Abstract
STIM1 and Orai represent the key components of Ca2+ release-activated Ca2+ channels. Activation of Orai channels requires coupling of the C terminus of STIM1 to the N and C termini of Orai. Although the latter appears to be central in the interaction with STIM1, the role of the N terminus and particularly of the conserved region close to the first transmembrane sequence is less well understood. Here, we investigated in detail the functional role of this conserved region in Orai3 by stepwise deletions. Molecular determinants were mapped for the two modes of Orai3 activation via STIM1 or 2-aminoethoxydiphenyl borate (2-APB) and for current gating characteristics. Increasing N-terminal truncations revealed a progressive decrease of the specific fast inactivation of Orai3 concomitant with diminished binding to calmodulin. STIM1-dependent activation of Orai3 was maintained as long as the second half of this conserved N-terminal domain was present. Further truncations abolished it, whereas Orai3 stimulation via 2-APB was partially retained. In aggregate, the N-terminal conserved region plays a multifaceted role in Orai3 current gating with distinct structural requirements for STIM1- and 2-APB-stimulated activation.
Keywords: Calcium, Calcium-binding Proteins, Calcium Channels, Ion Channels, Membrane Proteins, Signal Transduction, Orai3, STIM1, Activation, Fast Inactivation
Introduction
Ca2+ is a multifunctional messenger that regulates a variety of processes like muscle contraction, gene expression, proliferation, cell growth, and cell death. A major route for Ca2+ entry across the cell membrane is represented by the Ca2+ release-activated Ca2+ (CRAC)5 channel (1–4). STIM1 and Orai proteins have been identified as the key components of the CRAC signaling machinery. STIM1, which is located in the endoplasmic reticulum membrane, acts as a Ca2+ sensor and as a transducer that triggers CRAC channel activation (5–7). Upon depletion of endoplasmic reticulum Ca2+ stores, STIM1 multimerizes and redistributes into punctae in close proximity to the plasma membrane (6, 8, 9). Here it co-localizes with the CRAC channel component Orai, culminating in channel activation followed by Ca2+ influx (7, 8, 10).
The Orai family comprises three members (Orai1–3), all of which form highly Ca2+-selective channels upon coexpression with STIM1; however, they differ in fast Ca2+-dependent inactivation and in their responses to 2-APB (3, 11–14). Fast inactivation of CRAC channels is mediated by the cooperative interplay of several structures within Orai proteins, the CRAC modulatory domain of STIM1, and via calmodulin (CaM) binding to the Orai N terminus (3, 11, 14–19). Low concentrations of 2-APB (<10 μm) enhance store-operated Orai1 currents (20) together with an increased open probability (18). High 2-APB concentrations (≥50 μm) completely or partially inhibit STIM1-mediated Orai1 or Orai2 currents, respectively (12, 21), whereas Orai3 is robustly stimulated by 2-APB independently of STIM1 and Ca2+ store depletion (12, 21–23). In addition, 2-APB alters the ion selectivity of Orai3 channels by increasing their pore size from ∼3.8 Å to more than 5.34 Å (22). Lee et al. (11) have additionally demonstrated that an Orai3 chimera containing an Orai1 C terminus could be similarly activated by 2-APB like wild-type Orai3.
Store-dependent coupling of STIM1 and Orai is predominantly mediated by interaction of their C termini (10, 24) and involves a conformational transition within the STIM1 C terminus (25, 26). Although the role of the Orai C terminus in this coupling process has been extensively studied, much less is known about the function of the Orai N terminus. So far it has been hypothesized that a fully conserved N-terminal Orai region (e.g. aa 73–90 in Orai1 and aa 48–65 in Orai3) is essential for STIM1-dependent stimulation (19, 27–31). It has been reported recently that a single lysine within this conserved region (K85E in Orai1 and K60E in Orai3) represents a critipcal residue for store-operated activation (31). This conserved region includes a binding site for CAD, a small Orai-activating STIM1 C-terminal fragment that interacts with higher affinity also with the C terminus of Orai1 (30). Furthermore, an increased hydrophobicity at the N terminus/membrane interface as present in the SCID mutant Orai1 R91W impairs gating (19). Fast inactivation of Orai1 requires the interaction of CaM with the conserved N-terminal region (16). Hence, multiple residues in the N terminus of Orai1 are involved in the gating of the channel.
Here, we examined a series of N-terminal deletion mutants to pinpoint those residues within the N-terminal conserved region of Orai3 that are essential for the gating by STIM1 and 2-APB. Electrophysiological analysis of the Orai3 mutants revealed that increasing N-terminal truncations progressively interfered with fast inactivation, then with STIM1-mediated activation, and finally with 2-APB stimulation.
MATERIALS AND METHODS
Molecular Biology
Human STIM1 (hereafter referred to as STIM1; GenBankTM accession number NM_003156) N-terminally ECFP- and EYFP-tagged was kindly provided by T. Meyer's laboratory, Stanford University. STIM1 C terminus (aa 233–685) was cloned into the T/A site of pcDNA3.1V5 His TOPO (Invitrogen) by PCR and subcloned into pECFP-C1 and pEYFP-C1 (Clontech) via the restriction sites KpnI and XbaI. By introducing a stop codon at amino acid position 475, pECFP-C1 STIM1 233–474 was constructed.
Human Orai3 (Orai3; GenBank accession number NM_152288) was kindly provided by L. Birnbaumer (NIEHS, National Institutes of Health, Research Triangle Park, NC). Orai3 was cloned into the pEYFP-C1 (Clontech) expression vector via its BamHI and XbaI restriction sites also for N-terminal labeling. All Orai3 N-terminal deletion mutants (Orai3 ΔN1–46, ΔN1–47, ΔN1–51, ΔN1–53, ΔN1–55, ΔN1–57, ΔN1–58, ΔN1–60, ΔN1–62, and ΔN1–63) were cloned via PCR into the T/A site of pcDNA3.1V5 His TOPO vector (Invitrogen) and recloned into pEYFP-C1 (Clontech) expression vector via its BamHI and XbaI restriction sites. For the generation of pEYFP-C1 Orai3 R52A/R53A and pEYFP-C1 Orai3 ΔN1–51 R52A/R53A mutants, pEYFP-C1 Orai3 and pEYFP-C1 Orai3 ΔN1–51 were used as templates, respectively. Suitable primers exchanged the corresponding codons from CGC (Arg) to GCC (Ala). YFP-C1 Orai3 served as a template for the generation of pEYFP-C1 Orai3 ΔR52 ΔR53 and pEYFP-C1 Orai3 R47Q. For the generation of the latter, primers were used that exchanged the corresponding codon from CGG (Arg) to CAG (Gln). In the case of pEYFP-C1 Orai3 ΔR52 ΔR53, primers were designed to delete the corresponding codons CGC (Arg) and CGC (Arg). pEYFP-C1 Orai3 ΔN1–55 and pEYFP-C1 Orai3 ΔN1–57 served as templates for the generation of pEYFP-C1 Orai3 ΔN1–55 L285S/L292S and pEYFP-C1 Orai3 ΔN1–57 L285S/L292S mutants, respectively. Suitable primers exchanged the corresponding codons from CTG (Cys) to TCG (Ser). Orai3 ΔN1–51 and Orai3 ΔN1–53 chimeras both containing the C terminus of Orai1 were cloned via splicing by overlap extension into pcDNA3.1V5 His TOPO vector (Invitrogen) and via the internal restriction sites BamHI and XbaI into the pEYFP-C1 (Clontech) expression vector for N-terminal fluorescence labeling.
C-terminally enhanced GFP-labeled human CRACR2A and mouse CRACR2B were kindly provided by the lab of Yousang Gwack, University of California. cDNA was subcloned into pEYFP/pECFP-C1 vectors (Clontech) via their internal restriction sites BglII and EcoRI. All clones were confirmed by sequence analysis.
Peptide-CaM Binding PAGE Shift Assay
Calmodulin from bovine testes (300 pmol; Sigma) was incubated for 30 min at 21 °C with a variable molar excess of peptides aa 47–65, aa 52–6, or aa 52–65 R52A/R53A (Peptide 2.0 Inc.; derivatized on the additional C-terminal cysteine with biotin-PEG6-maleimide to prevent disulfide formation; see BIAcore experiments) in incubation buffer (150 mm NaCl, 5 mm HEPES, pH 7.3) together with either 2 mm EGTA or 2 mm Ca2+. Electrophoresis was carried out in 12% polyacrylamide gels in the same buffers under non-denaturing conditions. The gels were stained with Coomassie Blue. Peptides display a highly positive charge at neutral pH (pI between 9.2 and 10.5) and thus have the tendency to migrate to the cathode, which is located on the top of the gel. In the case of an interaction of CaM with a positively charged peptide, this complex would have a larger size and a lower net negative charge than free CaM, resulting in slower migration toward the anode on the bottom of the gel.
Electrophysiology
Electrophysiological recordings comparing characteristics of two to three constructs were carried out in a paired comparison on the same day. Expression patterns and levels of the various constructs were carefully monitored by confocal fluorescence microscopy and were not significantly changed by the introduced mutations. Electrophysiological experiments were performed at 20–24 °C using the patch clamp technique in the whole cell recording configuration. For STIM1/Orai as well as STIM1 C terminus/Orai current measurements, voltage ramps were usually applied every 5 s from a holding potential of 0 mV, covering a range of −90 to +90 mV over 1 s. The internal pipette solution for passive store depletion contained 3.5 mm MgCl2, 145 mm cesium methanesulfonate, 8 mm NaCl, 10 mm HEPES, 20 mm EGTA, pH 7.2. The 100 nm Ca2+ containing intracellular solution included 3.5 mm MgCl2, 145 mm cesium methanesulfonate, 8 mm NaCl, 10 mm HEPES, 10 mm EGTA, 4.3 mm CaCl2, pH 7.2. Extracellular solution consisted of 145 mm NaCl, 5 mm CsCl, 1 mm MgCl2, 10 mm HEPES, 10 mm glucose, 10 mm CaCl2, pH 7.4.
Confocal Fluorescence Microscopy
Confocal microscopy for co-localization experiments was performed similarly as described (32). In brief, a QLC100 Real-Time Confocal System (VisiTech International) connected to two Photometrics CoolSNAPHQ monochrome cameras (Roper Scientific) and a dual port adapter (dichroic, 505lp; cyan emission filter, 485/30 nm; yellow emission filter, 535/50 nm; Chroma Technology Corp.) was used for recording fluorescence images. This system was attached to an Axiovert 200M microscope (Zeiss) in conjunction with an argon ion multiwavelength (457, 488, and 514 nm) laser (Spectra Physics). The wavelengths were selected by an Acousto Optical Tuneable Filter (VisiTech International). MetaMorph 5.0 software (Universal Imaging Corp.) was used to acquire images and to control the confocal system. Illumination times for CFP and YFP images that were recorded with a minimum delay consecutively were about 900 ms.
BIAcore
Specific binding of calmodulin to N-terminal Orai peptides was measured by surface plasmon resonance in a BIAcore X setup at 25 °C. The Orai peptides were custom synthesized (Peptide 2.0 Inc.) with an extra cysteine on their C terminus (as amide with ammonia). The C-terminal cysteine was coupled to maleimide-PEG8-biotin6 using a slight excess of tris(2-carboxyethyl)phosphine for in situ reduction of potentially oxidized peptides as well as 1 mm EDTA and an argon atmosphere to prevent reoxidation. The biotin-PEG-peptide conjugates were purified by reversed phase chromatography, and the structures were confirmed by MALDI-TOF mass spectrometry. The biotin-PEG-peptide conjugates were bound to streptavidin-functionalized surface plasmon resonance chips by injection into flow cell 1 but only after the streptavidin molecules in flow cell 2 (control cell) had been blocked with an excess of free biotin (see legends to supplemental Figs. 4 and 5). The chips were superfused with HEPES buffer (150 mm NaCl, 5 mm HEPES, pH 7.3) at a constant rate of 20 μl/min. Binding of CaM was tested by injecting the protein (2 μm) in HEPES buffer containing either 2 mm CaCl2 or 2 mm EGTA and recording the surface plasmon resonance angle in both flow cells.
RESULTS
Deletion of Whole N Terminus of Orai3 Disrupts Its Activation
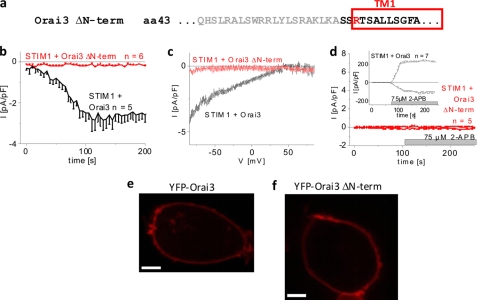
Because Orai3 channels can be stimulated by store depletion as well as 2-APB, we utilized both modes of activation to investigate the role of the N terminus in the Orai3 gating machinery. Initially, we generated an N-terminally YFP-labeled Orai3 deletion mutant lacking the complete N terminus (Orai3 ΔN-term = ΔN1–63) (Fig. 1a). Coexpression of CFP-STIM1 and YFP-Orai3 ΔN-term revealed a lack of current activation following passive store depletion by 20 mm EGTA (Fig. 1, b and c) in close analogy to previous reports with the corresponding Orai1 ΔN1–88 deletion mutant (10, 27). The Orai3-specific 2-APB stimulation was similarly disrupted by the full N-terminal truncation (Fig. 1d). Using confocal fluorescence microscopy, we observed a comparable level of plasma membrane expression for both the wild type (Fig. 1e) and the Orai3 N-terminal deletion mutant (Fig. 1f). Moreover, the deletion mutant showed the same ability to homomultimerize as deduced from FRET measurements of cells expressing YFP- and CFP-labeled Orai3 ΔN-term (data not shown). Hence, the full N terminus of Orai3 or at least a portion of it is indispensable for both STIM1- and 2-APB-mediated Orai3 activation.
FIGURE 1.
Deletion of whole N terminus (aa 1–63) of Orai3 disrupts its activation. a, amino acid sequence of Orai3 ΔN-term deletion mutant. Gray letters represent amino acids within the N terminus of Orai3 that were deleted. b, time course of whole cell inward currents at −74 mV activated by passive store depletion of HEK 293 cells coexpressing CFP-STIM1 and YFP-Orai3 ΔN-term in comparison with wild-type YFP-Orai3 (p < 0.01). c, current/voltage relationship corresponding to b. d, time course of whole cell currents at −74 and +74 mV of HEK 293 cells coexpressing CFP-STIM1 and YFP-Orai3 ΔN-term upon perfusion with 75 μm 2-APB (p < 0.01). Confocal fluorescence images from a representative cell expressing wild-type YFP-Orai3 (e) and YFP-Orai3 ΔN-term (f) are shown. Bars in fluorescence images correspond to 5 μm. pF, picofarad. Error bars represent S.E.
Progressive Truncations until Residues 48–51 Differentially Affect Activation and Inactivation of Orai3 Currents
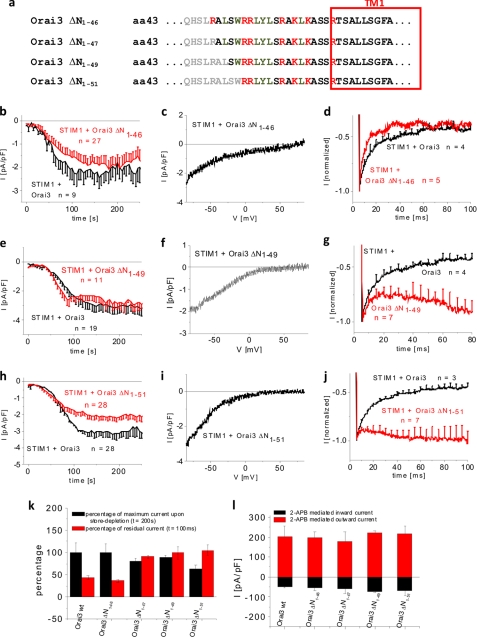
The Orai3 N terminus contains a region (aa 48–65) proximal to the first transmembrane domain that is fully conserved among all Orai proteins (supplemental Fig. 1). An N-terminal deletion until aa 73 in Orai1 (corresponding to aa 48 in Orai3) maintains store-operated activation but to a reduced extent (27). Hence, it has been hypothesized that the fully conserved region is essential for Orai1 activation (27, 29). We examined this hypothesis for Orai3 by generating a series of N-terminal deletion mutants and analyzing their overall gating behavior in terms of activation and inactivation using either STIM1 or 2-APB as the activating stimulus. In a first step, we investigated Orai3 N-terminal deletion mutants lacking 46, 47, 49, and 51 amino acids (Orai3 ΔN1–46, ΔN1–47, ΔN1–49, and ΔN1–51) (Fig. 2a). Although the Orai3 ΔN1–46 and ΔN1–47 mutants still contain the full conserved region (supplemental Fig. 1), the Orai3 ΔN1–49 and ΔN1–51 mutants lack the first two (Ala48-Leu49) and four (Ala48-Leu49-Ser50-Trp51) amino acids of the conserved domain. Orai3 ΔN1–46, ΔN1–47, and ΔN1–49 displayed store-operated activation to an extent comparable with wild type, and the Orai3 ΔN1–51 mutant exhibited slightly reduced currents (Fig. 2, b, e, and k, and supplemental Fig. 3a). All four deletion mutants showed similar current/voltage characteristics (Fig. 2, c, f, and i, and supplemental Fig. 3b). Confocal fluorescence microscopy indicated comparable plasma membrane localization of all four Orai3 deletion mutants (supplemental Fig. 2, a–h). The smaller current size of Orai3 ΔN1–51 was probably not due to reduced plasma membrane expression as 75 μm 2-APB stimulated the four deletion mutants to levels similar to that of wild-type Orai3 (Fig. 2l and supplemental Fig. 3g) consistent with their overall comparable plasma membrane expression. Analysis of the inactivation behavior revealed that the Orai3 ΔN1–46 mutant displayed strong, fast inactivation during a voltage step to −90 mV equivalent to wild-type Orai3 (Fig. 2d and supplemental Fig. 3j), whereas the shorter truncation mutants exhibited currents with substantial attenuation of inactivation (Fig. 2, g, j, and k, and supplemental Fig. 3, c and k). Furthermore, deletion of the arginine Arg47 significantly reduced inactivation, and continuative truncations until Trp51 led to further gradual loss of fast inactivation. A comparison of maximum current densities reached at 200 s and inactivation represented by normalized residual currents at 100 ms (Fig. 2k) showed no general correlation between the current amplitudes and the extent of inactivation among these mutants. In summary, N-terminal aa 47–51 represent key residues for fast inactivation of Orai3 truncation mutants but are not essential for Orai3 activation via STIM1 or 2-APB.
FIGURE 2.
Store-operated activation is retained upon deletion of first four amino acids of N-terminal conserved region of Orai3. a, amino acid sequence of Orai3 N-terminal deletion mutants. Gray letters represent amino acids within the N terminus of Orai3 that were deleted. Shown are times course of whole cell inward currents at −74 mV activated by passive store depletion of HEK 293 cells coexpressing CFP-STIM1 with YFP-Orai3 ΔN1–46 (b), YFP-Orai3 ΔN1–49 (e), and YFP-Orai3 ΔN1–51 (h) in comparison with wild-type YFP-Orai3. c, f, and i, current/voltage relationship corresponding to b, e, and h, respectively. The mean fast inactivation for 100 ms upon voltage steps from a holding potential of 0 to −90 mV of normalized current traces of CFP-STIM1-mediated YFP-Orai3 ΔN1–46 (d), YFP-Orai3 ΔN1–49 (g), and YFP-Orai3 ΔN1–51 (j) currents is shown. k, block diagram opposing the percentage of maximal store-operated currents (t = 200 s) against the percentage of residual currents after 100 ms upon application of a voltage step to −90 mV of Orai3 and Orai3 deletion mutants (for Orai3 ΔN1–46, p > 0.05 for store-operated currents at t = 200 s as well as residual currents of inactivation at t = 100 ms; for Orai3 ΔN1–47, ΔN1–49, and ΔN1–51, p > 0.05 for store-operated currents at t = 200 s but p < 0.05 for residual currents of inactivation at t = 100 ms) mentioned in this figure. l, block diagram of whole cell inward (−74 mV) and outward (+74 mV) currents of HEK 293 cells coexpressing CFP-STIM1 with YFP-Orai3 ΔN1–46, YFP-Orai3 ΔN1–47, YFP-Orai3 ΔN1–49, and YFP-Orai3 ΔN1–51 upon perfusion with 75 μm 2-APB (for inward as well as outward currents at t = 100 s upon application of 2-APB, p > 0.05). pF, picofarad. Error bars represent S.E.
Diminished Inactivation of Orai3 N-terminal Deletion Mutants Correlates with Reduced Calmodulin Binding
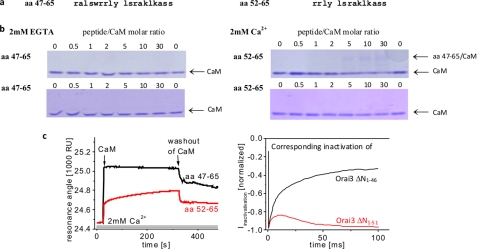
Mullins et al. (16) have reported that disruption of the CaM-binding domain within Orai1 N terminus by single point mutations results in reduced fast inactivation. We have recently demonstrated that CaM dynamically interacts in a Ca2+-dependent manner with the N terminus of Orai1 as well as Orai3 (14). The CaM-binding domain is located in the aa 67–82 stretch in Orai1 that corresponds to aa 43–62 in Orai3 (supplemental Fig. 1). This region overlaps with the N-terminal region (aa 48–65 in Orai3) that is conserved among all Orai isoforms. In an attempt to relate the reduction of inactivation of the Orai3 ΔN1–47 and shorter deletion mutants to a reduced CaM binding, we carried out native PAGE shift assays of CaM in the presence of the respective peptides. The two synthetic peptides, aa 47–65 and aa 52–65 (Fig. 3a), were designed to mimic CaM binding of the corresponding Orai3 ΔN1–46 and Orai3 ΔN1–51 channels that exhibited distinct fast inactivation. PAGE shift assays were carried out under non-denaturing conditions following preincubation of CaM with increasing amounts of peptides either in the absence (2 mm EGTA) or in the presence (2 mm Ca2+) of Ca2+. In the absence of Ca2+, the CaM bands remained comparably intense for both peptides independently of the peptide/CaM ratio as expected for the absence of peptide-CaM interaction (16) (Fig. 3b, left). In the presence of Ca2+, the CaM band displayed a progressive reduction of intensity, starting at a ratio of 5 for aa 47–65/CaM (Fig. 3b, upper right), together with the appearance of a second, weak band that likely corresponds to the peptide-CaM complex. The shorter peptide, aa 52–65, displayed almost no reduction of the CaM band with increasing peptide/CaM ratios (Fig. 3b, lower right). To quantitatively address the interaction of CaM with these two peptides, we additionally carried out BIAcore experiments. For this purpose, the C terminus of the peptides was extended with a hexa(ethylene glycol) linker carrying a terminal biotin group. The biotinylated peptide was immobilized on a streptavidin-functionalized sensor chip in flow cell 1, whereas the streptavidin molecules in the parallel flow cell 2 were blocked with free d-biotin. Both flow cells were perfused in series with buffer containing 2 mm EGTA or 2 mm Ca2+ before injecting 2 μm CaM either in Ca2+-free buffer or in Ca2+ buffer, respectively.
FIGURE 3.
Interaction of Orai3 N terminus with CaM. a, amino acid sequence of two Orai3 N-terminal peptides, aa 47–65 and aa 52–65. b, PAGE shift assay demonstrating Ca2+ dependence of CaM interaction with peptides aa 47–65 and aa 52–65 in 2 mm EGTA and 2 mm Ca2+ media at different peptide/CaM ratios. c, BIAcore sensorgrams displaying binding of CaM to peptide aa 47–65 compared with that of aa 52–65 in the presence of 2 mm Ca2+. On the right side, inactivation profiles of Orai3 ΔN1–46 and Orai3 ΔN1–51 are shown in accordance with the respective sensorgrams. RU, resonance units.
Consistent with Park et al. (30) and results obtained by the PAGE shift assays above, BIAcore experiments revealed no CaM binding for any peptide in the presence of EGTA (data not shown). In a 2 mm Ca2+-containing buffer, however, we observed a stronger as well as faster binding of CaM to the longer peptide (450 resonance units) compared with the shorter peptide (207 resonance units) (Fig. 3c, left). Thus, the distinct Ca2+-dependent CaM binding capabilities of the aa 47–65 and aa 52–65 peptides correlated well with the degree of inactivation of the Orai3 ΔN1–46 and Orai3 ΔN1–51 channels, respectively (Fig. 3c, right). Presumably, the progressive deletion of the Orai3 N terminus from aa 46 to 51 gradually decreases CaM binding, which is reflected in a successive attenuation of Orai3 inactivation.
Arginines at Positions 52/53 Exert Inhibitory Effect on Orai3 Gating
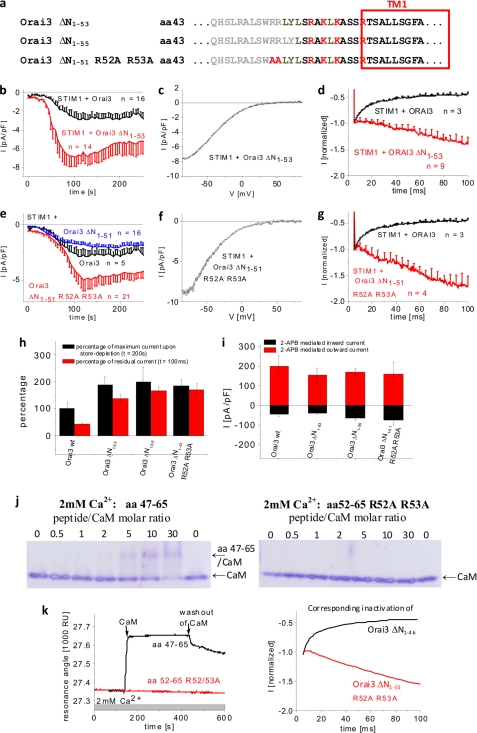
Mapping of the remaining residues in the conserved Orai3 N-terminal region was continued by generating further Orai3 deletion mutants, Orai3 ΔN1–53 and ΔN1–55 (Fig. 4a). The Orai3 ΔN1–53 lacked the two positively charged arginines Arg52/53, and the Orai3 ΔN1–55 additionally lacked the subsequent two hydrophobic amino acids. Both mutants exhibited well preserved store-operated activation with current densities that were more than 2-fold increased compared with that of Orai3 wild-type (Fig. 4, b and h, and supplemental Fig. 3d). Confocal fluorescence microscopy images displayed plasma membrane localization for Orai3 ΔN1–53 and Orai3 ΔN1–55 equivalent to wild-type Orai3 (supplemental Fig. 2). Accordingly, 75 μm 2-APB stimulated Orai3 ΔN1–53 and ΔN1–55 currents to levels similar to that of wild-type Orai3 (Fig. 3i and supplemental Fig. 3h) consistent with their similar levels of plasma membrane expression. Strikingly, their current/voltage (I/V) relationships displayed U-shaped characteristics at negative potentials (Fig. 4c and supplemental Fig. 3e) that were not visible for Orai3 inward currents. Voltage steps for both Orai3 ΔN1–53 and ΔN1–55 channels revealed substantial current potentiation without any Ca2+-dependent inactivation over the first 100 ms (Fig. 4, d and g, and supplemental Fig. 3, f and l). The current potentiation of these mutants likely resulted in the U-shaped I/V relationship and the increased maximum currents, suggesting a profound gating effect.
FIGURE 4.
Two arginines exert inhibitory effect on Orai3 gating. a, amino acid sequence of Orai3 N-terminal deletion mutants. Gray letters represent amino acids within the N terminus of Orai3 that were deleted. Shown are time courses of whole cell inward currents at −74 mV activated by passive store depletion of HEK 293 cells coexpressing CFP-STIM1 with YFP-Orai3 ΔN1–53 (b) and YFP-Orai3 ΔN1–51 R52A/R53A (e) in comparison with wild-type Orai3 (b) and in comparison with both wild-type Orai3 and YFP-Orai3 ΔN1–51 (e). c and f, current/voltage relationships corresponding to b and e, respectively. The mean fast inactivation for 100 ms upon voltage steps from a holding potential of 0 to −90 mV of normalized current traces of CFP-STIM1-mediated YFP-Orai3 ΔN1–53 (d) and YFP-Orai3 ΔN1–51 R52A/R53A (g) currents in comparison with wild-type YFP-Orai3. h, block diagram opposing the percentage of maximal store-operated currents (t = 200 s) against the percentage of residual currents after 100 ms upon application of a voltage step to −90 mV of Orai3 and the Orai3 deletion mutants indicated (p < 0.01 for store-operated currents at t = 200 s as well as residual currents of inactivation at t = 100 ms). i, block diagram of whole cell inward (−74 mV) and outward (+74 mV) currents of HEK 293 cells coexpressing CFP-STIM1 with YFP-Orai3 ΔN1–53, YFP-Orai3 ΔN1–55, and YFP-Orai3 ΔN1–51 R52A/R53A upon perfusion with 75 μm 2-APB (for inward as well as outward currents at t = 100 s upon application of 2-APB, p > 0.05). j, PAGE shift assay demonstrating Ca2+ dependence of CaM interaction with peptides aa 47–65 and aa 52–65 R52A/R53A in 2 mm Ca2+ medium at different peptide/CaM ratios. k, BIAcore sensorgrams displaying binding of CaM to peptide aa 47–65 compared with aa 52–65 R52A/R53A in the presence of 2 mm Ca2+. On the right side, inactivation profiles of Orai3 ΔN1–46 and Orai3 ΔN1–51 R52A/R53A are shown in accordance with the respective sensorgrams. pF, picofarad; RU, resonance units. Error bars represent S.E.
We hypothesized that the two arginines at positions 52 and 53 might affect Orai3 gating. Mutating these two arginines to alanines in the Orai3 ΔN1–51 R52A/R53A mutant indeed increased currents 2–3-fold (Fig. 4, e and h) and yielded a U-shaped I/V relationship (Fig. 4f) similar to that of Orai3 ΔN1–53. 2-APB currents of this double point mutant developed to levels comparable with wild-type Orai3 levels (Fig. 4i) consistent with similar plasma membrane expression (supplemental Fig. 2). In accordance with Orai3 ΔN1–53 and Orai3 ΔN1–55, Orai3 ΔN1–51 R52A/R53A displayed a pronounced potentiation during a voltage step to −90 mV (Fig. 4g and supplemental Fig. 3l). Thus, gating of the Orai3 ΔN1–51 R52A/R53A mutant (Fig. 4g) looked equivalent to that of Orai3 ΔN1–53/55, indicating a modulatory role of the two arginines Arg52-Arg53 in the gating of the Orai3 truncation mutant. The complete absence of fast inactivation in the Orai3 ΔN1–51 R52A/R53A or Orai3 ΔN1–53/55 mutant might be caused by abrogated CaM binding, which is still visible with the Orai3 ΔN1–51-mimicking peptide. Native PAGE shift assays with CaM and the peptide aa 52–65 R52A/R53A, corresponding to Orai3 ΔN1–51 R52A/R53A, did not display a change in CaM bands with increasing peptide/CaM ratio (Fig. 4j). Moreover, BIAcore experiments failed to detect significant binding of CaM to aa 52–65 R52A/R53A (Fig. 4k) contrary to the other two peptides mentioned in the previous section. In summary, ΔN1–53/55 deletions or mutation of Arg52-Arg53 within ΔN1–51 altered Orai3 gating by increasing maximum Orai3 currents and concomitantly reversed inactivation into potentiation. This potentiation was accompanied by a complete disruption of CaM binding.
N-truncated Orai3 Mutants with Potentiated Currents Exhibit Coupling Similar to STIM1
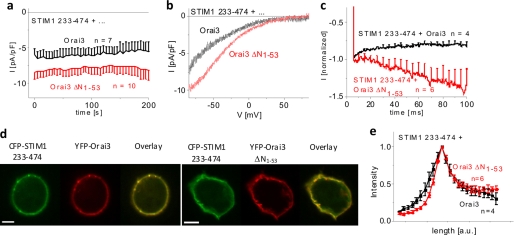
Because STIM1-mediated Orai3 ΔN1–53/55 currents were strongly increased, we examined the impact of the truncated N terminus on the coupling to STIM1. To reveal a potential change in affinity, we utilized the STIM1 233–474 cytosolic fragment as a surrogate for STIM1. Orai3 ΔN1–53/55 currents that were constitutively activated by STIM1 233–474 displayed enhanced current densities and slightly U-shaped I/V relationships (Fig. 5, a and b) as compared with wild-type Orai3. In line with the experiments with full-length STIM1, the slight inactivation of Orai3 currents changed into a strong potentiation with Orai3 ΔN1–53 when stimulated by STIM1 233–474 (Fig. 5c). Hence, the observed effects of STIM1 233–474 fragment were comparable with those obtained with full-length STIM1. The enhanced currents of Orai3 ΔN1–53/55 less likely resulted from increased interaction with STIM1 as co-localization and intensity profiles of STIM1 233–474 with wild-type Orai3 or Orai3 ΔN1–53 were similar (Fig. 5, d and e). Hence, we suggest that the observed biophysical characteristics of Orai3 ΔN1–53/55 result rather from an altered gating than from enhanced STIM1 binding.
FIGURE 5.
N-truncated Orai3 mutants with potentiated currents exhibit coupling similar to STIM1. a, time course of constitutive whole cell inward currents at −74 mV of HEK 293 cells coexpressing CFP-STIM1 233–474 with YFP-Orai3 ΔN1–53 in comparison with wild-type YFP-Orai3 (t = 0 s, p < 0.05). b, current/voltage relationships corresponding to a. c, mean fast inactivation for 100 ms upon voltage steps from a holding potential of 0 to −90 mV of normalized current traces of CFP-STIM1 233–474-mediated Orai3 ΔN1–53 currents compared with Orai3 currents (t = 0 s, p < 0.05). d, localization and overlay of CFP-STIM1 233–474 and YFP-Orai3 or YFP-Orai3 ΔN1–53. e, intensity plots representing the localization of STIM1 233–474 across the cell when coexpressed with Orai3 ΔN1–53 compared with wild-type Orai3. Bars in fluorescence images correspond to 5 μm. pF, picofarad; a.u., arbitrary units. Error bars represent S.E.
Continuative N-terminal Truncations Impair STIM1-mediated Activation of Orai3
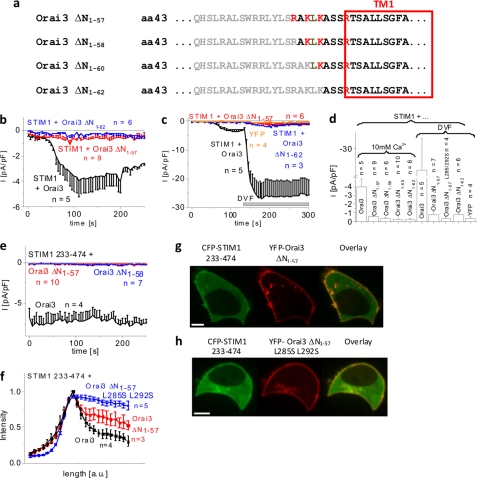
Deletion of nearly half of the N-terminal conserved region (aa 48–55) in Orai3 maintained store-operated activation and additionally affected current density and inactivation. To pinpoint the region within the remaining N terminus of Orai3 essential for STIM1-dependent activation, we carried out further truncations, generating Orai3 ΔN1–57, Orai3 ΔN1–58, Orai3 ΔN1–60, and Orai3 ΔN1–62 deletion mutants (Fig. 6a). All of these mutants still exhibited plasma membrane localization as judged from confocal fluorescence microscopy images (supplemental Fig. 2). Quantitative evaluation of membrane fluorescence intensities exemplarily compared for Orai3 wild type and Orai3 ΔN1–47 and Orai3 ΔN1–57 mutants only revealed a slight reduction by about 30% for Orai3 ΔN1–57 (supplemental Fig. 2i). However, all of the truncation mutants from Orai3 ΔN1–57 to Orai3 ΔN1–62, when coexpressed with STIM1, completely lacked store-operated activation (Fig. 6, b and d, and supplemental Fig. 3i), suggesting that these deletions fully abolished function but not plasma membrane localization. In support of this hypothesis, the application of divalent cation-free solution, which typically amplified wild-type Orai3 currents ∼8-fold, resulted in no significant current increases with Orai3 ΔN1–57 and ΔN1–62 mutants (Fig. 6, c and d). As additional controls, the Orai3 ΔN1–57 L285S/L292S mutant with disrupted STIM1 coupling (24) or YFP-expressing cells displayed similar, marginal increases of the currents in divalent cation-free solution (Fig. 5d). The shorter C-terminal STIM1 fragment 233–474 (Fig. 6e), which typically exhibits enhanced interaction with Orai proteins and efficient stimulation of Orai currents (28, 30, 33), also failed to evoke significant currents from these Orai3 deletion mutants. Therefore, we further examined whether the abolished store-operated currents might result from a lack of coupling of the Orai3 ΔN1–57 and ΔN1–62 mutants with STIM1. Co-localization experiments and density profiles showed a weaker interaction of STIM1 233–474 with Orai3 ΔN1–57 than with wild-type Orai3 that was still stronger than with the STIM1-disrupted coupling mutant Orai3 ΔN1–57 L285S/L292S (Figs. 6, f–h, and 5d) (24). Hence, Orai3 channel activation requires, in addition to STIM1 coupling to its C terminus, the presence of an additional N-terminal region upstream to aa 56 as an essential molecular determinant for STIM1-mediated Orai3 current activation.
FIGURE 6.
N-terminal deletions until position 57 or further significantly diminish store-operated Orai3 activation. a, amino acid sequence of Orai3 N-terminal deletion mutants. Gray letters represent amino acids within the N terminus of Orai3 that were deleted. b, time course of whole cell inward currents at −74 mV activated by passive store depletion of HEK 293 cells coexpressing CFP-STIM1 with YFP-Orai3 ΔN1–57 and YFP-Orai3 ΔN1–62 in comparison with wild-type YFP-Orai3. c, time course of whole cell inward currents at −74 mV upon perfusion with divalent cation-free (DVF) solution of HEK 293 cells expressing YFP or coexpressing CFP-STIM1 with YFP-Orai3, YFP-Orai3 ΔN1–57, or YFP-Orai3 ΔN1–62. d, block diagram comparing maximal currents in 10 mm Ca2+ of HEK cells coexpressing CFP-STIM1 and YFP-Orai3, YFP-Orai3 ΔN1–57, YFP-Orai3 ΔN1–58, YFP-Orai3 ΔN1–60, or YFP-Orai3ΔN1–62 (p < 0.05) as well as in divalent cation-free (DVF) solution of HEK 293 cells expressing YFP or coexpressing CFP-STIM1 and YFP-Orai3, YFP-Orai3 ΔN1–57, YFP-Orai3 ΔN1–57 L285S/L292S, or YFP-Orai3 ΔN1–62 (p < 0.05). e, time course of whole cell inward currents at −74 mV activated by passive store depletion of HEK 293 cells coexpressing CFP-STIM1 233–474 with YFP-Orai3 ΔN1–57 and YFP-Orai3 ΔN1–58 in comparison with wild-type YFP-Orai3 (p < 0.05). f, intensity plots representing the localization of CFP-STIM1 233–474 across the cell when coexpressed with YFP-Orai3 ΔN1–57 L285S/L292S compared with YFP-Orai3 ΔN1–57 and wild-type Orai3. g and h, localization and overlay of CFP-STIM1 233–474 and YFP-Orai3, YFP-Orai3 ΔN1–57, or YFP-Orai3 ΔN1–57 L285S/L292S. Bars in fluorescence images correspond to 5 μm. pF, picofarad. Error bars represent S.E.
Orai3 Mutants Lacking Store-operated Activation Are Still Stimulated by 2-APB
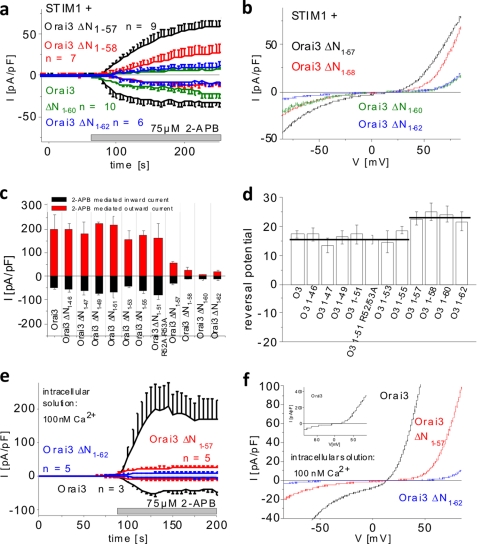
Because truncations beyond residue 57 in the Orai3 N terminus abolished store-operated activation, we examined their impact on stimulation by 2-APB. All four deletion mutants, i.e. Orai3 ΔN1–57, ΔN1–58, ΔN1–60, and ΔN1–62, that lacked store-operated activation still displayed a current response to 2-APB characterized by double rectifying current/voltage relationships (Fig. 7, a and b). The mean current densities, however, exhibited significantly reduced levels compared with wild-type Orai3 with the outward currents being more strongly diminished than the inward currents (Fig. 7, b and c). Close inspection of the I/V relationship revealed a right-shifted reversal potential in comparison with wild-type Orai3 determined at maximum 2-APB currents (Fig. 7d). The deletion mutants were also stimulated by 2-APB without coexpression of STIM1 (data not shown). To eliminate a potential effect of endogenous STIM1 contributing to current stimulation, we additionally performed experiments with a 100 nm Ca2+ intracellular solution to prevent store depletion. Under these conditions, the 2-APB-activated currents of Orai3, Orai3 ΔN1–57, and Orai3 ΔN1–62 developed to an extent similar to that in the presence of passive store depletion (Fig. 7, e and f). Thus, the second half (aa 56–62) of the conserved region within the Orai3 N terminus is sufficient for 2-APB activation. However, additional amino acids before residue 57 are required for maximum stimulation of Orai3 currents via 2-APB.
FIGURE 7.
Amino acids 58–62 of Orai3 N terminus are sufficient for 2-APB activation that is independent of STIM1. a, time course of whole cell currents at −74 and +74 mV of HEK 293 cells coexpressing CFP-STIM1 with YFP-Orai3 ΔN1–57, YFP-Orai3 ΔN1–58, YFP-Orai3 ΔN1–60, or YFP-Orai3 ΔN1–62 upon perfusion with 75 μm 2-APB (p < 0.05). b, current/voltage relationships corresponding to a. Block diagrams comparing inward and outward currents upon 2-APB stimulation (c) and reversal potential of double rectifying 2-APB currents of all Orai3 (O3) deletion mutants (Orai3 ΔN1–46–Orai3 ΔN1–55, p > 0.05; Orai3ΔN1–57–Orai3 ΔN1–62, p < 0.05) (d). e, time course of whole cell currents at −74 and +74 mV of HEK cells coexpressing CFP-STIM1 and YFP-Orai3, YFP-Orai3 ΔN1–57, or YFP-Orai3 ΔN1–62 in the presence of 100 nm Ca2+ in the intracellular solution and upon perfusion with 75 μm 2-APB (2-APB currents of respective Orai3 proteins in the presence of 10 mm EGTA compared with 100 nm Ca2+ intracellular solution, p > 0.05 at t = 100 s upon addition of 2-APB). f, current/voltage relationship corresponding to e. pF, picofarad. Error bars represent S.E.
DISCUSSION
The present study highlights the versatile role of the N-terminal conserved region (aa 48–65) in the gating of Orai3 channels. Distinct molecular determinants were identified for Orai3 activation by STIM1 and 2-APB. Moreover, fast inactivation of Orai3 channels mediated by CaM binding was reversed into strong potentiation upon partial truncation of the conserved N-terminal region.
Utilizing a detailed series of N-terminal truncation mutants of Orai3, we confined the minimal essential domain for STIM1-mediated activation to the second half (aa 56–65) of the N-terminal conserved region of Orai3. Further truncations beyond aa 57 resulted in Orai3 channels that were completely deprived of store-dependent activation. Lis et al. (31) have recently reported that an Orai3 K60E mutant similarly fails to be activated upon store depletion. Nevertheless, both the Orai3 ΔN1–57 and Orai3 K60E (31) mutants were still able to couple to STIM1 233–474 and CAD, respectively, used as surrogates of STIM1. Co-localization and density profiles of Orai3 ΔN1–57 and STIM1 233–474 indicated a reduced overall affinity compared with the fully functional Orai3 ΔN1–53. This weaker binding apparently reflected the contribution of the N terminus for STIM1 binding despite the predominant role of the Orai C terminus in mediating STIM1/Orai coupling (10, 24). This finding is in accordance with the co-immunoprecipitation studies of Lis et al. (31) that showed retained but weaker binding of CAD to an N-terminal peptide mimicking Orai3 K60E versus Orai3. Thus, the N-terminal region downstream of aa 55 and particularly of Lys60 (31) plays a key role in transducing the coupling with STIM1 into Orai3 gating and current activation.
Although N-terminal truncation until aa 57 completely abolished store-operated activation, shorter truncations up to residue 51 maintained STIM1-dependent activation similar to wild-type Orai3. However, truncations within the stretch of residues from 51 to 55 increased current activation and altered gating characteristics of Orai3 channels. This effect less likely resulted from enhanced STIM1 binding to respective truncation mutants (Fig. 5, d and e) but rather resulted from an alteration of Orai3 gating with the change from fast inactivation into a profound potentiation phase. Here, the two arginines within this N-terminal stretch substantially affected Orai3 gating.
The unique fast inactivation of Orai3 required the whole conserved region (aa 48–65) of the Orai3 N terminus together with Arg47. This region functions as a CaM-binding domain in Orai1 fast inactivation (16). In Orai3, CaM plays a similar role in fast inactivation as the reduction of CaM binding with increasing truncations correlated well with diminished fast inactivation (see Fig. 3). Hence, significantly reduced inactivation of Orai3 ΔN1–47/49/51 most likely reflects a concomitant decrease of CaM binding affinity as demonstrated here by in vitro binding assays. Point mutations disrupting CaM binding in Orai1 (16) were analogously introduced in Orai3 (Orai3 A48E and Orai3 W51E). We observed a similar loss of inactivation in accordance with the impaired fast inactivation of ΔN1–47/49/51 (data not shown), further underlining the involvement of CaM in Orai3 fast inactivation. Loss of CaM binding accompanied abrogation of fast inactivation of Orai3 ΔN1–53/55. Here, we identified the two arginines 52/53 in the center of the conserved region as further determinants for CaM binding and inactivation. Their deletion or mutation to alanines completely eliminated both CaM binding and fast inactivation. In addition, the Orai3 ΔN1–53/55 and Orai3 ΔN1–51 R52A/R53A mutants exhibited a pronounced potentiation and an increase in maximum current levels that exhibited U-shaped I/V characteristics. These effects might reflect an additional gating process.
Recently, a CRAC regulator protein (CRACR2A/B) has been identified to facilitate and stabilize STIM1-Orai complex formation (34). Its proposed interaction site on the Orai N terminus overlaps with that of CaM and can be disrupted in Orai1 by a double point mutation, K85A/K87A (34). To determine whether this protein is additionally involved in the regulation of Orai3 via its N terminus, we carried out knockdown as well as overexpression studies. Utilizing siRNA, suppression of endogenous CRACR2A but not of CRACR2B in HEK cells significantly inhibited STIM1/Orai1 current activation (supplemental Fig. 6a). However, neither STIM1/Orai3 currents nor their inactivation was affected by knockdown or overexpression of CRACR2A or CRACR2B (supplemental Fig. 6, b–e). Under the above conditions, Orai3 ΔN1–53 current characteristics were also not altered (supplemental Fig. 6, f–i). Hence, an involvement of CRACR2A/2B in regard to the changes of gating characteristics of wild type and Orai3 truncated forms is very unlikely.
We demonstrated that Orai3 channels required a minimal N-terminal segment starting from aa 56/57 to retain STIM1-dependent activation as well as 2-APB stimulation. 2-APB-stimulated outward currents were activated with some delay compared with inward currents (data not shown) as reported previously (35). Upon further deletion of aa 57, Orai3 lost store-operated current activation. Stimulation by 2-APB was maintained although to a reduced extent. Consistent with Orai3 ΔN1–57, the Orai3 K60E mutant that fails to respond to store depletion similarly exhibits reduced 2-APB-stimulated currents (31). Furthermore, these reduced 2-APB currents displayed no delay in the activation of outward versus inward currents (data not shown). It might be hypothesized that the weaker binding of STIM1 to store-non-responsive N-terminal Orai3 truncation mutants might have allowed activation of inward and outward currents at a similar time course consistent with results of Yamashita et al. (35). As the truncation mutants that lost store-operated current activation concomitantly exhibited diminished 2-APB stimulation, we suggest that activations of Orai3 via STIM1 and 2-APB share some structural determinants. Moreover, the truncations might impose structural effects on the tetrameric channel, affecting distant conformational multidomain sites for 2-APB binding. We additionally observed for these 2-APB-responsive truncation mutants slightly right-shifted reversal potentials as compared with 2-APB-stimulated Orai3 wild-type currents. Thus, it is tempting to speculate that truncation of approximately two-thirds of the conserved N-terminal region might impose an allosteric effect upon the first transmembrane region, thereby altering channel pore conformation of the Orai3 ΔN1–57 (or shorter) mutants. In summary, we have pinpointed several key residues within the N-terminal conserved region of Orai3 that play an essential role in STIM1- as well as 2-APB-stimulated activation and current gating processes of Orai3.
Supplementary Material
Acknowledgments
We thank S. Buchegger for excellent technical assistance and Rainer Schindl for comments on the manuscript.
This work was supported in part by Austrian Science Fund (FWF) Projects P211925 (to K. G.) and P22565 (to C. R.) and Austrian Research Promotion Agency (FFG) Austrian Nanoscience Initiative VO104-08-BI Project 819703 NSI-NABIOS (to H. J. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–6.
P. Pollheimer, A. Scherflér, P. Wiesauer, C. Schwarzinger, and H. J. Gruber, manuscript in preparation.
- CRAC
- Ca2+ release-activated Ca2+
- 2-APB
- 2-aminoethoxydiphenyl borate
- CaM
- calmodulin
- aa
- amino acid(s)
- CAD
- CRAC activation domain
- CFP
- cyan fluorescent protein
- ECFP
- enhanced CFP
- EYFP
- enhanced YFP
- I/V
- current/voltage.
REFERENCES
- 1. Hoth M., Penner R. (1992) Nature 355, 353–356 [DOI] [PubMed] [Google Scholar]
- 2. Zweifach A., Lewis R. S. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 6295–6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Parekh A. B., Putney J. W., Jr. (2005) Physiol. Rev. 85, 757–810 [DOI] [PubMed] [Google Scholar]
- 4. Prakriya M., Lewis R. S. (2003) Cell Calcium 33, 311–321 [DOI] [PubMed] [Google Scholar]
- 5. Roos J., DiGregorio P. J., Yeromin A. V., Ohlsen K., Lioudyno M., Zhang S., Safrina O., Kozak J. A., Wagner S. L., Cahalan M. D., Veliçelebi G., Stauderman K. A. (2005) J. Cell Biol. 169, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luik R. M., Wu M. M., Buchanan J., Lewis R. S. (2006) J. Cell Biol. 174, 815–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liou J., Kim M. L., Heo W. D., Jones J. T., Myers J. W., Ferrell J. E., Jr., Meyer T. (2005) Curr. Biol. 15, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu M. M., Buchanan J., Luik R. M., Lewis R. S. (2006) J. Cell Biol. 174, 803–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baba Y., Hayashi K., Fujii Y., Mizushima A., Watarai H., Wakamori M., Numaga T., Mori Y., Iino M., Hikida M., Kurosaki T. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16704–16709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Muik M., Frischauf I., Derler I., Fahrner M., Bergsmann J., Eder P., Schindl R., Hesch C., Polzinger B., Fritsch R., Kahr H., Madl J., Gruber H., Groschner K., Romanin C. (2008) J. Biol. Chem. 283, 8014–8022 [DOI] [PubMed] [Google Scholar]
- 11. Lee K. P., Yuan J. P., Zeng W., So I., Worley P. F., Muallem S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14687–14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lis A., Peinelt C., Beck A., Parvez S., Monteilh-Zoller M., Fleig A., Penner R. (2007) Curr. Biol. 17, 794–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. DeHaven W. I., Smyth J. T., Boyles R. R., Putney J. W., Jr. (2007) J. Biol. Chem. 282, 17548–17556 [DOI] [PubMed] [Google Scholar]
- 14. Frischauf I., Schindl R., Bergsmann J., Derler I., Fahrner M., Muik M., Fritsch R., Lackner B., Groschner K., Romanin C. (2011) J. Biol. Chem. 286, 8577–8584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Srikanth S., Jung H. J., Ribalet B., Gwack Y. (2010) J. Biol. Chem. 285, 5066–5075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mullins F. M., Park C. Y., Dolmetsch R. E., Lewis R. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15495–15500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Litjens T., Harland M. L., Roberts M. L., Barritt G. J., Rychkov G. Y. (2004) J. Physiol. 558, 85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamashita M., Navarro-Borelly L., McNally B. A., Prakriya M. (2007) J. Gen. Physiol. 130, 525–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Derler I., Fahrner M., Carugo O., Muik M., Bergsmann J., Schindl R., Frischauf I., Eshaghi S., Romanin C. (2009) J. Biol. Chem. 284, 15903–15915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peinelt C., Lis A., Beck A., Fleig A., Penner R. (2008) J. Physiol. 586, 3061–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. DeHaven W. I., Smyth J. T., Boyles R. R., Bird G. S., Putney J. W., Jr. (2008) J. Biol. Chem. 283, 19265–19273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schindl R., Bergsmann J., Frischauf I., Derler I., Fahrner M., Muik M., Fritsch R., Groschner K., Romanin C. (2008) J. Biol. Chem. 283, 20261–20267 [DOI] [PubMed] [Google Scholar]
- 23. Zhang S. L., Kozak J. A., Jiang W., Yeromin A. V., Chen J., Yu Y., Penna A., Shen W., Chi V., Cahalan M. D. (2008) J. Biol. Chem. 283, 17662–17671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Frischauf I., Muik M., Derler I., Bergsmann J., Fahrner M., Schindl R., Groschner K., Romanin C. (2009) J. Biol. Chem. 284, 21696–21706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Korzeniowski M. K., Manjarrés I. M., Varnai P., Balla T. (2010) Sci. Signal. 3, ra82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Muik M., Fahrner M., Schindl R., Stathopulos P., Frischauf I., Derler I., Plenk P., Lackner B., Groschner K., Ikura M., Romanin C. (2011) EMBO J. 30, 1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Z., Lu J., Xu P., Xie X., Chen L., Xu T. (2007) J. Biol. Chem. 282, 29448–29456 [DOI] [PubMed] [Google Scholar]
- 28. Yuan J. P., Zeng W., Dorwart M. R., Choi Y. J., Worley P. F., Muallem S. (2009) Nat. Cell Biol. 11, 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fahrner M., Muik M., Derler I., Schindl R., Fritsch R., Frischauf I., Romanin C. (2009) Immunol. Rev. 231, 99–112 [DOI] [PubMed] [Google Scholar]
- 30. Park C. Y., Hoover P. J., Mullins F. M., Bachhawat P., Covington E. D., Raunser S., Walz T., Garcia K. C., Dolmetsch R. E., Lewis R. S. (2009) Cell 136, 876–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lis A., Zierler S., Peinelt C., Fleig A., Penner R. (2010) J. Gen. Physiol. 136, 673–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singh A., Hamedinger D., Hoda J. C., Gebhart M., Koschak A., Romanin C., Striessnig J. (2006) Nat. Neurosci. 9, 1108–1116 [DOI] [PubMed] [Google Scholar]
- 33. Muik M., Fahrner M., Derler I., Schindl R., Bergsmann J., Frischauf I., Groschner K., Romanin C. (2009) J. Biol. Chem. 284, 8421–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Srikanth S., Jung H. J., Kim K. D., Souda P., Whitelegge J., Gwack Y. (2010) Nat. Cell Biol. 12, 436–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yamashita M., Somasundaram A., Prakriya M. (2011) J. Biol. Chem. 286, 9429–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.