Abstract
Chemical modulators of autophagy provide useful pharmacological tools for examination of autophagic processes, and also may lead to new therapeutic agents for diseases in which control of cellular sequestration and degradation capacity are beneficial. We have identified that timosaponin A-III (TAIII), a medicinal saponin reported to exhibit anticancer properties and improve brain function, is a pronounced activator of autophagy. In this work, the salient features and functional role of TAIII-induced autophagy were investigated. In TAIII-treated cells, autophagic flux with increased formation of autophagosomes and conversion into autolysosomes is induced in association with inhibition of mammalian target of rapamycin activity and elevation of cytosolic free calcium. The TAIII-induced autophagy is distinct from conventional induction by rapamycin, exhibiting large autophagic vacuoles that appear to contain significant contents of endosomal membranes and multivesicular bodies. Furthermore, TAIII stimulates biosynthesis of cholesterol, which is incorporated to the autophagic vacuole membranes. The TAIII-induced autophagic vacuoles capture ubiquitinated proteins, and in proteasome-inhibited cells TAIII promotes autophagy of aggregation-prone ubiquitinated proteins. Our studies demonstrate that TAIII induced a distinct form of autophagy, and one of its pharmacological actions is likely to enhance the cellular quality control capacity via autophagic clearance of otherwise accumulated ubiquitinated protein aggregates.
Keywords: Autophagy, Calcium, Cholesterol, Drug Action, mTOR, Proteasome, Protein Degradation, Ubiquitination
Introduction
Autophagy, or precisely referred to macroautophagy, is a distinct lysosomal degradation pathway for intracellular materials (1, 2). Although the ubiquitin proteasome system (UPS)2 usually degrades short-lived soluble proteins, autophagy complements UPS to degrade long-lived proteins, protein aggregates, or organelles (3–5). Autophagy is essentially a regulated intracellular vacuolization process consisting of two key steps: formation of autophagosomes that capture the cytosolic substrates, and fusion of autophagosomes and lysosomes to form autolysosomes in which the autophagic content is degraded. The membrane trafficking processes in autophagy are specifically controlled by proteins encoded by highly conserved autophagy related genes, ATGs, as well as the signaling lipid phosphatidylinositol 3-phosphate that recruits effector proteins to the membranes of autophagic vacuoles (6–8).
A major function of autophagy is to recycle free amino acids, lipids, and carbohydrates for resynthesis of biomolecules or energy production, particularly under starvation conditions. In this regard, autophagy was thought to be a nonspecific process involving random sequestration and degradation of cytoplasmic components, but it is now evident that cargo can also be selectively incorporated into autophagosomes (9). Emerging evidence has shown that autophagy may play a role in selective sequestration and degradation of aggregated protein complexes and/or damaged organelles, thereby serving as a cellular quality control system for elimination of defective macromolecules. Thus, impairment of autophagy has been linked to degenerative diseases and carcinogenesis in which pathological molecules build up (10). On the other hand, induction of autophagy is proposed as an attractive therapeutic strategy for diseases in which an enhanced cellular sequestration and degradation capacity is beneficial (11). Thus, a number of small molecule-based autophagy inducers, including drugs already in clinical uses (12, 13) or those from natural products (14, 15), have been identified with the purpose for treating or preventing degenerative diseases.
In the course of screening chemical modulators of autophagy, we have identified timosaponin A-III (TAIII) as a robust inducer of autophagy (16). TAIII is a spirostanol saponin consisting of a galactose-glucose disaccharide moiety attached to the C3 position of the aglycone sarsasapogenin (supplemental Fig. S1). TAIII is isolated from the medicinal herb Anemarrhena asphodeloides, which is used as an antipyretic, anti-inflammatory, antidiabetic, and antidepressive agent in traditional Chinese medicine (17, 18). Experimental evidence shows that purified timosaponins or the medicinal fractions containing timosaponins exhibit various pharmacological properties, including improvement of learning and memory in subjects with dementia (17, 18). We have reported the anticancer activities of TAIII, and demonstrated that TAIII induces autophagy, which plays a cytoprotective function (16). Recently, TAIII was also shown to be preferentially toxic to breast cancer cell lines but not non-transformed cells (19).
In the view that autophagy induction is a characteristic action of TAIII, we have identified several novel features of the autophagy induced by this compound. Herein we report that TAIII induced a distinct form of autophagy with the involvement of multivesicular bodies, cholesterol biosynthesis, mTOR, and calcium signaling. The pharmacological significance of TAIII-induced autophagy is linked to degradation of ubiquitinated proteins and may play a role in clearance of otherwise accumulated aggregation-prone proteins.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents
Expression plasmids for tfLC3 (20) and mRFP-LC3 were gifts of Dr. Tamotsu Yoshimori. Ubiquitin-GFP (21) and YFP-CL1 (22) were obtained from Addgene. Timosaponin AIII was from Wako Biochemicals. Bafilomycin A1 was obtained from Sigma. BODIPY FL-pepstatin A, Oregon Green 488-labeled phosphatidylethanolamine, and Fluo-4 were obtained from Invitrogen. Mevastatin, BAPTA-AM, MG132, and antibodies to ubiquitin (FK2) were from Enzo Life Science. Filipin was from Cayman Chemical. p62 antibody was from BD Bioscience. LC3 antibody was from Abgent. mTOR, p70 S6 kinase and phospho-p70 S6 kinase (threonine 389), 4E-BP1, and GAPDH antibodies were from Cell Signaling. ULK1 antibody was from Santa Cruz Biotechnology.
Cells and Transfection
HeLa cells were maintained in minimum essential medium supplemented with 10% fetal bovine serum and 2 mm glutamine. HeLa cells stably expressing tfLC3, mRFP-LC3, Ub-GFP, or YFP-CL1 were established by transfecting cells with the corresponding constructs using Lipofectamine 2000 (Invitrogen) and selected with 1 mg/ml of G418. HeLa cells expressing YFP-CL1 reporter were further selected for inducible expression of the reporter protein in the presence of MG132. Cells were grown to near confluence before experiments. 0.1% DMSO was added to the cells as vehicle control.
Immunofluorescence Staining
Cells were fixed with 3% paraformaldehyde in PBS for 10 min, permeabilized with 50 μg/ml of digitonin for 5 min, and blocked with 1% BSA in PBS. The fixed cells were incubated with primary antibodies at 1 μg/ml for 1 h followed with appropriate Alexa Fluor fluorescence dye-conjugated antibodies (Invitrogen) at 1 μg/ml for 1 h. The stained cells were visualized with a Zeiss fluorescence microscope.
Filipin Staining
Cells were fixed with 3% paraformaldehyde in PBS for 10 min, incubated with 5 μg/ml of filipin for 30 min, and then examined with a fluorescence microscope.
BODIPY FL-Pepstatin A Labeling
Cells were incubated with 1 μm BODIPY-pepstatin in culture medium at 37 °C for 1 h, washed with PBS, and then examined with a fluorescence microscope.
Oregon Green 488-conjugated Phosphatidylethanolamine (PE) Labeling
Cells were incubated with 3 μm Oregon Green 488-PE in culture medium for 1 h at 4 °C to allow labeling at the plasma membrane. The medium was removed and replaced with medium containing TAIII or DMSO vehicle and incubated for a further 18 h.
Fluorescence Image Analysis
The degree of colocalization of two independent fluorescent images were analyzed by Colocalizer Pro software to obtain the Pearson correlation coefficient (Rr). Rr values from 0.5 to 1 indicates significant colocalization.
Transmission Electron Microscopy (TEM)
Cells were washed, trypsinized, and fixed with 2.5% glutaraldehyde in 0.1 m sodium cacodylate buffer, pH 7.4, at 4 °C overnight. After washing, cells were post-fixed with 1% osmium tetroxide and embedded in Polybed resin. Ultrathin sections were doubly stained with uranyl acetate and lead citrate and analyzed with 208S Philips TEM.
Intracellular Free Calcium Measurement
After TAIII treatment, cells were incubated with 1 μm Fluo-4 in culture medium at 37 °C for 30 min, washed, and examined with a fluorescence microscope. The relative intracellular free calcium concentration was determined by measuring the cell-associated Fluo-4 fluorescence using a plate reader.
Immunoblot Analysis
Cells were lysed with buffer (50 mm Tris-Cl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with protease inhibitor. Equal amount of proteins (20 μg) was resolved by SDS-PAGE and transferred onto PVDF membrane. The membrane was blocked with Tris-buffered saline containing 0.1% Tween 20 and 3% BSA and then incubated with primary antibodies at 4 °C overnight, followed with appropriate secondary antibodies for 2 h. The immunoreactivities were detected using enhanced chemiluminescence reagents (GE Healthcare). Protein signals were quantitated using Image J software.
cDNA Microarray Analysis
HeLa cells were treated with 10 μm TAIII or 0.1% DMSO for 6 h. The total RNA was isolated by TRIzol reagent (Invitrogen) followed by clean-up with the RNeasy kit (Qiagen). cDNA microarray analysis was performed with the Affymetrix Human Genome U133 Plus 2.0 GeneChip and hybridization data were analyzed using the Affymetrix Expression Console software (Genome Research Centre, The University of Hong Kong). The regulated genes (with more than 2-fold differences in expression) were listed under supplemental Table S3.
RESULTS
TAIII Induces Autophagic Flux
We have previously demonstrated that TAIII induces autophagy using TEM, immunoblot for microtubule-associated protein 1 light chain 3 (LC3) expression, and immunofluorescence detection of GFP-LC3 punctates (16). Nonetheless, these assays are instructive of formation of autophagosomes without a direct indication of autophagic flux involving the eventual merging of the autophagosomal and lysosomal pathways (23, 24). In this work, an experimental system using a tandem fluorescent mRFP-GFP-LC3 expression construct (tfLC3) was employed for monitoring the TAIII-induced autophagic flux (20). Cells transfected with tfLC3 would show characteristic fluorescent punctate upon induction of autophagy. Because GFP fluorescence is quenched at the acidic environment, whereas mRFP fluorescence is insensitive to pH changes, tfLC3 associated with neutral compartments show both green and red fluorescence, whereas those associated with acidic compartments appear as localized red but not green fluorescent signals. Thus monitoring the tfLC3 fluorescence allows distinguishing of the neutral autophagosomes and more mature, acidified autophagic vacuoles such as autolysosomes. Fig. 1 shows the time course of autophagy induction by TAIII using HeLa cells stably transfected with tfLC3. Treatment of the tfLC3 expressing cells with TAIII for 3–6 h (Fig. 1, A and B) resulted in a significant increase in green/red fluorescent LC3 dots indicative of autophagosomes, and an even more dramatic increase in red-only LC3 dots, presumably being mature acidic autophagic vacuoles. Prolonged treatment for 18 h (Fig. 1B and Fig. 2A) resulted in predominant formation of acidic autophagic vacuoles.
FIGURE 1.
Induction of autophagy by TAIII as revealed by tfLC3 imaging. A, HeLa cells expressing tfLC3 were treated with 10 μm TAIII or DMSO vehicle (control) and then examined with fluorescence microscope. Scale bars, 10 μm. B, quantitation of the percentage of cells showing tfLC3 dots after TAIII treatment. For cells expressing the tfLC3 dots, the fractions of yellow spots (autophagosomes) and orange spots (autolysosomes) in the merged images were determined. At least 100 cells were counted. C, HeLa cells were treated with 10 μm TAIII, 200 nm rapamycin (rpm), 5 nm bafilomycin A1 or their combination as indicated for 8 h. Immunoblot analysis of the cell lysates was performed to examine the levels of LC3-II and p62.
FIGURE 2.
TAIII-induced autophagic vacuoles exhibit features of multivescicular bodies. A, HeLa cells expressing tfLC3 were treated with 10 μm TAIII or 200 nm rapamycin for 18 h and then examined with a fluorescence microscope. In TAIII-treated cells, locations of the tfLC3-associated autophagic vacuoles were also recognized under light microscope. Scale bars, 5 μm. B, HeLa cells expressing mRFP-LC3 were incubated with Oregon Green 488-labeled phosphatidylethanolamine (OG-PE) for 1 h to label the plasma membrane (shown in the inset in the upper left micrograph). Medium was removed and replaced with medium containing 10 μm TAIII or 200 nm rapamycin and incubation was continued for 18 h. Cells were then examined with fluorescence microscope. Portions of TAIII-treated cells were enlarged to show features of autophagic vacuoles. Scale bars, 20 μm.
If the autophagic vacuoles induced by TAIII treatment represent steady state autophagic activities, blocking the intermediate steps should result in accumulation of the premature form of vacuoles. Bafilomycin A1 is an inhibitor of vacuolar H+-ATPase that prevents acidification of autophagosomes and impairs autophagosome-lysosome fusion (25). It was found that cells treated with TAIII and bafilomycin A1 resulted in accumulation of green/red fluorescent punctates, presumably being autophagosomes (supplemental Fig. S2). Thus, the autophagic vacuoles eventually formed after TAIII treatment were predominantly mature vacuoles that have been fused with acidic lysosomes. The TAIII-induced autophagic vacuoles could also be labeled by fluorophore-conjugated pepstatin A, which binds to lysosomal hydrolyase cathepsins (supplemental Fig. S3, Pearson coefficient of colocalization, Rr = 0.8960). Furthermore, immunoblot analysis indicates that TAIII induced expression of LC3-II, which can be further enhanced in the presence of bafilomycin A1 (Fig. 1C). TAIII treatment also reduced the autophagy-specific substrate p62, and such a reduction was abrogated by bafilomycin A1 treatment (Fig. 1C). Fluorescence microscopy reveals that the immunoreactivity of p62 was colocalized with the TAIII-induced autophagic vacuoles (supplemental Fig. S4), and this was much more apparent when TAIII-induced autophagy was intercepted by bafilomycin A1. Collectively, TAIII induces functional autophagic flux and degradation.
TAIII Induces Formation of Large Autophagic Vacuoles Apparently Containing Endosomal Membranes and Multivesicular Bodies
When TAIII-induced autophagy is compared with that by a conventional inducer like rapamycin (Fig. 2A), a striking difference is that the TAIII-induced autophagic vacuoles were much larger in size (diameter >1 μm) than those mediated by rapamycin treatment, and even recognizable under phase-contrast light microscope. Furthermore, we have observed that in HeLa cells expressing tfLC3, some luminal LC3 dots were frequently found inside the TAIII-induced vacuoles. These led us to postulate that the TAIII-induced autophagic vacuoles characteristically constitute specialized structures such as multivesicular bodies (MVB)/late endosomes, which harbor smaller vesicles derived from perimeter membranes (26). Thus, we labeled the MVB in mRFP-LC3-transfected HeLa cells with Oregon Green 488-labeled PE and traced its association with autophagic vacuoles according to a previously described procedure (27, 28) (Fig. 2B). The fluorophore-conjugated phosphatidylethanolamine is initially incorporated to the plasma membrane, efficiently internalized via endocytosis, and targeted to the MVBs. In TAIII-treated cells, intense Oregon Green 488-PE labeling was significantly colocalized with the mRFP-LC3-labeled autophagic vacuoles (Rr = 0.6481). Closer examination of these vacuoles shows the presence of smaller luminal Oregon Green 488-labeled PE vesicles, resembling the expected MVB appearance. Coincidently, ultrastructural examination of TAIII-treated cells by TEM (supplemental Fig. S5) also shows that vacuolar structures of 1–5 μm diameter were formed with luminal vesicle-like structures (29). In comparison with rapamycin-treated cells (Fig. 2B), Oregon Green 488-PE labeling was much less conspicuous and was not found co-localized with mRFP-LC3-labeled autophagic vacuoles (Rr = 0.483).
TAIII Induces Autophagy in Concomitance with Up-regulation of Cholesterol Biosynthesis
We have performed cDNA microarray analysis to identify possible genetic changes involved in the mechanism of action of TAIII (supplemental Tables S1–S3). Treatment of HeLa cells with TAIII for 6 h resulted in ∼800 up-regulated and ∼200 down-regulated transcripts. Notably, a number of transcripts encoding proteins functioning at different hierarchy levels of autophagy were significantly up-regulated (supplemental Table S1) (30). These include proteins involved in formation of autophagic vacuoles such as unc-51-like kinase (ULK1), ATG2 autophagy-related 2 homolog B (ATG2B), microtubule-associated protein 1 light chain 3β (MAP1LC3B), phosphoinositide 3-kinase, catalytic, α-peptide (PIK3C2A), and phosphoinositide kinase FYVE finger containing (PIKFYVE). The simultaneous up-regulation of autophagy-related genes suggests autophagic activities are perhaps coordinated by a specific transcriptional mechanism.
Through the use of the DAVID functional annotation tool (31), a number of significant pathways modulated by TAIII are revealed (supplemental Table S2). The most prominent TAIII-induced transcriptional change is an increase in the mRNA encoding enzymes for cholesterol biosynthesis, including the rate-limiting enzyme 3-hydroxy-3-methylglutaryl coenzyme A reductase (supplemental Tables S1 and S2). The induction of the mRNA of several key cholesterogenic enzymes by TAIII was confirmed by RT-PCR (supplemental Fig. S6). Conventional induction of autophagy by rapamycin was not associated with marked changes in some cholesterogenic enzyme expressions (supplemental Fig. S7). To investigate the specific role of cholesterol in TAIII-induced autophagy, the localization of cholesterol-enriched compartments was examined by filipin staining of tfLC3 cells. In untreated (Fig. 3A) as well as in rapamycin-treated cells (supplemental Fig. S8), filipin-stained cholesterol was predominantly localized at the cell surface plasma membrane. In TAIII-treated cells, additional cholesterol was colocalized with the autophagic vacuoles, with most localized at the membranes lining the vacuoles (Fig. 3A). In cells treated with TAIII and bafilomycin A1 (Fig. 3B), the autophagy was clamped at the autophagosomal stage and the filipin-stained cholesterol was even more explicitly colocalized with the autophagosomes (Rr = 0.9051). When cholesterol biosynthesis was inhibited by mevastatin, an inhibitor of 3-hydroxy-3-methylglutaryl-coenzyme A reductase, the filipin-stained vacuolar cholesterol was markedly diminished (Fig. 3C). Furthermore, mevastatin treatment inhibited maturation of autophagosomes to acidic autolysosomes, as indicated by increased colocalization of GFP and RFP signals of tfLC3 (Rr = 0.9407) (Fig. 3C). Mevastatin treatment also induced accumulation of the autophagic substrate p62 in TAIII-treated cells (supplemental Fig. S9). These results suggest that TAIII induces the up-regulation of biosynthesis of cholesterol, which impacts on the autophagy process.
FIGURE 3.
TAIII-induced autophagy and cholesterol localization. A, HeLa cells expressing tfLC3 were treated with 10 μm TAIII or DMSO vehicle (control) for 18 h. Cells were fixed and stained with 5 μg/ml of filipin, and then examined with fluorescence microscope. Portions of TAIII-treated cells were enlarged to show features of autophagic vacuoles. B, HeLa cells expressing tfLC3 were treated with 10 μm TAIII and 5 nm bafilomycin A1. C, HeLa cells expressing tfLC3 were treated with 1 μm mevastatin for 24 h and then 10 μm TAIII for further 18 h. Scale bars, 20 μm.
TAIII Activates Autophagy via Inhibition of mTOR Signaling
Suppression of mTOR signaling is an important pathway of autophagy induction (32). In this work, involvement of the mTOR pathway in TAIII-induced autophagy was examined (Fig. 4). Immunoblot analysis of the mTOR substrate reveals that TAIII treatment markedly inhibited p70 S6 kinase (p70 S6K) phosphorylaton at threonine 389 and induced dephosphorylation of 4E-BP1, of which both are experimental readouts of inhibition of rapamycin-sensitive mTORC1 complex kinase activity. In one model of mTOR-regulated autophagy, mTORC1 can suppress autophagy through inhibitory phosphorylation of autophagy-initiating kinase ULK1 (the mammalian orthologue of ATG1) (32). When mTOR activity is inhibited upon nutrient starvation, ULK1 is relived from the inhibition and hypophosphorylated, thereby initiating autophagy. Fig. 4 shows that TAIII treatment resulted in a significant increase in ULK1 mobility in immunoblot analysis, indicative of dephosphorylation of the protein as previously reported (33, 34).
FIGURE 4.
TAIII inhibits mTOR signaling. HeLa cells were treated with 10 μm TAIII, 200 nm rapamycin or DMSO vehicle (control) for 1, 4, and 8 h and the phosphorylation of mTOR substrate (p70 S6K, 4E-BP1, and ULK1) and the levels of autophagy marker proteins (LC3-II and p62) were analyzed by immunoblot. Dotted lines were positioned to indicate the changes in mobility of the proteins (4E-BP1 and ULK1) as a result of phosphorylation events. C, control; T, TAIII; R, rapamycin.
TAIII-induced Autophagy Is Associated with Cytosolic Calcium Mobilization
In literature, TAIII was shown to increase cytosolic calcium levels in vascular endothelial cells and smooth muscle cells in buffered salt medium (35). This lead us to investigate TAIII-induced calcium mobilization in relationship to autophagy. Fig. 5A shows that treatment of cultured HeLa cells with TAIII elicited a rapid and sustained elevation of intracellular free calcium levels as measured by the calcium probe Fluo-4. Imaging calcium localization reveals that TAIII induced an increase in Fluo4 fluorescence not only in cytoplasm but also remarkably in the mRFP-LC3-labeled autophagic vacuoles (Fig. 5B). To examine the role of calcium in TAIII-induced autophagy, BAPTA-AM was used to buffer the intracellular calcium levels. Treatment of cells with BAPTA-AM was found to markedly inhibit TAIII-induced formation of autophagic vacuoles in tfLC3 cells (Fig. 5C), as well as the TAIII-induced LC3-II expression (Fig. 5D). BAPTA-AM treatment did not affect TAIII-induced S6K and ULK1 dephosphorylation, and even itself exerted inhibition on S6K phosphorylation (similar to previously reported (36). Thus there is no correlation between the TAIII-induced cytosolic calcium increase and inhibition of mTOR signaling, suggesting calcium may act downstream of mTOR in the autophagy induction pathway.
FIGURE 5.
TAIII-induced autophagy and cellular calcium mobilization. A, HeLa cells were treated with 10 μm TAIII and the cytosolic calcium levels were analyzed with calcium probe Fluo-4. Left panel, fluorescence microscopic image of Fluo-4-loaded cells treated with 10 μm TAIII or DMSO vehicle (control) for 30 min. Right panel, time course of changes of relative Fluo-4 fluorescence (TAIII/control) of TAIII-treated cells. B, HeLa cells expressing mRFP-LC3 were treated with 10 μm TAIII or DMSO vehicle (control) for 18 h. Cells were then loaded with Fluo-4 and examined with fluorescence microscope. C, HeLa cells expressing tfLC3 were treated with 20 μm BAPTA-AM or left untreated for 1 h, followed by treatment with 10 μm TAIII for 6 h. Scale bars, 20 μm. D, HeLa cells were treated as in C and LC3-II, p-S6K (Thr-389), and ULK1 expression was determined by immunoblot.
TAIII-induced Autophagic Vacuoles Capture Ubiquitinated Proteins
Autophagy was previously considered to be a bulk lysosome-dependent degradation system with little specificity, but cumulative evidence indicates that there is specific, autophagic degradation of ubiquitinated proteins, whose accumulation is implied in a number of degenerative diseases. Thus, whether ubiquitinated proteins are substrates of TAIII-induced autophagy was investigated. mRFP-LC3 expressing HeLa cells were transfected with GFP-tagged ubiquitin (ubiquitin-GFP), which has been previously demonstrated to conjugate cellular protein substrates, thereby allowing microscopic tracing of the ubiquitinated proteins (21). Fig. 6 shows that in TAIII-treated cells, there was a small amount of ubiquitin-GFP-labeled structures colocalized with the induced mRFP-LC3-labeled autophagic vacuoles. When TAIII-induced autophagy was inhibited by bafilomycin A1, mRFP-LC3-labeled vacuoles were accumulated and all of these were found colocalized with ubiquitin-GFP-labeled materials. Taken together, these results suggest that TAIII-induced autophagy capture ubiquitinated proteins and deliver them to lysosomes for degradation.
FIGURE 6.
TAIII-induced autophagic vacuoles capture ubiquitinated proteins. HeLa cells expressing ubiquitin-GFP were transiently transfected with mRFP-LC3. Cells were then treated with DMSO vehicle, 10 μm TAIII, or 10 μm TAIII and 5 nm bafilomycin A1 for 18 h, and then examined with a fluorescence microscope. Scale bars, 20 μm.
TAIII Induces Autophagy of Ubiquitinated Protein Aggregates Resulting from Proteasome Inhibition
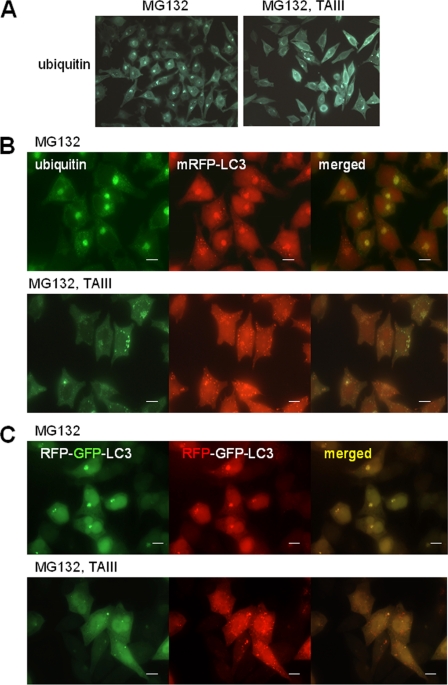
We have investigated the effect of TAIII-induced autophagy on the ubiquitinated protein expression after proteasomal degradation was inhibited. Fig. 7A shows that in cells treated with proteasome inhibitor MG132, ubiquitinated proteins were accumulated and part of them were presented as perinuclear inclusions. Exposure of these proteasome-inhibited cells to TAIII resulted in a significant reduction of ubiquitinated protein inclusions (Fig. 7A, right panel). In mRFP-LC3 expressing cells, the ubiquitinated protein inclusions were colocalized with mRFP-LC3 (Fig. 7B). Treatment of these cells with TAIII resulted in a decrease in the protein inclusions together with an increase in mRFP-LC3-labeled autophagic vacuoles. The TAIII-induced autophagy in the presence of proteasome inhibitor was also examined using tfLC3 expressing cells (Fig. 7C). In cells treated with MG132, inclusion bodies labeled with tfLC3 showing both mRFP and GFP fluorescence were obvious, suggesting that these LC3 harboring inclusions were relatively neutral compartments. Importantly, TAIII treatment induced a significant increase in the red-only LC3-labeled vacuoles, revealing that TAIII induced autophagic flux during the proteasome inhibition.
FIGURE 7.
TAIII-induced autophagy is associated with reduced ubiquitinated protein deposition upon proteasome inhibition. A, HeLa cells were treated with 2.5 μm MG132 for 14 h, followed by treatment with 10 μm TAIII or DMSO vehicle (control) for 8 h. Cells were fixed and stained with antiubiquitin antibody, and then examined with a fluorescence microscope. B, HeLa cells expressing mRFP-LC3 were treated as in A, fixed, and stained with anti-ubiquitin antibody. C, HeLa cells expressing tfLC3 were treated as in A to show induction of autophagy. Scale bars, 20 μm.
We have also investigated the effect of TAIII treatment on the expression of a YFP reporter for the ubiquitination substrate YFP-CL1 (22) (Fig. 8). CL1 is a hydrophobic peptide and conveys proteasome-dependent degradation when fused to YFP. The YFP-CL1 fusion protein resembles misfolded proteins, serving as a model protein for studies of disease-linked, aggregation-prone proteins (37, 38). In HeLa cells stably transfected with YFP-CL1, the reporter protein was subjected to proteasomal degradation and expressed at very low levels. When the cells were treated with proteasome inhibitor MG132, YFP-CL1 was accumulated and formed perinuclear inclusions (Fig. 8A) (22), which were found colocalized with transfected mRFP-LC3 (Fig. 8B). When these cells were treated with TAIII, there was an induction of autophagic vacuoles exhibiting mRFP-LC3 fluorescence, concomitant with a significant reduction of cells with YFP-CL1 inclusions (Fig. 8, A and B). Immunoblot analysis also shows that TAIII treatment resulted in a reduction of accumulated YFP-CL1, which could be reversed by co-treatment with the autophagosome-lysosome fusion inhibitor bafilomycin A1 (Fig. 8C). Furthermore, the levels of the adaptor protein and autophagic substrate p62 were also reduced upon TAIII treatment, and such p62 loss could be recovered in the presence of bafilomycin A1. The LC3-II expression was stimulated in the MG132-treated cells, as previously demonstrated (39, 40). Importantly, LC3-II expression was markedly increased in the presence of bafilomycin A1 and this was further augmented in TAIII-treated cells, suggesting that TAIII enhanced autophagy in the proteasome-inhibited cells. Collectively, these results support that TAIII treatment could mediate autophagic degradation of ubiquitinated aggregation-prone proteins accumulated upon impairment of UPS.
FIGURE 8.
TAIII-induced autophagy is associated with reduction of an aggregation-prone UPS reporter substrate. A, HeLa cells with YFP-CL1 reporter were treated with 2.5 μm MG132 for 14 h to induce accumulation of YFP-CL1, followed by treatment with 10 μm TAIII or DMSO vehicle (control) for 8 h, and then examined with fluorescence microscope. B, HeLa cells with YFP-CL1 reporter were transiently transfected with mRFP-LC3. The transfected cells were then treated as in A. Scale bars, 20 μm. C, HeLa cells with YFP-CL1 reporter were treated with 2.5 μm MG132 for 14 h to induce accumulation of YFP-CL1, followed by treatment with DMSO vehicle (control) or 10 μm TAIII in the presence or absence of 5 nm bafilomycin A1 for 8 h. Immunoblot analysis of the cell lysates was performed to examine the levels of YFP-CL1, p62, and LC3-II. The relative densitometric values of the protein signals normalized against GAPDH values are indicated. The values of the protein signals of the sample treated with MG132 alone are set at unity.
DISCUSSION
Modulation of autophagy is a potential therapeutic strategy for treatment of cancer and degenerative disorders. We have identified TAIII as a chemical inducer of autophagy and characterized its special autophagy-inducing properties. Our data reveal that TAIII induces robust autophagic responses involving initial autophagosome formation and merging of the lysosomal pathway with formation of autolysosomes, the eventual end point for autophagic degradation (Fig. 1). The TAIII-induced autophagy flux is not a generic response of saponins as a panel of structurally similar saponins has been found incapable of inducing significant formation of autophagic vacuoles.3
A striking feature of TAIII-induced autophagy is the formation of large acidic, cathespins containing autophagic vacuoles (autolysosomes) upon prolonged drug treatment; some of which are even observable using light microscopy (Fig. 2A). These autophagic vacuoles were apparently distinct from those formed during early time course of TAIII treatment or those induced by conventional autophagy inducers such as rapamycin or starvation (Fig. 2A). We speculate that these large vacuoles may signify a high autophagy-activating capacity of TAIII. When the plasma membranes were labeled with fluorophore-conjugated phosphatidylethanolamine and their cellular distribution was followed (Fig. 2B), there was significant colocalization of the fluorescent label with the TAIII-induced autophagic vacuoles. Although the fluorescent label may redistribute in various cellular compartments in addition to endosomes after internalization, the possibility exists that TAIII may affect the endosomal pathway, allowing the autophagosomes to have a higher chance to contain endosomal membranes. The phosphatidylethanolamine labeling experiments also provide evidence that the TAIII-induced autophagic vacuoles may have an origin from MVB (Fig. 2B and supplemental Fig. S5), a form of late endosome that harbors smaller vesicular structures derived from perimeter plasma membranes (26). Emerging evidence indicates that MVB can be incorporated into amphisomes, and eventually mature to autolysosomes after lysosomal fusion (26). Importantly, it has been shown that alteration in MVB biogenesis blocks the degradation of protein aggregates associated with neurodegeneration through autophagy (41, 42), indicating that coupling of MVB and autophagy is necessary to allow an efficient autophagic degradation.
TAIII induces autophagy in association with increased cholesterol incorporation into the autophagic vacuoles (Fig. 3A). Notably, transcriptional profiling data shows that up-regulation of mRNA levels for cholesterol biosynthetic enzymes is the most prominent feature of TAIII-induced gene expression (supplemental Table S1–S3). A link between TAIII-induced autophagy and cholesterol is revealed by the observation that an inhibitor of cholesterol biosynthesis, mevastatin, decreased vacuole-associated cholesterol and blocked the maturation of autophagic vacuoles (Fig. 3C). King et al. (19) have also shown that TAIII induced cholesterol biosynthesis in breast cancer cell lines but no functional role was assigned. Our experiments herein independently show this in HeLa cells but, in addition, demonstrate that augmented cholesterol biosynthesis has a significant impact on TAIII-induced autophagy. The incorporation of cholesterol into autophagic vacuoles is dynamic, as locking autophagy at the autophagosomal stage by bafilomycin A1 leads to a more significant accumulation of cholesterol in the resulting premature autophagic vacuoles (Fig. 3B). It has been shown in earlier TEM studies that cholesterol is detected in the membrane of the autophagic vacuoles (43), and our microscopic images also provide evidence for marked enrichment of cholesterol on autophagic vacuole membranes in TAIII-treated cells. It is conceivable that the high cholesterol content in the autophagic vacuole membranes support autophagic degradation, as cholesterol is required to maintain the integrity of the vacuolar membrane, preventing leakage of lysosomal enzymes and death-inducing factors. Sterols can fill the spaces between the acyl chains of phospholipids in the hydrophobic portion of the membrane bilayer, thereby conferring rigidity and decreasing permeability. It is noteworthy that tamoxifen, a known autophagy inducer in breast cancer cells, also activates autophagy with sterol accumulation (44).
The present studies demonstrated the involvement of mTOR in the signaling of the TAIII-induced autophagy (Fig. 4). mTOR activation stimulates cell growth and proliferation, and inhibits autophagy (32). Inhibition of mTOR is one of the conventional signals leading to autophagy induction under starvation conditions, which can be mimicked by rapamycin treatment. A model of the control of autophagy by mTOR has recently emerged, with the mTORC1 complex being the suppressor of autophagy through a direct interaction with the ULK1-Atg13-FIP200 complex, which are phosphorylation substrates of mTOR (32). Induction of autophagy can be mediated by inhibition of mTOR activity, resulting in activation of the ULK1 complex, which is the most upstream component of core autophagy machinery. In the present work, TAIII is shown to inhibit mTOR kinase activity (p70 S6K and 4E-BP1 phosphorylation) and induce dephosphorylation of ULK1. Thus TAIII-induced autophagy is apparently mediated at least in part by an mTOR/ULK1-regulated mechanism.
TAIII also induces an immediate and sustained increase of cytosolic calcium concentration, which appears to modulate autophagy independently or act downstream to mTOR (Fig. 5). A number of calcium mobilizing agents were shown to affect autophagy. Seglen and colleagues (45) first reported that autophagy depends on cytosolic calcium levels. Extended mechanistic studies on the role of calcium in autophagy have just emerged albeit with conflicting conclusions. Changes in cytosolic calcium have been shown to regulate the induction of autophagy via calmodulin-dependent protein kinase kinase, AMP-activated protein kinase, and mTOR (46). Recently, AMP-activated protein kinase-independent induction of autophagy by cytosolic calcium increase has also been demonstrated (47). Nonetheless, it has been demonstrated that calcium channel blockers induce autophagy in neuronal cells, and an increase in cytosolic calcium is suggested to be inhibitory to autophagic flux (48). Thus, it still remains to be elucidated how autophagy is exactly controlled by the complex dynamics of spatial and temporal calcium signals. In the present study, it is noteworthy that in TAIII-treated cells, autophagic flux could occur even in the presence of cytosolic calcium elevation. Furthermore, buffering the cytosolic calcium with BAPTA-AM abrogated the initial formation of autophagic vacuoles induced by TAIII (Fig. 5C). These findings are consistent with those previous studies suggesting that autophagy is dependent on mobilization of cytosolic calcium and the presence of calcium in some intracellular compartments (45–47). We have also shown that a salient feature of TAIII-induced autophagy is the intralumenal accumulation of calcium ions in autophagic vacuoles (Fig. 5B). Similar observations on the calcium accumulation in MVB-associated autophagic vacuoles in human erythroleukemia cells has been reported (28). Intravesicular accumulation of calcium and local calcium release is linked to intracellular fusion and exocytotic processes (49). Thus the possibility exists that fusion events in the autophagy pathway are controlled by the localized calcium concentration in autophagic vacuoles (8, 26). It has been demonstrated that autophagic vacuoles and lysosomal compartments are equipped with several calcium channels that may mediate local calcium fluxes (50).
The second part of our work concerns the functional role of TAIII-induced autophagy. We have demonstrated that the TAIII-induced autophagic vacuoles harbor ubiquitinated proteins (Fig. 6). Inhibition of TAIII-induced autophagy at the lysosomal fusion step by bafilomycin A1 resulted in accumulation of ubiquitinated materials in the autophagic vacuoles. Increasing evidence indicates that ubiquitinated proteins are substrates for autophagic degradation (51, 52). The selective autophagic turnover of ubiquitinated proteins may be mediated by p62 (53, 54). p62 binds both to ubiquitinated proteins and LC3, thereby serving as an “autophagic receptor.” One current model suggests that p62 binds ubiquitinated proteins, promoting oligomerization to form aggregates that are captured in autophagic vacuoles via its affinity with LC3 (9). Furthermore, it has also been demonstrated that when autophagy is blocked at the lysosomal fusion stage, p62 accumulates and binds ubiquitinated proteins, thereby preventing their delivery to and degradation by the proteasome (55).
The ubiquitinated proteins targeted for autophagy may include misfolded, aggregation-prone proteins or damaged organelles that accumulate during cellular stress. Accumulation of misfolded protein aggregates is often linked to various pathological conditions. Although the role of autophagy in these conformational diseases remains to be clarified, enhancement of autophagic activities is proposed to exhibit a beneficial effect on elimination of disease-associated protein aggregates (11). We have also examined whether TAIII-induced autophagy may impact on the accumulation of ubiquitinated proteins in the UPS-impaired cells (Figs. 7 and 8). In the presence of proteasome inhibitor, cells accumulate ubiquitinated proteins. Part of them are present as misfolded protein aggregates that eventually deposit in the perinuclear region as inclusion bodies. When the UPS-impaired cells were treated with TAIII, autophagy was induced with a marked decrease in ubiquitinated protein deposits. These observations have been obtained from experiments on both endogenous ubiquitinated proteins (Fig. 7) as well as on a UPS reporter that models aggregation-prone proteins (Fig. 8). We reason that TAIII-induced autophagy mediates degradation of aggregation-prone proteins formed upon UPS inhibition, resulting in less protein aggregates accumulated as inclusions. It is noteworthy that localized deposition of aggregated proteins not only signifies a preceding accumulation of denatured or damaged proteins particularly in conditions when protein degradation (UPS or autophagy) is limiting, but may also represent a sequestration mechanism for disposal of toxic aggregates (56–61). Thus, protein inclusions may also be subjected to autophagic clearance, which may be enhanced by TAIII treatment. Further work is clearly required to elucidate the spatial and temporal regulation of aggregation-prone protein degradation.
The pronounced induction of autophagic flux with enhanced autophagosome formation and autophagosomes-lysosomes fusion is a characteristic action of TAIII and may in part account for its versatile reported pharmacological actions (16–19). For example, the autophagy-inducing properties of TAIII may render protein degradation predominant over synthesis, thereby compromising the growth of rapidly proliferating cancer cells. Although prolonged TAIII treatment results in anticancer activities, evidence exists that TAIII induced early autophagy, which is cytoprotective in nature (16, 19). Also of particular interest is the reported claims on the effects of timosaponins, particularly TAIII, on the amelioration of learning and memory deficits associated with dementia (17, 18), a pathological condition in which autophagy in neural cells may impact. TAIII is thus on the emerging list of compounds that exhibit antiproliferative and cytoprotective properties attributable to autophagy (14, 62–64), and its application in relationship to autophagy activation warrants further investigation.
Supplementary Material
Acknowledgment
We thank Dr. Tamotsu Yoshimori for providing the LC3 expression plasmids.
This work was supported by Area of Excellence Scheme AoE/P10/01, established under the University Grants Committee and Department of Health of Hong Kong Special Administrative Region, People's Republic of China.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S9.
C. N. Lok, L. K. Sy, and C. M. Che, unpublished data.
- UPS
- ubiquitin proteasome system
- TAIII
- timosaponin AIII
- LC3
- microtubule-associated protein 1 light chain 3
- tfLC3
- tandem fluorescent mRFP-GFP-LC3
- PE
- phosphatidylethanolamine
- MVB
- multivesicular bodies
- mTOR
- mammalian target of rapamycin
- ULK1
- unc-51-like kinase 1
- 4E-BP1
- translation initiation factor 4E-binding protein 1
- p70 S6K
- p70 S6 kinase
- DMSO
- dimethyl sulfoxide
- TEM
- transmission electron microscopy
- BAPTA-AM
- 1,2-bis(2-aminophenoxyl)ethane-N,N,N′,N′-tetraacetic acid acetoxymethyl ester.
REFERENCES
- 1. Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ravikumar B., Sarkar S., Davies J. E., Futter M., Garcia-Arencibia M., Green-Thompson Z. W., Jimenez-Sanchez M., Korolchuk V. I., Lichtenberg M., Luo S., Massey D. C., Menzies F. M., Moreau K., Narayanan U., Renna M., Siddiqi F. H., Underwood B. R., Winslow A. R., Rubinsztein D. C. (2010) Physiol. Rev. 90, 1383–1435 [DOI] [PubMed] [Google Scholar]
- 3. Nedelsky N. B., Todd P. K., Taylor J. P. (2008) Biochim. Biophys. Acta 1782, 691–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lamark T., Johansen T. (2010) Curr. Opin. Cell Biol. 22, 192–198 [DOI] [PubMed] [Google Scholar]
- 5. Ding W. X., Yin X. M. (2008) Autophagy 4, 141–150 [DOI] [PubMed] [Google Scholar]
- 6. Xie Z., Klionsky D. J. (2007) Nat. Cell Biol. 9, 1102–1109 [DOI] [PubMed] [Google Scholar]
- 7. Tooze S. A., Yoshimori T. (2010) Nat. Cell Biol. 12, 831–835 [DOI] [PubMed] [Google Scholar]
- 8. Burman C., Ktistakis N. T. (2010) FEBS Lett. 584, 1302–1312 [DOI] [PubMed] [Google Scholar]
- 9. Kirkin V., McEwan D. G., Novak I., Dikic I. (2009) Mol. Cell 34, 259–269 [DOI] [PubMed] [Google Scholar]
- 10. Knaevelsrud H., Simonsen A. (2010) FEBS Lett. 584, 2635–2645 [DOI] [PubMed] [Google Scholar]
- 11. Rubinsztein D. C., Gestwicki J. E., Murphy L. O., Klionsky D. J. (2007) Nat. Rev. Drug Discov. 6, 304–312 [DOI] [PubMed] [Google Scholar]
- 12. Zhang L., Yu J., Pan H., Hu P., Hao Y., Cai W., Zhu H., Yu A. D., Xie X., Ma D., Yuan J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19023–19028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sarkar S., Perlstein E. O., Imarisio S., Pineau S., Cordenier A., Maglathlin R. L., Webster J. A., Lewis T. A., O'Kane C. J., Schreiber S. L., Rubinsztein D. C. (2007) Nat. Chem. Biol. 3, 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vingtdeux V., Giliberto L., Zhao H., Chandakkar P., Wu Q., Simon J. E., Janle E. M., Lobo J., Ferruzzi M. G., Davies P., Marambaud P. (2010) J. Biol. Chem. 285, 9100–9113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sarkar S., Davies J. E., Huang Z., Tunnacliffe A., Rubinsztein D. C. (2007) J. Biol. Chem. 282, 5641–5652 [DOI] [PubMed] [Google Scholar]
- 16. Sy L. K., Yan S. C., Lok C. N., Man R. Y., Che C. M. (2008) Cancer Res. 68, 10229–10237 [DOI] [PubMed] [Google Scholar]
- 17. Li T. J., Qiu Y., Yang P. Y., Rui Y. C., Chen W. S. (2007) Neurosci. Lett. 421, 147–151 [DOI] [PubMed] [Google Scholar]
- 18. Lee B., Jung K., Kim D. H. (2009) Pharmacol. Biochem. Behav. 93, 121–127 [DOI] [PubMed] [Google Scholar]
- 19. King F. W., Fong S., Griffin C., Shoemaker M., Staub R., Zhang Y. L., Cohen I., Shtivelman E. (2009) PLoS One 4, e7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kimura S., Noda T., Yoshimori T. (2007) Autophagy 3, 452–460 [DOI] [PubMed] [Google Scholar]
- 21. Dantuma N. P., Groothuis T. A., Salomons F. A., Neefjes J. (2006) J. Cell Biol. 173, 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Menéndez-Benito V., Verhoef L. G., Masucci M. G., Dantuma N. P. (2005) Hum. Mol. Genet. 14, 2787–2799 [DOI] [PubMed] [Google Scholar]
- 23. Klionsky D. J., Abeliovich H., Agostinis P., Agrawal D. K., Aliev G., Askew D. S., Baba M., Baehrecke E. H., Bahr B. A., Ballabio A., Bamber B. A., Bassham D. C., Bergamini E., Bi X., Biard-Piechaczyk M., Blum J. S., Bredesen D. E., Brodsky J. L., Brumell J. H., Brunk U. T., Bursch W., Camougrand N., Cebollero E., Cecconi F., Chen Y., Chin L. S., Choi A., Chu C. T., Chung J., Clarke P. G., Clark R. S., Clarke S. G., Clavé C., Cleveland J. L., Codogno P., Colombo M. I., Coto-Montes A., Cregg J. M., Cuervo A. M., Debnath J., Demarchi F., Dennis P. B., Dennis P. A., Deretic V., Devenish R. J., Di Sano F., Dice J. F., Difiglia M., Dinesh-Kumar S., Distelhorst C. W., Djavaheri-Mergny M., Dorsey F. C., Dröge W., Dron M., Dunn W. A., Jr., Duszenko M., Eissa N. T., Elazar Z., Esclatine A., Eskelinen E. L., Fésüs L., Finley K. D., Fuentes J. M., Fueyo J., Fujisaki K., Galliot B., Gao F. B., Gewirtz D. A., Gibson S. B., Gohla A., Goldberg A. L., Gonzalez R., González-Estévez C., Gorski S., Gottlieb R. A., Häussinger D., He Y. W., Heidenreich K., Hill J. A., Høyer-Hansen M., Hu X., Huang W. P., Iwasaki A., Jäättelä M., Jackson W. T., Jiang X., Jin S., Johansen T., Jung J. U., Kadowaki M., Kang C., Kelekar A., Kessel D. H., Kiel J. A., Kim H. P., Kimchi A., Kinsella T. J., Kiselyov K., Kitamoto K., Knecht E., Komatsu M., Kominami E., Kondo S., Kovács A. L., Kroemer G., Kuan C. Y., Kumar R., Kundu M., Landry J., Laporte M., Le W., Lei H. Y., Lenardo M. J., Levine B., Lieberman A., Lim K. L., Lin F. C., Liou W., Liu L. F., Lopez-Berestein G., López-Otín C., Lu B., Macleod K. F., Malorni W., Martinet W., Matsuoka K., Mautner J., Meijer A. J., Meléndez A., Michels P., Miotto G., Mistiaen W. P., Mizushima N., Mograbi B., Monastyrska I., Moore M. N., Moreira P. I., Moriyasu Y., Motyl T., Münz C., Murphy L. O., Naqvi N. I., Neufeld T. P., Nishino I., Nixon R. A., Noda T., Nürnberg B., Ogawa M., Oleinick N. L., Olsen L. J., Ozpolat B., Paglin S., Palmer G. E., Papassideri I., Parkes M., Perlmutter D. H., Perry G., Piacentini M., Pinkas-Kramarski R., Prescott M., Proikas-Cezanne T., Raben N., Rami A., Reggiori F., Rohrer B., Rubinsztein D. C., Ryan K. M., Sadoshima J., Sakagami H., Sakai Y., Sandri M., Sasakawa C., Sass M., Schneider C., Seglen P. O., Seleverstov O., Settleman J., Shacka J. J., Shapiro I. M., Sibirny A., Silva-Zacarin E. C., Simon H. U., Simone C., Simonsen A., Smith M. A., Spanel-Borowski K., Srinivas V., Steeves M., Stenmark H., Stromhaug P. E., Subauste C. S., Sugimoto S., Sulzer D., Suzuki T., Swanson M. S., Tabas I., Takeshita F., Talbot N. J., Tallóczy Z., Tanaka K., Tanaka K., Tanida I., Taylor G. S., Taylor J. P., Terman A., Tettamanti G., Thompson C. B., Thumm M., Tolkovsky A. M., Tooze S. A., Truant R., Tumanovska L. V., Uchiyama Y., Ueno T., Uzcátegui N. L., van der Klei I., Vaquero E. C., Vellai T., Vogel M. W., Wang H. G., Webster P., Wiley J. W., Xi Z., Xiao G., Yahalom J., Yang J. M., Yap G., Yin X. M., Yoshimori T., Yu L., Yue Z., Yuzaki M., Zabirnyk O., Zheng X., Zhu X., Deter R. L. (2008) Autophagy 4, 151–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mizushima N., Yoshimori T., Levine B. (2010) Cell 140, 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klionsky D. J., Elazar Z., Seglen P. O., Rubinsztein D. C. (2008) Autophagy 4, 849–950 [DOI] [PubMed] [Google Scholar]
- 26. Fader C. M., Colombo M. I. (2009) Cell Death Differ. 16, 70–78 [DOI] [PubMed] [Google Scholar]
- 27. Savina A., Furlán M., Vidal M., Colombo M. I. (2003) J. Biol. Chem. 278, 20083–20090 [DOI] [PubMed] [Google Scholar]
- 28. Fader C. M., Sánchez D., Furlán M., Colombo M. I. (2008) Traffic 9, 230–250 [DOI] [PubMed] [Google Scholar]
- 29. Fawcett D. W. (1981) in The Cells, pp. 487–510, W. B. Saunders, Philadelphia, PA [Google Scholar]
- 30. Behrends C., Sowa M. E., Gygi S. P., Harper J. W. (2010) Nature 466, 68–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang, da W., Sherman B. T., Lempicki R. A. (2009) Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 32. Jung C. H., Ro S. H., Cao J., Otto N. M., Kim D. H. (2010) FEBS Lett. 584, 1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ganley I. G., Lam du H., Wang J., Ding X., Chen S., Jiang X. (2009) J. Biol. Chem. 284, 12297–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., Guan J. L., Oshiro N., Mizushima N. (2009) Mol. Biol. Cell 20, 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang G. J., Lin L. C., Chen C. F., Cheng J. S., Lo Y. K., Chou K. J., Lee K. C., Liu C. P., Wu Y. Y., Su W., Chen W. C., Jan C. R. (2002) Life Sci. 71, 1081–1090 [DOI] [PubMed] [Google Scholar]
- 36. Gulati P., Gaspers L. D., Dann S. G., Joaquin M., Nobukuni T., Natt F., Kozma S. C., Thomas A. P., Thomas G. (2008) Cell Metab. 7, 456–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Link C. D., Fonte V., Hiester B., Yerg J., Ferguson J., Csontos S., Silverman M. A., Stein G. H. (2006) J. Biol. Chem. 281, 1808–1816 [DOI] [PubMed] [Google Scholar]
- 38. Bence N. F., Sampat R. M., Kopito R. R. (2001) Science 292, 1552–1555 [DOI] [PubMed] [Google Scholar]
- 39. Iwata A., Riley B. E., Johnston J. A., Kopito R. R. (2005) J. Biol. Chem. 280, 40282–40292 [DOI] [PubMed] [Google Scholar]
- 40. Ding W. X., Ni H. M., Gao W., Yoshimori T., Stolz D. B., Ron D., Yin X. M. (2007) Am. J. Pathol. 171, 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerød L., Fisher E. M., Isaacs A., Brech A., Stenmark H., Simonsen A. (2007) J. Cell Biol. 179, 485–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee J. A., Beigneux A., Ahmad S. T., Young S. G., Gao F. B. (2007) Curr. Biol. 17, 1561–1567 [DOI] [PubMed] [Google Scholar]
- 43. Eskelinen E. L. (2005) Autophagy 1, 1–10 [DOI] [PubMed] [Google Scholar]
- 44. de Medina P., Silvente-Poirot S., Poirot M. (2009) Autophagy 5, 1066–1067 [DOI] [PubMed] [Google Scholar]
- 45. Gordon P. B., Holen I., Fosse M., Røtnes J. S., Seglen P. O. (1993) J. Biol. Chem. 268, 26107–26112 [PubMed] [Google Scholar]
- 46. Høyer-Hansen M., Bastholm L., Szyniarowski P., Campanella M., Szabadkai G., Farkas T., Bianchi K., Fehrenbacher N., Elling F., Rizzuto R., Mathiasen I. S., Jäättelä M. (2007) Mol. Cell 25, 193–205 [DOI] [PubMed] [Google Scholar]
- 47. Grotemeier A., Alers S., Pfisterer S. G., Paasch F., Daubrawa M., Dieterle A., Viollet B., Wesselborg S., Proikas-Cezanne T., Stork B. (2010) Cell. Signal. 22, 914–925 [DOI] [PubMed] [Google Scholar]
- 48. Sarkar S., Rubinsztein D. C. (2008) Mol. Biosyst. 4, 895–901 [DOI] [PubMed] [Google Scholar]
- 49. Luzio J. P., Bright N. A., Pryor P. R. (2007) Biochem. Soc. Trans. 35, 1088–1091 [DOI] [PubMed] [Google Scholar]
- 50. Martina J. A., Lelouvier B., Puertollano R. (2009) Traffic 10, 1143–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lenk S. E., Dunn W. A., Jr., Trausch J. S., Ciechanover A., Schwartz A. L. (1992) J. Cell Biol. 118, 301–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim P. K., Hailey D. W., Mullen R. T., Lippincott-Schwartz J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20567–20574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 54. Ichimura Y., Kumanomidou T., Sou Y. S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., Komatsu M. (2008) J. Biol. Chem. 283, 22847–22857 [DOI] [PubMed] [Google Scholar]
- 55. Korolchuk V. I., Mansilla A., Menzies F. M., Rubinsztein D. C. (2009) Mol. Cell 33, 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fortun J., Dunn W. A., Jr., Joy S., Li J., Notterpek L. (2003) J. Neurosci. 23, 10672–10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wong E. S., Tan J. M., Soong W. E., Hussein K., Nukina N., Dawson V. L., Dawson T. M., Cuervo A. M., Lim K. L. (2008) Hum. Mol. Genet. 17, 2570–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Salomons F. A., Menéndez-Benito V., Böttcher C., McCray B. A., Taylor J. P., Dantuma N. P. (2009) Mol. Cell. Biol. 29, 1774–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mitra S., Tsvetkov A. S., Finkbeiner S. (2009) Autophagy 5, 1037–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Balgi A. D., Fonseca B. D., Donohue E., Tsang T. C., Lajoie P., Proud C. G., Nabi I. R., Roberge M. (2009) PLoS One 4, e7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gies E., Wilde I., Winget J. M., Brack M., Rotblat B., Novoa C. A., Balgi A. D., Sorensen P. H., Roberge M., Mayor T. (2010) PLoS One 5, e14410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Puissant A., Robert G., Fenouille N., Luciano F., Cassuto J. P., Raynaud S., Auberger P. (2010) Cancer Res. 70, 1042–1052 [DOI] [PubMed] [Google Scholar]
- 63. Aoki H., Takada Y., Kondo S., Sawaya R., Aggarwal B. B., Kondo Y. (2007) Mol. Pharmacol. 72, 29–39 [DOI] [PubMed] [Google Scholar]
- 64. Herman-Antosiewicz A., Johnson D. E., Singh S. V. (2006) Cancer Res. 66, 5828–5835 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.