Abstract
Neuroinflammation and traumatic brain injury involve activation of inflammatory cells and production of local pro-inflammatory mediators that can amplify tissue damage. Using LC-UV-MS-MS-based lipidomics in tandem with functional screening at the single-cell level in microfluidic chambers, we identified a series of novel bioactive oxygenated docosahexaenoyl ethanolamide- (DHEA) derived products that regulated leukocyte motility. These included 10,17-dihydroxydocosahexaenoyl ethanolamide (10,17-diHDHEA) and 15-hydroxy-16(17)-epoxy-docosapentaenoyl ethanolamide (15-HEDPEA), each of which was an agonist of recombinant CB2 receptors with EC50 3.9 × 10−10 and 1.0 × 10−10 m. In human whole blood, 10,17-diHDHEA and 15-HEDPEA at concentrations as low as 10 pm each prevented formation of platelet-leukocyte aggregates involving either platelet-monocyte or platelet-polymorphonuclear leukocyte. In vivo, 15-HEDPEA was organ-protective in mouse reperfusion second organ injury. Together these results indicate that DHEA oxidative metabolism produces potent novel molecules with anti-inflammatory and organ-protective properties.
Keywords: Chemotaxis, Fatty Acid, Inflammation, Leukocyte, Neutrophil, Cannabinoids, Novel Bioactive Products, Tissue Injury
Introduction
Neuroinflammation and local pro-inflammatory mediators are associated with neurodegenerative diseases as well as traumatic brain injury (1). In both scenarios, treatment with docosahexaenoic acid (DHA)2 reduces inflammation and local tissue injury. For example, DHA reduces the damage from impact acceleration injury and reduces β-amyloid precursor, a marker of axonal injury in vivo relevant in traumatic brain injury (2). Also, DHA reduces ischemic stroke in rats via production of neuroprotectin D1, which acts on leukocytes and reduces leukocyte infiltration and leukocyte-mediated tissue damage and regulates NF-κB (3). Neuroprotectin D1 stimulates neuronal stem cell differentiation (4) and has potent anti-inflammatory and proresolving actions in several in vivo disease models (5–7). D series resolvins are biosynthesized from DHA in brain tissue and resolving inflammatory exudates (7, 8). Resolvin D1 and resolvin D2 display potent stereoselective actions that are anti-inflammatory and proresolving, reduce pain signaling, and act in the pico-to-nanomolar range in vivo, a dose range where DHA itself displays no demonstrable action (9–11). Hence, the metabolome and metabolic fate of DHA is of interest in the resolution of pain, inflammation, and tissue injury.
Another metabolic fate of DHA in brain is conversion to docosahexaenoyl ethanolamide (DHEA), which is thought to be produced by the same pathway as N-acyl-arachidonoyl-ethanolamide (AEA, anandamide) (12). DHEA is directly related to dietary intake of DHA and is enriched in brain tissue at levels comparable with AEA (13). AEA is an endocannabinoid that regulates neurofunctions and the immune system via cannabinoid (CB) 1 and 2 receptors (14–17). Because AEA undergoes oxidative metabolism to bioactive molecules (16, 18), we addressed whether the beneficial actions of DHA treatment, for example, protection against brain injury (2), can be regulated in part by conversion of DHEA to bioactive products.
Herein we report on the DHEA metabolome with identification of novel potent bioactive molecules that are organ-protective in vivo. These novel bioactive products from DHEA were identified using LC-MS-MS-based lipidomics in tandem with functional single-cell screening in newly engineered microfluidic chambers and in vivo systems. These new bioactive products from DHEA may underlie some of the beneficial effects of DHA administration.
EXPERIMENTAL PROCEDURES
Materials
LC grade solvents were purchased from Fisher Scientific. Phenomenex Luna C18 (150 mm × 2 mm × 5 μm) column and Strata-X solid phase extraction columns were purchased from Phenomenex (Torrance, CA). Soybean lipoxygenase, human hemoglobin, human serum albumin, d4-MeOH, and Hanks' balanced salt buffer were purchased from Sigma-Aldrich. BSTFA was purchased from Pierce. P-selectin was purchased from R&D Systems (Minneapolis, MN). Phycoerythrin-conjugated mouse anti-human CD62P and FITC-CD41 anti-human were purchased from BD Biosciences. FITC-conjugated mouse anti-human CD14, mouse anti-human CD16, Cy5-conjugated mouse anti-human CD3, and mouse anti-human CD20 were all purchased from Pharmingen. 17-Hydroxydocosahexaenoic acid (17-HDHA) standard was prepared from DHA and soybean lipoxygenase (8, 19). Docosahexaenoyl ethanolamide (DHEA) was custom synthesized by Dr. Piomelli's group at University of California, Irvine or purchased, as was 15(S)-HETE ethanolamide from Cayman Chemical (Ann Arbor, MI).
Animals
All animals used in the present study were male FVB mice (Charles River Laboratories) that were 6–8 weeks old (weighing 20–25 g). They were maintained in a temperature- and light-controlled environment and had unlimited access to water and food (laboratory standard rodent diet 5001 (Lab Diet)), containing 1.5% eicosapentaenoic acid, 1.9% DHA of total fatty acids. Experiments were performed in accordance with the Harvard Medical School Standing Committee on Animals guidelines for animal care (Protocol 02570).
Reverse Phase-HPLC
Liquid chromatographic analyses and separations were performed using an Agilent 1100 series high performance liquid chromatography (HPLC) system (Agilent, Santa Clara, CA) equipped with G1379A degasser, G1312A binpump, and G1315B UV diode array detector. HPLC analyses were carried out using a Phenomenex C18 column (150 mm × 2 mm × 5 μm) with the mobile phase of 0.2 ml/min flow rate (methanol:water, 70:30 v/v from 0 to 18 min, then ramped to 100% methanol from 18 to 35 min). Compound isolations/purifications were carried out using a Beckman ODS column (10 mm × 250 mm × 5 μm) with the mobile phase flow rate at 4 ml/min (methanol:water, 70: 30 v/v from 0 to 18 min, then ramped to 100% methanol from 18 to 35 min).
Lipidomics MS-MS Analysis
Sample analyses were carried out using a mass spectrometer (Qstar XL quadrupole TOF hybrid mass spectrometer; Applied Biosystems, Foster City, CA) equipped with two Shimadzu LC20AD HPLC pumps (Shimadzu, Columbia, MD) and an Agilent G1315B UV diode array detector (Agilent). For routine analyses, samples were extracted using C-18 cartridge as in Ref. 19 and injected to a Phenomenex C18 column (150 mm × 2 mm × 5 μm), and the mobile phase (methanol:water; 70:30 v/v from 0 to 18 min, then ramped to 100% methanol from 18 to 35 min) was eluted at a 0.2 ml/min flow rate and UV detector-scanned from 200 to 400 nm before samples entered the MS-MS. GC-MS analysis was carried out as in Ref. 9. Samples were injected in 2.5 μl of hexane.
Preparation of Oxygenated DHEA Products
DHEA (12.5 mg) was suspended in 0.05 m borate buffer (250 ml, pH = 9.3) at 4 °C, and 160 kilounits of soybean LOX3 (type VI, 640 kilounits total, 701 kilounits/mg of protein, 3.6 mg of protein/ml) was added at 0, 2, 4, and 6 min. The incubation was monitored using a UV spectrometer (Agilent). Incubations were treated with NaBH4 before extraction two times with 200 ml of ether. The organic layers were combined, washed twice with 100 ml of double distilled H2O, taken to dryness under nitrogen flow, and subjected to preparative HPLC isolation monitoring online UV at 235, 245, and 270 nm for isolation of 4,17-diHDHEA, 7,17-diHDHEA, and 14,17-diHDHEA, respectively. The corresponding fraction was collected, dried under nitrogen, and resuspended in methanol. Preparation of each compound was confirmed using GC-MS or LC-MS-MS before further investigation.
Preparation of HEDPEA
Human hemoglobin (400 mg) was added to 17-hydroperoxydocosahexaenoyl ethanolamide (17-HpDHEA) (2.75 mg), suspended in 25 ml of phosphate buffer (0.1 m, pH 7.3, 37 °C), and vortexed (5 min). The incubations were carried out at 37 °C for 6 min and then diluted with double distilled H2O to 100 ml and extracted twice with 150 ml of ether. The organic layer was combined and washed twice with 100 ml of double distilled H2O. The crude product was taken to dryness under nitrogen flow and then isolated by preparative HPLC isolation. The fractions were isolated and collected, monitoring UV absorbance at 215 nm. Each fraction was collected, taken to dryness under nitrogen flow, and either subjected to LC-MS-MS and/or NMR analysis or derivatized with BSTFA and then subjected to GC-MS analysis.
Receptor-Ligand Interactions
Receptor activation with the CB2 β-arrestin system was carried out essentially as in Refs. 20 and 21. HEK cells stably overexpressing human CB2 receptor tagged with Pro-Link and Enzyme Acceptor-labeled β-arrestin (Discoverx, Fremont CA) were plated at 20,000 cells/well of a 96-well plate. Forty-eight hours after plating, cells were incubated with compounds at concentrations from 1 pm to 100 nm for 1 h in serum-free DMEM at 37 °C. Ligand-receptor interaction was determined by measuring chemiluminescence using the PathHunter EFC detection kit (Discoverx), generated upon coupling of the Enzyme Acceptor-labeled β-arrestin with the Pro-Link-tagged receptor, with a plate reader (Envision, PerkinElmer Life Sciences).
PMN Isolation and Incubations
Human whole blood was collected (Brigham and Women's Hospital Protocol 88-02642), and PMNs were isolated as in Refs. 8, 9, and 11. PMNs (2 × 106) suspended in 1 ml of Dulbecco's PBS+/+ with 0.2% bovine serum albumin (Sigma) were incubated with 5 μg of HPLC-isolated 17-HpDHEA or DHEA, alone or with zymosan A (100 μg/ml) for 30 min at 37 °C, and incubations were stopped with 2 volumes of ice-cold methanol. The mixture was kept in −20 °C for at least 2 h to precipitate proteins and then taken for C18 solid phase extraction and analysis.
Leukocyte Chemotaxis Screening of DHEA Metabolites with Microfluidic Chamber
The fabrication and surface modification of the microfluidic devices were prepared as in Refs. 9 and 22. Whole blood (5–10 μl) diluted in Hanks' balanced salt buffer (1:10, v/v) was introduced into the chemotaxis chamber via a cell inlet, and neutrophils were captured along the chamber via P-selectin tethering. Next, the transversal gradient of IL-8 (0–10 nm) was introduced to the chemotaxis chamber. After 15 min, novel DHEA metabolites (at a uniform concentration) were introduced to the chemotaxis chamber from the second gradient generator network, and 10 nm IL-8 gradient was maintained. Single-cell neutrophil chemotaxis was recorded using microscopy (Nikon, Eclipse E600) equipped with a video camera (Diagnostic, RT Slider) and subject to analysis using the ImageJ software (9).
PAF-stimulated Platelet-Leukocyte Aggregate Formation
Whole blood was incubated with either vehicle or HPLC-isolated 10,17-diHDHEA or 15-HEDPEA (0.01–100 nm) for 15 min at 37 °C with intermittent mixing. Vehicle or PAF (100 nm, PAF C-16, Cayman Chemical, Ann Arbor, MI) was added for another 30 min at 37 °C with intermittent mixing. Incubation was stopped by ice-cold red blood cell lysis buffer (10 min at 4 °C). Cells were collected using centrifugation (210 × g, 5 min, 4 °C) and then fixed with 3% formalin (15 min, 4 °C). Cells were stained with FITC-anti-human CD41 (1:100, v/v) and phycoerythrin-anti-human-CD62P (1:100, v/v) for 20 min at 4 °C and were analyzed using flow cytometry and the CellQuest software as in Ref. 23. Cellular composition within whole blood was determined by forward and side scattering as well as cell-specific markers, anti-human-CD41 for platelets, anti-human-CD14 for monocytes, and anti-human-CD16 for neutrophils.
Second Organ Reperfusion Injury
Murine hind limb vascular occlusion second organ lung reperfusion injury was performed using 6–8-week-old FVB male mice and carried out as in Ref. 24.
Statistical Analysis
The significance of difference between groups was evaluated using the two-tailed Student's t test. p values of less than 0.05 were considered to be statistically significant.
RESULTS
Functional Metabolomics
LC-UV-MS-MS Identification of 17-HDHEA from Brain
To investigate the potential endogenous generation of DHEA-derived bioactive products, mouse brain was harvested and subjected to solid phase extraction (19), and the resulting methyl formate fractions were taken for LC-UV-MS-MS-based metabolomics. Tandem mass fragmentations and online UV spectrum with characteristic λmax at 237 nm are consistent with the proposed structure as shown in Fig. 1B, inset. Because of the lack of suitable functional groups for direct efficient ionization and analysis of 17-hydroxy-4Z,7Z,10Z,13Z,15E,19Z-docosahexaenoylethanolamide (17-HDHEA), its acetate adduct m/z 446 = [M+CH3COOH-H] was targeted for analysis. The major tandem mass ions were assigned as following: m/z 386 = [M-H], 368 = [M-H-H2O], 281 = [299-H2O]. The m/z 288 is consistent with fragmentation at Position 17 (see Table 1 for numbering) (Fig. 1B). Because of the limited quantities of endogenous 17-HDHEA produced in brain tissue, further analyses and in vitro enzymatic preparation were carried out by incubating DHEA with 15-LOX followed by reduction with NaBH4 (see “Experimental Procedures”). Endogenous 17-HDHEA and the enzymatically prepared compound in vitro gave essentially the same LC retention times and tandem mass fragmentations using LC-MS-MS (see supplemental Fig. 1). To assess their production by human and mouse tissues, DHEA was also incubated with isolated human PMN or whole mouse brain because DHEA is enriched in this tissue. LC-MS-MS-based targeted lipidomics indicated the production of a novel series of oxygenated DHEA (Table 1).
FIGURE 1.
Identification of hydroxydocosahexaenoyl ethanolamide and functional screening of DHEA brain metabolome. A, online UV. B, tandem mass spectrum of 17-HDHEA. Inset shows fragment assignments for HDHEA mass spectrum. amu, atomic mass units. C, representative average of PMN directional migration velocity in 0–10 nm IL-8 gradient (μm/min) before and after metabolite mixtures were infused to the chamber. The mixtures were isolated from mouse brain homogenate incubated with 5 μg of DHEA. Error bars represent migration distance ± S.D. for mean of 26 single PMN (n = 3 separate donors). *, p < 0.01 for IL-8 versus brain extracts.
TABLE 1.
Structures, LC-MS and GC-MS fragmentations, and UV λ max for novel DHEA metabolites identified using mediator-based lipidomics
† Stereochemistries shown are tentative assignments.
Decoding Metabolomics Using Microfluidic Chambers
In parallel to structure elucidation, chemotactic screening of HPLC-isolated DHEA metabolites obtained from mouse brain was carried out by utilizing microfluidic chamber (Fig. 1C). After IL-8 (0–10 nm gradient) was introduced to the main channel of the microfluidic device, P-selectin tethered leukocytes rapidly migrated along the IL-8 chemotactic gradient at an average rate of 2.3 μm/min. After 15 min, the mixture of metabolites was infused into the microfluidic main channel while an IL-8 (0–10 nm) gradient was maintained (Fig. 1C, left panel). Human PMN chemotaxis was dramatically reduced (p < 0.01) upon the addition of the brain metabolite mixture, whereby average human PMN chemotaxis velocity dropped from 2.3 to ∼0.7 μm/min (Fig. 1C, middle panel). This decrease in chemotaxic velocity was maintained even after the gradient was switched back to IL-8. These results indicated that the brain metabolites contained bioactive components that stopped PMN chemotaxis.
LC-UV-MS-MS and GC-MS-based Metabolomics of DHEA
Results from this screening uncovered that at least one bioactive product was present among the mixture of DHEA metabolites; thus, we pursued the metabolic fates of DHEA and 17-HpDHEA/17-HDHEA identified in mouse brain (Fig. 1B) using LC/UV/MS/MS-based lipidomics. As with 17-HDHEA, acetate adducts of potential DHEA-derived metabolites [M+CH3COOH-H] were targeted for tandem mass analysis (Table 1). These results demonstrated the presence and production of novel products in the DHEA metabolome.
Incubations of isolated human PMNs with DHEA or 17-HpDHEA led to the generation of 17-HDHEA, 4,17-diHDHEA, 10,17-diHDHEA, and 15-HEDPEA. Human hemoglobin, which can be liberated upon tissue damage (25), was incubated with 17-HpDHEA and gave 13-HEDPEA and 15-HEDPEA as prominent products, as well as 17-HDHEA (Table 1). Mouse brain homogenates with DHEA also produced 17-HDHEA and 4,17-diHDHEA as major products with smaller amounts of 7,17-diHDHEA, 10,17-diHDHEA, and 15-HEDPEA. The online UV and tandem mass spectra for 4,17-diHDHEA are shown in Fig. 2, A and B. The adduct parent ion, the analyte parent ion, and the ions resulting from neutral loss are m/z 462 = [M+CH3COOH-H], 402 = [M-H], 384 = [M-H-H2O], and 366 = [M-H-2H2O], which are common signature ions for all dihydroxy-containing DHEA products. The ions m/z 333, 315 = [333-H2O], 304, and 286 = [304-H2O] were assigned as diagnostic ions for fragmentations at Position 17. Fragmentations at Position 4 can lead to m/z 144, 257, and 239 = [257-H2O]. Its UV spectrum displayed characteristic maximum absorbance at 238 nm, which was consistent with the presence of two separated conjugated diene structures in this compound. As shown in Fig. 2, C and D, diagnostic ions m/z 304, 286 = [304-H2O]; 184 and 156 corresponded to the fragmentations at Positions 7 and 17 of 7,17-diHDHEA respectively (see Table 1 for numbering). The UV spectrum of the compound displayed maximum absorbance, λmax, at 246 nm (26), consistent with the presence of two diene structures separated by a methylene group. For 10,17-diHDHEA, m/z 333, 315 = [333-H2O], 304, 286 = [304-H2O], and 196 came from fragmentations at Positions 10 and 17 as shown in Fig. 2, E and F. The presence of a conjugated triene structure in 10,17-diHDHEA was confirmed by the characteristic UV spectrum with λmax at 270 nm. Tandem mass spectrum of 13-HEDPEA is shown in supplemental Fig. 2A with signature fragmentation ions m/z 320, 304, 286 = [304-H2O], and 236. GC/MS was also utilized for additional structural analysis with 13-HEDPEA and 15-HEDPEA that confirmed the original tandem MS assignments shown in supplemental Fig. 2, C and D. The C-value for 13-HEDPEA was determined as 32.1 ± 0.2 (supplemental Fig. 2E), and for 15-HEDPEA, it was determined as 33.7 ± 0.2 (supplemental Fig. 2F).
FIGURE 2.
DHEA metabolome via LC-UV-MS-MS-based lipidomics. A–F, online UV, fragment assignments shown in inset, and tandem mass spectra of 4,17-diHDHEA, 7,17-diHDHEA, and 10,17-diHDHEA. amu, atomic mass units.
To determine concentrations, as well as to further confirm structures, HPLC-isolated 13-HEDPEA and 15-HEDPEA were characterized using proton NMR (1H NMR). The chemical shift assignments are shown in supplemental Table 2, a and b, respectively. For 15-HEDPEA, the proton at Position 15 (H-15) displayed two distinct chemical shifts, which will be discussed later. Because of the limited amounts of materials and the lack of informative UV chromophores present in these compounds, NMR spectroscopy was also used for quantitation using 17-hydroxydocosahexaenoic acid (17-HDHA) as an internal standard with known concentrations. The NMR quantitated compounds were then used as standards for HPLC quantitation monitoring UV chromatogram at 210 nm or LC-tandem mass profiling (see “Experimental Procedures” for further details).
Human PMN Single-cell Chemotactic Functional Screening
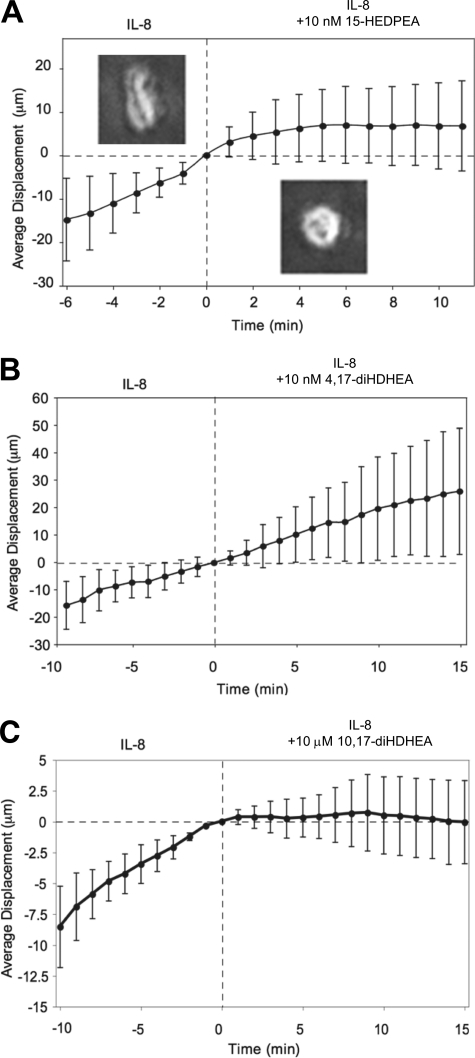
HPLC-isolated dioxygenated DHEA products were individually screened for direct PMN actions using microfluidic chambers. Infusion of isolated 15-HEDPEA at 10 nm to the main channel stimulated changes in morphology and chemotaxis of PMN in the IL-8 gradient and stopped further PMN migration after ∼4 min (Fig. 3A). For direct comparison, PMN chemotaxis velocity did not change with time with the IL-8 gradient (supplemental Fig. 3A). At 10 nm, 4,17-diHDHEA (Fig. 3B), 7,17-diHDHEA, or 10,17-diHDHEA did not significantly regulate chemotaxis (supplemental Fig. 3, B and C), whereas at higher concentrations, e.g. 10 μm, 10,17-diHDHEA rapidly stopped PMN chemotaxis (Fig. 3C). These results indicate that 15-HEDPEA is the most potent of this series in regulating human PMN shape change and motility.
FIGURE 3.
Microfluidic chamber-based screening of human PMN chemotaxis with DHEA-derived products. A–C, representative average PMN directional migration displacement against 0–10 nm IL-8 gradient from original positions (in μm) before and after HPLC-isolated DHEA-derived products were individually infused to the chambers. Insets in A show morphology of PMN before (left side) and after (right side) exposure to 10 nm 15-HEDPEA (average of 23–30 PMN in each panel). Error bars represent migration distance ± S.D. for mean of 26 single PMN (n = 3 separate donors).
Cannabinoid Receptor Activation
Because AEA exerts a wide range of bioactions via activating cannabinoid receptor(s) (14, 27), we therefore next tested whether DHEA, 10,17-diHDHEA, or 15-HEDPEA also activated CB receptors. To this end, we used recombinant human CB receptors overexpressed in a β-arrestin system as described under “Experimental Procedures.” AEA was used for direct comparison as a known agonist. Fig. 4 shows the dose response of CB1 and CB2 with each compound. Activation of CB2 by AEA gave EC50 ∼1.1 × 10−10 m and activation by DHEA gave EC50 9.8 × 10−9 m (Fig. 4B). For comparison, EC50 values for metabolically oxygenated products, 10,17-diHDHEA and 15-HEDPEA, were 3.9 × 10−10 and 1.0 × 10−10 m, respectively (Fig. 4, C and D). These results demonstrate that enzymatic oxidation products from DHEA are activators of CB2 receptors and that 10,17-diHDHEA and 15-HEDPEA also activated CB1 receptors but required much higher concentrations (Fig. 4A). By comparison, 15(S)-HETE ethanolamide, the oxygenated product of AEA, did not stimulate CB2 receptors in this dose range (Fig. 4B). CB2 receptor-ligand interactions were confirmed with the dose response of CB2-specific antagonist AM630. When incubated with GPCR CB2 overexpressed cells, AM630 inhibited activation stimulated with 15-HEDPEA (10 nm) and AEA (10 nm), used here for a known positive and direct comparison (Fig. 4, E and F). AM630 also inhibited GPCR CB2 interaction with 10,17-diHDHEA at higher concentration (n = 3, data not shown).
FIGURE 4.
GPCR CB1 and CB2 are activated by DHEA-derived products. HEK cells overexpressing CB1 or CB2 in a β-arrestin system were incubated with the indicated concentrations of compounds for 1 h in serum-free DMEM at 37 °C. Ligand receptor interactions were determined by increases in chemiluminescence generated upon interaction of the Enzyme Acceptor-labeled β-arrestin with the Pro-Link-tagged receptor (see “Experimental Procedures”). A–D, dose-response activation of GPCR CB1 (A) and GPCR CB2 (B–D) with the indicated compounds. RLU, relative luminescence units; 15(S)-HAEA, 15(S)-HETE ethanolamide. E and F, CB2 receptor-ligand interactions were confirmed with the dose response of the CB2-specific inhibitor AM630 co-incubated with GPCR CB2 overexpressed cell activation stimulated with 15-HEDPEA (10 nm) (E) and AEA (10 nm) (F) as positive control. See under “Results” for more details. Results are mean ± S.E. (n = 3–5).
DHEA Products Reduced Platelet-Leukocyte Aggregate Formation in Human Whole Blood
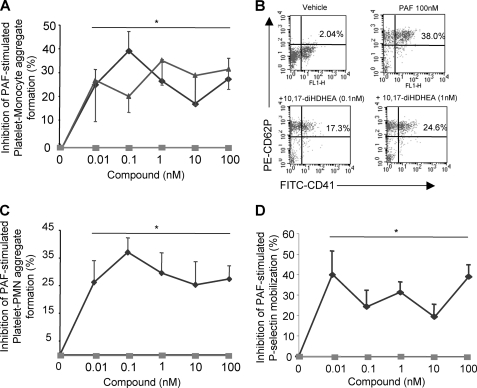
Platelet-leukocyte interactions play important roles in hemostasis, thrombosis, and inflammation (for recent review, see Ref. 28 and references within). At concentrations as low as 10 pm, 10,17-diHDHEA or 15-HEDPEA decreased PAF- (100 nm) stimulated platelet-monocyte aggregate formation in human whole blood by ∼30% (Fig. 5, A and B). The inhibitory action of 10,17-diHDHEA displayed a bell-shaped dose response and reached maximum reduction at ∼40% with 100 pm. Formation of PMN-platelet aggregates with PAF (100 nm) was also inhibited by 10,17-diHDEA at concentrations as low as 10 pm, as was the surface expression of P-selectin on platelets in whole blood (Fig. 5D). By comparison, the precursor DHEA (unoxidized) was not active in this dose range (Fig. 5, A and B).
FIGURE 5.
In human whole blood, 10,17S-diHDHEA and 15-HEDPEA each block PAF-stimulated platelet-leukocyte aggregate formation. Human whole blood stimulated with PAF (100 nm) was incubated with 10,17-diHDHEA, 15-HEDPEA, or DHEA for 30 min at 37 °C. The incubation was stopped via ice-cold RBC lysis buffer. The majority of RBCs were removed, and the remaining cells were labeled. Platelet-leukocyte aggregate formation or P-selectin mobilization was analyzed using FACS (see “Experimental Procedures”). A–D, dose-response inhibition of platelet-monocyte aggregate formation (A), representative dot plot of platelet-monocyte aggregates (B), platelet-PMN aggregate formation (C), and platelet P-selectin mobilization (D), with the indicated products (diamonds, 10,17-diHDHEA; triangles, 15-HEDPEA; squares, DHEA). PE, phycoerythrin. Results are mean ± S.E. of n = 5–6 donors. *, p < 0.05 when compared with vehicle treatment.
Organ Protection in Ischemia/Reperfusion Injury
Because 15-HEDPEA displayed potent bioactions with human PMN at the single-cell level (Fig. 3) and in human whole blood (Fig. 5), we next questioned whether it had protective actions in vivo in murine hind limb ischemia (1 h) and second organ reperfusion (2 h) injury (24). Indeed, following reperfusion, 15-HEDPEA significantly reduced lung PMN accumulation in mice and associated lung injury at 1 μg/mouse (supplemental Fig. 4) (∼50% reduction when compared with vehicle; p < 0.05).
DISCUSSION
Although AEA functions as a cannabinoid receptor agonist and its metabolism is well appreciated (12, 14–16, 27), the roles of DHEA and its metabolome are of interest because DHA treatment reduces traumatic brain injury (2) and is the precursor to potent proresolving mediators, including the resolvins and protectins (1, 7, 8). In the present study, we identified HDHEA in mouse brain, which provided the basis for further investigation of 17-HDHEA and 17-HpDHEA metabolic fates and potential biological impact of DHEA metabolism. Given the lack of functional groups for efficient ionization via electrospray ionization, direct analysis/detection of DHEA or its oxygenated metabolites with LC-MS-MS was impeded with low sensitivity. To this end, their acetate adducts, [M+CH3COOH-H], were targeted for analysis, which proved to be a useful alternative strategy employed in the present investigation. In terms of both detection limits and tandem mass fragmentation patterns, these oxygenated DHEA acetate adducts were comparable with those of the corresponding free acid-derived products.
Because AEA is a reported substrate for murine leukocyte type 12/15-LOX, reticulocyte type 15-LOX, and soybean 15-LOX to generate 15-hydroperoxyarachidonoyl ethanolamide (18), we rationalized 17-HDHEA as the reduced hydroxyl group containing the product of 15-lipoxygenase-like enzyme with DHEA. This hypothesis proved consistent with LC-MS-MS mass analysis of the reduced product obtained from incubation of DHEA with soy bean 15-LOX, which essentially showed the same LC retention time, tandem mass fragmentation patterns as well as online UV spectrum with endogenous 17-HDHEA (Fig. 1).
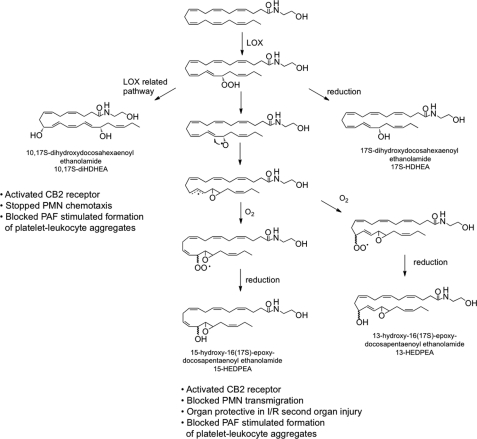
To determine 17-HpDHEA/17-HDHEA metabolic fates, LC-MS-MS-based lipidomic investigation led to identification of a series of novel oxygenated products listed in Table 1. Incubation of either 17-HpDHEA or DHEA with human PMN or mouse brain also gave a novel series of dioxygenated products, such as 4,17-diHDHEA, 7,17-diHDHEA, 10,17-diHDHEA, as well as 15-HEDPEA (Table 1). From DHA, some of these products are biosynthesized in inflammatory exudates, namely resolvin D5 (7,17-dihydroxydocosahexaenoic acid; 7,17-diHDHA) and resolvin D6 (4,17-dihydroxydocosahexaenoic acid; 4,17-diHDHA) (8), as well as the double dioxygenation product 10,17-dihydroxydocosahexaenoic acid (10,17-diHDHA), an isomer of neuroprotectin D1 (6). Hence, their ethanolamide counterparts were identified in the present study. In addition, incubation of 17-HpDHEA with hemoglobin generated two major hepoxilin-like structures (29), 13-HEDPEA and 15-HEDPEA. Given that hepoxilin diastereomer mixtures are generated from hemoglobin or hemin (29), it was of interest whether this was the case for 17-HpDHEA-derived compounds. To this end, NMR chemical shift of H-18 of isolated 13-HEDPEA displayed two distinguishable peaks at 4.23 and 4.45 ppm, and chemical shift of H-13 of isolated 13-HEDPEA showed broad peaks of ∼3.9 ppm (supplemental Table 2, a and b), which strongly suggested the presence of diastereomers. To determine the biosynthetic mechanism of formation of 13-HEDPEA and 15-HEDPEA from hemoglobin and 17-HpDHEA, incubations were also carried out in 18O water. Tandem mass analysis of these incubation products indicated that 18O was not incorporated within these products (data not shown), which suggested that the oxygen source of hydroxyl group could be attributed to atmospheric O2.
Combining results from our lipidomic analyses and the mechanisms proposed for phytooxylipin and hepoxilin biosynthesis (30), the pathways for novel oxygenated DHEA products are proposed in Fig. 6. In this scheme, DHEA is first converted to 17-HpDHEA mediated by 15-LOX. Then 17-HpDHEA is partially reduced to the oxide radical (Fig. 6) by hemoglobin, which reacts with the vicinal double bond at the 16-position to yield the 16(17)-epoxide radical. Non- or low stereospecific addition of oxygen to the intermediate leads to formation of two types of peroxide radical diastereomers. Further reduction can generate 13-HEDPEA and 15-HEDPEA. Alternatively, 17-HpDHEA can undergo reduction to yield 17-HDHEA. Of interest, hemoglobin interactions with 17-HpDHEA yield approximately equal amounts of 13-HEDPEA and 15-HEDPEA; for comparison, incubation of PMN with DHEA or HpDHEA generated predominantly 15-HEDPEA (Table 1 and supplemental Table 1), suggesting the presence of a distinct 15-HEDPEA synthase in human PMN.
FIGURE 6.
Proposed DHEA metabolome and bioactive products. See “Discussion” for details. I/R, ischemia/reperfusion.
In view of the requirement for methodology development for functional screening to keep up with the rapid expansion of modern metabolomics, microfluidic chambers were coupled in tandem for screening the chemotactic activity of human PMN with the novel DHEA-derived products. Given the ∼1-μl volume of the assay chamber, only small amounts of materials were required for these analyses. Results from the screening reported in Fig. 3 indicated that 15-HEDPEA (10 nm) effectively stopped PMN chemotaxis stimulated with IL-8 gradient. Microfluidic chamber-based screening of human PMN chemotaxis offers several advantages that include: (a) the small amounts needed in the ∼1-μl3 chamber, (b) capture of human leukocytes in less than 5 min when compared with several hours (2–3) of isolation using density gradient, and (c) video documentation of single PMN responses (9). Hence, the present results further demonstrate microfluidic chamber-based functional screening as an effective novel approach to decode rare and transient functional metabolites.
AEA exerts a wide range of functions via binding to CB receptors (14–17). However, its DHA metabolite DHEA displays only moderate affinity to CB1 receptor (Ki value of 324 nm versus 40 nm for AEA) (31). To investigate the biological implications of DHEA metabolic oxidation in terms of activating CB receptors, we assessed two of the major PMN products, 10,17-diHDHEA and 15-HEDPEA, using CB2-β-arrestin ligand systems. The EC50 for the novel DHEA-derived products, 10,17-diHDHEA and 15-HEDPEA, were 3.9 × 10−10 and 1.0 × 10−10 m, respectively, similar to that of AEA (Fig. 4). For comparison, the EC50 for DHEA was 9.8 × 10−9 m, ∼2 orders of magnitude higher. 10,17-diHDHEA and 15-HEDPEA also activated CB1, as shown in Fig. 4A. Ligand-CB2 interactions were confirmed using the specific CB2 antagonist AM630 (Fig. 4, E and F). Additional molecular targets of AEA are the vanilloid receptors (TRPV1) in addition to the cannabinoid receptors, which required micromolar range for activity (32). Our results indicated that metabolic oxygenation of DHEA produces novel CB agonists with enhanced potencies that are in the nanomolar range.
Because the production of certain N-acyl ethanolamide is enhanced during stroke (33), it was of interest to investigate biological functions of DHEA and its metabolites in platelet-leukocyte aggregate formation in human whole blood. Platelet-leukocyte aggregate formation is a component of many vascular diseases, stroke, diabetes, and hypertension (28). Specifically, increased platelet-leukocyte aggregates were suggested as an early marker for acute myocardial infarction and are increasingly regarded as a cardiovascular risk factor (34). Also, patients with elevated circulating platelet-monocyte aggregates may reflect a pro-atherogenic phenotype (35). The presence of platelet-leukocyte aggregates stimulates production of pro-inflammatory cytokines, such as IL-1β, IL-8, MCP-1, MIP-1b, PAF, and matrix metalloproteinase, as well as procoagulant tissue factors (for recent review, see Ref. 28). For these reasons, the formation of platelet-leukocyte aggregates is targeted for therapeutic intervention (for reviews, see Refs. 28 and 36). Our lipidomics investigation indicated that 10,17-diHDHEA and 15-HEDPEA were two major DHEA-derived products produced by isolated human PMN. Thus the actions of these compounds were assessed in PAF-stimulated platelet-leukocyte aggregate formation. Both 10,17-diHDHEA and 15-HEDPEA were potent signals and, at concentrations as low as 10 pm, each decreased 100 nm PAF-stimulated platelet-monocyte aggregate formation ∼30% in human whole blood (Fig. 5A). The 10,17-diHDHEA also decreased PAF-stimulated platelet-PMN aggregates by 25–35% (Fig. 5B). For comparison, the precursor DHEA did not significantly inhibit formation of platelet-leukocyte aggregates within this dose range (Fig. 5, A and B).
Formation of platelet-leukocyte aggregates depends mostly on the activation of platelets (37). Along these lines, 10,17-diHDHEA (10 pm to 100 nm) blocked P-selectin surface expression of PAF-stimulated platelets (Fig. 5C), suggesting that 10,17-diHDHEA actions were at least partially achieved via reductions in P-selectin mobilization and surface appearance-related platelet activation. Our results demonstrate that DHEA metabolic oxygenation generated potent molecules that reduce platelet activation and platelet-leukocyte aggregate formation in human whole blood.
Ischemia/reperfusion or reflow injury is the major cause of organ injury after myocardial infarction, stroke, surgery, and organ transplantation injury and involves platelet and PMN activation (24). In this setting, neutrophils play critical roles in the initiation of reperfusion or reflow injury and in consequent tissue damage. Hence, the prevention of PMN activation or accumulation in ischemia organ reduces tissue injury after reperfusion (24, 38). The present results obtained from chemotaxis screening might serve as useful benchmarks for searching/selecting potential protective mediators for ischemia/reperfusion injury. In this regard, 15-HEDPEA, which effectively stopped PMN chemotactic migration, was next evaluated in the mouse ischemia/reperfusion second organ injury initiated by hind limb occlusion. Indeed, 15-HEDPEA at 1 μg/mouse was organ-protective, decreasing PMN infiltration in lung by ∼50%. It is noteworthy that aberrant and excessive leukocytic infiltration is also associated with other diseases, including arthritis and psoriasis (39, 40). Of interest, Kim et al. (41) recently reported that DHEA promotes development of hippocampal neurons.
In summation, lipidomic investigation of DHEA functional metabolome uncovered a series of novel oxygenated products that 1) are potent CB2 agonists, 2) regulate single-cell PMN chemotactic responses, 3) modulate platelet-leukocyte interaction in whole blood, and 4) are organ-protective. In view of the role of lipid mediators in inflammation and its resolution as well as hemostasis(7), the present new DHEA metabolome documented herein may serve as a counter-regulatory system in neural tissues and those rich in DHEA as well as in administration of DHA (42) to regulate leukocyte-mediated tissue damage.
Supplementary Material
Acknowledgments
We thank Mary H. Small for skillful manuscript preparation and Timothy F. Porter for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01NS067686 (to C. N. S.), RC2AT005909 (to C. N. S.), and R01DE019938 (to C. N. S. and D. I.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2, a and b, and Figs. 1–4.
LOX abstracts hydrogen and inserts molecular oxygen in a stereoselective reaction with 1,4-cis-pentadiene units present in polyunsaturated fatty acids.
- DHA
- docosahexaenoic acid
- BSTFA
- N,O-bis-(trimethylsilyl)-trifluoroacetamide
- AEA
- anandamide (arachidonoylethanolamide)
- DHEA
- docosahexaenoylethanolamide
- 4,17-diHDHEA
- 4,17-dihydroxydocosa-5,7Z,10Z,13Z,15,19Z-hexaenoylethanolamide
- 7,17-diHDHEA
- 7,17-dihydroxydocosa-4Z,8E,10Z,13Z,15,19Z-hexaenoylethanolamide
- 10,17-diHDHEA
- 10,17-dihydroxydocosa-4Z,7Z,11,13Z,15,19Z-hexaenoylethanolamide
- 17-HDHEA
- 17-hydroxydocosa-4Z,7Z,10Z,13Z,15,19Z-hexaenoylethanolamide
- 13-HEDPEA
- 13-hydroxy-16(17)-epoxydocosa-4Z,7Z,10Z,14,19Z-pentaenoylethanolamide
- 15-HEDPEA
- 15-hydroxy-16(17)-epoxydocosa-4Z,7Z,10Z,13Z,19Z-pentaenoylethanolamide
- 17-HpDHEA
- 17-hydroperoxydocosahexaenoyl ethanolamide
- LOX
- lipoxygenase
- CB
- cannabinoid
- PMN
- polymorphonuclear leukocytes
- GPCR
- G-protein-coupled receptor
- PAF
- platelet-activating factor.
REFERENCES
- 1. Arnason B. G. (ed). (2010) The Brain and Host Defense, Elsevier, San Diego, CA [Google Scholar]
- 2. Bailes J. E., Mills J. D. (2010) J. Neurotrauma 27, 1617–1624 [DOI] [PubMed] [Google Scholar]
- 3. Marcheselli V. L., Hong S., Lukiw W. J., Tian X. H., Gronert K., Musto A., Hardy M., Gimenez J. M., Chiang N., Serhan C. N., Bazan N. G. (2003) J. Biol. Chem. 278, 43807–43817 [DOI] [PubMed] [Google Scholar]
- 4. Yanes O., Clark J., Wong D. M., Patti G. J., Sánchez-Ruiz A., Benton H. P., Trauger S. A., Desponts C., Ding S., Siuzdak G. (2010) Nat. Chem. Biol. 6, 411–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hassan I. R., Gronert K. (2009) J. Immunol. 182, 3223–3232 [DOI] [PubMed] [Google Scholar]
- 6. Bazan N. G., Calandria J. M., Serhan C. N. (2010) J. Lipid Res. 51, 2018–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Serhan C. N., Chiang N., Van Dyke T. E. (2008) Nat. Rev. Immunol. 8, 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Serhan C. N., Hong S., Gronert K., Colgan S. P., Devchand P. R., Mirick G., Moussignac R. L. (2002) J. Exp. Med. 196, 1025–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kasuga K., Yang R., Porter T. F., Agrawal N., Petasis N. A., Irimia D., Toner M., Serhan C. N. (2008) J. Immunol. 181, 8677–8687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu Z. Z., Zhang L., Liu T., Park J. Y., Berta T., Yang R., Serhan C. N., Ji R. R. (2010) Nat. Med. 16, 592–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Spite M., Norling L. V., Summers L., Yang R., Cooper D., Petasis N. A., Flower R. J., Perretti M., Serhan C. N. (2009) Nature 461, 1287–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. De Petrocellis L., Melck D., Bisogno T., Di Marzo V. (2000) Chem. Phys. Lipids 108, 191–209 [DOI] [PubMed] [Google Scholar]
- 13. Berger A., Crozier G., Bisogno T., Cavaliere P., Innis S., Di Marzo V. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6402–6406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Devane W. A., Hanus L., Breuer A., Pertwee R. G., Stevenson L. A., Griffin G., Gibson D., Mandelbaum A., Etinger A., Mechoulam R. (1992) Science 258, 1946–1949 [DOI] [PubMed] [Google Scholar]
- 15. Di Marzo V., Melck D., Bisogno T., De Petrocellis L. (1998) Trends Neurosci. 21, 521–528 [DOI] [PubMed] [Google Scholar]
- 16. Kozak K. R., Marnett L. J. (2002) Prostaglandins Leukot. Essent. Fatty Acids 66, 211–220 [DOI] [PubMed] [Google Scholar]
- 17. Pavlopoulos S., Thakur G. A., Nikas S. P., Makriyannis A. (2006) Curr. Pharm. Des. 12, 1751–1769 [DOI] [PubMed] [Google Scholar]
- 18. Ueda N., Yamamoto K., Yamamoto S., Tokunaga T., Shirakawa E., Shinkai H., Ogawa M., Sato T., Kudo I., Inoue K., Takizawa H., Nagano T., Hirobe M., Matsuki N., Saito H. (1995) Biochim. Biophys. Acta 1254, 127–134 [DOI] [PubMed] [Google Scholar]
- 19. Yang R., Chiang N., Oh S. F., Serhan C. N. (2011) Curr. Protoc. Immunol., Suppl. 94, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Olson K. R., Eglen R. M. (2007) Assay Drug Dev. Technol. 5, 137–144 [DOI] [PubMed] [Google Scholar]
- 21. Krishnamoorthy S., Recchiuti A., Chiang N., Yacoubian S., Lee C. H., Yang R., Petasis N. A., Serhan C. N. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1660–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Irimia D., Liu S. Y., Tharp W. G., Samadani A., Toner M., Poznansky M. C. (2006) Lab Chip 6, 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dona M., Fredman G., Schwab J. M., Chiang N., Arita M., Goodarzi A., Cheng G., von Andrian U. H., Serhan C. N. (2008) Blood 112, 848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Qiu F. H., Wada K., Stahl G. L., Serhan C. N. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4267–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kumar V., Fausto N., Abbas A. (2004) Robbins and Cotran Pathologic Basis of Disease, 7th Ed., Saunders Elsevier, Philadelphia [Google Scholar]
- 26. Tjonahen E., Oh S. F., Siegelman J., Elangovan S., Percarpio K. B., Hong S., Arita M., Serhan C. N. (2006) Chem. Biol. 13, 1193–1202 [DOI] [PubMed] [Google Scholar]
- 27. Felder C. C., Briley E. M., Axelrod J., Simpson J. T., Mackie K., Devane W. A. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 7656–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van Gils J. M., Zwaginga J. J., Hordijk P. L. (2009) J. Leukoc. Biol. 85, 195–204 [DOI] [PubMed] [Google Scholar]
- 29. Pace-Asciak C. R., Reynaud D., Demin P., Nigam S. (1999) Adv. Exp. Med. Biol. 447, 123–132 [PubMed] [Google Scholar]
- 30. Rowley F. A., Kuhn H., Schewe T. (eds). (1998) Eicosanoids and Related Compounds in Plants and Animals, Portland Press, London [Google Scholar]
- 31. Sheskin T., Hanus L., Slager J., Vogel Z., Mechoulam R. (1997) J. Med. Chem. 40, 659–667 [DOI] [PubMed] [Google Scholar]
- 32. Zygmunt P. M., Petersson J., Andersson D. A., Chuang H., Sørgård M., Di Marzo V., Julius D., Högestätt E. D. (1999) Nature 400, 452–457 [DOI] [PubMed] [Google Scholar]
- 33. Franklin A., Parmentier-Batteur S., Walter L., Greenberg D. A., Stella N. (2003) J. Neurosci. 23, 7767–7775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Furman M. I., Barnard M. R., Krueger L. A., Fox M. L., Shilale E. A., Lessard D. M., Marchese P., Frelinger A. L., 3rd, Goldberg R. J., Michelson A. D. (2001) J. Am. Coll. Cardiol. 38, 1002–1006 [DOI] [PubMed] [Google Scholar]
- 35. Sarma J., Laan C. A., Alam S., Jha A., Fox K. A., Dransfield I. (2002) Circulation 105, 2166–2171 [DOI] [PubMed] [Google Scholar]
- 36. Weyrich A. S., Zimmerman G. A. (2004) Trends Immunol. 25, 489–495 [DOI] [PubMed] [Google Scholar]
- 37. Rinder H. M., Bonan J. L., Rinder C. S., Ault K. A., Smith B. R. (1991) Blood 78, 1730–1737 [PubMed] [Google Scholar]
- 38. Kilgore K. S., Todd R. F., Lucchesi B. R. (1999) Reperfusion Injury, Lippincott, Williams and Wilkins, Philadelphia [Google Scholar]
- 39. Preissner W. C., Schröder J. M., Christophers E. (1983) Br. J. Dermatol. 109, 1–8 [DOI] [PubMed] [Google Scholar]
- 40. Nishiura H., Shibuya Y., Matsubara S., Tanase S., Kambara T., Yamamoto T. (1996) J. Biol. Chem. 271, 878–882 [DOI] [PubMed] [Google Scholar]
- 41. Kim H. Y., Moon H. S., Cao D., Lee J., Kevala K., Jun S. B., Lovinger D. M., Akbar M., Huang B. X. (2011) Biochem. J. 435, 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Calder P. C. (2010) Proc. Nutr. Soc. 69, 565–573 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.