Abstract
Heat shock factor Hsf1 is involved in the regulation of a variety of cellular processes including heat shock response, development and differentiation, aging, and tumorigenesis. Hsf1 transcriptional activity is tightly controlled through phosphorylation, sumoylation, and acetylation, and through association with a number of regulatory proteins. However, regulation of Hsf1 protein stability or turnover remains unknown. We have identified a novel Hsf1-interacting protein, FILIP-1L, that was found to bind to Hsf1 through yeast two-hybrid screening. FILIP-1L encodes multiple isoforms spanning from 711 to 1135 amino acid residues. FILIP-1L contains four coiled-coil and two N-terminal leucine zipper domains. Ectopic expression of FILIP-1L reduces the expression of the Hsf1 protein because FILIP-1L promotes Hsf1 ubiquitination and degradation through the ubiquitin-proteasome system, leading to a reduction in Hsf1-mediated transcription. FILIP-1L, Hsf1, and the ubiquitin-binding domain of HhR23A, a receptor that transports polyubiquitinated proteins to the 19 S proteasome subunit targeting them for degradation, are found in a complex. This indicates that FILIP-1L is a potential adaptor that is involved in the Hsf1 degradation pathway. Taken together, our results indicate that FILIP-1L interacts with Hsf1, controlling its stability and thus modulating the heat shock response. These data indicate a novel function for FILIP-1L and a pathway for Hsf1 degradation through the ubiquitin-proteasome system.
Keywords: Adaptor Proteins, Cellular Regulation, Protein Degradation, Transcription Factors, Ubiquitination, FILIP-1L, Hsf1, UPS, hHR23
Introduction
The heat shock response is controlled by a family of heat shock transcription factors (Hsfs)2 that consist of Hsf-1, -2, -3, and -4, whose expression and activities are differentially regulated (1–3). Structurally Hsfs contain an N-terminal winged helix turn helix DNA-binding domain and an N-terminal hydrophobic heptad repeat (HR-A/B) that mediates trimerization of the Hsfs (1, 4). Hsfs also contain a regulatory domain, a C-terminal hydrophobic region (HR-C), and a transactivation domain. Within the Hsfs, only Hsf4 lacks the C-terminal hydrophobic region (1, 3, 5). The homology of Hsfs at the protein level is ∼40%, and this defines their unique biological function in cells. In mammalian cells, although the inter-relationship between Hsfs is complex, Hsf2 and Hsf4, which are expressed in specific tissues, mainly contribute to cellular differentiation (1, 6–10). In contrast, Hsf1 is expressed in all tissues and is involved in many cellular processes, and it is the main regulator of the heat shock response. Hsf1 is activated by heat shock and plays a complex function during early development, as well as during tumorigenesis as it potentiates p53-mediated tumorigenesis, and can impact the myelination process in the CNS (11–14).
Transcriptional activation of Hsf1 is complex, and its activity is controlled at the post-translational level by phosphorylation, acetylation, and sumoylation (1, 15–19). These modifications can modulate Hsf1 transcriptional activity through impacting the structure of Hsf1 and its interaction with other proteins. In addition to the above modifications, Hsf1 activity is also regulated through interactions with a number of other proteins, including Hsp70, Hsp90 (1, 20, 21), Hsbp1 (22), CHIP (23), RalBP1 (24), Daxx (25), MTA1 (26), 14-3-3 (27), and Cdc20 (28). These proteins either exert negative regulation on Hsf1 transcriptional activity (e.g. Hsbp1), participate in the activation of Hsf1 (e.g. CHIP, Daxx), or confer a regulatory function toward Hsf1 in a specific signaling pathway (RalBP1, MTA1). Hsf1 is expressed in a monomeric form and is in complexes with Hsp90. Upon heat shock, Hsf1 is dissociated from the Hsp90 complexes (20, 24), trimerizes, and is translocated into the nucleus and binds to the promoter of Hsps and other downstream target genes, driving transcription. Hsf1 also interacts with Hsp70, and this suppresses its transcriptional activity (29). The interaction of Hsf1 with Cdc20 inhibits the interaction of Cdc20 with Cdc27, phosphorylation of Cdc27, and the ubiquitination activity of anaphase-promoting complex (28). Interaction of Hsf1 with 14-3-3 at 37 °C leads to cytoplasmic sequestration and repression of Hsf1 activity. Ral-binding protein-1 (RalBP1) forms a complex with Hsf1 and Hsp90 under physiological growth conditions, and regulates the Hsf1-mediated heat shock response in the Ral-dependent signaling pathway (24). The phosphorylated Hsf1 has been shown to undergo ubiquitin-mediated degradation via the SCFβ-TrCP pathway in mitotic cells (28). However, the mechanism underlying Hsf1 protein turnover in interphase cells and during the heat shock response has not been elucidated.
In the present study, we report another novel Hsf1-associating protein named filamin interacting protein 1-like (FILIP-1L) whose biological function is presently unclear. FILIP-1L is up-regulated in response to angiogenic inhibitors (30). Overexpression of FILIP-1L in cells leads to an increase in cellular apoptosis and inhibition of cell growth and migration (31). We show that FILIP-1L forms complexes with Hsf1 and Hsp72 and ectopic expression of FILIP-1L reduces Hsf1 protein leading to inhibition of Hsf1-mediated transcription. FILIP-1L expression in cells leads to an increase in Hsf1 ubiquitination and its recruitment to the complexes with the UBA domain of hHR23A protein. We anticipate that hHR23A is a ubiquitin receptor involved in proteasomal degradation, promoting Hsf1 degradation through the UPS.
EXPERIMENTAL PROCEDURES
Cell Culture
H1299 (human small cell lung carcinoma), HEK293 (human embryonic kidney epithelial), and C2C12 (murine myoblast) cell lines were maintained in Dulbecco's minimal essential medium (DMEM) supplemented to 10% fetal bovine serum (FBS) and streptomycin/penicillin. Heat shock treatment was performed in a temperature-controlled circulating water bath.
Yeast Two-hybrid System
For yeast two-hybrid screening, the CytoTrap Yeast Two-hybrid System (Stratagene, La Jolla, CA) was used as we have previously reported (24). A fragment of human Hsf1 (amino acid residues 1–378) was subcloned into the pSos vector and used as bait. A human heart tissue cDNA library that had been subcloned into the pMyr vector was used to find Hsf1-interacting proteins. Ten μg of the bait construct and 10 μg of the cDNA library were cotransfected into the competent yeast cdc25H strain. The transformed yeast were plated in SD (uracil/leucine-deficient) glucose plates and allowed to grow at 25 °C for 48–72 h. The yeast colonies that appeared on the plates were replica-plated on the SD/galactose plates and SD/glucose plates, and plates were incubated at 37 °C for 10 days. The positive colonies were cultured in 5 ml of SD/glucose broth and cultured at 25 °C. The plasmid DNA was extracted and used to transform Escherichia coli DH5α and then selected on chloramphenicol-containing agar plates. The plasmids pMyr and pSos encoding Maf B transcription factor were supplied by the manufacturer and were used as a positive control (24). The isolated plasmids were verified by DNA sequencing.
Plasmids
For pSos-Hsf1, Hsf1 cDNA encoding 1–378 amino acids was subcloned into the pSos vector (24). For pcDNA3-FLAG-FILIP-1L and pcDNA3-HA-FILIP-1L (both FLAG and HA tags located at the N terminus of FILIP-1L cDNA), the intact FILIP-1L cDNA (amino acid residues 1–893) was amplified by PCR from plasmid pMyr-FILIP-1L and subcloned into pcDNA3 at the BamHI and XhoI restriction enzyme sites. For plasmid pcDNA3-FLAG-FILIP-1L-N, amino acid residues 1 to 230 of FILIP-1L were released from pcDNA3-FLAG-FILIP-1L using restriction enzymes BamHI and EcoRI and the fragment was subcloned into the same restriction enzyme sites of pcDNA3. For pEBG-Hsf1 mutants, pEBG-Hsf1 amino acid residues 1–80, 1–161, 1–196, 1–278, 1–378, and 379–529 were constructed by inserting the appropriate Hsf1 fragments into the BamHI and NotI restriction enzyme sites of the pEBG vector. Similar strategies were used to generate pEBG-FILIP-1L (amino acid residues 1–893) and pEBG-FILIP-1L-N (amino acid residues 1–288) expression vectors. The plasmids pcDNA3-His-Hsf1 containing human Hsf1 and pcDNA3-HA-ubiquitin were gifts from Drs. R. Morimoto (Northwestern University, Evanston, IL) and T. Kamitani (Georgia Health Sciences University), respectively. All the constructs were generated by PCR and confirmed by DNA sequencing.
Immunoblotting, Immunoprecipitation, and GST Pull-down Assays
For immunoblotting, cells were lysed in 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Nonidet P-40, and protease inhibitor mixtures (Roche Applied Science). Equal amounts of proteins (30 μg) were subjected to SDS-PAGE and immunoblotting (24). For immunoprecipitation, 48 h following transfection of appropriate vectors, cells were lysed in buffer. 800 μg of cell lysate was pre-cleared using 20 μl of a 50% slurry of Protein A-Sepharose 4B beads. 2–3 μg of primary antibody was added to the pre-cleared cell lysate and incubated overnight at 4 °C. Cell lysates were then incubated with 40 μl of protein A beads for 2 h. Protein A beads containing protein complexes were centrifuged, rinsed four times with lysis buffer, and boiled in SDS sample buffer in preparation for immunoblotting. For the in vivo GST pulldown assays, cells were co-transfected with appropriate plasmids together with recombinant constructs of pEBG encoding full-length or truncated mutants. 48 h after transfection, cells were lysed and 700 μg of soluble protein fractions were incubated with 30 μl of glutathione-Sepharose 4B beads for 16 h. The beads were rinsed four times with lysis buffer. Samples were subjected to immunoblotting analyses. For the in vitro GST pulldown assays, cell lysates containing overexpressed protein were incubated with bacterially purified GST or GST-conjugated purified protein overnight at 4 °C. The GST and GST fusion proteins were immobilized with glutathione-Sepharose 4B beads (24). After rinsing, beads were placed in SDS sample buffer and boiled for 10 min, and samples were analyzed by immunoblotting using the appropriate antibody. Antibodies were from the following sources: β-actin and FLAG, Sigma; ubiquitin and HA, Santa Cruz Biotechnology; Hsf1, Cell Signaling; and Hsp72 and Hsp27, Stressgen.
For in vivo ubiquitination reactions, HEK293 cells were transiently transfected with empty vector or with appropriate plasmids. Cells were left untreated or treated with proteasome inhibitors and lysed in Nonidet P-40 lysis buffer containing a protease inhibitor mixture. The proteins were pulled down and, after rinsing with high salt buffer containing Tris-HCl, pH 7.4, 0.4 NaCl, and 2 mm DTT, the immunoprecipitated complexes were immunoblotted using specific antibody as indicated in the figure legends.
Talon Beads Pulldown Assay
HEK293 cells were transiently cotransfected with appropriate plasmids using Trans IT-LT (Mirus). After 48 h, cells were treated with 10 μm MG132 for 6 h and lysed in denaturing lysis buffer containing 6 m guanidinium HCl, 100 mm Na2HPO4, 150 mm NaCl. Cell lysates were incubated with 30–40 μl of taylon beads for 1 h and beads were rinsed three times in 8 m urea buffer and 2 times with PBS. The proteins were eluted in SDS sample buffer and analyzed by immunoblotting.
Immunofluorescent Analyses
The empty vector containing the enhanced yellow fluorescent protein (EYFP) or construct expressing FILIP-1L-EYFP was transiently transfected into H1299 cells. Cells were left untreated or treated at 43 °C for 1 h and left to recover at 37 °C for 30 min. Cells were rinsed with PBS and fixed in 4% paraformaldehyde for 15 min at 25 °C and then permeabilized with 0.5% Triton X-100 in PBS (PBST) for 5 min at 4 °C. Cells were rinsed three times and blocked in 5% BSA in PBST buffer for 1 h and incubated with the primary antibody for 1 h at 25 °C. After three rinses, cells were incubated with secondary antibody conjugated with Texas Red for 1 h (17). After three rinses, nuclei were stained with DAPI in PBS buffer. Cells were analyzed using a Zeiss Axio-Imager Fluorescence microscope.
Statistical Analyses
All experiments were performed at least three times. Data are presented as mean ± S.D. Statistical significance between experimental groups was assessed using Student's t test and p < 0.05 was considered significant.
RESULTS
Hsf1 Interacts with FILIP-1L
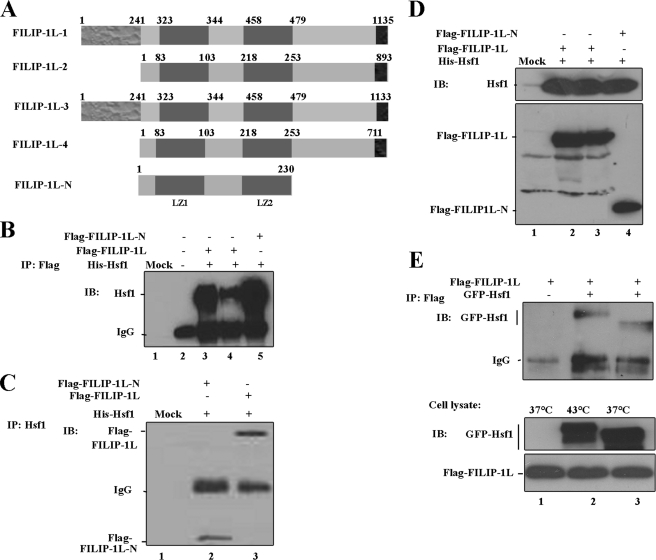
Using the human Hsf1 cDNA as a bait, we screened a human heart cDNA library using the Cyto-Trap Yeast Two-hybrid screening system (24). A novel Hsf1-interacting protein whose sequence was identical to FILIP-1L mRNA version 2 (also known as down-regulated ovary cancer protein 1 (DOC1), GenBankTM designation number NP_055705) was identified (Fig. 1A). FILIP-1L version 1 encodes a 1135-amino acid residue protein and has two leucine zipper domains at the N-terminal region (amino acid residues 323–344 and 458–479) and four coiled-coiled domains spanning amino acids 78 to 781. Amino acid residues 250–760 are homologous to the N-terminal of the Structural Maintenance of Chromosomes, which is a chromosome segregation ATPase, and amino acid residues 875–1115 are homologous to BLLF1 (herpesvirus major outer envelope glycoprotein Gp350/220) (Fig. 1A). FILIP-1L also encodes a potential nuclear localization domain at amino acid residues 168–183. FILIP-1L has been reported to possess four versions (Fig. 1A). The last seven amino acid residues of versions 1 and 4 are 1128VEPLLLPH1135 and 704VEPLLLPH711, respectively, and that of versions 2 and 3 are 1129SNIYN1133 and 889SNIYN893, respectively. The significance of these isoforms in cellular function is not known.
FIGURE 1.
Hsf1 interacts with FILIP-L1 in mammalian cells. A, schematic representation of different FILIP-1L isoforms. FILIP-1L-2 was isolated by the yeast two-hybrid system to interact with Hsf1. FILIP-1L-N is not an isoform but was constructed to determine the minimal domain of the FILIP-1L interaction with Hsf1. B, interaction of FLAG-FILIP-1L with Hsf1. H1299 cells were mock transfected (lane 2) or co-transfected with His-Hsf1 and FLAG-FILIP-1L, or FLAG-FILIP-1L-N (amino acid residues 1–230) (lanes 3–5). Cells were left at 37 °C (lanes 2, 3, and 5) or were heated at 43 °C for 1 h (lane 4). Complexes of Hsf1 and FILIP-1L or FILIP-1L-N were co-immunoprecipitated (IP) using antibody to FLAG and were detected using antibody to His-Hsf1. Location of the IgG band is also indicated. Lane 1 represents pre-cleared protein A. C, co-immunoprecipitation of FLAG-FILIP-1L or FLAG-FILIP-1L-N (amino acid residues 1–230) with Hsf1 using antibody to Hsf1. Transient transfections were carried out as in B. FILIP-1L or FILIP-1L-1N were co-immunoprecipitated using antibody to Hsf1 and immunoblotted using antibody to FLAG. Lane 1 represents pre-cleared protein A; lane 2 represents cells transiently transfected with His-Hsf1 and FLAG-FILIP-1L-N. Lane 3 represents cells transiently transfected with His-Hsf1 and FLAG-FILIP-1L. D, immunoblot analyses showing the expression levels of ectopically expressed His-Hsf1, FLAG-FILIP-1L, and FLAG-FILIP-1L-N (amino acid residues 1–230) in the cell lysates used in panels B and C for co-immunoprecipitation studies. E, FLAG-FILIP-1L interacts with GFP-Hsf1 under both physiological growth conditions and following heat shock. H1299 cells were transiently transfected with FLAG-FILIP-1L alone (lane 1) or cotransfected with FLAG-FILIP-1L and GFP-Hsf1 fusion protein (lanes 2 and 3). Cells were left at 37 °C (lanes 1 and 3) or treated with heat shock at 43 °C for 1 h (lane 2). The GFP-Hsf1 and FLAG-FILIP-1L complexes were co-immunoprecipitated using antibody to FLAG. Immunoblotting (IB) was performed using antibody to GFP. Lower panels represent expression of GFP-Hsf1 and FLAG-FILIP-1L in the cell lysate.
The interaction between Hsf1 and FILIP-1L was examined in mammalian cells using immunoprecipitation assays. H1299 cells were either mock-transfected, or transiently co-transfected with plasmids encoding His-Hsf1 together, with either plasmids encoding FLAG-FILIP-1L version 2 (thereafter called FILIP-1L) or FLAG-FILIP-1L-N (encoding amino acid residues 1–230) (Fig. 1, A and B). Cells were left untreated or were heated at 43 °C for 1 h to activate Hsf1. As indicated in Fig. 1B, Hsf1 was co-immunoprecipitated by both FLAG-FILIP-1L and FLAG-FILIP-1L-N under physiological growth conditions (lanes 3 and 5). However, the interaction of Hsf1 with FLAG-FILIP-1L was reduced when cells were treated at 43 °C (Fig. 1B, lane 4). No Hsf1 was co-immunoprecipitated in the pre-cleared beads (Fig. 1B, lane 1), or in mock-transfected cells incubated with FLAG antibody (Fig. 1B, lane 2). FLAG-FILIP-1L and FLAG-FILIP-1L-N were also co-immunoprecipitated with His-Hsf1, using antibody to Hsf1 (Fig. 1C, lanes 2 and 3). These results demonstrate that the leucine zipper region of the N-terminal fragment of FILIP-1L (encoded by FILIP-1N) is required for the interaction with Hsf1 (Fig. 1, A-C). Immunoblot analyses showing the expression levels of FLAG-FILIP-1L, FLAG-FILIP-1L-N, and His-Hsf1 in the H1299 cells are presented in Fig. 1D. Comparable results were also obtained when expression plasmids encoding GFP-Hsf1 and FLAG-FILIP-1L were co-expressed in cells and complexes were pulled down from untreated (37 °C) or heated (43 °C) cells using antibody to FLAG (Fig. 1E). Levels of GFP-Hsf1 and FLAG-FILIP-1L in cell lysates are indicated in the lower panel of Fig. 1E. These results indicate that Hsf1 interacts with FILIP-1L in cultured mammalian cells.
N-terminal Hydrophobic Region of Hsf1 Interacts with FILIP-1L
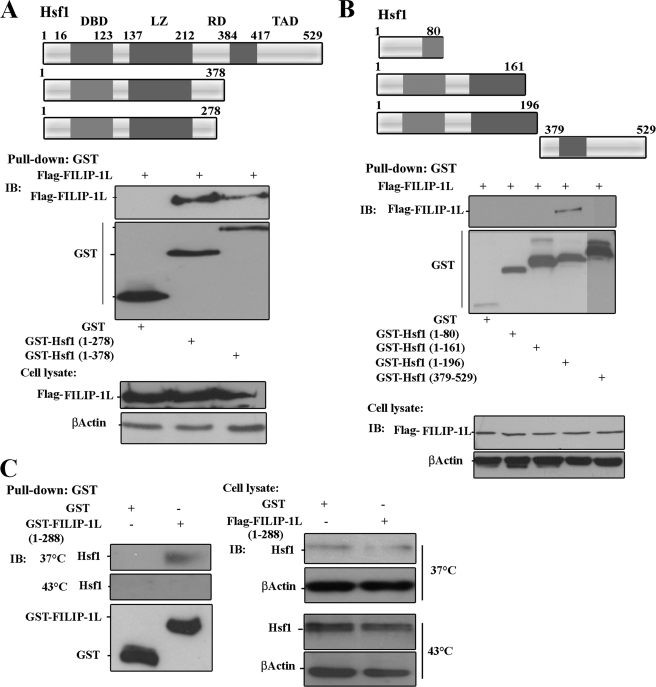
We next performed an in vivo GST pulldown assay to determine the domains of Hsf1 that associate with FILIP-1L. Thus, plasmids containing FLAG-FILIP-1L were cotransfected into H1299 cells with constructs expressing GST alone, GST-Hsf1(1–378), or GST-Hsf1(1–278). As indicated in Fig. 2A, FLAG-FILIP-1L was co-immunoprecipitated together with GST-Hsf1(1–278) and GST-Hsf1(1–378), but not with GST alone. These results suggest that the FILIP-1L-interacting region is located between amino acid residues 1 and 278 of the Hsf1 protein, which contains the Hsf1 DNA-binding domain, N-terminal hydrophobic domain, and a portion of the regulatory domain. To more precisely determine the domain of Hsf1 that interacts with FILIP-1L, we co-transfected H1299 cells with constructs encoding FLAG-FILIP-1L together with GST alone, or GST-Hsf1 containing amino acid residues 1–80, 1–161, 1–196, or 378–529. As presented in Fig. 2B, we found that FLAG-FILIP-1L could be pulled down with GST-Hsf1 amino acid residues 1–196, but not with 1–80, 1–161, or 378–529, or with GST alone. The results indicate that Hsf1 amino acid residues 1–196, which contain the full-length N-terminal hydrophobic region, is the FILIP-1L-interacting domain (Fig. 2B). The expression of plasmids encoding FLAG-FILIP-1L, GST, and GST-Hsf1 mutant proteins in H1299 cells were detected by immunoblot analyses using antibodies to FLAG, GST, or β-actin (Fig. 2, A and B, lower panels). Using a comparable approach, we determined that GST-FILIP-1L amino acid residues 1–288, which contain two leucine zipper domains, could pull down endogenous Hsf1 protein under normal physiological growth conditions, but not when cells were exposed to heat shock (Fig. 2C). The right panel in Fig. 2C shows the expression of Hsf1 in the cell lysate. Results presented in Fig. 2C further support that the N-terminal leucine zipper domain of FILIP-1L is the Hsf1-interacting domain.
FIGURE 2.
Hsf1 N-terminal hydrophobic domain interacts with FILIP-1L leucine zipper domain. A and B, top panels represent schematic drawings of the GST-Hsf1 mutants. DBD represents DNA-binding domain; LZ, leucine zipper domain; RD, regulatory domain; TAD, transactivation domain. H1299 cells were transiently co-transfected with expression vectors containing FLAG-FILIP-1L and GST alone, or with GST-Hsf1 deletion mutants. The GST and GST-Hsf1 mutants were pulled-down with glutathione-Sepharose 4B beads. Pulldown materials were immunoblotted using antibody to FLAG or GST. The lower panels in A and B represent immunoblots of the overexpressed FLAG-FILIP-1L and β-actin in the cell lysates. C, GST-FILIP-1L can pull down endogenous Hsf1 under physiological growth conditions. H1299 cells were transfected with GST alone, or GST-FILIP-1L (amino acid residues 1–288). Cells were left at 37 °C or heated at 43 °C for 1 h. Endogenous Hsf1 was co-immunoprecipitated with GST-FILIP-1L using glutathione-Sepharose 4B beads. The immunoprecipitated materials were immunoblotted using antibody to Hsf1 and GST. The right panel represents the immunoblot (IB) analyses of cell lysate showing endogenous Hsf1 and β-actin.
FILIP-1L Forms a Complex with Hsf1 and Hsp72
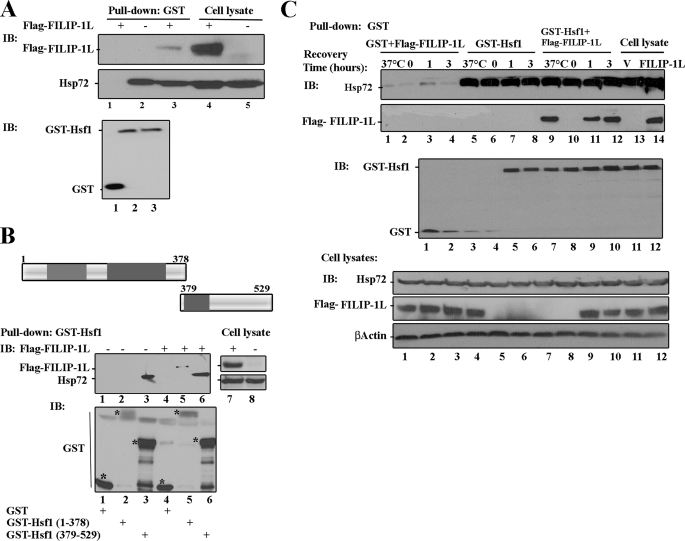
In mammalian cells, Hsf1 exists in a monomeric form through its interactions of the N- and C-terminal hydrophobic domains (4). Hsf1 has also been shown to form heterocomplexes with Hsp72. Hsp72 interacts with the C-terminal hydrophobic region of Hsf1 and inhibits Hsf1 transcriptional activity under physiological growth conditions. Because we showed that FILIP-1L can interact with the N-terminal hydrophobic repeats of Hsf1 under physiological growth conditions, we tested whether FILIP-1L·Hsf1 complexes also contain Hsp72. To this end, we performed an in vivo GST pulldown assay. The constructs expressing GST or GST-Hsf1 amino acid residues 1–529, 1–378, or 379–529 were transiently transfected alone, or co-transfected with plasmids containing FLAG-FILIP-1L into HEK293 cells. As indicated in Fig. 3A, GST-Hsf1 amino acid residues 1–529 could be co-immunoprecipitated with FLAG-FILIP-1L and Hsp72 (Fig. 3A, lanes 2 and 3). In contrast, GST-Hsf1(1–378) could only be co-immunoprecipitated with FLAG-FILIP-1L but not Hsp72 (Fig. 3B, compare lane 2 with lane 5). However, GST-Hsf1 amino acid residues 379–529 could pull down Hsp72, but not FLAG-FILIP-1L (Fig. 3B, lanes 3 and 6). No interactions between GST, Hsp72, and FILIP-1L could be detected (Fig. 3, A, lane 1, and B, lanes 1 and 4). These results indicate that FILIP-1L and Hsp72 associate with different domains of the Hsf1 protein. Expression levels of GST or GST-Hsf1 mutants in the cell lysates are presented in Fig. 3, A and B, lower panels. Expression levels of FLAG-FILIP-1L and Hsp72 in the cell lysates are also presented. These data demonstrate that full-length Hsf1 binds both Hsp72 and FILIP-1L in one complex.
FIGURE 3.
FILIP-1L is in Hsf1 and Hsp72 complexes. A, full-length Hsf1 forms complexes with Hsp72 and FLAG-FILIP-1L. HEK293 cells were transiently transfected with GST and FLAG-FILIP-1L (lane 1), GST-Hsf1 alone (lane 2), or GST-Hsf1 and FLAG-FILIP-1L (lane 3). The endogenous Hsp72 and FLAG-FILIP-1L that were pulled down with GST-Hsf1 were immunoblotted (IB) using antibodies against FLAG, Hsp72, or GST. The overexpressed FLAG-FILIP-1L and endogenous Hsp72 in the cell lysates are indicated in lanes 4 and 5. B, top panel represents a schematic drawing of Hsf1 mutant constructs. Lower panel represents immunoblotting analyses showing FLAG-FILIP-1L and Hsp72 interacting with different domains of Hsf1. HEK293 cells were transiently transfected with expression vector encoding GST (lane 1), GST-Hsf1 (amino acid residues 1–378) (lane 2), GST-Hsf1 (amino acid residues 379–529) (lane 3), co-transfected with FLAG-FILIP-1L and GST or GST-Hsf1 mutants (lanes 4–6). The endogenous Hsp72 or overexpressed FLAG-FILIP-1L was pulled down and immunoblotted using the corresponding antibodies. The expression of Hsp72 and FLAG-FILIP-1L in cell lysates is presented in lanes 7 and 8. The expression levels of the pulled down GST or GST-Hsf1 mutants are indicated in the lower panel. C, regulation of Hsf1, FLAG-FILIP-1L, and Hsp72 complexes by heat shock. HEK293 cells were transiently transfected with GST and FLAG-FILIP-1L (lanes 1–4), GST-Hsf1 alone (lanes 5–8), or GST-Hsf1 and FLAG-FILIP-1L (lanes 9–12). Cells were left at 37 °C (lanes 1, 5, and 9) or heated at 43 °C for 1 h, and then allowed to recover at 37 °C for 0 (lanes 2, 6, and 10), 1 (lanes 3, 7, and 11), or 3 h (lanes 4, 8, and 12). The GST and GST-Hsf1 pulled down endogenous Hsp72, or ectopically expressed FLAG-FILIP-1L were immunoblotted using their corresponding antibodies. Expression of GST and GST-Hsf1 are indicated in the middle panel. Lanes 13 and 14 of the upper panel show expression of Hsp72 and FLAG-FILIP-1L in the cell lysates. V indicates pcDNA3 vector alone. Levels of Hsp72, FLAG-FILIP-1L, and β-actin in cell lysates are indicated in the lower panels.
To determine whether the association between Hsf1, Hsp72, and FILIP-1L is regulated by heat shock, cells were cotransfected with GST plus GST-FILIP-1L (Fig. 3C, lanes 1-4), GST-Hsf1 (Fig. 3C, lanes 5–8), or co-transfected with GST-Hsf1 plus FLAG-FILIP-1L (Fig. 3C, lanes 9–12) and left at 37 °C, or were heated at 43 °C for 1 h, and then incubated at 37 °C for a recovery period of 0, 1, or 3 h. As indicated in Fig. 3C, lanes 5–8, GST-Hsf1 could pull down Hsp72 under both physiological growth conditions and after cells were exposed to heat shock and left to recover from heat shock exposure. Data show that exposure of the HEK293 cells to heat shock does not significantly alter the interaction between Hsf1 and Hsp72, and that Hsp72 forms a heterodimer with Hsf1 even under physiological growth conditions. Hsp72 and Hsf1 continue to interact following exposure of the cells to heat shock and during the recovery from heat shock. This is because HEK293 cells express the inducible Hsp72 constitutively. Surprisingly, GST-Hsf1, which was shown to interact with FILIP-1L under physiological growth conditions, could not pull down FLAG-FILIP-1L immediately following exposure of the cells to 43 °C heat shock (Fig. 3C, compare lanes 9 and 10). However, Hsf1 and FLAG-FILIP-1L began to interact again during the 1- and 3-h recovery period following heat shock (Fig. 3C, lanes 11 and 12). Cells expressing GST plus FLAG-FILIP-1L did not show any interaction with Hsp72 under any condition (Fig. 3C, lanes 1–4). Expression of GST and GST-Hsf1 are presented in Fig. 3C, middle panel. Expression of Hsp72 and FLAG-FILIP-1L in the cell lysates are presented in Fig. 3C. These data further demonstrate that the interaction between Hsf1 and FILIP-1L occurs predominantly under physiological growth conditions. Under comparable conditions, neither GST protein nor GST-Hsf1 could immunoprecipitate Hsp90 (data not shown). This is consistent with previous reports that intracellular association between Hsf1 and Hsp90 requires use of a protein cross-linker (20, 24).
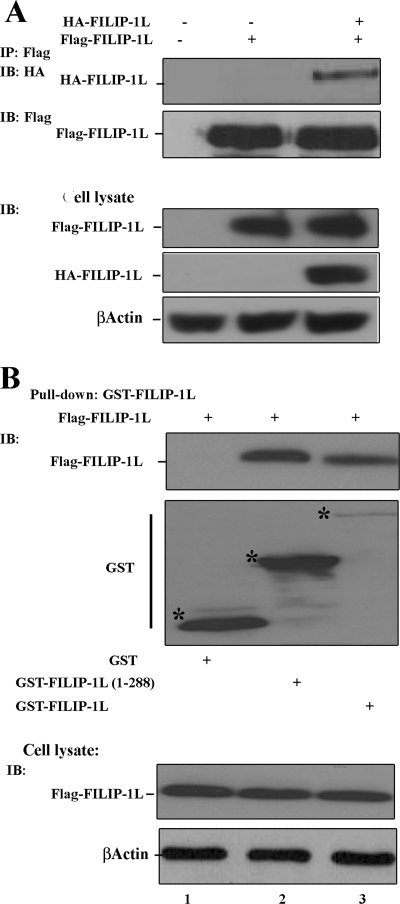
FILIP-1L Exists as a Dimer or Oligomer
To examine whether intracellular FILIP-1L exists as a monomer or oligomer, we performed immunoprecipitation and in vivo pulldown assays. Data show that FLAG-FILIP-1L could be co-immunoprecipitated with HA-FILIP-1L (Fig. 4A). Furthermore, GST-FILIP-1L or GST-FILIP-1L amino acid residues 1–288 could pull down the FLAG-FILIP-1L protein (Fig. 4B). Expression levels of FLAG-FILIP-1L and β-actin in the cell lysate are presented as control. These results demonstrate that the FILIP-1L protein is expressed in cells as either a dimer or oligomer.
FIGURE 4.
FILIP-1L in cells exist as dimer or oligomer. A, HEK293 cells transiently transfected with FLAG-FILIP-1L alone (lane 2) or cotransfected with HA-FILIP-1L and FLAG-FILIP-1L (lane 3). HA-FILIP-1L was immunoprecipitated (IP) using antibody to FLAG and the immunoprecipitated materials were immunoblotted (IB) using antibodies to HA (upper panel) or FLAG (lower panel). The expression of HA-, FLAG-FILIP-1L, and β-actin in cell lysates is also presented. Lane 1 represents negative control where cell lysate expressing empty vector was incubated with anti-FLAG antibody. B, GST-FILIP-1L pulls down FLAG-FILIP-1L. HEK293 cells were transiently transfected with FLAG-FILIP-1L and GST (lane 1) or FLAG-FILIP-1L with GST-FILIP-1L or GST-FILIP-1L(1–288) (lanes 2 and 3). Cell extracts were then used in pull-down experiments using GST. The pull down materials were immunoblotted using FLAG-FILIP-1L and GST. Lower panels show expression of FLAG-FILIP-1L and β-actin in the cell lysates.
FILIP-1L Expression in Cells Reduces Hsf1 Expression
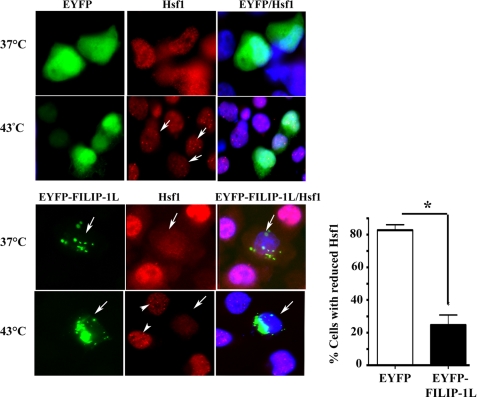
In the human tumor cell lines, Hsf1 is normally localized in both the cytoplasm and nucleus under physiological growth conditions. Following heat shock, Hsf1 translocates into the nucleus and forms nuclear stress granules and drives transcription (17). To examine whether FILIP-1L is colocalized with endogenous Hsf1 following heat shock, cells were transiently transfected with EYFP-N1 empty vector or plasmids encoding EYFP-FILIP-1L. Cells were left untreated or heated at 43 °C for 1 h. As presented in Fig. 5, upper panels, expression of EYFP does not affect formation of Hsf1 stress granules, which form upon exposure of the cells to heat stress. Surprisingly, we found that ectopic expression of EYFP-FILIP-1L reduced the expression of endogenous Hsf1 in cells that were untreated or cells that were heated at 43 °C for 1 h (Fig. 5, lower panels, arrows). Note that Hsf1 normally form stress granules in cells exposed to heat shock (Fig. 5, lower panels, arrowheads). Ectopic expression of FILIP-1L also interfered with the formation of Hsf1 stress granules following heat shock. Quantification of cells that express FILIP-1L and show reduced Hsf1 are presented in the Fig. 5, right panel. As indicated in Fig. 5, lower panels, EYFP-FILIP-1L is expressed in both the cytoplasm and nucleus and often as granules; the reason for this is unclear. These structures do not colocalize with Grp75, a mitochondrial protein, Grp78, an ER resident protein, and Lamp1, a lysosomal marker (data not shown).
FIGURE 5.
Hsf1 protein level is reduced in the presence of FILIP-1L. Upper panels, H1299 cells were transiently transfected with expression plasmids containing EYFP alone. Cells were left at 37 °C or heated at 43 °C for 1 h. Expression of EYFP was detected using fluorescence microscopy. The level of Hsf1 was detected using antibody to Hsf1. Arrows show cells containing Hsf1 that forms stress granules following heat shock. Lower panels, H1299 cells were transiently transfected with expression plasmids containing EYFP-FILIP-1L. Cells were left at 37 °C or heated at 43 °C for 1 h. Expression of EYFP-FILIP-1L was detected by fluorescence microscopy. The level of Hsf1 was detected using antibody to Hsf1. Arrows show cells expressing EYFP-FILIP-1L and Hsf1 in the same cell. Arrowheads show Hsf1 nuclear granules in cells that do not express EYFP-FILIP-1L. Quantification of the percent number of cells expressing EYFP-FILLIP-1L with reduced Hsf1 levels compared with cells expressing EYFP is presented in the right panel. Bars are mean ± S.D. Statistical significance is indicated (*, p < 0.05).
FILIP-1L Represses Hsf1 Transcriptional Activity
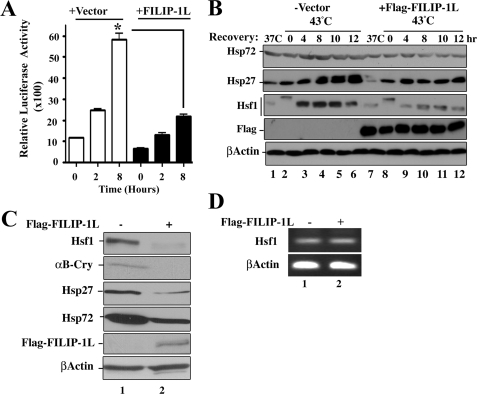
One function of Hsf1 in mammalian cells is the enhanced expression of Hsps following exposure of the cells to stress conditions (1, 4, 13). The data presented in Fig. 5 indicated that FILIP-1L expression in cells leads to reduction in Hsf1 protein and its ability to form stress granules in the nuclei following stress, suggesting that FILIP-1L may exert a negative regulatory effect on Hsf1 transcriptional activity. To this end, we performed reporter assays using plasmids containing the Hsp70 promoter fused to the luciferase (hsp70-luciferase) reporter gene. Cells were transiently transfected with plasmids containing hsp70-luciferase alone, or co-transfected with expression plasmids containing FLAG-FILIP-1L. Cells were left untreated or treated at 43 °C for 1 h, and left to recover at 37 °C for 2 or 8 h to allow luciferase expression. As the data in Fig. 6A indicate, hsp70 promoter-driven luciferase activity was significantly reduced in cells expressing FILIP-1L under both physiological growth conditions and following exposure of the cells to heat shock.
FIGURE 6.
Inhibition of Hsf1 transcriptional activity by FILIP-1L. A, HEK293 cells were transiently transfected with expression constructs containing Hsp70-luciferase plus pcDNA3-β-galactosidase (as an internal control) and Vector alone, or cotransfected with Hsp70-luciferase, pcDNA3-β-galactosidase, and FLAG-FILIP-1L. Cells were left at 37 °C or heated at 43 °C for 1 h and left to recover at 37 °C for 0, 2, and 8 h. Luciferase activity was determined and normalized to β-galactosidase expression levels. Bars are mean ± S.D. Statistical significance is indicated for 8 h post-heat treatment (*, p < 0.02). B, FILIP-1L reduces heat-induced Hsp expression. HEK293 cells were transiently transfected with empty vector (lanes 1–6) or with constructs expressing FLAG-FILIP-1L (lanes 7–12). After 48 h, cells were left at 37 °C (lanes 1 and 7) or treated at 43 °C for 1 h and then left to recover at 37 °C for 0 (lanes 2 and 8), 4 (lanes 3 and 9), 8 (lanes 4 and 10), 10 (lanes 5 and 11), or 12 h (lanes 6 and 12). The expression of Hsp72, Hsp27, Hsf1, and β-actin were determined by immunoblotting using the indicated antibodies. C, ectopic expression of FILIP-1L reduces the level of Hsf1 and its downstream target genes. C2C12 cells were infected using empty retroviral vector (lane 1) or vector expressing FLAG-FILIP-1L (lane 2). Immunoblotting experiments were performed using antibodies to detect Hsf1, αB-Crystallin (αB-Cry), Hsp27, Hsp72, or FLAG. β-Actin represents loading control. D, ectopic expression of FILIP-1L does not affect Hsf1 mRNA expression levels. C2C12 cells were infected using empty retroviral vector (lane 1) or vector expressing FLAG-FILIP-1L (lane 2). After 48 h, total RNA was isolated and Hsf1 and β-actin mRNA expression in the presence or absence of FLAG-FILIP-1L were determined using semi-quantitative RT-PCR.
To confirm whether Hsp expression is affected in cells expressing FLAG-FILIP-1L, we performed immunoblotting analyses to determine whether ectopic expression of FLAG-FILIP-1L could reduce the expression level of Hsf1 target genes (hsp72 and hsp27). HEK293 cells were transiently transfected with plasmids encoding FLAG-FILIP-1L. Cells were then heated at 43 °C for 1 h and allowed to recover at 37 °C for 0 to 12 h, and the expression levels of Hsp72 and Hsp27 were determined by immunoblotting. As indicated in Fig. 6B, the expression of these two Hsps was significantly lower (2-fold) when cells expressed FLAG-FILIP-1L. The results of immunoblotting analysis using Hsf1 antibody indicate that FILIP-1L expression in cells reduces the level of Hsf1 expression (Fig. 6B). Data in Fig. 6, C and D, also indicate that ectopic expression of FLAG-FILIP-1L reduces the levels of Hsf1, αB-crystallin, Hsp27, and Hsp72 at the level of translation by 2–4-fold, whereas the level of Hsf1 mRNA was not affected. Taken together, the above data indicate that FILIP-1L inhibits Hsf1 transcriptional activity by altering the level of Hsf1 protein.
FILIP-1L Promotes Hsf1 Polyubiquitination
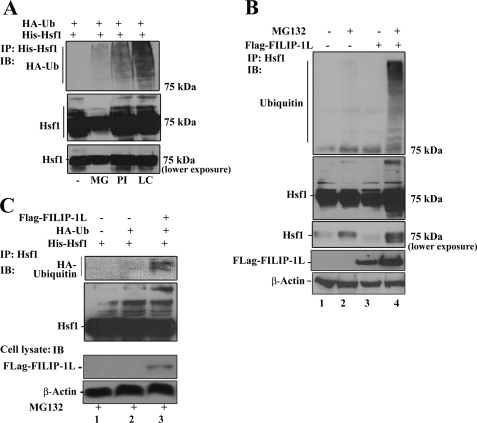
Our data indicate that ectopic expression of FLAG-FILIP-1L reduces Hsf1 expression, suggesting that FILIP-1L may regulate Hsf1 protein stability. To elucidate whether FILIP-1L can mediate Hsf1 degradation through polyubiquitination and degradation through the UPS, we first determined whether Hsf1 protein could be polyubiquitinated. HEK293 cells were transiently transfected with expression vectors encoding HA-ubiquitin and His-Hsf1. Cells were then left untreated or were treated with proteasome inhibitors MG132, proteasome inhibitor 1, or lactocystein. Hsf1 was immunoprecipitated using antibody to Hsf1 followed by immunoblotting using antibody to ubiquitin. As presented in Fig. 7A, Hsf1 could be polyubiquitinated. The proteasome inhibitor lactocystein more effectively blocked the degradation of polyubiquitinated Hsf1. The level of Hsf1 in the cell lysate is presented in Fig. 7A, lower panels. To investigate whether FILIP-1L can modulate the levels of Hsf1 polyubiquitination, cells were transfected with empty vector (Fig. 7B, lanes 1 and 2) or plasmids containing FLAG-FILIP-1L (Fig. 7B, lanes 3 and 4), and cells were left untreated (Fig. 7B, lanes 1 and 3) or were treated with MG132 (Fig. 7B, lanes 2 and 4). Endogenous Hsf1 was immunoprecipitated using antibody to Hsf1 followed by immunoblotting using antibody to ubiquitin. Data show that FLAG-FILIP-1L can enhance Hsf1 polyubiquitination in the presence of MG132. To further confirm whether FILIP-1L can modulate the Hsf1 polyubiquitination state, we performed a Talon pulldown assay. HEK293 cells were transiently transfected with expression vectors encoding His-Hsf1 alone or co-transfected with His-Hsf1 plus plasmids containing HA-ubiquitin or His-Hsf1 plus HA-ubiquitin and FLAG-FILIP-1L. Cells were treated with MG132. His-Hsf1 was pulled down with Talon beads followed by immunoblotting analyses using anti-HA-ubiquitin antibody. As indicated in Fig. 7C, FILIP-1L could enhance His-Hsf1 polyubiquitination even under denaturation conditions (Fig. 7C, lane 3). Taken together, our results demonstrate that FILIP-1L promotes Hsf1 polyubiquitination.
FIGURE 7.
FILIP-1L mediates Hsf1 degradation through the UPS. A, the Hsf1 protein level is regulated by ubiquitination. HEK293 cells were co-transfected with plasmids containing His-Hsf1 and HA-ubiquitin (HA-Ub). Cells were left untreated or were treated with 10 μm MG123 (MG), 1 μg/ml of proteasome inhibitor (PI), or 5 μm lactocystein (LC) for 6 h. His-Hsf1 was immunoprecipitated (IP) and the immunoprecipitated materials were subjected to immunoblotting (IB) analyses using antibody to HA-ubiquitin. Lower panels show the expression level of Hsf1 in the cell lysates. B, FILIP-1L enhances Hsf1 ubiquitination. HEK293 cells were transiently transfected with empty vector (lanes 1 and 2) or plasmids containing FLAG-FILIP-1L (lanes 3 and 4). Cells were then left untreated (lanes 1 and 3), or treated with 10 μm MG132 for 6 h (lanes 2 and 4). Endogenous Hsf1 was immunoprecipitated using antibody to Hsf1 and complexes were subjected to immunoblotting analyses using antibody to ubiquitin. Lower panels show the level of expression of Hsf1, FLAG-FILIP-1L, and β-actin in the cell lysates. C, determination of FILIP-1L-mediated Hsf1 ubiquitination using the nickel pulldown assay. Cells were transiently transfected with plasmids containing His-Hsf1 alone (lane 1), or cotransfected with plasmids containing HA-ubiquitin (lane 2), or HA-ubiquitin and FLAG-FILIP1L (lane 3). Cells were treated with 10 μm MG132 for 6 h and lysed in denaturing lysis buffer. The His-Hsf1 was pulled down using nickel beads, and the pulldown materials were subjected to immunoblotting using antibody to HA (upper panel) or Hsf1 (lower panel). Expression of FLAG-FILIP-1L and β-actin in the cell lysate is indicated.
FILIP-1L Binds to the UBA Domain of HhR23 and Hsf1
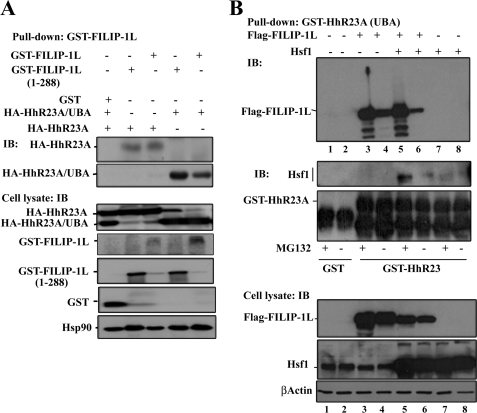
Ubiquitinated proteins are transported to the 19 S proteasome through their association with ubiquitin receptor proteins such as HhR23A (yeast homolog of Rad23A) (32, 33). Within its protein structure, HhR23A contains two ubiquitin associating UBA domains at its C-terminal domain, and the ubiquitin-like (UBL) sequences at its N-terminal domain. To elucidate whether FILIP-1L-mediated proteasome degradation of Hsf1 is regulated by adaptor protein HhR23A, we performed an in vitro GST pulldown assay. HEK293 cells were transiently transfected with GST, GST-FILIP-1L amino acid residues 1–893 or 1–288, and HA-HhR23A or HA-HhR23A UBA domains. GST pulldown materials were immunoblotted using antibody to HA. Data presented in Fig. 8A indicate that both fragments of FILIP-1L interact with full-length HA-HhR23A and its UBA domains. The lower panels of Fig. 8A show expression of overexpressed proteins in the cell lysate. These data indicate that the FILIP-1L leucine zipper domains interact with the UBA domain of HhR23A. To examine whether Hsf1, FILLIP-1L, and HhR23A are in the same complex, we performed an in vitro GST pulldown assay. HEK293 cells were transiently transfected with empty vector (Fig. 8B, lanes 1 and 2), FLAG-FILIP1L alone (Fig. 8B, lanes 3 and 4), or His-Hsf1 alone (Fig. 8B, lanes 7 and 8) or cotransfected with FLAG-FILIP-1L plus His-Hsf1 (Fig. 8B, lanes 5 and 6). Cells were left untreated or were treated with MG132. Cell lysates were prepared and incubated with bacterially purified protein GST-HhR23A (containing only the two UBA domains) (34). As the data in Fig. 8 indicate, FILIP-1L can be pulled down using GST-HhR23A (Fig. 8B, lanes 3 and 4). Furthermore, both FILIP-1L and Hsf1 could be pulled down using GST-HhR23A (Fig. 8B, lanes 5 and 6). No Hsf1 or FILIP-1L could be pulled down using GST alone (Fig. 8B, lanes 1 and 2). In addition, HhR23A alone (without FILIP-1L) could not pull down Hsf1 significantly (Fig. 8B, lanes 7 and 8). In all cases, treatment of cells with MG132 enhanced the interaction of HhR23A with FILIP-1L or Hsf1. The levels of overexpressed His-Hsf1 and FLAG-FILIP-1L in cell lysates are indicated in Fig. 8B, lower panels. Taken together, our data indicate that FILLIP-1L brings Hsf1 and HhR23A together into the same complexes.
FIGURE 8.
FILIP-1L and Hsf1 form complexes with HhR23A. A, FILIP-1L interacts with the UBA domain of HhR23A in an in vivo pulldown assay. HEK293 cells were transiently transfected with the indicated expression vectors (+). After 48 h, the GST pulldown assay was performed, and immunoprecipitated materials were used in immunoblotting (IB) using the HA antibody. Expression levels of HA-HhR23A and its UBA domain, GST-FILIP-1L, and GST alone are presented in the cell lysates (lower panels). The expression level of Hsp90 is presented as loading control. B, FILIP-1L interacts with Hsf1 and the HhR23A UBA domain in an in vitro pulldown assay. HEK293 cells were transiently transfected with the indicated expression vectors (+). Cells were untreated or treated with 10 μm MG132 for 6 h. GST-HhR23A was used in pulldown assays and the level of immunoprecipitated materials was detected using antibodies to FLAG and Hsf1. The level of purified GST and the GST-HhR23A UBA domain are indicated. Lower panels show the expression of FLAG-FILIP-1L and Hsf1 in the cell lysates. β-Actin is the loading control. Lanes 1–8 indicates the same groups in the upper and lower panels.
DISCUSSION
Hsf1 transcription factor activity is tightly regulated; however, whether Hsf1 is degraded during its cycles of activation and inactivation remain poorly understood. One study has found that in cells undergoing mitosis, Hsf1 can be ubiquitinated and degraded by the SCFβ-TriCP complex (28). Ubiquitination and degradation of Hsf1 by the SCFβ-TriCP complex only occurred during mitosis when the phosphorylated Hsf1 at serine 216 was released from the Cdc20 complex. Hsf1 was then found in SCFβ-TrCP complexes, ubiquitinated, and degraded (28). Another report indicates that anaphase-promoting complex/cyclosome (APC/C) ubiquitin E3 ligase mediates the ubiquitination and degradation of another Hsf family member, Hsf2 through substrate recognition by both Cdc20 and Cdh1 (5). Degradation of Hsf2 was observed during the heat shock response. Comparable interactions or ubiquitination of Hsf1 was not detected (5).
In the study reported here, we have found that a protein named FILIP-1L interacts with Hsf1. The function of FILIP-1L has not clearly been characterized. FILIP-1L encodes coiled-coiled, leucine zipper, and ATPase domains and exists as multiple isoforms in many cell types (30, 31). Ectopic expression of FILIP-1L results in the inhibition of cellular proliferation, migration, and apoptosis. Furthermore, endothelial cells treated with angiogenic inhibitors up-regulate FILIP-1L expression. FILIP-1L expression in tumor vasculature reduces tumor cell proliferation in vivo (31). FILIP-1L has been identified to be absent in ovarian cancer cells, but present in normal epithelial cells and was named down-regulated ovarian cancer protein 1 or DOC1 (35). We present evidence that Hsf1 interacts with FILIP-1L in the yeast two-hybrid system as well as in mammalian cells. The N-terminal fragment of FILIP-1L containing the leucine zipper domain interacts with the N-terminal hydrophobic region of Hsf1 that is required for its trimerization and transcriptional activity. Interestingly, the Hsf1 and FILIP-1L interaction, which also contains Hsp72 in the complexes leads to polyubiquitination and degradation of Hsf1 protein. Cells co-expressing Hsf1 and FILIP-1L exhibit reduction in the Hsf1 protein level, and inhibition of Hsf1 stress granule formation following exposure to heat shock. Ectopic expression of full-length FILIP-1L most efficiently degraded the endogenous Hsf1 protein or Hsf1 protein containing short tags. Additionally, FILIP-1L expression in cells also inhibits Hsf1-mediated transcriptional activity. Because the FILIP-1L protein appears to lack an E3 ligase domain and we were unable to show E3 ligase activity of FILIP-1L using an in vitro transcription/translation system (data not shown), we performed experiments to determine whether FILIP-1L acts as a scaffold mediating ubiquitination and degradation of Hsf1. To determine whether there could be additional proteins that may be involved in the transfer of Hsf1 to the proteasome, we used full-length hHR23A (hPlic-1, Chap1/hPlic-2) as well as the HhR23A UBA domain, which is a known ubiquitin receptor (33, 34). We were able to detect Hsf1, FILIP-1L, and the hHR23A UBA domain in the same complex, suggesting that the hHR23A UBA domain could be an intermediate molecule delivering the ubiquitinated Hsf1 to the UPS. HhR23A was originally identified as a protein that has UBA and UBL domains in yeast (33). The UBL domain is recognized by S5a, which is a subunit of the proteasome. Based on the domains present in the ubiquitin receptors, they most likely transport ubiquitinated proteins to the proteasome. HhR23A (Rad23) has been identified to bind the E6-AP ubiquitin ligase, and it has been shown to target the polyubiquitinated p53 and Png1 proteins to the UPS. The UBA domain of hHR23A binds the ubiquitinated p53, protecting the molecule from deubiquitination, whereas the UBL domain of hHR23A transfers the ubiquitinated p53 to the UPS. Png1, which is a deglycosylating enzyme, is able to transfer ubiquitinated proteins to the UPS (36). The speculation is that the substrate specificity of Rad23A or other ubiquitin-binding proteins may be defined by other adaptor proteins. We can envision that FILIP-1L may be such an adaptor protein for the transport of ubiquitinated Hsf1 to the UPS system. As to what is the exact function of FILIP-1L, we show that hHR23A does not directly interact with Hsf1 but that Hsf1, through its interaction with the adaptor protein FILIP-1L, is transported to the UPS via the HhR23A UBA domain.
In conclusion, we have identified the adaptor protein FILIP-1L to interact with Hsf1 and facilitate its ubiquitination and degradation. FILIP-1L and Hsf1 complexes are transported to the proteasome, likely via Class 1 UBA domain containing ubiquitin receptors such as hHR23A.
Acknowledgment
We thank Dr. Tetsu Kamitani for the plasmid encoding HA-ubiquitin and discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants CA062130 and CA132640 (to N. F. M.).
- Hsf
- heat shock transcription factor
- FILIP-1L
- filamin A-interacting protein 1-like
- UPS
- ubiquitin-proteosome system.
REFERENCES
- 1. Morimoto R. I. (1998) Genes Dev. 12, 3788–3796 [DOI] [PubMed] [Google Scholar]
- 2. Kroeger P. E., Sarge K. D., Morimoto R. I. (1993) Mol. Cell. Biol. 13, 3370–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nakai A., Tanabe M., Kawazoe Y., Inazawa J., Morimoto R. I., Nagata K. (1997) Mol. Cell. Biol. 17, 469–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu C. (1995) Annu. Rev. Cell Dev. Biol. 11, 441–469 [DOI] [PubMed] [Google Scholar]
- 5. Ahlskog J. K., Björk J. K., Elsing A. N., Aspelin C., Kallio M., Roos-Mattjus P., Sistonen L. (2010) Mol. Cell. Biol. 30, 5608–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kallio M., Chang Y., Manuel M., Alastalo T. P., Rallu M., Gitton Y., Pirkkala L., Loones M. T., Paslaru L., Larney S., Hiard S., Morange M., Sistonen L., Mezger V. (2002) EMBO J. 21, 2591–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sistonen L., Sarge K. D., Phillips B., Abravaya K., Morimoto R. I. (1992) Mol. Cell Biol. 12, 4104–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Min J. N., Zhang Y., Moskophidis D., Mivechi N. F. (2004) Genesis 40, 205–217 [DOI] [PubMed] [Google Scholar]
- 9. Wang G., Ying Z., Jin X., Tu N., Zhang Y., Phillips M., Moskophidis D., Mivechi N. F. (2004) Genesis 38, 66–80 [DOI] [PubMed] [Google Scholar]
- 10. Fujimoto M., Izu H., Seki K., Fukuda K., Nishida T., Yamada S., Kato K., Yonemura S., Inouye S., Nakai A. (2004) EMBO J. 23, 4297–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Min J. N., Huang L., Zimonjic D. B., Moskophidis D., Mivechi N. F. (2007) Oncogene 26, 5086–5097 [DOI] [PubMed] [Google Scholar]
- 12. Dai C., Whitesell L., Rogers A. B., Lindquist S. (2007) Cell 130, 1005–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Y., Huang L., Zhang J., Moskophidis D., Mivechi N. F. (2002) J. Cell Biochem. 86, 376–393 [DOI] [PubMed] [Google Scholar]
- 14. Homma S., Jin X., Wang G., Tu N., Min J., Yanasak N., Mivechi N. F. (2007) J. Neurosci. 27, 7974–7986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang X., Khaleque M. A., Zhao M. J., Zhong R., Gaestel M., Calderwood S. K. (2006) J. Biol. Chem. 281, 782–791 [DOI] [PubMed] [Google Scholar]
- 16. Westerheide S. D., Anckar J., Stevens S. M., Jr., Sistonen L., Morimoto R. I. (2009) Science 323, 1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dai R., Frejtag W., He B., Zhang Y., Mivechi N. F. (2000) J. Biol. Chem. 275, 18210–18218 [DOI] [PubMed] [Google Scholar]
- 18. Hietakangas V., Anckar J., Blomster H. A., Fujimoto M., Palvimo J. J., Nakai A., Sistonen L. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hilgarth R. S., Murphy L. A., Skaggs H. S., Wilkerson D. C., Xing H., Sarge K. D. (2004) J. Biol. Chem. 279, 53899–53902 [DOI] [PubMed] [Google Scholar]
- 20. Zou J., Guo Y., Guettouche T., Smith D. F., Voellmy R. (1998) Cell 94, 471–480 [DOI] [PubMed] [Google Scholar]
- 21. Akerfelt M., Morimoto R. I., Sistonen L. (2010) Nat. Rev. Mol. Cell Biol. 11, 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Satyal S. H., Chen D., Fox S. G., Kramer J. M., Morimoto R. I. (1998) Genes Dev. 12, 1962–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dai Q., Zhang C., Wu Y., McDonough H., Whaley R. A., Godfrey V., Li H. H., Madamanchi N., Xu W., Neckers L., Cyr D., Patterson C. (2003) EMBO J. 22, 5446–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu Y., Mivechi N. F. (2003) J. Biol. Chem. 278, 17299–17306 [DOI] [PubMed] [Google Scholar]
- 25. Boellmann F., Guettouche T., Guo Y., Fenna M., Mnayer L., Voellmy R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4100–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khaleque M. A., Bharti A., Gong J., Gray P. J., Sachdev V., Ciocca D. R., Stati A., Fanelli M., Calderwood S. K. (2008) Oncogene 27, 1886–1893 [DOI] [PubMed] [Google Scholar]
- 27. Wang G., Zhang J., Moskophidis D., Mivechi N. F. (2003) Genesis 36, 48–61 [DOI] [PubMed] [Google Scholar]
- 28. Lee Y. J., Kim E. H., Lee J. S., Jeoung D., Bae S., Kwon S. H., Lee Y. S. (2008) Cancer Res. 68, 7550–7560 [DOI] [PubMed] [Google Scholar]
- 29. Shi Y., Mosser D. D., Morimoto R. I. (1998) Genes Dev. 12, 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tandle A. T., Mazzanti C., Alexander H. R., Roberts D. D., Libutti S. K. (2005) Cytokine 30, 347–358 [DOI] [PubMed] [Google Scholar]
- 31. Kwon M., Hanna E., Lorang D., He M., Quick J. S., Adem A., Stevenson C., Chung J. Y., Hewitt S. M., Zudaire E., Esposito D., Cuttitta F., Libutti S. K. (2008) Cancer Res. 68, 7332–7341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Léon S., Haguenauer-Tsapis R. (2009) Exp. Cell Res. 315, 1574–1583 [DOI] [PubMed] [Google Scholar]
- 33. Dantuma N. P., Heinen C., Hoogstraten D. (2009) DNA Repair 8, 449–460 [DOI] [PubMed] [Google Scholar]
- 34. Hurley J. H., Lee S., Prag G. (2006) Biochem. J. 399, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mok S. C., Wong K. K., Chan R. K., Lau C. C., Tsao S. W., Knapp R. C., Berkowitz R. S. (1994) Gynecol. Oncol. 52, 247–252 [DOI] [PubMed] [Google Scholar]
- 36. Kim I., Ahn J., Liu C., Tanabe K., Apodaca J., Suzuki T., Rao H. (2006) J. Cell Biol. 172, 211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]