting. Expression was induced by 1 μg/ml tetracycline.
SDS-PAGE and Western Blotting
Cells were grown and transfected or induced in 6-well dishes and harvested after 18–24 h unless stated otherwise. Cells were rinsed twice with PBS and lysed in Laemmli sample buffer (60 mm Tris, pH 6.8, 4% SDS, 5% (w/v) glycerol). The cell lysates were then sonicated on ice and centrifuged to remove insoluble material. The resulting supernatants were collected, and the protein concentration was determined using a Bio-Rad protein assay kit according to the manufacturer's protocol. Approximately, 50 mm DTT and 0.05% bromphenol blue were added to the samples. Samples were incubated at 95 °C for 5 min or 70 °C for 10 min, and 25 μg of total protein was loaded onto 8, 10, 12, or 15% polyacrylamide gels containing 0.1% SDS. Gels were then transferred to 0.45 μm polyvinylidene difluoride membrane (Millipore) and immunoblotted using an enhanced chemiluminescence system (13). For quantification of Western blots, data are expressed graphically and represent the relative band intensity for each component detected following correction of any background. Signals were quantified using Gbox software (Syngene), and several different exposure times were used to ensure that each image analyzed gave a linear response.
Statistical Analysis
Statistical analysis (two-sample t test) was carried out using SPSS 10.1 software. For all other experimental procedures, see the supplemental material.
RESULTS
Expression and Localization of DC2 and KCP2 in Mammalian Tissues
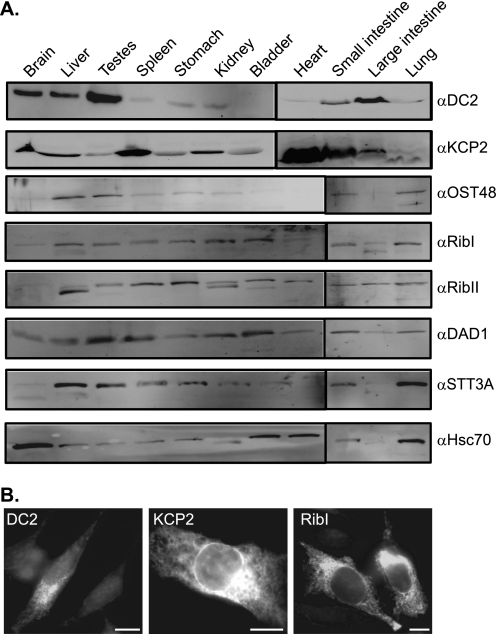
DC2 and KCP2 were previously identified as two subunits of the OST in proliferating animal cells (12), but whether DC2 and KCP2 are expressed in differentiated tissues of multicellular organisms and, if so, where is not known. To address these questions, we analyzed protein extracts from 11 different adult mouse tissues by immunoblotting with antibodies to several core subunits of the human OST (Fig. 1A). To determine the tissue distribution of DC2 and KCP2 relative to the other OST subunits, antibodies were raised to the C terminus of DC2 and KCP2. We observed the presence of DC2 in the mouse brain, liver, and small and large intestines with enrichment in the testes (Fig. 1A, DC2 panel). Similarly, DAD1 was enriched in the testes (Fig. 1A, compare DC2 and DAD1 panel). Conversely, KCP2 was ubiquitously expressed throughout all tissues with enrichment in the heart (Fig. 1A, KCP2 panel). The other OST subunits appeared to be ubiquitously expressed and were found in most tissues (Fig. 1A, compare panels OST48–STT3A). Interestingly, we detected faster migrating bands of OST48, ribophorin I and II, and Hsc70, which are indicative of other isoforms present in different tissues (14). Next, we were interested in studying the cellular localization of DC2 and KCP2 either by overexpressing in cells or staining for endogenous levels of the protein. The subcellular distribution of DC2 and KCP2 in HeLa cells was examined by indirect immunofluorescence microscopy (Fig. 1B). The expression of DC2 and KCP2 showed a reticular pattern typical of the ER (Fig. 1B). Similar results were obtained either by the overexpression of ribophorin I by observing the endogenous levels (Fig. 1B) or costaining with the ER markers Sec61β or ribophorin I, which colocalized with DC2 and KCP2 (data not shown). Hence, we reconfirm that DC2 and KCP2 are localized to the ER compartment of the cell. Most noticeably, it is clear that KCP2 can be found ubiquitously expressed throughout all tissues, whereas DC2 is confined to certain tissues analyzed so far.
FIGURE 1.
Expression and localization of DC2 and KCP2 in mammalian tissues. A 24-week-old male mouse was sacrificed, and organs were removed and lysed in lysis buffer as described in the supplemental material. A, equal amounts of protein from 10,000 × g supernatant fractions from different mouse tissue extracts were separated by SDS-PAGE and analyzed by immunoblotting by using antibodies against DC2, KCP2, OST48, ribophorin I (RibI), ribophorin II (RibII), DAD1, STT3A, and Hsc70. Differential mobilities of OST48, ribophorin I, ribophorin II, STT3A, and Hsc70 may represent tissue-specific isoforms. B, HeLa cells were transiently transfected with DC2 (left), KCP2 (middle), or ribophorin I (right) and stained for DC2, KCP2, and ribophorin I, followed by an FITC-conjugated secondary antibody. Bar, 20 μm.
DC2 and KCP2 Are Dispensable for N-Glycosylation Activity of the OST Complex
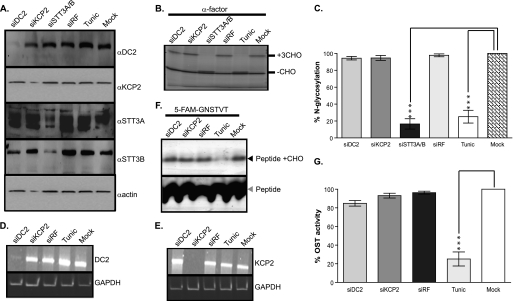
We had previously studied the role of ribophorin I and the catalytic subunits, STT3A and STT3B, using RNA interference (RNAi) to tease out their function and mechanism of action in N-glycosylation (10). First of all, HeLa cells were treated with RNA duplexes specific for DC2, KCP2, and STT3A/B mRNAs, and cells were then analyzed for levels of these subunits. Three days after siRNA treatment, Western blotting revealed that cellular levels of DC2, KCP2, and STT3A/B were specifically reduced to 20% or less of those seen in control cells (Fig. 2A, compare lanes 1–3 with lane 6 for each of the products indicated). In contrast, the levels of other proteins were similar, as evidenced by other subunits of the OST complex and the amount of β-actin (Fig. 2A, lanes 1–6, α-actin panel). Cells treated with a non-functional siRNA showed no such reduction in protein levels (Fig. 2A, lane 4, see DC2, KCP2, STT3A, and STT3B panels), showing that the losses observed were not due to pleotropic effects resulting from transfection. Having established that we could knock down DC2 and KCP2 using siRNA treatment, next we addressed the question of whether DC2 and KCP2 were required for efficient N-glycosylation of glycoproteins. We had previously established an assay to analyze the effect of OST subunit knockdown upon the modification of a glycoprotein (9, 10, 13). Briefly, this assay utilizes cells RNAi-treated in combination with an in vitro readout of N-glycosylation efficiency using semipermeabilized cells (9). Using this assay, we analyzed the soluble glycoprotein α-factor, the precursor for α-mating factor from Saccharomyces cerevisiae, with a cleavable, N-terminal signal sequence and three N-glycosylation sites. We synthesized α-factor using rabbit reticulocyte lysate supplemented with semipermeabilized HeLa cells that had been treated with DC2, KCP2, or STT3A/B siRNA duplexes (Fig. 2B). First, when we analyzed cells pretreated with the antibiotic tunicamycin that inhibits synthesis of the lipid-linked oligosaccharide precursor or STT3A/B siRNA duplexes, we observed a dramatic reduction in N-glycosylation to less than 30% (Fig. 2, B (compare lanes 3, 5, and 6) and C). However, when we analyzed both DC2 and KCP2 depletion upon N-glycosylation of α-factor, it was immediately apparent that there was no significant effect (Fig. 2, B (compare lanes 1, 2, and 6) and C). This in vitro N-glycosylation assay relies upon the protein sequence and its transport across the ER membrane, and the effect of knockdown can be affected by the substrate presented to the catalytic core (10). To address the function of DC2 and KCP2 in the OST complex, we decided to use a previously established fluorescent peptide assay (17) as an alternative way to measure the OST activity of semipermeabilized cells. This assay is similar to the tripeptide assay, which uses an iodinated peptide acceptor (Nα-Ac-Asn-Tyr-Thr-NH2) and was important in the identification of the functional OST complex (18–20). This assay differs from the first assay using semipermeabilized cells and in vitro translation in that the tripeptides are passively transported into the ER and represent a minimal substrate for the OST that is unaffected by either folding status or substrate being presented to the catalytic core. The fluorescent peptide assay uses an N-terminally fluorescence-labeled (5-carboxyfluorescein) hexapeptide sequence that contains the tripeptide motif required for N-glycosylation (5-carboxyfluorescein-Gly-Asn-Ser-Thr-Val-Thr-NH2, where the tripeptide motif is underlined). First, we used RT-PCR and showed that mRNA levels of DC2 and KCP2 were specifically reduced to 20% or less of that in control cells after 72 h (Fig. 2, D and E, compare lanes 1 and 2 with lane 5 for both DC2 and KCP2, respectively). Conversely, the levels of GAPDH were mainly unaffected (Fig. 2, D and E, lanes 1–5, GAPDH panels), and a non-functional siRNA also had no effect (Fig. 2, D and E, lane 3, see DC2 and KCP2 panels), confirming that the losses were not due to pleiotropic effects. When we performed the N-glycosylation assay using the fluorescent peptide as a substrate, we observed a dramatic reduction in N-glycosylation in the HeLa cells pretreated with tunicamycin (Fig. 2, F (lanes 4 and 5) and G). Likewise, depletion of DC2 and KCP2 led to no significant effect upon N-glycosylation (Fig. 2, F (lanes 1, 2, and 5) and G).
FIGURE 2.
DC2 and KCP2 are dispensable for N-glycosylation activity of the OST complex. A, HeLa cells treated with siRNA duplexes for DC2 (siDC2), KCP2 (siKCP2), STT3A and STT3B (siSTT3A/B), or a siRisc-free control (siRF) or mock-transfected (Mock). HeLa cells were also incubated overnight with 20 μg/ml tunicamycin (Tunic). Western blots were performed after treatments with antibodies against DC2, KCP2, STT3A, STT3B, and β-actin. B, S. cerevisiae α-factor was synthesized as a radiolabeled polypeptide using a rabbit reticulocyte lysate system supplemented with semipermeabilized HeLa cells prepared 72 h after transfection with siRNAs specific for the mRNAs encoding DC2 (lane 1), KCP2 (lane 2), or STT3A/B (lane 3), or a non-functional control siRNA (siRF) (lane 4) or following mock transfection (lane 6). As a positive control for loss of N-glycosylation, HeLa cells were incubated with 20 μg/ml tunicamycin overnight prior to isolation on day 2 (lane 5). The resulting glycosylated (+CHO) and non-glycosylated (−CHO) polypeptides are shown. C, the relative proportion of glycosylated polypeptide was calculated for each sample and expressed as a percentage of the total protein recovered. The values expressed in the graph are the mean ± S.E. (error bars) of three independent experiments. Levels of N-glycosylation that differ from the mock-treated control with a significance of p < 0.001 (***) are indicated by asterisks. D and E, HeLa cells treated with siRNA duplexes for DC2 (siDC2) and KCP2 (siKCP2) or a siRisc-free control (siRF) or mock-transfected (Mock). HeLa cells were also incubated overnight with 20 μg/ml tunicamycin (Tunic). RT-PCR was performed on RNA isolated after the above treatments with primers specific for DC2 and GAPDH (D) or KCP2 and GAPDH (E). F, 5-carboxyfluorescein-GNSTVT peptide was added to the reaction mix containing free lipid-linked oligosaccharide donor, LLO buffer supplemented with semipermeabilized HeLa cells prepared 72 h after transfection with siRNAs specific for the mRNAs encoding DC2 (lane 1) or for KCP2 (lane 2) or a non-functional control siRNA (siRF) (lane 3) or following mock transfection (lane 5). As a positive control for loss of N-glycosylation, HeLa cells were incubated with 20 μg/ml tunicamycin overnight prior to isolation on day 3 (lane 4). The reaction was separated through a 15–25% gradient gel and analyzed using LAS3000 using a blue LED. The non-modified peptide (Peptide) and resulting glycosylated peptide products (Peptide +CHO) are shown. G, the resulting peptide products and the proportion of N-glycosylated products obtained after various treatments are indicated as described above.
DC2 and KCP2 Affect APP Processing
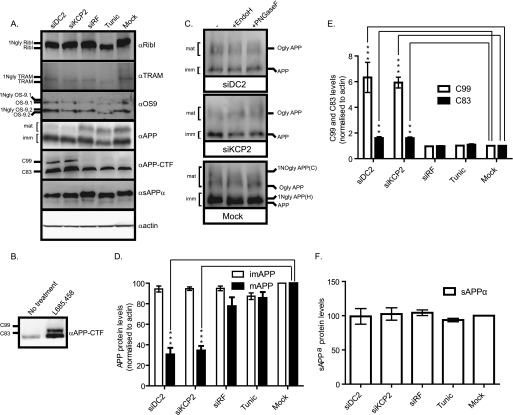
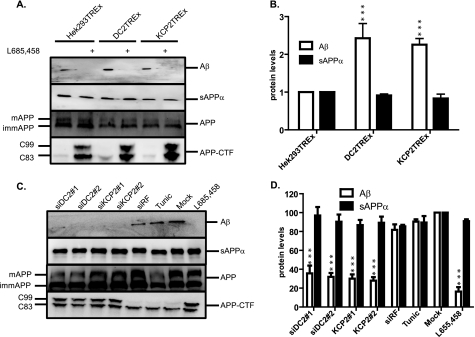
To investigate further the global effect of DC2 and KCP2 knockdown, we analyzed by Western blot the endogenously expressed proteins found in HeLa cells and transiently expressed the full-length version of APP (APP695). The first three proteins that we analyzed are examples of glycoproteins: the subunit of the OST, ribophorin I, a subunit associated with the Sec61 complex; the translocating chain-associating membrane (TRAM) protein; and the subunit implicated in the ER-associated degradation pathway, OS-9. Treatment with tunicamycin clearly shows that these proteins are N-glycosylated, as indicated by the faster migrating band (Fig. 3A, RibI, TRAM, and OS-9 panels, lanes 4 and 5). Next, we analyzed the DC2- and KCP2-depleted samples and observed no effect upon N-glycosylation of all three glycoproteins (Fig. 3A, RibI, TRAM, and OS-9 panels, lanes 1, 2, and 5). In the same experiment, we also blotted for the other components of the OST, DAD1 and OST48, which are not glycoproteins. We observe no effect of DC2 and KCP2 depletion upon DAD1 and OST48 protein levels (data not shown). Western analysis using an antibody specific for the C terminus of APP recognizes a number of specific APP products that were observed (see Fig. 3A, lane 5, αAPP panel). In particular, the products that were observed included two major bands corresponding, respectively, to (i) a lower molecular weight form corresponding to immature APP (∼90 kDa) that was generated after core N-glycosylation at the ER, carries only high mannose form N-glycans, and was sensitive to both endoglycosidase H and peptide:N-glycosidase F treatments (Fig. 3, A (lane 5, αAPP panel) and C (Mock panel)) and (ii) a higher molecular weight form corresponding to mature APP (∼100 kDa), which was O-glycosylated, carries complex N-linked sugars that were both added to APP during transit in the Golgi complex, and was sensitive solely to peptide:N-glycosidase F treatment (Fig. 3, A (lane 5, αAPP panel) and C (Mock panel)). In addition to these products, we also detected the C-terminal fragments (CTFs) C99 and C83 migrating below ∼15 kDa that are generated after either β- or α-secretase cleavage of APP, respectively (Fig. 3A, lane 5, αAPP-CTF panel). HeLa cells treated with tunicamycin affected the migration of both mature and immature APP products due to the block of N-glycosylation, but the ratio of mature and immature APP levels was not significantly affected (Fig. 3, A (compare lanes 4 and 5, αAPP panel) and D). Likewise, when HeLa cells were treated with DC2 and KCP2 RNA duplexes showed that N-glycosylation of both APP products (immature and mature) was blocked and insensitive to both endoglycosidase H and peptide:N-glycosidase F treatment (Fig. 3, A (compare lanes 1, 2, 4, and 5, αAPP panel) and C (siDC2 and siKCP2 panels)). In addition, DC2 and KCP2 depletion resulted in a significant accumulation of both C99 and C83 products similar to that observed when the γ-secretase is chemically inhibited with L-685,458 (see Fig. 3, A (compare lanes 1 and 2 with lane 5), B (αAPP-CTF panels), and E). At the same time, mature APP levels were reduced considerably by 69.4% (p < 0.001) for DC2 and 65.5% (p < 0.001) for KCP2 (see Fig. 3, A (compare lanes 1 and 2 with lane 5, αAPP panel) and D). This reduction in mature APP levels was not due to a block in secretion because the levels of sAPPα secreted into the media were unaffected (Fig. 3, A (compare lanes 1–5, sAPPα panel) and F). These results also show that α-secretase cleavage undergone by ADAM10 was unaffected by DC2 and KCP2 knockdown. Interestingly, there was no significant effect upon the ratio of mature and immature APP products when HeLa cells were treated with tunicamycin, indicating that N-glycosylation per se was not responsible for the reduction in mature APP that we observe with DC2 and KCP2 depletion (Fig. 3, A (compare lanes 4 and 5, αAPP panel), C, and D). The non-targeting siRNA, Risc-free (siRF) served as an internal control for these experiments; there was no effect upon the cellular levels of APP products with this treatment (Fig. 3, A (compare lanes 3 and 5, αAPP panel), C, and D). There are a number of other substrates that undergo γ-secretase cleavage. Notch protein is cleaved by a number of proteases at different locations in the cell referred to as S1, S2, and S3 cleavages (reviewed in Ref. 21). S1 cleavage in the extracellular domain of Notch occurs constitutively in the trans-Golgi network and is mediated by a furin-like convertase; reassembly of the fragments creates a heterodimeric Notch receptor at the cell surface. This event has been most closely characterized with respect to mammalian Notch, but there is evidence that fly Notch is similarly processed. S2 cleavage by a disintegrin/metalloprotease (tumor necrosis factor-α converting enzyme (TACE) in vertebrates) occurs in response to ligand binding and releases the majority of the extracellular domain. The resulting membrane-anchored fragment, referred to as the Notch extracellular truncation, is subject to intramembranous S3 cleavage by γ-secretase, which finally releases the Notch intracellular domain fragment. First, in order to study the effect of DC2 and KCP2 upon γ-secretase (S3) cleavage of Notch, we depleted both DC2 and KCP2 levels by siRNA treatment for 2 days and then transiently transfected human Notch 1 into HeLa cells. On day 3, to analyze γ-secretase cleavage of Notch 1, we stimulated γ-secretase cleavage by the addition of 5 mm EDTA for 15 min at 37 °C (22) and isolated cell extracts. We performed Western analysis with antibodies against DC2 and KCP2 to verify the efficiency of knockdown, more than 80% depletion (see supplemental Fig. S1A). We detected Notch 1 expression using an antibody recognizing the full-length protein (NotchFL) (supplemental Fig. S1A). We observed that the Notch intracellular domain fragment generated after γ-secretase cleavage can be blocked by the addition of the γ-secretase inhibitor (L-685,458) but was unaffected by both DC2 and KCP2 depletion (supplemental Fig. S1, A and B). These results confirm that DC2 and KCP2 may alter how the γ-secretase sees its substrates and could change the location of cleavage in the cell, and this could be unique to APP. Our results show that DC2 and KCP2 depletion specifically affect APP glycosylation and processing. In addition, the above findings suggest that depleting DC2 and KCP2 could affect components involved in the APP processing pathway.
FIGURE 3.
DC2 and KCP2 depletion affects APP processing. A, media and lysates of HeLa cells treated with siRNA duplexes for DC2 (siDC2) and KCP2 (siKCP2) or a siRisc-free control (siRF) or mock-transfected (Mock). HeLa cells were also incubated overnight with 20 μg/ml tunicamycin (Tunic). For APP analysis, HeLa cells were transfected with APP695, and after 5 h, the medium was replaced with fresh OptiMEM, and cells were incubated for 18 h. The following day, the medium was collected and concentrated by TCA precipitation with a carrier protein, and cell lysates were prepared. Western analysis was performed for ribophorin I (RibI), TRAM, and OS-9 (two isoforms present, OS-9.1 and OS9.2). In most cases, the presence and number (x) of N-linked glycans present on a particular protein is indicated by the suffix xNgly. Shown is Western analysis with APP recognizing the C terminus of APP and various isoforms of APP including immature (∼110 kDa; imm) and mature (∼125 kDa; mat). Two CTFs, C99 and C83, resulted from proteolytic cleavage of APP. The analysis of actin levels confirmed that similar levels of protein were present in each lane. In addition, HeLa cells treated with or without the γ-secretase inhibitor, L-685,458, are shown (B). C, lysates treated with siRNA duplexes for DC2 and KCP2 or mock-transfected and transfected with APP695 were treated with or without endoglycosidase H (EndoH) and peptide:N-glycosidase F (PNGaseF). For APP, three populations of glycoproteins reflecting products with high mannose type N-linked glycan (1NglyAPP(H)), O-linked glycans (OglyAPP), and both O-linked and complex N-glycans (1NOglyAPP(C)) were detected. D–F, graphs showing the density analysis of APP (D), C83 and C99 (E), and sAPPα (F). Data in D and E were normalized by dividing the density of the APP, C83, and C99 bands by the density of the β-actin band. D and F, data were represented as percentage change compared with the mock controls. E, data were represented as -fold increase over the mock control. **, p < 0.01; ***, p < 0.001. Error bars, S.E.
DC2 and KCP2 Knockdown Affects the γ-Secretase Complex
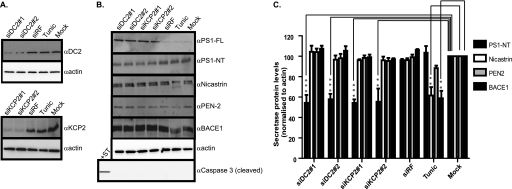
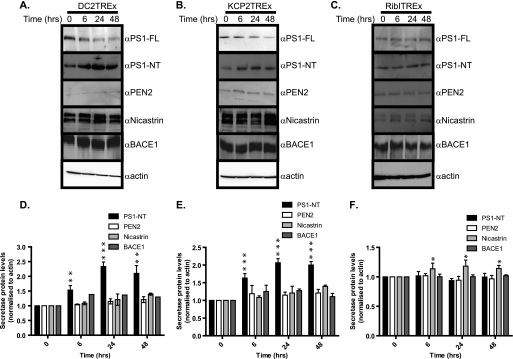
The site of cleavage is of clinical relevance because Aβ peptides (Aβ 39–43) derived from β-secretase and γ-secretase cleavage of APP are major components of the amyloid plaques that are a characteristic brain lesion of individuals with Alzheimer disease. The pepsin-like aspartyl protease β-secretase (BACE1) cleaves the ectodomain of APP, releasing the soluble protein sAPPβ (23–25). Alternative cleavage by a metalloprotease called α-secretase (ADAM10) leads to proteolysis within the Aβ region and produces a slightly longer soluble protein sAPPα (26, 27). After ectodomain cleavage, γ-secretase cuts the remaining membrane-bound stub to release the APP intracellular domain as well as either Aβ (via the β-secretase pathway) or the N-terminally truncated peptide p3 (via the α-secretase pathway) (28). The failure to remove Aβ peptides from the body, in particular Aβ42 peptides, may result in the formation of amyloid plaques, a neuropathological hallmark of Alzheimer disease. To establish which pathway of APP processing is affected by DC2 and KCP2 depletion, we analyzed cell lysates isolated after DC2 and KCP2 knockdown for cellular levels of various components of the γ- and β-secretases. We observed more than 80% depletion of the cellular protein levels of both DC2 and KCP2 after 3 days of knockdown (Fig. 4A, see DC2 and KCP2 panels). Similarly, we observed a reduction in mature APP products for both siRNAs against DC2 and KCP2. Next, we blotted for components of the γ-secretase (PS1, nicastrin, and PEN-2), the complex responsible for the final stage of APP processing generating Aβ peptides. When we analyzed the effect of DC2 and KCP2 depletion upon these components, we observed that the amount of N-terminal fragments of PS1 was reduced for DC2 and KCP2 knockdowns by more than 42% (p < 0.001) and 44.6% (p < 0.01), respectively (Fig. 4, B and C). PS1 can undergo endoproteolysis by an unidentified aspartyl protease called the “presenilinase” or self-cleavage assisted by PEN2, resulting in cleavage of PS1 within a cytosolic loop between transmembrane domains 6 and 7, releasing N- and C-terminal fragments found in the active γ-secretase complex along with nicastrin, PEN-2, and APH-1 (7, 29). In contrast, we observe no significant effect upon endoproteolysis of PS1 when N-glycosylation was blocked, indicating that N-glycosylation was not responsible for the reduction in endoproteolysis of PS1 (Fig. 4, B and C). Similarly, we observe no effect or only a minor effect upon the γ-secretase subunit, PEN2, or the β-secretase component, BACE1, upon inhibition of N-glycosylation (Fig. 4, B and C). Nevertheless, both nicastrin and BACE1 protein levels were significantly affected by tunicamycin treatment by 44.5 and 41% (p < 0.01), respectively (Fig. 4, B and C).
FIGURE 4.
DC2 and KCP2 knockdown affects PS1. A, lysates of HeLa cells treated with siRNA duplexes for DC2 (siDC2#1 and siDC2#2) and KCP2 (siKCP2#1 and siKCP2#2) or a siRisc-free control (siRF) or mock-transfected (Mock). HeLa cells were also incubated overnight with 20 μg/ml tunicamycin (Tunic) or for 6 h with 1 μm staurosporine. Shown is Western analysis with rabbit polyclonal antibodies raised against DC2 and KCP2 (αDC2 and αKCP2) and β-actin. B, HeLa lysates prepared as above were used for Western analysis using rabbit polyclonal antibodies against PS1 detecting full-length (PS1-FL) and PS1 N-terminal fragment (PS1-NT) nicastrin and PEN-2 components of the γ-secretase complex and the β-secretase, BACE1. In addition, caspase 3 cleavage was analyzed with the staurosporine (+ST) control, which induces caspase 3 cleavage, leading to apoptosis. The analysis of β-actin levels confirmed that similar levels of protein were present in each lane. C, the percentage of secretase protein levels for PS1-NT, nicastrin, PEN2, and BACE1 remaining calculated as a ratio of band intensity compared with the mock minus background intensity as an average from three independent experiments with error bars representing S.E. Levels of protein that differ from the mock-treated control with a significance of p < 0.01 (**) and p < 0.001 (***) are indicated by asterisks.
A number of reports have suggested a role for caspase activation and apoptosis in Alzheimer disease (30, 31). In the same experiments, we also addressed the question of whether DC2 and KCP2 depletion resulting in a perturbation of PS1 endoproteolysis could be due to an alteration in the cellular apoptotic response. When HeLa cells were treated with the proapoptotic drug staurosporine for 8 h, we observed a product of caspase 3 cleavage (Fig. 4B, Caspase panel, +ST). We observed no caspase 3 cleavage products for both DC2 and KCP2 knockdown, indicating that the effect upon PS1 endoproteolysis that we observed was not due to caspase activation resulting in apoptosis.
Overexpression of DC2 and KCP2 Increases Active γ-Secretase
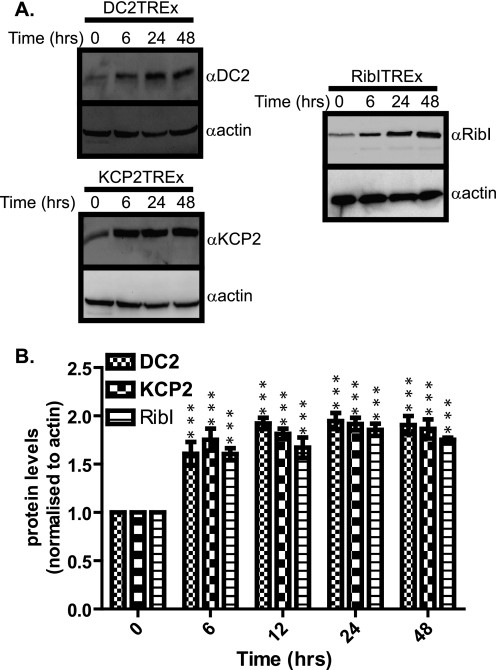
In order to understand the effect of DC2 and KCP2 knockdown upon APP processing, we investigated the consequence of overexpression using the Flp-In T-REx system (Invitrogen) to generate stable mammalian cell lines exhibiting tetracycline-inducible expression of DC2, KCP2, and ribophorin I as a control from a specific genomic location. We cloned the cDNAs of DC2, KCP2, and ribophorin I into pcDNA5/FRT/TO mammalian inducible expression vector and stably expressed these genes in an HEK293T-REx-inducible cell line. When we treated the stable cell lines with 1 μg/ml tetracycline, we observed an induction of DC2, KCP2, and ribophorin I expression, 1.61-, 1.75-, and 1.6-fold (p < 0.001) after a 6-h treatment, respectively (Fig. 5, A and B). The peak of expression for DC2, KCP2, and ribophorin I, 1.95-, 1.92-, and 1.86-fold (p < 0.001), respectively, was attained after 24 h of treatment and leveled out by 48 h (Fig. 5, A and B). Next, we performed the same time course of induction over 48 h and isolated samples at specific time points postinduction (i.e. 0, 6, 24, and 48 h) and performed Western blots to determine the effects of DC2 and KCP2 overexpression upon the β- and γ-secretases. We found that the overexpression of DC2 and KCP2 leads to a significant increase in PS1-NT protein levels that peaks at 2.33-fold for DC2 and 2.1-fold for KCP2 after 24 h of treatment, confirming our previous observations (Fig. 6, A–C and D–E, respectively). In contrast, there was no significant effect of DC2 and KCP2 overexpression upon the other subunits of the γ- and β-secretases, PEN2, nicastrin, and BACE1 protein levels (Fig. 6, A, B, D, and E). Likewise, we observe no effect of ribophorin I overexpression upon PS1-NT protein levels (Fig. 6, C and F). However, we did observe a small significant increase, 1.2-fold (p < 0.05), peaking at 24 h for nicastrin protein levels with ribophorin I overexpression (Fig. 6, C and F). These results suggest that DC2 and KCP2 have a more specific and distinct role in APP maturation because overexpression of other ER chaperones (BIP, calnexin ERdj3, and ERdj4) inhibits the maturation of APP (32, 33). The overexpression of DC2 and KCP2 results in the reverse effect upon PS1 endoproteolysis. Generally, there is a correlation between increasing protein expression of DC2 and KCP2 and an increase in PS1 endoproteolysis. It is presumed that this leads to more active complex being available to cleave APP. In addition when another subunit of the OST complex, ribophorin I, is overexpressed, we observed no effect upon PS1 endoproteolysis. In addition, our results confirm that the overexpression of both DC2 and KCP2 enhances endoproteolysis of PS1, and this effect was unique to these OST subunits.
FIGURE 5.
Inducible expression of DC2, KCP2, and ribophorin I. A, stable HEK293T-REx cell lines DC2 (DC2T-REx), KCP2 (KCP2T-REx), and ribophorin I (RibIT-REx) were incubated with 1 μg/ml tetracycline over a 48-h time course. Lysates were isolated at 0, 6, 24, and 48 h postinduction and analyzed by Western blot for DC2, KCP2, ribophorin I, and β-actin. B, the protein levels of DC2, KCP2, and ribophorin I were quantified and normalized against the loading control, β-actin, and indicated as a -fold increase over time (the value at the start (0 h) is 1). Levels of protein that increased at specific time points over a 48-h period that were significantly higher p < 0.001 (***)) are indicated. Error bars, S.E.
FIGURE 6.
DC2 and KCP2 affect the γ-secretase. A–C, lysates isolated for DC2T-Rex, KCP2T-Rex, and RibIT-REx at 0, 6, 24, and 48 h postinduction were analyzed by Western blot for PS1-FL, PS1-NT, PEN-2, nicastrin, BACE1, and β-actin. D–F, the -fold increase of PS1-NT, nicastrin, PEN2, and BACE1 calculated as a ratio of band intensity compared with the mock minus background intensity as an average from three independent experiments with error bars representing S.E. Levels of protein that differ from the mock-treated control with a significance of p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***) are indicated by asterisks.
DC2 and KCP2 Alter Aβ Production
We sought to investigate whether this effect upon PS1 endoproteolysis has any further involvement in the amyloidogenic processing pathway, in particular in the production of Aβ. Using a well characterized γ-secretase cleavage assay, we examined whether DC2 and KCP2 could influence the cleavage of the full-length version of APP (APP695) construct by γ-secretase. This assay makes use of the APP695 plasmid, with an intact γ-secretase cleavage site that is expressed in HEK293T-REx, DC2T-REx, and KCP2T-REx stable cell lines. Next, we performed an induction time course as before over 24 h except we isolated both cell lysates (for 0, 6, and 24 h) and medium samples (for 6 and 24 h). Afterward, we analyzed by Western blot the medium samples using an Aβ-specific antibody, W02, that recognizes all Aβ products and cell lysates for APP expression levels. We observed a significant dramatic increase in the amount of Aβ products secreted into the media after 24-h induction of DC2 (2.43-fold (p < 0.001)) and KCP2 (2.26-fold (p < 0.001)) compared with HEK293T-REx (Fig. 7, A (Aβ panel) and B). We can confirm that the Aβ products secreted into the media are γ-secretase products because we can block γ-secretase cleavage by use of the inhibitor L-685,458 (Fig. 7A, Aβ panel, compare lanes 1–6). We observed no significant effect of DC2 and KCP2 induction upon sAPPα products secreted into the media detected with the W02 antibody that recognizes APP products cleaved by the α-secretase as judged by no perturbation of products (Figs. 7, A (sAPPα panel) and B). In addition, we performed a similar set of experiments to analyze the effect of knocking down DC2 and KCP2 upon Aβ generation. We performed the knockdown of DC2 and KCP2 on day 0, followed by the expression of APP695 on day 1, replaced the medium on day 2, and harvested the cell lysates and medium on day 3 for analysis. Strikingly, we found that both DC2 and KCP2 knockdown results in a significant dramatic decrease in Aβ protein levels to 35.6, 32, 30.1, and 28% (p < 0.001) for siDC2#1, siDC2#2, siKCP2#1, and siKCP2#2, respectively (Fig. 7, C and D). We observe no significant effect when we knockdown DC2 and KCP2 upon sAPPα production (Fig. 7, C and D). We can conclude from these results that DC2 and KCP2 can act and enhance specifically γ-secretase cleavage of APP.
FIGURE 7.
DC2 and KCP2 stimulate Aβ production. A, HEK293T-REx cell lines DC2 (DC2T-REx), KCP2 (KCP2T-REx), or mock (HEK293T-REx) were transfected with full-length APP695 (APP) prior to overexpression of DC2 and KCP2 by the induction with tetracycline. γ-Secretase cleavage was inhibited by the addition of 1 μm L-685,458. The medium and cell lysate samples were collected at 24 h postinduction. The medium samples were TCA-precipitated with a carrier protein and analyzed by Western blot using antibodies specific against soluble αAPP and Aβ peptides. Cell lysates were analyzed using the C-terminal antibody against APP for APP and APP-CTF. B, the amount of Aβ peptide and sAPPα present in the medium samples represented as -fold increase at 24 h was calculated. The data shown represent an average from three independent experiments with the S.E. (error bars). Levels of Aβ peptide and sAPPα that differ from the control (HEK293T-REx) with a significance of p < 0.001 (***) are indicated by asterisks. C, both the medium and cell lysate samples of HeLa cells treated with siRNA duplexes for DC2 (siDC2#1 and siDC2#2) and KCP2 (siKCP2#1 and siKCP2#2) or a siRisc-free control (siRF) or mock-transfected (Mock) followed by transfection with full-length APP695 (APP) and Western analysis. D, the percentage of Aβ peptide and sAPPα present in the medium samples at 24 h was calculated. The data shown represent an average from three independent experiments with the S.E. Levels of Aβ peptide and sAPPα that differ from the control (Mock) with a significance of p < 0.001 (***) are indicated by asterisks.
DC2 and KCP2 Interact with the γ-Secretase Complex
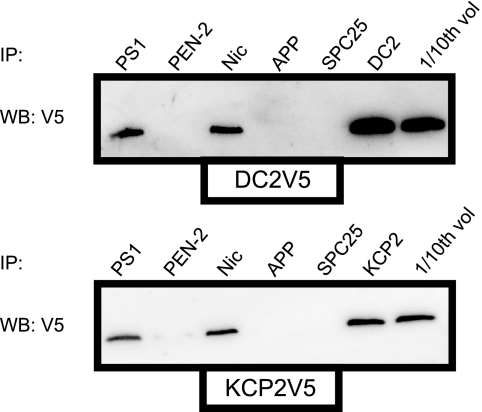
To assess the possibility that DC2 and KCP2 could associate with the γ-secretase complex, we carried out co-immunoprecipitation analysis of extracts of SH-SY5Y cells in the presence of transiently transfected DC2 and KCP2 with a V5 tag. Extracts were immunoprecipitated with antibodies to PS1, PEN2, nicastrin, APP, and DC2 or KCP2, as well as with a non-related serum against the signal peptidase complex subunit 25 (SPC25). These immunoprecipitates were analyzed by Western blot with V5 antibody (Fig. 8). Both DC2 and KCP2 appear to be able to be co-precipitated with some of the components of the γ-secretase complex, PS1 and nicastrin. Likewise, we detected for immunoprecipitates of PS1 and nicastrin but not PEN2 when we immunoprecipitated with V5 and blotted for PS1, PEN2, and nicastrin (data not shown). Because we could detect for interactions of DC2 and KCP2 with some of the components of the γ-secretase complex, we were interested in asking the question of whether depletion of DC2 and KCP2 could disrupt the cellular localization of the γ-secretase complex. In order to study the effect of knockdown on the cellular localization of the γ-secretase, we first knocked down DC2 and KCP2 in SH-SY5Y cells on day 0 followed by the expression of wild-type PS1 on day 2 and immunofluorescence on day 3. We performed a triple immunofluorescence staining with antibodies against PS1 (localized to the ER and Golgi complex), Sec61β (localized to the ER and the ER-Golgi intermediate compartment) and p58 (localized to the Golgi complex) (34–36). We observed that PS1 in SH-SY5Y cells was mainly localized to the ER with a reticular and perinuclear pattern and Golgi complex similar to previously reported localization studies (supplemental Fig. S2, see Mock panel) (37, 38). Brefeldin A treatment prevents anterograde traffic from the ER to the Golgi, resulting in retrograde transport of proteins back from the Golgi to the ER (39). Brefeldin A treatment blocked PS1 transport to the Golgi and caused retrograde transport of p58 because both appeared as a reticular pattern, indicating a block in transport and retention at the ER (supplemental Fig. S2, BFA panel). However, SH-SY5Y cells transiently transfected with siRNAs against DC2 or KCP2 and stained for PS1 display similar staining compared with the mock (supplemental Fig. S2, siDC2 and siKCP2 panels). These findings suggest that DC2 and KCP2 are unable to affect the cellular localization of the γ-secretase complex, whereas DC2 and KCP2 may interact with γ-secretase subcomplexes containing PS1 and nicastrin that could be formed in the immature γ-secretase complex before PEN2 is able to associate with the complex in the final stages of assembly.
FIGURE 8.
DC2 and KCP2 interact with the γ-secretase complex. SH-SY5Y cells were transfected with V5-tagged DC2 and KCP2 in a mammalian expression vector as shown (DC2V5 and KCP2V5 panels). Cells were harvested 18 h after transfection and lysed in immunoprecipitation buffer containing 1% CHAPSO, and associated cellular components were then recovered by immunoisolation using antibodies specific for PS1, PEN-2, nicastrin (Nic), APP, SPC25, DC2, or KCP2. The resulting material was analyzed by Western blotting with monoclonal antibody specific for the V5 epitope tag. A fraction of the total cellular extract from the transfected cells (about one-tenth that was used for immunoprecipitation) was included as a control (1/10th vol). IP, immunoprecipitation; WB, Western blot.
DISCUSSION
Sequential cleavage of APP is a prerequisite to Aβ generation because cleavage by β-secretase directly contributes to Aβ generation, whereas cleavage by α-secretase prevents Aβ formation. The mechanisms that regulate APP cleavage are not well defined but include trafficking factors and cell signaling pathways. Moreover, recent studies increasingly indicate that intracellular APP trafficking as a mechanism to regulate APP proteolytic cleavage by the secretases and, in turn, the amount of APP processing (40, 41). This study identifies DC2 and KCP2 as novel proteins modulating the APP pathway through APP cleavage by the γ-secretase and Aβ generation.
Our hypothesis is that the majority of eukaryotic OST subunits are not required for the core catalytic activity of the complex but perform other distinct roles in N-glycosylation and quality control at the ER membrane. Our working model predicts that DC2 and KCP2 play a role in the APP processing pathway either by (i) interacting with the unidentified aspartyl protease called the “presenilinase” at the ER or by altering the self-cleavage PS1 or (ii) by affecting the assembly of the γ-secretase complex. Our data indicate first that DC2 and KCP2 plays a minor role in N-glycosylation, affecting APP but not all glycoproteins, including those associated with the APP processing pathway, such as nicastrin and BACE1. The reduction in APP N-glycosylation seems unlikely to directly affect APP cleavage because a block of N-glycosylation by tunicamycin treatment did not perturb APP maturation. Indeed, it has been previously shown that APP cleavage is unaffected in cells expressing a APP mutant lacking N-glycosylation sites (42). Second, DC2 and KCP2 depletion leads to an accumulation of C-terminal fragments of APP, C99 and C83, as a result of less active γ-secretase complex being available. Finally, we show that Aβ production is altered in a DC2 and KCP2 knock-out model while the overexpression of DC2 and KCP2 enhances γ-secretase activity associated with PS1.
Little was known until now about the elusive subunits of the mammalian OST, DC2 and KCP2. The C terminus of DC2 is weakly homologous, sharing less than 10 and 18% homology with the yeast subunits, Ost3p and Ost6p, respectively. It is believed that Ost3/6p are present in two subcomplexes that associate either with Sbh1p or Sbh2p, respectively, forming two structurally different translocons (43, 44). Recently, Ost3/6p have been shown to contain thioredoxin-like folds, which would enable the oxidation folding of glycoproteins (45). It remains to be elucidated if this folding event occurs immediately after translocation of a nascent chain into the ER or before presentation of a substrate to the OST catalytic core. We can rule out DC2 having this role in the OST because first DC2 is a weak homologue and lacks a CXXC active site motif that is essential for oxidoreduction activity. Second, other subunits of the human OST, IAP and N33, share more homology to Ost3/6p, which could perform oxidative folding in the OST. Remarkably, KCP2 has no homologue in yeast that has been identified.
A major consensus indicates that presenilins along with nicastrin, PEN2, and Aph1 form the catalytic core of the γ-secretase complex. These four proteins are necessary and sufficient for γ-secretase activity in vitro. Nicastrin and Aph1 are thought to form a stable subcomplex prior to presenilin and/or PEN2 recruitment. A PEN2-PS1 subcomplex has been reported, and it appears that the docking site for PEN2 in the active complex is within TMD4 of PS1 (46–49). The minimal content requires only PS1 and PEN2 that is sufficient to stimulate and activate PS1 (7). Interestingly, a study identified Rer1p, a protein that retrieves proteins from the ER and cis-Golgi, as a novel limiting factor that negatively regulates the assembly of the complex by binding to nicastrin and competing with Aph1 (50). A number of studies increasingly imply that the assembly of the γ-secretase complex is a result of recycling of its components from the ER to the Golgi (51). However, the endoproteolysis of PS1 most likely occurs in a more acidic compartment, such as the cis-Golgi complex that extends the half-life of PS1 because the FL-PS1 is turned over in less than 1 h (52–55). These studies highlight the importance of quality control mechanisms that exist at the ER and Golgi in the regulation of the γ-secretase complex. DC2 and KCP2 may regulate the final stages of assembly by enabling the recruitment of PEN2 to the subcomplex facilitating transit beyond the ER and early biosynthetic compartment.
The regulation and maturation of the activity of the γ-secretase complex could possibly be maintained by the G-protein scaffold protein, tetraspanin (15). Interestingly, this same study pulled out interactions of PS1 with components of the OST, in particular OST48 and ribophorin II. In our earlier studies, we became interested in the role of ribophorin I in the OST after we had characterized and identified cross-links of amyloid precursor protein (βAPP), a fragment of APP that is generated by β-secretase cleavage to ribophorin I (11). This association of ribophorin I with APP was independent of glycosylation status and could be observed both in vitro and in vivo (11). In addition, STT3B, a component of the OST complex, has been linked to macroautophagy of the ATPase subunit, VOa1, and remarkably requires PS1 interaction for efficient autophagy. Surprisingly, PS1 knock-out impairs N-glycosylation and targeting of VOa1 to autophagosomes, leading to an accumulation of autophagic vacuoles observed in several neurodegenerative diseases. This provides us with supporting evidence linking the role of the OST complex to Alzheimer disease (16). Our results indicate for the first time that the OST complex, in particular the subunits DC2 and KCP2, can interact with the γ-secretase components and regulate γ-secretase-directed processing of APP as a consequence of modulation of the PS1 endoproteolytic cleavage. We therefore can hypothesize that the role of DC2 and KCP2 in PS1 endoproteolysis could occur by two ways: (i) by the recruitment of PS1/nicastrin via DC2 and/or KCP2 at the ER, allowing the binding of PEN2 to the complex facilitating the intracleavage of PS1, or (ii) by DC2 and/or KCP2 acting as an accessory to the assembly of the γ-secretase complex. It would be interesting to generate a KO animal model to determine if DC2 and KCP2 are important both for development and for reducing Aβ load in the brain.
In conclusion, our studies define an essential role for DC2 and KCP2 in the maturation and processing of APP. We have demonstrated that DC2 and KCP2 are localized to the ER compartment and are dispensable for N-glycosylation and OST function. We have clearly shown that DC2 and KCP2 modulate γ-secretase cleavage of APP through their ability to control PS1 endoproteolysis. Our future studies will seek to build upon this new knowledge, facilitating the identification of the elusive protease, the “presenilinase,” and to determine the molecular basis of DC2 and KCP2 function at the ER.
Supplementary Material
Acknowledgments
We are eternally grateful to all of our colleagues who have contributed reagents and advice toward this project. We thank Professor Stephen High and Dr. Peristera Roboti (University of Manchester) for the generation of antibodies and for valuable discussions and Dr. Eric Pinaud (Université de Limoges) and Professor Guylène Page (Université de Poitiers) for advice during the preparation of the manuscript.
This project was supported by a BQR Actions Internationales (Region Limousin) (to C. M. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- AD
- Alzheimer disease
- APP
- amyloid precursor protein
- ER
- endoplasmic reticulum
- OST
- oligosaccharyltransferase
- TRAM
- translocating chain-associating membrane
- CTF
- C-terminal fragment.
REFERENCES
- 1. Hardy J., Selkoe D. J. (2002) Science 297, 353–356 [DOI] [PubMed] [Google Scholar]
- 2. Haass C. (2004) EMBO J. 23, 483–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Price D. L., Sisodia S. S., Borchelt D. R. (1998) Science 282, 1079–1083 [DOI] [PubMed] [Google Scholar]
- 4. Sisodia S. S., St George-Hyslop P. H. (2002) Nat. Rev. Neurosci. 3, 281–290 [DOI] [PubMed] [Google Scholar]
- 5. Laudon H., Hansson E. M., Melén K., Bergman A., Farmery M. R., Winblad B., Lendahl U., von Heijne G., Näslund J. (2005) J. Biol. Chem. 280, 35352–35360 [DOI] [PubMed] [Google Scholar]
- 6. Thinakaran G., Borchelt D. R., Lee M. K., Slunt H. H., Spitzer L., Kim G., Ratovitsky T., Davenport F., Nordstedt C., Seeger M., Hardy J., Levey A. I., Gandy S. E., Jenkins N. A., Copeland N. G., Price D. L., Sisodia S. S. (1996) Neuron 17, 181–190 [DOI] [PubMed] [Google Scholar]
- 7. Ahn K., Shelton C. C., Tian Y., Zhang X., Gilchrist M. L., Sisodia S. S., Li Y. M. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 21435–21440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kelleher D. J., Gilmore R. (2006) Glycobiology 16, 47R–62R [DOI] [PubMed] [Google Scholar]
- 9. Wilson C. M., High S. (2007) J. Cell Sci. 120, 648–657 [DOI] [PubMed] [Google Scholar]
- 10. Wilson C. M., Roebuck Q., High S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 9534–9539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wilson C. M., Kraft C., Duggan C., Ismail N., Crawshaw S. G., High S. (2005) J. Biol. Chem. 280, 4195–4206 [DOI] [PubMed] [Google Scholar]
- 12. Shibatani T., David L. L., McCormack A. L., Frueh K., Skach W. R. (2005) Biochemistry 44, 5982–5992 [DOI] [PubMed] [Google Scholar]
- 13. Wilson C. M., High S. (2010) Methods Mol. Biol. 619, 389–402 [DOI] [PubMed] [Google Scholar]
- 14. George M., Ying G., Rainey M. A., Solomon A., Parikh P. T., Gao Q., Band V., Band H. (2007) BMC Cell Biol. 8, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wakabayashi T., Craessaerts K., Bammens L., Bentahir M., Borgions F., Herdewijn P., Staes A., Timmerman E., Vandekerckhove J., Rubinstein E., Boucheix C., Gevaert K., De Strooper B. (2009) Nat. Cell Biol. 11, 1340–1346 [DOI] [PubMed] [Google Scholar]
- 16. Lee J. H., Yu W. H., Kumar A., Lee S., Mohan P. S., Peterhoff C. M., Wolfe D. M., Martinez-Vicente M., Massey A. C., Sovak G., Uchiyama Y., Westaway D., Cuervo A. M., Nixon R. A. (2010) Cell 141, 1146–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kohda D., Yamada M., Igura M., Kamishikiryo J., Maenaka K. (2007) Glycobiology 17, 1175–1182 [DOI] [PubMed] [Google Scholar]
- 18. Kelleher D. J., Kreibich G., Gilmore R. (1992) Cell 69, 55–65 [DOI] [PubMed] [Google Scholar]
- 19. Kelleher D. J., Gilmore R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4994–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kelleher D. J., Karaoglu D., Mandon E. C., Gilmore R. (2003) Mol. Cell 12, 101–111 [DOI] [PubMed] [Google Scholar]
- 21. Kopan R., Ilagan M. X. (2009) Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rand M. D., Grimm L. M., Artavanis-Tsakonas S., Patriub V., Blacklow S. C., Sklar J., Aster J. C. (2000) Mol. Cell. Biol. 20, 1825–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hussain I., Powell D., Howlett D. R., Tew D. G., Meek T. D., Chapman C., Gloger I. S., Murphy K. E., Southan C. D., Ryan D. M., Smith T. S., Simmons D. L., Walsh F. S., Dingwall C., Christie G. (1999) Mol. Cell Neurosci. 14, 419–427 [DOI] [PubMed] [Google Scholar]
- 24. Vassar R., Bennett B. D., Babu-Khan S., Kahn S., Mendiaz E. A., Denis P., Teplow D. B., Ross S., Amarante P., Loeloff R., Luo Y., Fisher S., Fuller J., Edenson S., Lile J., Jarosinski M. A., Biere A. L., Curran E., Burgess T., Louis J. C., Collins F., Treanor J., Rogers G., Citron M. (1999) Science 286, 735–741 [DOI] [PubMed] [Google Scholar]
- 25. Sinha S., Anderson J. P., Barbour R., Basi G. S., Caccavello R., Davis D., Doan M., Dovey H. F., Frigon N., Hong J., Jacobson-Croak K., Jewett N., Keim P., Knops J., Lieberburg I., Power M., Tan H., Tatsuno G., Tung J., Schenk D., Seubert P., Suomensaari S. M., Wang S., Walker D., Zhao J., McConlogue L., John V. (1999) Nature 402, 537–540 [DOI] [PubMed] [Google Scholar]
- 26. Jorissen E., Prox J., Bernreuther C., Weber S., Schwanbeck R., Serneels L., Snellinx A., Craessaerts K., Thathiah A., Tesseur I., Bartsch U., Weskamp G., Blobel C. P., Glatzel M., De Strooper B., Saftig P. (2010) J. Neurosci. 30, 4833–4844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuhn P. H., Wang H., Dislich B., Colombo A., Zeitschel U., Ellwart J. W., Kremmer E., Rossner S., Lichtenthaler S. F. (2010) EMBO J. 29, 3020–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. De Strooper B., Vassar R., Golde T. (2010) Nat. Rev. Neurol. 6, 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tolia A., De Strooper B. (2009) Semin. Cell Dev. Biol. 20, 211–218 [DOI] [PubMed] [Google Scholar]
- 30. Holtzman D. M., Deshmukh M. (1997) Nat. Med. 3, 954–955 [DOI] [PubMed] [Google Scholar]
- 31. Xie Z., Romano D. M., Kovacs D. M., Tanzi R. E. (2004) J. Biol. Chem. 279, 34130–34137 [DOI] [PubMed] [Google Scholar]
- 32. Kudo T., Okumura M., Imaizumi K., Araki W., Morihara T., Tanimukai H., Kamagata E., Tabuchi N., Kimura R., Kanayama D., Fukumori A., Tagami S., Okochi M., Kubo M., Tanii H., Tohyama M., Tabira T., Takeda M. (2006) Biochem. Biophys. Res. Commun. 344, 525–530 [DOI] [PubMed] [Google Scholar]
- 33. Hoshino T., Nakaya T., Araki W., Suzuki K., Suzuki T., Mizushima T. (2007) Biochem. J. 402, 581–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim T. W., Pettingell W. H., Jung Y. K., Kovacs D. M., Tanzi R. E. (1997) Science 277, 373–376 [DOI] [PubMed] [Google Scholar]
- 35. Greenfield J. J., High S. (1999) J. Cell Sci. 112, 1477–1486 [DOI] [PubMed] [Google Scholar]
- 36. Katayama T., Imaizumi K., Sato N., Miyoshi K., Kudo T., Hitomi J., Morihara T., Yoneda T., Gomi F., Mori Y., Nakano Y., Takeda J., Tsuda T., Itoyama Y., Murayama O., Takashima A., St George-Hyslop P., Takeda M., Tohyama M. (1999) Nat. Cell Biol. 1, 479–485 [DOI] [PubMed] [Google Scholar]
- 37. Busciglio J., Hartmann H., Lorenzo A., Wong C., Baumann K., Sommer B., Staufenbiel M., Yankner B. A. (1997) J. Neurosci. 17, 5101–5107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kovacs D. M., Fausett H. J., Page K. J., Kim T. W., Moir R. D., Merriam D. E., Hollister R. D., Hallmark O. G., Mancini R., Felsenstein K. M., Hyman B. T., Tanzi R. E., Wasco W. (1996) Nat. Med. 2, 224–229 [DOI] [PubMed] [Google Scholar]
- 39. Lippincott-Schwartz J., Yuan L. C., Bonifacino J. S., Klausner R. D. (1989) Cell 56, 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Y. W., Thompson R., Zhang H., Xu H. (2011) Mol. Brain 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sannerud R., Annaert W. (2009) Semin. Cell Dev. Biol. 20, 183–190 [DOI] [PubMed] [Google Scholar]
- 42. Påhlsson P., Spitalnik S. L. (1996) Arch. Biochem. Biophys. 331, 177–186 [DOI] [PubMed] [Google Scholar]
- 43. Yan A., Lennarz W. J. (2005) Glycobiology 15, 1407–1415 [DOI] [PubMed] [Google Scholar]
- 44. Chavan M., Chen Z., Li G., Schindelin H., Lennarz W. J., Li H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8947–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schulz B. L., Stirnimann C. U., Grimshaw J. P., Brozzo M. S., Fritsch F., Mohorko E., Capitani G., Glockshuber R., Grütter M. G., Aebi M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 11061–11066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Prokop S., Haass C., Steiner H. (2005) J. Neurochem. 94, 57–62 [DOI] [PubMed] [Google Scholar]
- 47. Watanabe N., Tomita T., Sato C., Kitamura T., Morohashi Y., Iwatsubo T. (2005) J. Biol. Chem. 280, 41967–41975 [DOI] [PubMed] [Google Scholar]
- 48. Kim S. H., Sisodia S. S. (2005) J. Biol. Chem. 280, 1992–2001 [DOI] [PubMed] [Google Scholar]
- 49. Francis R., McGrath G., Zhang J., Ruddy D. A., Sym M., Apfeld J., Nicoll M., Maxwell M., Hai B., Ellis M. C., Parks A. L., Xu W., Li J., Gurney M., Myers R. L., Himes C. S., Hiebsch R., Ruble C., Nye J. S., Curtis D. (2002) Dev. Cell 3, 85–97 [DOI] [PubMed] [Google Scholar]
- 50. Spasic D., Raemaekers T., Dillen K., Declerck I., Baert V., Serneels L., Füllekrug J., Annaert W. (2007) J. Cell Biol. 176, 629–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. De Strooper B., Annaert W. (2010) Annu. Rev. Cell Dev. Biol. 26, 235–260 [DOI] [PubMed] [Google Scholar]
- 52. Ratovitski T., Slunt H. H., Thinakaran G., Price D. L., Sisodia S. S., Borchelt D. R. (1997) J. Biol. Chem. 272, 24536–24541 [DOI] [PubMed] [Google Scholar]
- 53. Thinakaran G., Harris C. L., Ratovitski T., Davenport F., Slunt H. H., Price D. L., Borchelt D. R., Sisodia S. S. (1997) J. Biol. Chem. 272, 28415–28422 [DOI] [PubMed] [Google Scholar]
- 54. Zhang J., Kang D. E., Xia W., Okochi M., Mori H., Selkoe D. J., Koo E. H. (1998) J. Biol. Chem. 273, 12436–12442 [DOI] [PubMed] [Google Scholar]
- 55. Campbell W. A., Iskandar M. K., Reed M. L., Xia W. (2002) Biochemistry 41, 3372–3379 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.