Abstract
The interaction between the orphan nuclear receptor FTZ-F1 (Fushi tarazu factor 1) and the segmentation gene protein FTZ is critical for specifying alternate parasegments in the Drosophila embryo. Here, we have determined the structure of the FTZ-F1 ligand-binding domain (LBD)·FTZ peptide complex using x-ray crystallography. Strikingly, the ligand-binding pocket of the FTZ-F1 LBD is completely occupied by helix 6 (H6) of the receptor, whereas the cofactor FTZ binds the co-activator cleft site of the FTZ-F1 LBD. Our findings suggest that H6 is essential for transcriptional activity of FTZ-F1; this is further supported by data from mutagenesis and activity assays. These data suggest that FTZ-F1 might belong to a novel class of ligand-independent nuclear receptors. Our findings are intriguing given that the highly homologous human steroidogenic factor-1 and liver receptor homolog-1 LBDs exhibit sizable ligand-binding pockets occupied by putative ligand molecules.
Keywords: Development, Drosophila, Nuclear Receptors, Protein Structure, X-ray Crystallography, Fushi Tarazu Factor 1
Introduction
FTZ-F1 (Fushi tarazu factor 1)is a member of the orphan nuclear receptor (NR)3 family, NR5A3, which interacts with the homeodomain protein FTZ to define alternate parasegmental regions in the Drosophila embryo (1–4). FTZ-F1 was originally isolated as a putative transcriptional activator of the ftz (fushi tarazu) gene (5). In blastoderm stage embryos, FTZ-F1 is uniformly distributed, whereas FTZ is expressed in seven stripes representing even-numbered parasegments (6). Both proteins are required for the formation of these even-numbered parasegments (7).
FTZ-F1 has two conserved structural domains: a DNA-binding domain consisting of two zinc fingers and a putative ligand-binding domain (LBD) at the C terminus. Sequence alignments with other NRs and secondary structure predictions suggest that the FTZ-F1 LBD, like that of other NR proteins, is composed of 12 helices. Recent reports have also shown that transcriptional regulation by ligand-dependent NRs is achieved by interactions with cognate lipophilic ligands, which convert apo-NRs into the holo-conformation (8–14) and lead to transcriptional activation (15, 16) or repression (17, 18). These studies suggest that ligand binding to NR LBDs induces a conformational change in the LBD such that the AF-2 helix, which is the last α-helix at the C terminus, reorients its position to facilitate the binding of cofactors. However, FTZ-F1 has been considered an orphan NR because no cognate ligand has yet been identified. Therefore, the molecular mechanism governing FTZ-F1 activation and regulation has remained largely unknown.
Among the vertebrate NRs, those most similar to FTZ-F1 are steroidogenic factor-1 (SF-1/NR5A1) and liver receptor homologue-1 (LRH-1/NR5A2), which share ∼40% sequence identity with FTZ-F1 in the LBD. Recently, the structure of the mouse LRH-1 LBD has been solved, revealing that the AF-2 helix forms an active conformation despite the presence of a large and empty ligand-binding pocket (LBP) (19). Unlike mouse LRH-1, human LRH-1 requires the putative ligand phosphatidylinositol for optimum activity (20). Because FTZ-F1 constitutively interacts with its co-activator FTZ, we presume that its regulation mechanism in the absence of exogenous ligand might be similar to that of mouse LRH-1.
Although FTZ contains a highly conserved homeodomain that is required for DNA binding (21), FTZ is capable of performing some of its functions in the absence of its homeodomain through its interaction with FTZ-F1, showing that FTZ and FTZ-F1 cooperate to regulate target gene expression (22). FTZ, like most NR co-activators, interacts with FTZ-F1 through a conserved LXXLL motif (NR box). The LXXLL motif of the co-activator interacts with the NR LBD via LBD helices H3, H4, and H12 (AF-2). The AF-2 helix, which has a tendency to switch its position in a ligand-dependent manner, forms a crucial part of the interaction surface (23, 24). Previous reports have shown that FTZ-F1 contains a highly conserved AF-2 helix that is required for FTZ binding both in vitro and in vivo (22).
To gain functional insight into FTZ-F1·FTZ, we performed structural and biochemical studies on the FTZ-F1·FTZ complex. Here, we present the first atomic resolution structure of the FTZ-F1·FTZ complex derived from x-ray crystallography. Our structure demonstrates the detailed binding mode of the FTZ co-activator and surprisingly reveals that the LBP of the FTZ-F1 LBD is completely occupied by the hydrophobic residues of helix 6 (H6). In combination with results from mutagenesis experiments, our data suggest that helix 6 acts as a pseudoligand and plays a critical role in FTZ-F1 function.
EXPERIMENTAL PROCEDURES
Cloning of GST-FTZ Fusions and FTZ-F1 LBD
FTZ residues 88–119 and 88–136 were generated by PCR, and resulting PCR fragments were subcloned into pGEX4T-1 (GE Healthcare) to generate expression plasmids pGEX-4T-1-FTZ (residues 88–119) and pGEX-4T-1-FTZ (residues 88–136). Plasmids were transformed into Escherichia coli strain BL21 (DE3), grown in LB medium, and induced for 3 h with 1 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C. Cells were pelleted and lysed in Laemmli buffer. Plasmids used to express FTZ and FTZ(1–170) (pFNS) were described previously (22). The N-terminal start site of the LBD was delineated by aligning sequences of FTZ-F1 homologues with Drosophila melaganoster, Bombyx mori, and Tribolium castaneum FTZ-F1 proteins, as well as with mLRH-1, mSF-1, and the closest known Drosophila NR to FTZ-F1, DHR39, using the Clustal X program. The secondary structures of these proteins were predicted by the PHD program. Based on these results, the LBD expression sequence was cloned into the pET15(b) vector (Novagen).
Protein Preparation and Peptide Synthesis
The plasmid encoding the His-tagged Drosophila FTZ-F1 LBD (22) was transformed into E. coli BL21 (DE3), which were then induced with 0.25 mm isopropyl 1-thio-β-d-galactopyranoside and then grown at 25 °C for 6 h. Cell pellets were resuspended in lysis buffer (25 mm sodium phosphate, 300 mm NaCl, and 10 mm dithiothreitol, pH 7.0) and then disrupted by sonication. The soluble supernatant was collected by ultracentrifugation (140,000 × g for 30 min at 4 °C), and His-tagged FTZ-F1 LBD protein was purified on a cobalt TALON column (BD Biosciences). After removal of the His tag using bovine thrombin (Amersham Biosciences), the FTZ-F1 LBD protein was further purified by gel filtration on a Superdex 75 column (Amersham Biosciences) equilibrated with the following buffer: 150 mm NaCl, 25 mm Na2HPO4/NaH2PO4, pH 6.5, 5 mm dithiothreitol. To express SeMet-substituted FTZ-F1 LBD, FTZ-F1 LBD plasmid was transformed into E. coli B834(DE3) cells, and the protein was expressed in minimal media. A 19-residue FTZ peptide (FTZPEP) was synthesized (Anygen Co.), and the synthesized product was shown by NMR spectroscopy to be >98% pure.
Crystallization, Data Collection, and Structure Determination
Crystals of the FTZ-F1 LBD·FTZPEP complex were obtained at 17 °C from a 15 mg/ml solution using the microbatch method with an equal volume of reservoir buffer (1.3 m tri-basic sodium citrate, 0.1 m HEPES, pH 7.5). A crystal appeared after 1 day and then grew continuously to 0.6 × 0.1 × 0.05 mm in 3–5 days. The FTZ-F1 LBD·FTZPEP complex crystal contained one complex per asymmetry unit; the space group was P3(1)21. X-ray diffraction data were collected at the Pohang Accelerator Laboratory (6B1 beamline) at three wavelengths for multiwavelength anomalous dispersion at 100 K using mother liquor supplemented with 10% glycerol for cryoprotection. All diffraction data were processed and scaled using the HKL 2000 program package (25). The FTZ-F1 LBD·FTZPEP complex structure was solved by multiwavelength anomalous dispersion using the SOLVE (26) and RESOLVE programs (27). The initial model was refined using REFMAC5 (28, 29) and finally by CNS (version 1.1) (30) at 2.8 Å. All model building was performed using the program O (31). The quality of the model was analyzed using the program PROCHECK (32). Details of refinement statistics are summarized in Table 1.
TABLE 1.
Data collection and refinement statistics
| Peak | Edge | Remote | |
|---|---|---|---|
| Data collection | |||
| Space group | P3(1)21 | ||
| Resolution | 50-2.8 | 50-2.8 | 50-2.8 |
| Wavelength (Å) | 0.97922 | 0.97939 | 0.97158 |
| Unique reflection | 17,390 | 179,341 | 17,380 |
| Completeness (%) | 100.0 (99.9) | 99.7 (99.3) | 99.8 (99.7) |
| Rsym (%)a | 12.5 (42.3) | 11.3 (41.3) | 10.8 (41.6) |
| Average I/σ(I) | 19.08 (2.5) | 12.70 (1.7) | 14.30 (1.7) |
| Structure refinement | |||
| Resolution (Å) | 30-2.8 | ||
| Rwork (%)b | 21.7 | ||
| Rfree (%)c | 24.5 | ||
| r.m.s.d.d | |||
| Bond (Å) | 0.02 | ||
| Angle | 1.7° | ||
| Ramachandran plot (%)e | |||
| Most favored | 99.6 | ||
| Additional allowed | 0.4 | ||
| Disallowed | 0.0 | ||
a Rsym = Σ|Iobs − Iavg|/Iobs, where Iobs is the observed intensity of individual reflection and Iavg is the average over symmetry equivalents.
b Rcyst = Σ‖Fobs| − |Fcalc‖/Σ|Fobs| × 100 for 95% of recorded data.
c Rfree is the R-factor calculated by using 5% of the reflection data chosen randomly and omitted from the start of refinement.
d r.m.s.d., root mean square deviation.
e Data were calculated with the program PROCHECK.
Surface Plasmon Resonance
Association and dissociation rates of the FTZ-F1 LBD (coupled to a nitrilotriacetic acid sensor chip) and FTZPEP were measured with a BIAcore 3000 instrument (BIAcore Life Sciences, Uppsala, Sweden). The binding assay was performed with a constant flow rate of 10 μl/min at 25 °C with FTZPEP concentrations in the range of 15–200 μm. Surface plasmon resonance buffers and solutions were as follows: eluent buffer (10 mm HEPES, 150 mm NaCl, 50 μm EDTA, 0.005% Tween 20, pH 7.4); μ-dispensor buffer/HBS-EP buffer (10 mm HEPES, 150 mm NaCl, 3 mm EDTA, 0.005% Tween-20, pH 7.4); and regeneration solution (10 mm HEPES, 0.15 m NaCl, 0.35 mm EDTA, pH 8.3).
Isothermal Titration Calorimetry
Titration calorimetry experiments were performed using a VP-ITC titration calorimetric system (Microcal, Inc.). The solution in the calorimetric cell consisted of 0.04 mm FTZ-F1 LBD in 25 mm sodium phosphate (pH 6.5) and was titrated at 25 °C with 0.3 mm FTZPEP, dissolved in the same buffer, in a 250-μl injection syringe. The raw calorimetry data were collected and analyzed by Origin data analysis software (version 7.0; Microcal, Inc.). The binding isotherms were fitted to a one-site binding model, yielding values for the stoichiometry (N) of the interaction, the enthalpy of binding (ΔH), and the dissociation constant (Kd).
Construction of Gal4 DBD-FTZ-F1 LBD Fusion Proteins for Luciferase Assays
The pAc5.1-G4DBD vector, which contains the gene encoding the Gal4 DNA-binding domain followed by a multiple cloning site, was obtained from Prof. J. Choe (Korea Advanced Institute of Science and Technology). The wild-type FTZ-F1 LBD was cloned into the EcoRI/XbaI sites of this vector to create a Gal4 DBD-FTZ-F1 LBD fusion construct. FTZ-F1(Gly-6), with the 12 residues from Leu-901 to Leu-912 replaced by six glycine residues, was synthesized (Genscript Co.) and subcloned into EcoRI/XbaI sites of the same vector. Five mutant constructs, each replacing the five leucine residues in the H6 individually with alanine (L901A, L902A, L904A, L906A, and L912A), were also subcloned into the same EcoRI/XbaI sites of the vector.
S2 Cell Cultures and Luciferase Assays to Evaluate Transcriptional Activity of FTZ-F1 LBD
Drosophila S2 cells were cultured and transfected with DNA using the calcium phosphate precipitation method as described by the manufacturer's instructions (Invitrogen). A total of 5 μg of each DNA construct was used to transfect a 3-ml culture of S2 cells. An equal amount of the appropriate empty vector was transfected for a negative control in each case. Luciferase assays were performed using a kit as suggested by the kit manufacturer (Promega) using a luminometer (Turner Designs). Values were normalized for transfection efficiency by measuring the β-galactosidase activity from a co-transfected pActin-LacZ (Promega). Anti-G4DBD (sc-577) and anti-β-tubulin (sc-20852) antibodies were purchased from Santa Cruz Biotechnology.
RESULTS AND DISCUSSION
Minimal-binding Motif for FTZ and FTZ-F1 Interaction
To delineate the N-terminal start site of the FTZ-F1 LBD, we made secondary structure predictions based on sequence alignments of FTZ-F1 homologues. Leu-791 was chosen as the start site of the FTZ-F1 LBD based on the following results: 1) among the insect FTZ-F1 homologues used in the alignment, the residue immediately N-terminal to Leu-791 is not conserved; 2) the α-helical secondary structure is predicted to begin a few residues C-terminal to Leu-791; and 3) as shown by Krause and colleagues (22), this construct (Leu-791–Gly-1027) is soluble and sufficient for a interaction with FTZ in vitro. Minimal FTZ fragments sufficient for binding to FTZ-F1 were determined using a similar approach. Alignments of FTZ sequences from D. melanogaster, Drosophila hydei, and T. castaneum were submitted for secondary structure prediction (supplemental Fig. S1A). Based on these alignments, GST-FTZ fusion proteins of decreasing size spanning residues 88–119 were generated (supplemental Fig. S1B). FTZ fusion proteins were expressed in bacteria and used in far Western experiments with a radiolabeled FTZ-F1 LBD probe. All FTZ fragments tested bound to the FTZ-F1 LBD (supplemental Fig. S1C, lanes 2–5), whereas GST alone did not (supplemental Fig. S1C, lane 1). The amount of protein loaded is shown in the Coomassie-stained gel (supplemental Fig. S1C, lanes 6–10).
Determining Binding Affinity of FTZ for FTZ-F1 LBD
Surface plasmon resonance and isothermal titration calorimetry experiments were used to determine the binding affinity of the LXXLL motif, corresponding to residues Val-102–Leu-120 of FTZPEP, for the FTZ-F1 LBD. Binding to the FTZ-F1 LBD was observed upon injection of different concentrations of FTZPEP, from which a dissociation constant of 0.99 × 10−6 m was calculated (supplemental Fig. S2A). Isothermal titration calorimetry was also used to confirm FTZPEP binding to the FTZ-F1 LBD (supplemental Fig. S2B). Analysis of the data showed a 1:1 binding stoichiometry with a Kd value of 1.52 × 10−6 m and an apparent ΔH of −7.568 kcal/mol.
Structure of FTZ-F1·FTZPEP Complex
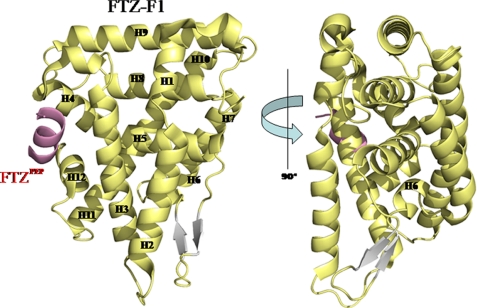
The crystal structure of the FTZ-F1 LBD·FTZPEP complex was solved using multiwavelength anomalous dispersion, and refined at 2.8 Å resolution. Parameters and properties of crystallographic refinements are summarized in Table 1. The overall structure of the FTZ-F1 LBD resembles the canonical fold of other reported NR LBDs in the NR5A family, with four α-helical sandwich layers containing 12 α-helices (Fig. 1). A well defined AF-2 helix (H12), which participates in LXXLL cofactor binding together with H3 and H4, is clearly observed (22). Residues from Ser-108 to Asn-115 in the FTZPEP are observed within the co-activator binding cleft site (Fig. 1). This form is similar to that of other cofactor-bound NRs, suggesting that the FTZ-F1 LBD adopts an active conformation in the presence of cofactor FTZPEP. FTZ-F1 LBD, like other NR5A family members, has an H2 helix that constitutes the fourth helical sandwich layer. It is known that the H2 helix in NR5A family NRs stabilizes the H3 and AF-2 helices for NR activation (19, 33). The structure of FTZ-F1·FTZPEP confirms that all NR5A members have this distinctive and unique H2 α-helix. The most distinctive feature of the FTZ-F1 LBD is an LBP. Surprisingly, the LBP of the FTZ-F1 LBD is occupied by helix 6 (Fig. 1).
FIGURE 1.
Crystal structure of the FTZ-F1 LBD in complex with its cofactor, FTZPEP. The ribbon representations of the FTZ-F1 LBD in two views are displaced by 90°. FTZPEP is shown in pink.
Molecular Interactions between FTZ-F1 LBD and FTZPEP
FTZPEP bound in the co-activator cleft is well stabilized by both electrostatic and hydrophobic interactions with residues in H3, H4, and AF-2 of the FTZ-F1 LBD. An extensive hydrogen-bonding network, referred to as a charge clamp, is observed (Fig. 2A). As with the SF-1 and LRH-1 LBDs, the FTZ-F1 LBD contains well conserved canonical residues required for co-activator binding (19, 20, 34, 35). Like the LRH-1·SF-1 LBD in complex with SHP and NcoA-2 cofactor peptides, FTZPEP also forms an amphipathic α-helix upon binding to the FTZ-F1 LBD. Residues Arg-849, Asp-860, and Glu-1019 of the FTZ-F1 LBD form hydrogen bonds with Leu-109, Arg-110, and Leu-112 of FTZPEP, respectively. Glu-1019 of H12 serves to stabilize FTZ, mainly via a hydrogen bond (Fig. 2A). The unique feature is an additional ionic interaction between Arg-110 of FTZ and Asp-860 in H4 of the FTZ-F1 LBD. This interaction is variable in both SF-1 and LRH-1 due to an amino acid substitution (Fig. 2B). The other stabilizing force comes from hydrophobic interactions between the cluster of side chains of Phe-842, Val-845, Val-859, Met-863, Leu-866, Leu-1016, and Met-1020 of the FTZ-F1 LBD, and hydrophobic residues of FTZPEP. Specifically, Leu-109 of FTZ interacts with residues Met-863 and Met-1020 of the FTZ-F1 LBD, whereas residues Leu-112 and Leu-113 of FTZ are embedded within the hydrophobic grooves formed by residues Val-859, Val-845, and Phe-842 in the FTZ-F1 LBD (Fig. 2A).
FIGURE 2.
The cofactor binding site of the FTZ-F1 LBD. A, left, the ribbon representation shows intermolecular interactions between the FTZ-F1 LBD and FTZPEP. The ribbon depiction of FTZPEP (pink) in the co-activator binding site of the FTZ-F1 LBD (yellow) is presented. Middle, the hydrophobic interaction patch between the FTZ-F1 LBD and FTZPEP is represented. Leucine resides Leu-109, Leu-112, and Leu-113 of FTZPEP are involved in hydrophobic contacts with the hydrophobic cluster formed by helices H3, H4, and H12 of the FTZ-F1 LBD. Right, the surface representation shows intermolecular interactions between the FTZ-F1 LBD and FTZPEP. All residues involved in interactions between the FTZ-F1 LBD and FTZPEP are represented by surface charge distribution. B, comparison of cofactor binding sites in proteins from NR5A family. Common charge clamps of FTZ-F1, LRH-1, and SF-1 LBD are shown, respectively. Residues in NR LBDs and in the cofactors are displayed in black and red, respectively.
Unique LBP of FTZ-F1
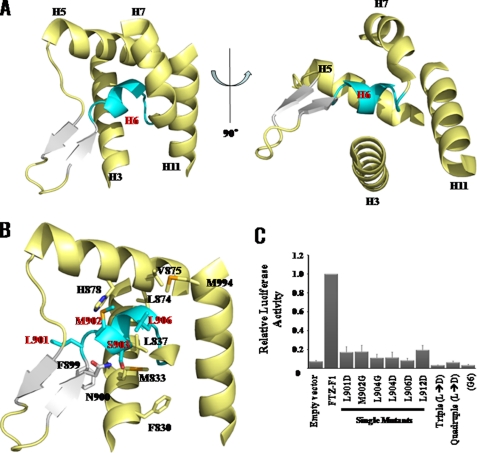
Most of the NR LBD structures characterized to date, including those of SF-1 and LRH-1, exhibit a well defined LBP. In the relatively few NR LBDs that do not contain such a pocket (e.g. the NR4A orphan receptors Nurr1 and DHR38), the ligand pocket is occupied by bulky and hydrophobic side chains of residues in surrounding α-helices (36, 37). The FTZ-F1 LBD structure, however, reveals an unusual LBP in which a single helix H6, comprising residues Leu-901–Leu-912, occupies the LBP, blocking the ligand from entering and occupying it (Fig. 3, A and B). Interactions between H6 and residues within the LBP are mainly stabilized by hydrophobic forces. These residues include Leu-901, Met-902, Leu-904, and Val-906 in the H6 and H9 helices, which form a hydrophobic patch (Fig. 3B). The N terminus of the loop is linked to short anti-parallel β-strands, which strongly interact at their turns with the loop between H2 and H3 through the following hydrogen bonds: Gln-897 NE2–Val-825 carbonyl oxygen, Asn-895 ND2–Val-825 carbonyl oxygen, Asn-895 ND2–Thr-821 carbonyl oxygen, and Asn-895 OD1–Thr-821 carbonyl oxygen. These hydrogen bonds play an important role in stabilizing the turn structure of the short anti-parallel β-strands. The structure of H6 is well supported through an extensive network of hydrogen bonds that includes bonding between the side chain of Ser-903 of H6 and Asn-900 of β2 (Fig. 3B).
FIGURE 3.
The novel LBP of the LBD of FTZ-F1. A, ribbon representations of the LBP of FTZ-F1 in two different views are displayed with H6 in cyan and with β-strands in white. B, detailed intramolecular interactions between residues of LBP (yellow) and H6 (cyan) are shown. C, activity assay of wild-type and mutant proteins of FTZ-F1 found H6 was essential for the transcriptional activity of FTZ-F1 because mutants of H6 significantly reduced transcriptional activity compared with wild type. Triple, L901D, L904D, and L9006D; Quadruple, 901D, L904D, L906D, and L912D; G6, 12 residues from approximately Leu-901–Leu-912 were replaced with six glycines.
Biological Role of Helix 6 of FTZ-F1
The role of H6 in the function of FTZ-F1 was confirmed by assaying transcriptional activity for FTZ-F1 in Drosophila S2 cells (Fig. 3C). Mutations in H6 residues decreased the transcriptional activity of FTZ-F1, and there was a correlation between the reduction in FTZ-F1 activity and position of the mutated residues (Fig. 3C). The results show that residues critical for transcriptional activity are located within the hydrophobic cluster in H6. The most profound effects were observed for the FTZ-F1 LBDL904D and FTZ-F1 LBDL906D mutants, which exhibited approximately a 90% decrease in transcriptional activity, suggesting that the hydrophobic patch formed by Leu-904 and Leu-906 in the LBP is essential for transcriptional activity. Consistent with this, the FTZ-F1 LBDL901D/L904D/L906D triple mutant lost virtually all transcriptional activity, as did the corresponding FTZ-F1(Gly-6) LBD mutant (Fig. 3C). Western blot analyses using an anti-G4DBD antibody showed that mutations in H6 of the FTZ-F1 LBD did not significantly affect protein expression, and CD spectra of mutants show similar folds to those observed in wild type (supplemental Fig. S3), indicating that the observed decrease in transcriptional activity is not due to structural changes in the mutant proteins. Taken together, these findings suggest that interactions between H6 and the residues of the LBP of FTZ-F1 are directly responsible for FTZ-F1 function.
Structural Comparison with LBDs of Other NRs
We performed structural superposition and sequence alignment of the FTZ-F1 LBD with the LRH-1 and SF-1 LBDs (supplemental Fig. S4 and Fig. 4A). The structure of the FTZ-F1 LBD shows two dramatic structural differences from other NRs: the position of the β-sheet and the orientation of the H6 helix (Fig. 4B). The anti-parallel β-sheet of the FTZ-F1 LBD is nearly parallel to H2 and H3, whereas those of the LRH-1 and SF-1 LBDs are oriented perpendicular to H2 and H3. This is due to the unique position of the H2 helix in the FTZ-F1 LBD, which is located in the LBP (Fig. 4B). As a result, the anti-parallel β-sheet is sterically hindered by the H2 helix, resulting in a rotation of the β-strands by 90° compared with those of the SF-1 and LRH-1 LBDs. Residues in the loop between the β-strands interact with residues in the loop between H2 and H3; forming hydrogen bonds between side chains of Asn-895 and Asn-897 with backbone atoms of the residues in the loop between H2 and H3. We propose that the amino acid sequence differences between the FTZ-F1 LBD and those of other NRs cause the conformational change of the β-sheet in FTZ-F1 (supplemental Fig. S4). Because H6 is linked to the β-sheet, this structural change in the β-sheet directly affects the H6 position.
FIGURE 4.
Differences in the position of helix 6 between the FTZ-F1, LRH-1, and SF-1 LBDs. A, differences in the amino acid sequence of H6 between the FTZ-F1, LRH-1, and SF-1 LBDs. B, position of H6 shown as a schematic. FTZ F1, LRH-1, and SF-1 LBD are represented as yellow, orange, and green.
Crystal structures of human SF-1 and LRH-1 LBDs show that the LBP of these NRs are occupied by free phospholipids (20, 33, 35). Interestingly, the H6 helix of the FTZ-F1 LBD is found in the position occupied by the phospholipid in the structure of the LRH-1 complex, suggesting that H6 could serve as a ligand substitute in place of a natural ligand or act as a molecular placeholder in the absence of a natural ligand (Fig. 5A). The hydrophobic leucine residues in H6 of FTZ-F1 LBD interact with hydrophobic residues in the LBP (Fig. 5A). The 2Fo − Fc omit map of H6 at 1.2σ shows the quality of the final electron density in the LBP (Fig. 5B). Furthermore, the relative orientation of H6 is quite different from that of the LRH-1 and SF-1 LBDs as shown in Fig. 5C. Although H6 interacts with most of the pocket-forming residues, it leaves some portion of the LBP unfilled (Fig. 5, A and C). Using the CASTp server (38), we calculated volumes of the LBP of the SF-1 and LRH-1 LBDs. They are 1749.7 Å3 and 2322.7 Å3, respectively, large enough to accommodate phospholipids. However, that of the FTZ-F1 LBD is calculated as a 764.1 Å3, which is a relatively small space to accommodate phospholipid molecules. Helix H6 is oriented away from LBP in the LRH-1 and SF-1 LBDs (Fig. 5C). In contrast, H6 of the FTZ-F1 LBD is located in the LBP (Fig. 5A).
FIGURE 5.
Structural comparison of the LBPs of FTZ-F1 and LRH-1. A, structures of both FTZ-F1 LBD (left) and hLRH-1 LBD/EPH (right) show that H6 (cyan) of FTZ-F1 LBD occupies the LBP similar that of EPH in the hLRH-1 LBD. B, the 2Fo − Fc omit map of the H6 of FTZ-F1 LBD is shown at 1.2σ. C, the LBP of the FTZ-F1 LBD is completely filled by H6 unlike in LRH-1 LBD and SF-1 LBD. Helix 6 is displayed in cyan. Positions of H6 in FTZ-F1 LBD, LRH-1 LBD bound to EPH, and SF-1 LBD bound to phosphotidylethanol are represented with surface figures.
The conformational uniqueness of the FTZ-F1 LBD can be compared with that of the T. castaneum USP (TcUSP) LBD (Fig. 6A) (39). Here, the loop blocks the LBP and functions like H6 of FTZ-F1 (Fig. 6B). However, the FTZ-F1 LBD has 12 α-helices and two β-strands with a canonical nuclear receptor topology unlike TcUSP. Another interesting fact is the position of the AF-2. The position of the AF-2 of the TcUSP LBD is different from that of the AF-2 in FTZ-F1 (Fig. 6, A and B).
FIGURE 6.
Structural comparison of the LBDs of FTZ-F1 and TcUSP. Ribbon representations of the FTZ-F1 (A) and TcUSP (B) LBD structures are displayed. H6, AF-2, and cofactor FTZPEP are shown in green, blue, and pink, respectively.
Implications for Biological Functions
The primary sequences of NR LBDs are relatively well conserved despite their different biological functions. The structures of the FTZ-F1 and hLRH-1 LBDs, like their primary sequences, superimpose very well, with an root mean square deviation value of 0.72 Å (Cα) for residues Leu-791–Arg-1025 of the FTZ-F1 LBD. The close alignment includes the AF-2 helix, which is in the proper orientation for cofactor binding. In both structures, an extended H2 helix stabilizes H3 in the absence of a ligand molecule, which in turn places the AF-2 helix in the active position (19). This orientation in the stabilized, active conformation appears to be a trait specific to the NR5A family. The structure of the FTZ-F1 LBD shows that H6 prevents binding of a ligand molecule in the LBP. These results suggest that the LBD of FTZ-F1 can adopt a fully active form in the absence of a ligand molecule by orienting its AF-2 helix.
The identification of cognate ligands has proven crucial in elucidating NR functions and enabling therapeutic drug development (40–42). However, the recent structural characterization of LBD structures of several orphan NRs, showing LBPs filled with NR side chains (19, 36, 37, 43), has raised the issue of whether these NRs actually have exogenous ligands. The discovery of phospholipid bound forms of the LRH-1 and SF-1 LBDs has suggested that these NRs, at least, may not be orphans and that they might have a ligand-dependent mode of action. On the other hand, mouse SF-1, in which a 2.7 Å salt bridge between Arg-314 (H5) and Glu-238 (H2) is disrupted with no effect on ligand binding, shows sharply diminished receptor activity (20), suggesting that NR5A receptors may exist in an “active conformation” at all times. Our structural and functional analyses of the FTZ-F1 LBD are consistent with this hypothesis. The novel use of an intramolecular helix to fill the LBP (Fig. 5C) together with additional structural modifications, such as the elongated H2, might indicate that the FTZ-F1 can form an active structure with FTZ co-activator in the absence of a ligand molecule, supporting the inclusion of FTZ-F1 in a new class of ligand-independent nuclear receptors.
Supplementary Material
Acknowledgment
We thank professor Kurt Wüthrich for fruitful discussion.
This work was supported by World Class University Program R33-2009-000-10123-0 (to W. L.) and National Creative Research Initiatives Center for Proteome Biophysics Grant 2008-0061984 (to I. C.) from the National Research Foundation/Ministry of Education, Science and Technology (MEST) of Korea. This work was also supported by National Research Foundation (NRF) of Korea through the National Core Research Center Grant R15-2004-024-00000-0 and the Midcareer Researcher Program through an NRF grant funded by the Korean government (MEST) (2009-0073145 and 2009-0084897; to H.-S. C.) and by Basic Science Research Program through the NRF of Korea funded by MEST (to K.-M. C.) (Grant 2011-0005000).
The atomic coordinates and structure factors (code 2XHS) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- NR
- nuclear receptor
- SF-1
- steroidogenic factor-1
- LRH-1
- liver receptor homologue-1
- AF-2
- activation function-2
- FTZPEP
- a 19-residue peptide containing the LXXLL motif that binds to FTZ-F1
- LBP
- ligand-binding pocket
- EPH
- l-α-phosphatidyl-β-oleoyl-γ-palmitoylphosphatidylethanolamine.
REFERENCES
- 1. Lavorgna G., Karim F. D., Thummel C. S., Wu C. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 3004–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ohno C. K., Petkovich M. (1993) Mechanisms of development 40, 13–24 [DOI] [PubMed] [Google Scholar]
- 3. Woodard C. T., Baehrecke E. H., Thummel C. S. (1994) Cell 79, 607–615 [DOI] [PubMed] [Google Scholar]
- 4. Struhl G. (1985) Nature 318, 677–680 [DOI] [PubMed] [Google Scholar]
- 5. Ueda H., Sonoda S., Brown J. L., Scott M. P., Wu C. (1990) Genes Dev. 4, 624–635 [DOI] [PubMed] [Google Scholar]
- 6. Carroll S. B., Scott M. P. (1985) Cell 43, 47–57 [DOI] [PubMed] [Google Scholar]
- 7. Wakimoto B. T., Turner F. R., Kaufman T. C. (1984) Dev. Biol. 102, 147–172 [DOI] [PubMed] [Google Scholar]
- 8. Beato M. (1985) Haematol. Blood Transfus. 29, 217–223 [DOI] [PubMed] [Google Scholar]
- 9. Blumberg B., Evans R. M. (1998) Genes Dev. 12, 3149–3155 [DOI] [PubMed] [Google Scholar]
- 10. Giguère V. (1999) Endocr. Rev. 20, 689–725 [DOI] [PubMed] [Google Scholar]
- 11. Kastner P., Mark M., Chambon P. (1995) Cell 83, 859–869 [DOI] [PubMed] [Google Scholar]
- 12. Mangelsdorf D. J., Evans R. M. (1995) Cell 83, 841–850 [DOI] [PubMed] [Google Scholar]
- 13. Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. (1995) Cell 83, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thummel C. S. (1995) Cell 83, 871–877 [DOI] [PubMed] [Google Scholar]
- 15. Bourguet W., Ruff M., Chambon P., Gronemeyer H., Moras D. (1995) Nature 375, 377–382 [DOI] [PubMed] [Google Scholar]
- 16. Renaud J. P., Rochel N., Ruff M., Vivat V., Chambon P., Gronemeyer H., Moras D. (1995) Nature 378, 681–689 [DOI] [PubMed] [Google Scholar]
- 17. Brzozowski A. M., Pike A. C., Dauter Z., Hubbard R. E., Bonn T., Engström O., Ohman L., Greene G. L., Gustafsson J. A., Carlquist M. (1997) Nature 389, 753–758 [DOI] [PubMed] [Google Scholar]
- 18. Shiau A. K., Barstad D., Loria P. M., Cheng L., Kushner P. J., Agard D. A., Greene G. L. (1998) Cell 95, 927–937 [DOI] [PubMed] [Google Scholar]
- 19. Sablin E. P., Krylova I. N., Fletterick R. J., Ingraham H. A. (2003) Mol. Cell 11, 1575–1585 [DOI] [PubMed] [Google Scholar]
- 20. Krylova I. N., Sablin E. P., Moore J., Xu R. X., Waitt G. M., MacKay J. A., Juzumiene D., Bynum J. M., Madauss K., Montana V., Lebedeva L., Suzawa M., Williams J. D., Williams S. P., Guy R. K., Thornton J. W., Fletterick R. J., Willson T. M., Ingraham H. A. (2005) Cell 120, 343–355 [DOI] [PubMed] [Google Scholar]
- 21. Desplan C., Theis J., O'Farrell P. H. (1988) Cell 54, 1081–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schwartz C. J., Sampson H. M., Hlousek D., Percival-Smith A., Copeland J. W., Simmonds A. J., Krause H. M. (2001) EMBO J. 20, 510–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feng W., Ribeiro R. C., Wagner R. L., Nguyen H., Apriletti J. W., Fletterick R. J., Baxter J. D., Kushner P. J., West B. L. (1998) Science 280, 1747–1749 [DOI] [PubMed] [Google Scholar]
- 24. Mak H. Y., Hoare S., Henttu P. M., Parker M. G. (1999) Mol. Cell. Biol. 19, 3895–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Otwinowski Z., Minor O.(1997) Methods in Enzymology 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26. Terwilliger T. C., Berendzen J. (1999) Acta Crystallogr. 55, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Terwilliger T. C. (2000) Acta Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29. Murshudov G. N., Vagin A. A., Lebedev A., Wilson K. S., Dodson E. J. (1999) Acta Crystallogr. 55, 247–255 [DOI] [PubMed] [Google Scholar]
- 30. Brunger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 31. Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A. 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 32. Laskowski R. A., Rullmann J. A., MacArthur M. W., Kaptain R., Thornton J. M. (1996) J. Biomol. NMR 4, 477–486 [DOI] [PubMed] [Google Scholar]
- 33. Li Y., Choi M., Cavey G., Daugherty J., Suino K., Kovach A., Bingham N. C., Kliewer S. A., Xu H. E. (2005) Mol. Cell 17, 491–502 [DOI] [PubMed] [Google Scholar]
- 34. Ortlund E. A., Lee Y., Solomon I. H., Hager J. M., Safi R., Choi Y., Guan Z., Tripathy A., Raetz C. R., McDonnell D. P., Moore D. D., Redinbo M. R. (2005) Nat. Struct. Mol. Biol. 12, 357–363 [DOI] [PubMed] [Google Scholar]
- 35. Wang W., Zhang C., Marimuthu A., Krupka H. I., Tabrizizad M., Shelloe R., Mehra U., Eng K., Nguyen H., Settachatgul C., Powell B., Milburn M. V., West B. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 7505–7510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baker K. D., Shewchuk L. M., Kozlova T., Makishima M., Hassell A., Wisely B., Caravella J. A., Lambert M. H., Reinking J. L., Krause H., Thummel C. S., Willson T. M., Mangelsdorf D. J. (2003) Cell 113, 731–742 [DOI] [PubMed] [Google Scholar]
- 37. Wang Z., Benoit G., Liu J., Prasad S., Aarnisalo P., Liu X., Xu H., Walker N. P., Perlmann T. (2003) Nature 423, 555–560 [DOI] [PubMed] [Google Scholar]
- 38. Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J. (2006) Nucleic Acids Res. 34, W116–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Iwema T., Billas I. M., Beck Y., Bonneton F., Nierengarten H., Chaumot A., Richards G., Laudet V., Moras D. (2007) EMBO J. 26, 3770–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chiang J. Y. (2005) Curr. Opin Investig. Drugs 6, 994–1001 [PubMed] [Google Scholar]
- 41. Pearce K. H., Iannone M. A., Simmons C. A., Gray J. G. (2004) Drug Discov. Today 9, 741–751 [DOI] [PubMed] [Google Scholar]
- 42. Kallenberger B. C., Love J. D., Chatterjee V. K., Schwabe J. W. (2003) Nat. Struct. Biol. 10, 136–140 [DOI] [PubMed] [Google Scholar]
- 43. Woo E. J., Jeong D. G., Lim M. Y., Jun Kim S., Kim K. J., Yoon S. M., Park B. C., Ryu S. E. (2007) J. Mol. Biol. 373, 735–744 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.