Abstract
Tumor suppressor protein p53, our most critical defense against tumorigenesis, can be made powerless by mechanisms such as mutations and inhibitors. Fortilin, a 172-amino acid polypeptide with potent anti-apoptotic activity, is up-regulated in many human malignancies. However, the exact mechanism by which fortilin exerts its anti-apoptotic activity remains unknown. Here we present significant insight. Fortilin binds specifically to the sequence-specific DNA binding domain of p53. The interaction of fortilin with p53 blocks p53-induced transcriptional activation of Bax. In addition, fortilin, but not a double point mutant of fortilin lacking p53 binding, inhibits p53-dependent apoptosis. Furthermore, cells with wild-type p53 and fortilin, but not cells with wild-type p53 and the double point mutant of fortilin lacking p53 binding, fail to induce Bax gene and apoptosis, leading to the formation of large tumor in athymic mice. Our results suggest that fortilin is a novel p53-interacting molecule and p53 inhibitor and that it is a logical molecular target in cancer therapy.
Keywords: Apoptosis, Carcinogenesis, Gene Regulation, p53, Protein-Protein Interactions, Fortilin
Introduction
Tumor suppressor protein p53 keeps us free of cancer when it is functional. Mice lacking p53 (p53−/−) spontaneously develop numerous neoplasms within 6 months (1). Mutated p53 genes are seen in more than 50% of all human cancers, making them the most frequently observed genetic derangement in human cancer (2). At a molecular level, the ability of p53 to eliminate cancerous cells relies on its ability to induce apoptosis, through either the transcriptional activation of proapoptotic genes such as Noxa (3), PUMA4 (4), and Bax (5) or the direct transcription-independent activation of Bax on mitochondria (6). Growing cancers manage to keep p53 in check either by mutating the p53 gene itself (7–9) or by expressing p53 inhibitors such as Mdm2 (9, 10).
The function of fortilin, a ubiquitous, highly conserved, 172-amino acid polypeptide also known as “translationally controlled tumor protein,” or TCTP, remained unknown (11, 12). Investigation in our laboratory and others showed that fortilin possesses potent anti-apoptotic activity (13–15). Fortilin is overexpressed in human cancers (16, 17), the depletion of which is associated with spontaneous death of cancerous cells (13, 18). Higher levels of fortilin are associated with more malignant cancer phenotypes (14). Although heterozygous fortilin-deficient mice (fortilin+/−) were normal in appearance and fertile, homozygous fortilin-deficient (fortilin−/−) mice were embryonically lethal around 3.5 days postcoitus due to massive apoptosis, as reported by our laboratory and others (19–21).
The mechanism by which fortilin functions as an anti-apoptotic molecule has been under robust investigation. First, based on the fact that fortilin physically interacts with myeloid cell leukemia protein-1 (MCL1), an anti-apoptotic Bcl-2 family member, it was suggested that fortilin stabilizes and exerts its anti-apoptotic activity through MCL1 (22). However, fortilin is capable of protecting cells from apoptosis in the absence of MCL1 (23), suggesting that fortilin is anti-apoptotic independently of MCL1. Secondly, fortilin functionally antagonizes Bax, a pro-apoptotic Bcl-2 family member (21), presumably by inserting itself into the mitochondrial membrane and preventing Bax from dimerizing within the membrane. However, there is no physical interaction demonstrable between fortilin and Bax (21), and it is still unclear how fortilin blocks Bax dimerization in the mitochondrial membrane. Thirdly, fortilin binds calcium (17, 24–28) and blocks calcium-dependent apoptosis (29). However, the strength of the binding of fortilin to calcium was moderate at most with a dissociation constant of 7.58–17.5 μm (29). Fourthly, fortilin binds and destabilizes transforming growth factor-β-stimulated clone-22 (TSC-22), a pro-apoptotic molecule, and protects cells against apoptosis (30). However, it remains unclear whether the direct interaction between fortilin and TSC-22 is required for the destabilization of TSC-22.
Fortilin is localized in not only the cytosol but also in the nucleus (13). The putative mechanisms described above by which fortilin functions anti-apoptotically do not necessarily explain the nuclear presence of fortilin. We speculated that fortilin might interact with one of the nuclear pro-apoptotic molecules and block its pathway, leading to apoptosis. Because p53 rapidly accumulates in the nucleus in response to DNA damage (31) and induces apoptosis through the transcriptional activation of pro-apoptotic genes including Bax (32), Noxa (3), and PUMA (4), we asked whether fortilin physically interacted with p53 and blocked the pro-apoptotic activity of p53. The data presented here support that fortilin specifically binds p53 and blocks the p53-induced transcriptional activation of Bax and resultant apoptosis. We propose that fortilin is a novel negative regulator of p53-mediated apoptosis.
EXPERIMENTAL PROCEDURES
The detailed experimental procedures are found in the supplemental Experimental Procedures.
Cell Culture and Cell Lines
All cell lines were maintained in high glucose Dulbecco's modified Eagle's medium (DMEM) and supplemented with 10% fetal bovine serum (FBS) at 37 °C in an atmosphere containing 5% CO2.
Biochemical Characterization of Fortilin-p53 Interaction
Western blot analyses (13), in vitro glutathione S-transferase (GST) pulldown assays (33), in vivo immunoprecipitation and Western blot analysis (22, 34), the immunocytochemical co-localization of fortilin and p53 (34), and the docking study (35) were performed as described previously.
Functional Characterization of Fortilin-p53 Interaction
The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (19), DNA fragmentation assay (19), and caspase 3 activation assay (13) were performed as described previously. Real-time quantitative reverse transcription-PCR, ELISA, and electrophoretic mobility shift assay (EMSA) were performed as described by us previously (36).
Nude Mouse Tumor Xenograft Assays
All mouse experiments were performed under the approved Institutional Animal Care and Use Committee (IACUC) as described previously (37).
Statistical Analyses
The degree of the spread of data was expressed by ± S.D. p < 0.05 was considered to be statistically significant.
RESULTS
Fortilin Specifically Interacts with p53
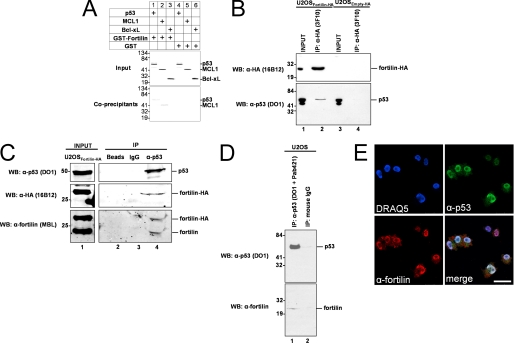
To test whether fortilin interacts with p53, we performed a standard GST pulldown assay, mixing [35S]methionine-labeled p53, MCL1 (known to interact with fortilin) or Bcl-xL (control) in separate reaction buffers containing either GST-fortilin or GST alone. MCL1 was co-precipitated by GST-fortilin (Fig. 1A, lane 2, Co-precipitants), but not by GST alone (lane 5, Co-precipitants). Bcl-xL was not co-precipitated by either GST-fortilin or GST alone (Fig. 1A, lanes 3 and 6, Co-precipitants). In this system, p53 was co-precipitated by GST-fortilin (Fig. 1A, lane 1, Co-precipitants), but not by GST alone (lane 4, Co-precipitants), suggesting the presence of specific interaction between purified fortilin and p53 in vitro.
FIGURE 1.
Fortilin specifically interacts with p53, a tumor suppressor protein. A, in vitro co-precipitation of p53 by fortilin in GST pulldown assay. Input, autoradiography of the 1/20 portion of binding mixture; Co-precipitants, 35S-labeled proteins co-precipitated by pulled down GST-fortilin or GST only. B, in vivo co-immunoprecipitation of native p53 by overexpressed HA-tagged fortilin using rat anti-HA antibody (3F10) and magnetic beads coated with anti-rat antibody. U2OSfortilin-HA, U2OS cells stably overexpressing HA-tagged fortilin; U2OSempty-HA, U2OS cells stably overexpressing human influenza HA tag; upper panel, Western blot (WB) analysis of HA-fortilin in the input and immunoprecipitated (IP) fractions; lower panels, Western blot analysis of p53 contained in input and co-immunoprecipitated fractions by DO1 anti-p53 antibody. C, in vivo reverse co-immunoprecipitation of native and HA-tagged fortilin by native p53 followed by Western blot analysis of p53 (top panel), HA tag (middle panel), and fortilin (bottom panel) in the input and immunoprecipitated fractions. D, in vivo reverse co-immunoprecipitation of native fortilin by native p53. Upper panel, Western blot analysis of immunoprecipitated p53; lower panel, Western blot analysis of co-immunoprecipitated fortilin. E, confocal co-localization of p53 with fortilin. UV-irradiated HeLa cells were stained with DRAQ5, anti-p53 (DO-1), and anti-fortilin (MBL International) antibodies. merge, a merged image of the above three images. Size bar = 50 μm.
U2OS cells harbor wild-type p53 (38). To validate the interaction between fortilin and p53 in vivo, we generated U2OS cells overexpressing hemagglutinin (HA)-tagged fortilin (U2OSfortilin-HA) or HA tag alone (U2OSEmpty-HA) and subjected the cleared total cell lysates from these cells to immunoprecipitation using anti-HA antibody (Fig. 1B, top panel, lanes 1 and 3). HA-tagged fortilin was successfully immunoprecipitated from the lysate from U2OSfortilin-HA, but not from U2OSEmpty-HA (Fig. 1B, top panel, lanes 2 and 4). In this system, the p53 signal was detectable only in the presence of successfully immunoprecipitated fortilin-HA (Fig. 1B, bottom panel, lanes 2 and 4). We then repeated the same experiment using a different anti-HA antibody preparation (rat 3F10 primary antibody and anti-rat IgG magnetic beads for Fig. 1B; 3F10 matrix beads for supplemental Fig. S1). In this system, the anti-HA antibody successfully immunoprecipitated fortilin-HA (supplemental Fig. S1, top panel, lane 4). The p53 signal was detectable in the presence of fortilin-HA (supplemental Fig. S1, middle and bottom panels, lane 4), but not in its absence (supplemental Fig. S1, middle and bottom panels, lane 3).
We then performed a reverse in vivo co-immunoprecipitation assay by equally dividing the cleared total cell lysates from U2OSfortilin-HA cells into three microcentrifuge tubes and incubating them with the bare agarose beads, beads coated with normal mouse IgG, or beads coated with anti-p53 antibody (FL-393AC). Beads coated with anti-p53 antibody, but not other types of beads, successfully immunoprecipitated native p53 (Fig. 1C, top panel, lane 4 versus lanes 2 and 3). In this system, both fortilin-HA (Fig. 1C, middle and bottom panels, lane 4) and native fortilin (Fig. 1C, bottom panel, lane 4) were co-immunoprecipitated by p53.
Finally, we performed the same in vivo reverse co-immunoprecipitation assay on cells expressing only native fortilin and p53. The equally divided aliquots of the cleared total cell lysates from wild-type U2OS cells were treated with either a mixture of DO1 and Pab421 antibodies or control mouse normal IgG. Native p53 was successfully immunoprecipitated by anti-p53 antibodies, but not by control IgG (Fig. 1D, top panel, lanes 1 and 2). In this system, the signal of native fortilin was detectable only in the presence of immunoprecipitated p53 (Fig. 1D, bottom panel, lane 1), but not in its absence (lane 2).
These data (Fig. 1, A–D) consisting of in vitro pulldown assays and in vivo forward and reverse immunoprecipitation Western blot assays, clearly suggest that fortilin specifically interacts with p53. To evaluate whether ultraviolet (UV) irradiation and resultant DNA damage affect the intensity of the fortilin-p53 interaction, we UV-irradiated U2OSfortilin-HA cells, immunoprecipitated HA-tagged fortilin, and evaluated the amount of p53 co-immunoprecipitated by fortilin-HA. UV irradiation increased p53 expression in a dose-dependent fashion (supplemental Fig. S2A, Input). More p53 was co-immunoprecipitated by fortilin-HA in the presence of mild to moderate UV irradiation (5–40 mJ/cm2), suggesting that UV irradiation leads to more fortilin-p53 interaction (supplemental Fig. S2, A and B). Further investigation is necessary to determine whether this is due to increased affinity between fortilin and p53 (such as through the post-translational modification of fortilin and/or p53) or increased availability of fortilin and/or p53 for each other.
To evaluate the spatial localization of fortilin in relation to that of p53, UV-irradiated HeLa cells were immunostained using anti-p53 (DO1) and anti-fortilin (MBL International) antibodies. Bound antibodies were detected by anti-mouse Alexa Fluor 488 and anti-rabbit rhodamine red-X secondary antibodies, respectively. The p53 signal was mostly localized in the nucleus, whereas the fortilin signal was present in both the nucleus and the cytosol, although a higher amount was found in the nucleus (Fig. 1E, DRAQ5, α-p53, and α-fortilin). The merged image suggests that the two molecules co-localize in the nucleus, but not in the nucleoli or cytosol (Fig. 1E, merge), suggesting that fortilin interacts with p53 in the nucleus.
Both N Terminus and C Terminus Portions of Fortilin Are Crucial for Its Interaction with p53
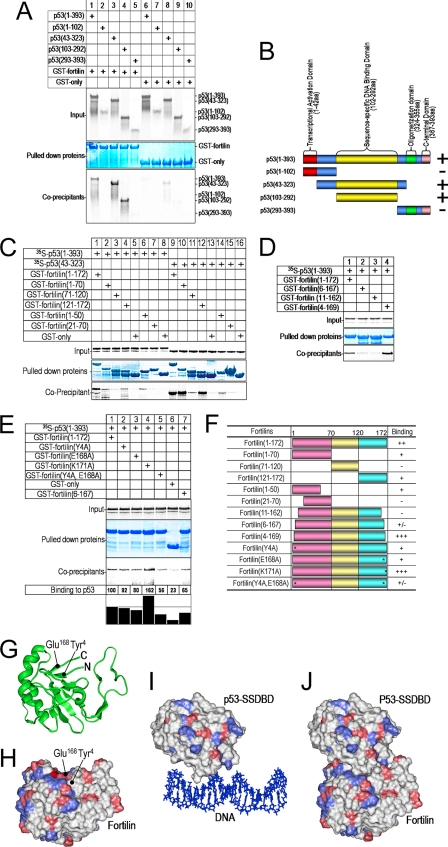
p53 is a 393-amino acid protein consisting of four major domains (Fig. 2B): 1) transcriptional activation domain (amino acids 1–42); 2) sequence-specific DNA binding domain (SSDBD, amino acids 102–292); 3) oligomerization domain (amino acids 324–355); and 4) C-terminal domain (amino acids 367–393) (39). Fig. 2, A and B, summarize an experiment in which we incubated the in vitro-translated wild-type p53 and its four deletion mutants in individual mixtures with either GST alone or GST-fortilin. Although GST alone failed to pull down wild-type or mutant p53 proteins (Fig. 2A, lanes 6–10), GST-fortilin pulled down both wild-type p53(1–393) and two mutants p53(43–323) and p53(103–292), but not p53(1–102) or p53(293–393). This suggests that fortilin binds to the SSDBD of p53 (Fig. 2, A and B). p53(1–102) migrated more slowly than p53(103–292), presumably because the rigidity of the proline-rich region between amino acid residues 50 and 85 of p53 contributed to anomalously slow migration in SDS-PAGE as reported previously (40). These data are also consistent with the in vivo data that fortilin interacted with wild-type p53 in U2OS cells, but not with a mutated p53 that contained only the half of the SSDBD in NCI-H1793 cells (supplemental Fig. S3).
FIGURE 2.
Fortilin binds the sequence-specific DNA binding domain through its N and C terminus ends. A, in vitro co-precipitation of p53 deletion mutants by fortilin in GST pulldown assay. Input, autoradiography of the 1/20 portion of binding mixture; Pulled down proteins, Coomassie Blue staining of GST-fusion proteins pulled-down by GST-Sepharose beads; Co-precipitants, 35S-labeled proteins co-precipitated by pulled down GST-fortilin or GST only. B, summary of interaction between fortilin and the wild-type and deletion mutants of p53. aa, amino acids. C and D, GST pulldown assay of fortilin deletion mutants. p53(1–393), full-length p53; p53(43–323), the SSDBD of p53. E, GST pulldown assay of fortilin point mutants. Binding to p53, a densitometric analysis of signal intensity of co-precipitants. F, identification of a fortilin double point mutant that lacks binding to p53. *, point mutation; −, no p53 binding; +/−, minimum p53 binding; +, decreased p53 binding; ++, p53 binding similar to wild-type fortilin; +++, greater p53 binding. The same GST pulldown assay was performed at least three times, and the data were found to be consistent among these experiments. G, the proximity of N and C terminus ends of fortilin to each other. H, the spatial representation of Tyr4 and Glu168 on the surface of fortilin molecule. Positively and negatively charged amino acid residues are colored blue and red, respectively. I, the SSDBD of p53 bound to DNA. J, docking of fortilin to the DNA binding surface of p53.
Fortilin comprises three domains with distinct hydrophilicities: domain 1 (amino acids 1–70), domain 2 (amino acids 71–120), and domain 3 (amino acids 121–172) (Fig. 2F) (13). To identify the regions of fortilin important for p53 binding, we first performed GST pulldown assays on deletion mutants of fortilin. Domains 1 and 3 bound p53 (Fig. 2, C, lanes 2 and 4, and F), but domain 2 did not (lane 3). Within domain 1, the N terminus 20 amino acids were required for p53 binding (Fig. 2, C, lanes 6 and 7, and F). p53(43–323), the SSDBD of p53, bound more strongly to fortilin and its mutants than did wild-type p53 (Fig. 2C, lanes 9–16 versus lanes 1–8).
To further evaluate the importance of the N and C termini of fortilin, we generated fortilin(11–162), fortilin(6–167), and fortilin(4–169). We subjected them to the same GST pulldown assay. Wild-type fortilin and fortilin(4–169) (Fig. 2D, lanes 1 and 4) bound p53. However, fortilin(6–167) in lane 2 or fortilin(11–162) in lane 3 did not bind p53, suggesting that the 4th and 5th amino acids from both ends of fortilin, but not the 1st through 3rd amino acids of fortilin, were critical for p53 binding (Fig. 2F).
We then mutated charged amino acids in these regions, generating fortilin(Y4A), fortilin(K171A), fortilin(E168A), and fortilin(Y4A,E168A). We subjected these fortilin mutants to the GST pulldown assay. Fortilin(K171A) bound more strongly to p53 than did wild-type fortilin (Fig. 2E, lane 4), a result consistent with our observation of fortilin(4–169) (Fig. 2D, lane 4). In addition, the binding of fortilin(Y4A) and fortilin(E168A) to p53 was weaker than that of wild-type fortilin (lanes 2 and 3 versus lane 1). Remarkably, fortilin(Y4A,E168A) showed little ability to bind p53 in this system (Fig. 2, E, lane 5, and F). Far-UV CD spectropolarimetric analyses (41, 42) showed that the secondary structures of these fortilin mutants are preserved (supplemental Fig. S4). Based on an analysis using published human fortilin NMR data (Protein Data Bank (PDB) ID: 2HR9), the 4th tyrosine (Tyr4) and 168th glutamate (Glu168) are located close to each other (Fig. 2G) on the surface of the fortilin molecule (Fig. 2H). An energy minimization procedure between the SSDBD of p53 (PDB ID: 1TUP, Fig. 2I) and the area of fortilin containing Tyr4 and Glu168 (Fig. 2H) yielded a very low intermolecular energy at −797 kcal/mol. This suggests highly stable binding between p53 and fortilin through the area containing Tyr4 and Glu168 (Fig. 2J).
Fortilin Blocks p53-dependent Apoptosis by Preventing the Transcriptional Activation of Bax Gene by p53
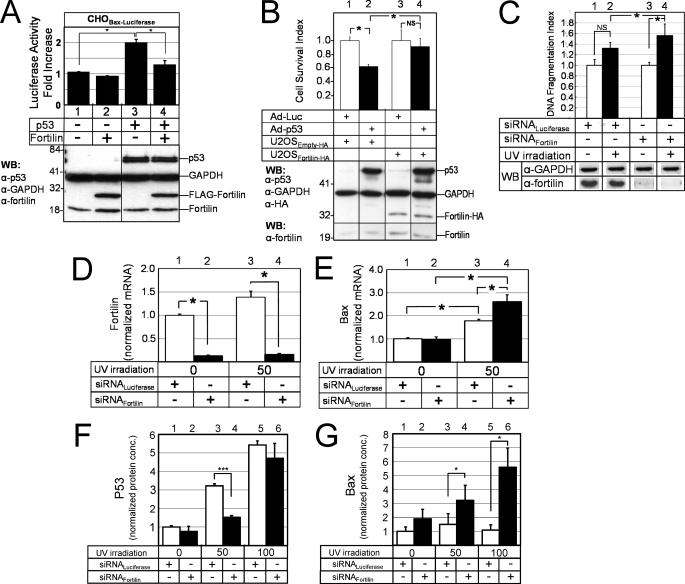
To evaluate the role of fortilin in the transcriptional activation of p53 target genes, we developed a unique luciferase assay system in which each CHO cell stably harbors a single copy of luciferase reporter plasmid containing the p53-responsive element (RE) of Bax gene (CHOBax-luciferase) (32). We transiently transfected the CHOBax-luciferase cells with p53 (or control) and with fortilin (or control) mammalian expression plasmids. Western blot analyses confirmed adequate expression of both p53 and fortilin proteins (Fig. 3A, WB). Background luciferase activity measured in CHOBax-luciferase cells was low in the absence of p53 either with or without fortilin overexpression (Fig. 3A, lanes 1 and 2, respectively). When p53 was overexpressed, luciferase activity increased more than 2-fold (lane 3), suggesting that p53 appropriately activated the p53-RE of the Bax promoter. In this system, co-overexpression of fortilin significantly reduced the activation by p53 of the p53-RE of the Bax promoter. (Fig. 3A, lane 4 versus lane 3), suggesting that fortilin blocked p53-induced transcriptional activation of Bax gene.
FIGURE 3.
Fortilin prevents p53 from transcriptionally activating Bax and blocks p53-dependent apoptosis. A, the blockade by fortilin of the transcriptional activation of Bax gene in luciferase assays (n = 3). CHOBax-Luciferase, CHO cells with the p53-RE of the Bax gene fused to the luciferase gene. WB, Western blot analysis. B, the inhibition by fortilin of Ad-p53-induced cell death in U2OS cells (n = 6) by the MTT assay. NS, not statistically significant. C, the protection by fortilin against UV-induced DNA fragmentation in U2OS cells (n = 3). siRNALuciferase, small interfering RNA against luciferase; siRNAFortilin, siRNA against fortilin. D and E, quantitative, real-time RT-PCR assay of fortilin (D) and Bax (E) message in UV-irradiated U2OS cells treated with either siRNALuciferase or siRNAFortilin (n = 4). F and G, ELISA of p53 (F) and Bax (G) protein concentrations (conc.) in UV-irradiated U2OS cells treated with either siRNALuciferase or siRNAFortilin (n = 4). *, p < 0.05; ***, p < 0.001.
To evaluate whether fortilin inhibited p53-induced cell death, we transduced an adenoviral vector that encoded p53 (Ad-p53) or luciferase (Ad-Luc) into U2OS cells stably overexpressing HA tag (U2OSEmpty-HA, control) or fortilin-HA (U2OSfortilin-HA). We assessed the survival of these cells using the MTT assay (19). The survival of U2OSEmpty cells significantly decreased when p53 was overexpressed by Ad-p53 (Fig. 3B, lanes 1 and 2). Strikingly, however, the survival of cells that overexpress fortilin was not affected by p53 overexpression (Fig. 3B, lanes 3 and 4). These data suggest that fortilin inhibited p53-induced cell death.
We next compared the susceptibilities of U2OS cells treated with siRNA against fortilin (siRNAFortilin) or siRNALuciferase to UV irradiation (43). In the presence of fortilin, UV irradiation did not significantly increase the DNA fragmentation of U2OS cells (Fig. 3C, lanes 1 and 2). In the absence of fortilin, however, UV irradiation drastically increased the DNA fragmentation (Fig. 3C, lanes 3 and 4), suggesting that fortilin protected the cells against UV-induced apoptosis.
Next, we quantified the Bax message levels within the UV-irradiated U2OS cells that were treated with either siRNAFortilin or siRNALuciferase, using a quantitative real-time RT-PCR assay. As expected, siRNAFortilin, but not siRNALuciferase, drastically decreased fortilin message levels in both UV-irradiated and control U2OS cells (Fig. 3D, lanes 2 and 4 versus lanes 1 and 3, respectively). In that system, Bax message levels were significantly higher in siRNAFortilin-treated cells than in siRNALuciferase-treated cells (Fig. 3E, lanes 3 versus 4), suggesting that the lack of fortilin led to the greater transcriptional activation of Bax gene in response to UV irradiation.
Next, we quantified p53 and Bax protein levels in response to UV irradiation using ELISA, in the presence and absence of fortilin. The intracellular p53 concentrations drastically increased as UV doses increased (Fig. 3F). In that system, Bax protein concentrations were significantly higher in siRNAFortilin-treated cells than in siRNALuciferase-treated cells, at both the 50-mJ/cm2 and the 100-mJ/cm2 UV doses (Fig. 3G), suggesting that fortilin prevented UV-induced p53 from increasing intracellular Bax concentrations. Considered together, these data suggest that fortilin prevents p53 from (a) transcriptionally activating Bax gene (Fig. 3, A, E, and G) and (b) inducing p53-dependent apoptosis in U2OS cells (Fig. 3, B and C).
To test whether the above observation is true in a different cell line, we stably knocked down p53 and/or fortilin using a lentiviral shRNA system in PMJ2-PC cells, a peritoneal macrophage cell line, and generated the four polyclonal cell lines, PMJ2-PCp53+fortilin+, PMJ2-PCp53+fortilin−, PMJ2-PCp53-fortilin+, and PMJ2-PCp53-fortilin− (supplemental Fig. S5A). These cell lines were subjected to UV irradiation, and the degree of DNA fragmentation and caspase 3 activity was quantified. Upon UV irradiation, all four cell lines exhibited a significant increase in DNA fragmentation (supplemental Fig. S5B, lanes 2, 4, 6, and 8 in comparison with supplemental Fig. S5B, lanes 1, 3, 5, and 7) and caspase 3 activities (supplemental Fig. S5C, lanes 2, 4, 6, and 8 in comparison with supplemental Fig. S5C, lanes 1, 3, 5, and 7). Strikingly, the lack of fortilin was associated with an ∼6-fold increase in DNA fragmentation (supplemental Fig. S5B, lane 2 versus lane 4) and 2.5-fold increase in caspase activities (supplemental Fig. S5C, lane 2 versus lane 4), only in the presence of p53 (supplemental Fig. S5, B and C, lanes 2 and 4) but not in the absence of p53 (supplemental Fig. S5, B and C, lanes 6 and 8). In other words, fortilin failed to protect cells against either DNA fragmentation or caspase 3 activation in the absence of p53 (supplemental Fig. S5, B and C, lanes 6 and 8). These data suggest that fortilin specifically blocks UV-induced, p53-dependent apoptosis through Bax.
The Binding of Fortilin to p53 Is Required for Fortilin to Prevent p53-dependent Apoptosis
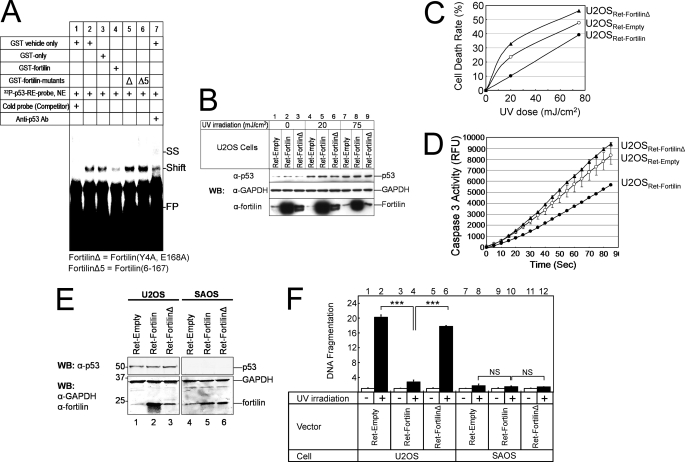
To evaluate whether fortilin is required to bind p53 to block the ability of p53 to function as a transcriptional activator, we developed an EMSA (36). In this study, we incubated p53-rich nuclear extracts from U2OS cells transduced by adenoviral vector containing p53 cDNA and 32P-labeled oligonucleotides representing the p53-RE (32P-p53-RE-probe) with one of the following before gel electrophoresis and autoradiography: vehicle, GST, GST-fortilin, GST-fortilin(Y4A,E168A) (Δ), or GST-fortilin(6–167) (Δ5). The 32P-p53-RE-probe and nuclear extracts of p53 combined generated a band (Fig. 4A, lane 2, Shift), which vanished in the presence of competitor or cold p53-RE-probe (lane 1). The addition of anti-p53 antibody caused formation of a probe-p53-antibody complex, which migrated even more slowly (lane 7, supershift (SS)). Here, GST-fortilin, but not GST alone, significantly reduced p53 binding to the 32P-p53-RE-probe (lane 3 versus lane 4). Strikingly, fortilin, but neither fortilin(Y4A,E168A) (Fig. 4A, lane 5, Δ) nor fortilin(6–167) (Fig. 4A, lane 6, Δ5), prevented p53 from binding to the 32P-p53-RE-probe. This suggests that fortilin is required to bind p53 to disrupt p53 binding to its target DNA sequence.
FIGURE 4.
Fortilin must bind p53 to block p53-dependent apoptosis. A, physical antagonism by fortilin of the binding of p53 to its consensus sequence in EMSA. Cold probe, p53-RE oligonucleotides without radiolabeling; Ab, antibody; FP, free probe; SS, supershift; p53-RE, a p53-responsive element; NE, nuclear extract; Δ, fortilin(Y4A,E168A) or fortilinΔ; Δ5 = fortilin(6–167). B, characterization of U2OSRet-Empty, U2OSRet-Fortilin, and U2OSRet-FortilinΔ cells. C, MTT cell death assay of UV-irradiated U2OSRet-Empty, U2OSRet-Fortilin, and U2OSRet-FortilinΔ cells (n = 3). Analysis of variance shows that all three curves are statistically significantly different from each other. D, caspase 3 activity assay of UV-irradiated U2OSRet-Empty, U2OSRet-Fortilin, and U2OSRet-FortilinΔ cells (n = 2). E, characterization of SAOSRet-Empty, SAOSRet-Fortilin, and SAOSRet-FortilinΔ cells. F, DNA fragmentation assay of UV-irradiated U2OS and SAOS cells. ***, p < 0.001 (n = 4).
Next, we infected U2OS cells with retroviral vector containing wild-type fortilin (Ret-fortilin), fortilin(Y4A,E168A) (Ret-fortilinΔ), or empty vector (Ret-empty, control). We then subjected them to UV radiation and performed both MTT and caspase 3 activity assays. Neither Ret-fortilin nor Ret-fortilinΔ contained epitope tags. Western blot analysis confirmed that both U2OSRet-fortilin and U2OSRet-fortilinΔ expressed more fortilins than did U2OSRet-Empty and that UV irradiation increased p53 expression in all three cell lines (Fig. 4B). In this system, the cell death rate was significantly lower for U2OSRet-fortilin cells than for U2OSRet-empty cells (Fig. 4C, p < 0.05 by analysis of variance). Remarkably, U2OSRet-fortilinΔ cells were more susceptible to UV irradiation than were U2OSRet-fortilin or U2OSRet-empty cells (Fig. 4C, p < 0.05 by analysis of variance). Consistently, U2OSRet-fortilinΔ cells had more caspase 3 activation than did U2OSRet-empty cells, whose caspase activity surpassed that of U2OSRet-fortilin (Fig. 4D, p < 0.05 by analysis of variance). These results, when taken together, suggest that fortilin is required to bind to p53 to block p53-mediated cell death (Fig. 4, C and D).
U2OS cells harbor wild-type p53 (38), whereas SAOS cells do not express p53 (44), offering a unique opportunity to evaluate the role of p53 and fortilin in UV-induced apoptosis. We first generated and characterized SAOSRet-Empty, SAOSRet-Fortilin, and SAOSRet-FortilinΔ (Fig. 4E) and subjected them and the three U2OS counterparts to UV irradiation and the DNA fragmentation assay. As expected, fortilin, but not fortilinΔ, protected U2OS cells against UV-induced DNA fragmentation (Fig. 4F, lanes 2, 4, and 6). Remarkably, however, fortilin did not protect SAOS cells any better than did fortilinΔ against UV-induced DNA fragmentation (Fig. 4F, lanes 8, 10, and 12). These results suggest that fortilin protected cells more than fortilinΔ only in the presence of wild-type p53 (Fig. 4F, lane 4 versus lane 6 and lane 10 versus lane 12). Also shown was that UV irradiation induced apoptosis predominantly through p53-dependent pathways in this system (Fig. 4F, lane 2 versus lane 8). Together, these data suggest that fortilin is required to bind p53 to block p53-dependent apoptosis.
Fortilin Negates the Tumor-suppressing Effects of p53 through Its Binding to p53 in Growing Tumors in Nude Mice
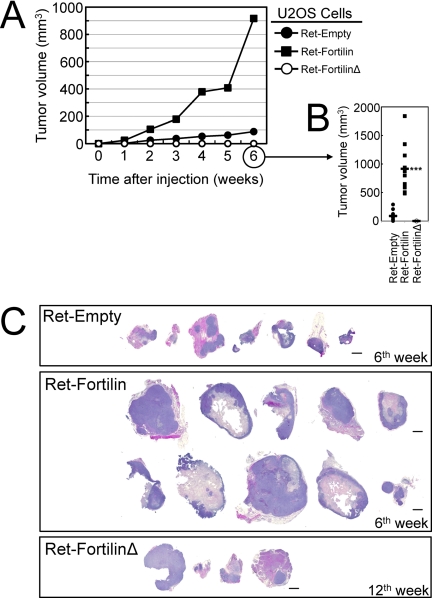
To determine whether fortilin facilitates tumor growth through its binding and inhibition of p53, we injected USOS and SAOS cells containing Ret-Empty, Ret-fortilin, or Ret-fortilinΔ into nude mice and followed the tumor growth (n = 10 each). All 10 nude mice injected with U2OSRet-fortilin and six injected with U2OSRet-Empty formed tumors at the 6th week where U2OSRet-fortilin formed larger tumors than did U2OSRet-Empty (Fig. 5, A–C). Strikingly, U2OSRet-fortilinΔ did not form any tumors at the 6th week (Fig. 5, A and B) with only four animals showing detectable tumors at the 12th week (Fig. 5C). None of the nude mice injected with SAOSRet-Empty, SAOSRet-Fortilin, or SAOSRet-FortilinΔ cells in Matrigel developed significant tumors at the 6th week, thus preventing us from assessing the effect of fortilin in tumor growth in the p53-null environment of SAOS at this time point (data not shown). At the 15th week, however, five, three, and four nude mice injected with SAOSRet-Empty, SAOSRet-Fortilin, and SAOSRet-FortilinΔ cells, respectively, exhibited significant tumors. The average tumor volumes from SAOSRet-Empty, SAOSRet-Fortilin, or SAOSRet-FortilinΔ cells were 235 ± 17.1, 23.3 ± 40.7, and 95.8 ± 154.9 mm3, respectively. Although the small sample size (n = 12 total) precluded an adequate statistical comparison on the tumor volumes among these three groups, these tumor materials were sufficient for and thus subjected to immunohistochemical evaluation as described below in Fig. 6. In summary, the U2OS cell data above (Fig. 5) strongly suggest that fortilin facilitates tumor growth through its interaction with and inhibition of p53.
FIGURE 5.
Tumor xenograft assays. A and B, tumor xenograft assay using U2OSRet-Empty, U2OSRet-Fortilin, and U2OSRet-FortilinΔ cells (n = 10). C, hematoxylin-eosin (H&E) staining of tumor xenografts harvested at the 6th week (U2OSRet-Empty and U2OSRet-Fortilin) and at the 12th week (U2OSRet-FortilinΔ). Size bar = 1 mm.
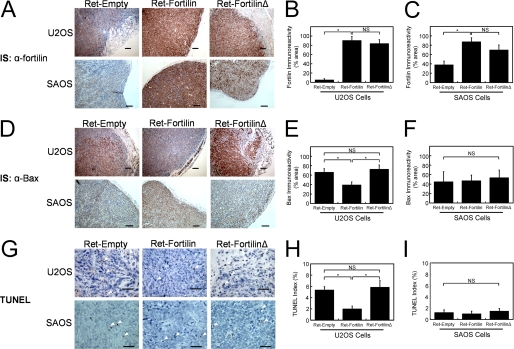
FIGURE 6.
Suppression of the induction of Bax and apoptosis by fortilin, but not by fortilinΔ. A–C, immunostaining of tumor xenografts by anti-fortilin antibody. IS, immunostaining; NS, not statistically significant. D–F, immunostaining of tumor xenografts by anti-Bax antibody (D) shows the inhibition of the induction of Bax gene by wild-type fortilin but not by fortilinΔ in the presence of wild-type p53 (E, U2OS). However, in the absence of wild-type p53, neither wild-type fortilin nor fortilin-Δ inhibited the induction of Bax. (F, SAOS). G–I, TUNEL staining of tumor xenografts (G) shows the inhibition of apoptosis by wild-type fortilin, but not by fortilinΔ in the presence of wild-type p53 (H). Again, in the absence of wild-type p53, neither wild-type fortilin nor fortilin-Δ inhibited apoptosis in the tumor (I). Immunoreactivity was quantified using SigmaScan Pro5 and expressed at the percentage of the diaminobenzidine-positive area within the region of interest. TUNEL indices were calculated at the number of TUNEL-positive cells divided by the number of total cells counted. Size Bars = 100 μm (A and D) and 50 μm (G). *, p < 0.05. The following numbers of tumor samples were harvested from the nude mice injected with U2OSRet-Empty, U2OSRet-fortilin, U2OSRet-fortilinΔ, SAOSRet-Empty, SAOSRet-Fortilin, and SAOSRet-FortilinΔ cells, respectively: n = 7, 10, 4, 5, 3, and 4.
Fortilin Decreases Bax Expression and Apoptosis in p53-positive Tumors
To test whether the presence of fortilin interfered with the ability of p53 to induce Bax and apoptosis within the growing tumors in the nude mouse model above, we immunostained the tissue with anti-fortilin (α-fortilin) and anti-Bax (α-Bax). We also performed TUNEL staining on these samples. Twelve (12) tumor samples from the nude mice injected with SAOS cells were also evaluated. Both U2OS and SAOS cells infected with Ret-fortilin and Ret-fortilinΔ showed the robust expression of these constructs in comparison with cells infected with the control vector (Ret-Empty) (Fig. 6, A–C). In this system, the immunoreactivity of Bax was significantly less in tumors from U2OSRet-fortilin than those from U2OSRet-fortilinΔ (Fig. 6, D, top panel, and E), whereas the immunoreactivity of Bax was identical among all the tumors from SAOSRet-Empty, SAOSRet-fortilin, and SAOSRet-fortilinΔ (Fig. 6D, bottom panel, and F). In addition, U2OSRet-fortilin, but not U2OSRet-fortilinΔ, had a significantly lower TUNEL index than did U2OSRet-Empty (Fig. 6, G, top panel, and H). Intriguingly, there was no difference in TUNEL indices of SAOSRet-Empty, SAOSRet-fortilin, and SAOSRet-fortilinΔ (Fig. 6, G, bottom panel, and I), which were uniformly lower than those of the U2OS cells. These data, when taken together, suggest that fortilin facilitates tumor growth by binding p53 and blocking its ability to induce Bax and apoptosis.
DISCUSSION
The current study for the first time provides a clear mechanistic insight as to exactly how fortilin protects cells against apoptosis. This is because we show here that fortilin, a unique anti-apoptotic molecule that does not resemble either Bcl-2 or inhibitor of apoptosis (IAP) family member proteins (13), specifically interacts with p53 (Fig. 1, A–E), prevents p53 from transcriptionally activating Bax gene (Figs. 3, A, E, and G, and 6, D and E) and inducing apoptosis (Fig. 3, B, C, supplemental Fig. S5, B and C), and sustains tumor growth in a whole animal (Fig. 5, A–C) by suppressing p53-dependent Bax induction and resultant apoptosis (Fig. 6, D–I). Intriguingly, in the absence of wild-type p53, fortilinΔ, which lacks p53 binding, behaved in the same fashion as wild-type fortilin, failing to protect cells from Bax induction or apoptosis (Figs. 4F and 6, D, F, G, and I), suggesting that fortilin is minimally involved in the non-p53 pathways. Immunocytochemical staining shows that the fortilin-p53 interaction likely takes place in the nucleus (Fig. 1E). The detailed analyses on deletion and point mutants of fortilin and p53 showed that the N and C terminus ends of fortilin participate in the binding to the SSDBD of p53 (Fig. 2, A–J). The ability of fortilin to inhibit the binding of p53 to its consensus sequence (Fig. 4A) and to block p53-mediated apoptosis was dependent on its binding to p53 (Fig. 4, B–F). We thus propose that fortilin is a key negative regulator of the p53-Bax apoptosis pathway. In a supplemental experiment, we evaluated the ability of fortilin to prevent p53 from inducing PUMA (4) and Noxa (3), p53-inducible genes. We found that fortilin inhibits the induction by p53 of Noxa, but not PUMA (supplemental Fig. S6). A possible differential inhibition by fortilin of p53 target genes, Bax and Noxa versus PUMA, is currently under investigation in our laboratory. In the final stage of our manuscript preparation, we were made aware of a study published on the interaction between fortilin and p53 where the authors used fortilin as bait, employed the yeast two-hybrid screening system, and identified a portion of p53 as a fortilin interacting molecule (45). Our work confirms the interaction and provides the totally new mechanistic insight as to how fortilin protects cells against p53-mediated apoptosis at all three levels, in vitro, in vivo, and whole animal.
p53, the guardian of the genome (46), is extensively regulated. In addition to the regulation by a number of post-translational modifications, methylation, acetylation, phosphorylation, ADP-ribosylation, ubiquitination, sumoylation, neddylation, and glycosylation (47), p53 is regulated by its physical association with p53 regulatory molecules. Although there have been over a dozen p53-interacting molecules reported in the literature, only five gene products are known to bind the SSDBD of p53 aside from fortilin. First, both 53BP1 and 53BP2, 1977 and 1134 amino acid polypeptides, respectively, bind the SSDBD (48) and stimulate p53-mediated transcriptional activation (49), playing a critical role in ionizing radiation-induced cell cycle arrest (50). Secondly, hematopoietic zing finger (Hzf), transcriptionally induced by p53, binds the SSDBD. This binding results in preferential transactivation of pro-arrest p53 target gene (p21) over its pro-apoptotic target genes (Bax) (51). Thirdly, Brn-3a, a Pit-Oct-Unc (POU) family transcriptional factor, binds the SSDBD of p53 and prevents p53 from transcriptionally activating Bax while facilitating p53 to induce p21CIP1/WAF1 (52, 53). Finally, apoptosis stimulating protein of p53 (ASPP) a protein homologous to 53BP2, also binds the SSDBD and enhances p53-induced apoptosis, but not cell cycle arrest (54). Further, fortilin is one of only three molecules that bind to p53 SSDBD and block Bax gene activation (other two are Hzf and Brn-3a). Notably, fortilin does not structurally resemble either Hzf or Brn-3a. Among the other p53 binding molecules, both Mdm2 (55) and Mdmx (56) bind the N terminus transactivation domains of p53, whereas YB-1 (57) and ATM and p53-associated KZNF protein (Apak) (58) both bind the C terminus of p53.
p53 is designed to play two important, but fundamentally opposing, roles in response to stress (59). p53, under the low, constitutive levels of DNA damage, induces cell cycle arrest through the transcriptional activation of p21CIP1/WAF1, allowing the cells to repair themselves. Upon the higher levels of DNA damage, on the contrary, p53 switches from promoting survival and repair to the induction of apoptosis (59, 60), allowing the organism to eliminate hopelessly damaged cells from itself. The exact mechanism by which p53 differentially activates the cell cycle regulatory versus the apoptosis pathway is unknown, although the partial and total acetylation of p53 by histone acetyltransferases may be involved in the activation of the cell cycle regulatory and pro-apoptotic genes, respectively (47). In addition, the molecules that bind the SSDBD of p53, such as Hzf (51), Brn-3a (52, 53), and ASPP (54), are shown to differentially regulate cell cycle progression and apoptosis. Although it is tempting to speculate that fortilin, an SSDBD binding molecule, also differentially regulates cell cycle progression and apoptosis, the role of fortilin in the regulation by p53 of cell cycle progression is currently unknown and under investigation.
Yang et al. (15) reported that fortilin interacts with Bcl-xL using both GST pulldown and co-immunoprecipitation assays. However, in our current assay (Fig. 1A) as well our previous work (22), fortilin binds MCL1, but not Bcl-xL. It is likely that fortilin interacts with p53 and MCL1 more strongly than with Bcl-xL and that our wash condition was too stringent to retain the interaction between fortilin and Bcl-xL.
The binding of Mdm2 to p53 (55) results in ubiquitination and proteasome-mediated degradation of p53, making the half-life of p53 very short (61–63) and intracellular levels of p53 very low. Cell stress such as DNA damage decreases the degree of sumoylation of Mdm2 and increases Mdm2 degradation, leading to the stabilization of p53 (64). Unlike Mdm2, neither fortilin overexpression nor depletion showed any consistent or detectable changes in the p53 levels by either Western blots or ELISA (Figs. 3, A, B, and F, and 4, B and E and supplemental Fig. S5A). The fortilin double point mutant identified in the current study (fortilinΔ) may prove to be a viable reagent as the role of fortilin in the stability of p53 is further investigated.
Our discovery of the physical and functional interaction between fortilin and p53 has significant clinical implications. First, targeting of the fortilin-p53 interaction by small molecules may result in the reactivation of p53 and the induction of apoptosis within cancer cells that harbor wild-type p53, although such strategies may not work in cancers that harbor a mutated p53. Such anti-fortilin small molecules may also be useful in further improving the response of cancer cells to chemotherapeutic agents, ionizing radiation, and Nutlin-3A (65) and MI-219 (66), small molecules that disrupt the p53-Mdm2 interaction. In addition, such anti-fortilin small molecules may be useful in the prevention and treatment of atherosclerosis refractory to 3-hydroxy-3-methyl-glutaryl-CoA reductase inhibitors (statins) because the lack of functioning p53, more specifically, macrophage p53 (67, 68), is associated with accelerated atherosclerosis (69).
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant HL68024 (to K. F.) from National Heart, Blood, and Lung Institute. This work was also supported by American Heart Association Grant 0540054N (to K. F.) and MacDonald General Research Fund Grant 04RDM013 (to K. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures and Figs. S1–S6.
- PUMA
- p53 up-regulated modulator of apoptosis
- MCL1
- myeloid cell leukemia protein
- TSC-22
- transforming growth factor-β-stimulated clone-22
- SSDBD
- the sequence-specific DNA binding domain
- RE
- responsive element
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- Ret
- retroviral
- Ad
- adenoviral
- Hzf
- hematopoietic zing finger.
REFERENCES
- 1. Donehower L. A., Harvey M., Slagle B. L., McArthur M. J., Montgomery C. A., Jr., Butel J. S., Bradley A. (1992) Nature 356, 215–221 [DOI] [PubMed] [Google Scholar]
- 2. Hainaut P., Soussi T., Shomer B., Hollstein M., Greenblatt M., Hovig E., Harris C. C., Montesano R. (1997) Nucleic Acids Res. 25, 151–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Oda E., Ohki R., Murasawa H., Nemoto J., Shibue T., Yamashita T., Tokino T., Taniguchi T., Tanaka N. (2000) Science 288, 1053–1058 [DOI] [PubMed] [Google Scholar]
- 4. Nakano K., Vousden K. H. (2001) Mol. Cell 7, 683–694 [DOI] [PubMed] [Google Scholar]
- 5. Vousden K. H. (2005) Science 309, 1685–1686 [DOI] [PubMed] [Google Scholar]
- 6. Chipuk J. E., Kuwana T., Bouchier-Hayes L., Droin N. M., Newmeyer D. D., Schuler M., Green D. R. (2004) Science 303, 1010–1014 [DOI] [PubMed] [Google Scholar]
- 7. Moll U. M., Erster S., Zaika A. (2001) Biochim. Biophys. Acta 1552, 47–59 [DOI] [PubMed] [Google Scholar]
- 8. Zalcenstein A., Stambolsky P., Weisz L., Müller M., Wallach D., Goncharov T. M., Krammer P. H., Rotter V., Oren M. (2003) Oncogene 22, 5667–5676 [DOI] [PubMed] [Google Scholar]
- 9. Vousden K. H., Prives C. (2005) Cell 120, 7–10 [DOI] [PubMed] [Google Scholar]
- 10. Martins C. P., Brown-Swigart L., Evan G. I. (2006) Cell 127, 1323–1334 [DOI] [PubMed] [Google Scholar]
- 11. Chitpatima S. T., Makrides S., Bandyopadhyay R., Brawerman G. (1988) Nucleic Acids Res. 16, 2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gross B., Gaestel M., Böhm H., Bielka H. (1989) Nucleic Acids Res. 17, 8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li F., Zhang D., Fujise K. (2001) J. Biol. Chem. 276, 47542–47549 [DOI] [PubMed] [Google Scholar]
- 14. Tuynder M., Susini L., Prieur S., Besse S., Fiucci G., Amson R., Telerman A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14976–14981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang Y., Yang F., Xiong Z., Yan Y., Wang X., Nishino M., Mirkovic D., Nguyen J., Wang H., Yang X. F. (2005) Oncogene 24, 4778–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chung S., Kim M., Choi W., Chung J., Lee K. (2000) Cancer Lett 156, 185–190 [DOI] [PubMed] [Google Scholar]
- 17. Arcuri F., Papa S., Carducci A., Romagnoli R., Liberatori S., Riparbelli M. G., Sanchez J. C., Tosi P., del Vecchio M. T. (2004) Prostate 60, 130–140 [DOI] [PubMed] [Google Scholar]
- 18. Gnanasekar M., Thirugnanam S., Zheng G., Chen A., Ramaswamy K. (2009) Int. J. Oncol 34, 1241–1246 [PubMed] [Google Scholar]
- 19. Koide Y., Kiyota T., Tonganunt M., Pinkaew D., Liu Z., Kato Y., Hutadilok-Towatana N., Phongdara A., Fujise K. (2009) Biochim. Biophys. Acta 1790, 326–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen S. H., Wu P. S., Chou C. H., Yan Y. T., Liu H., Weng S. Y., Yang-Yen H. F. (2007) Mol. Biol. Cell 18, 2525–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Susini L., Besse S., Duflaut D., Lespagnol A., Beekman C., Fiucci G., Atkinson A. R., Busso D., Poussin P., Marine J. C., Martinou J. C., Cavarelli J., Moras D., Amson R., Telerman A. (2008) Cell Death Differ. 15, 1211–1220 [DOI] [PubMed] [Google Scholar]
- 22. Zhang D., Li F., Weidner D., Mnjoyan Z. H., Fujise K. (2002) J. Biol. Chem. 277, 37430–37438 [DOI] [PubMed] [Google Scholar]
- 23. Graidist P., Phongdara A., Fujise K. (2004) J. Biol. Chem. 279, 40868–40875 [DOI] [PubMed] [Google Scholar]
- 24. Haghighat N. G., Ruben L. (1992) Mol. Biochem. Parasitol. 51, 99–110 [DOI] [PubMed] [Google Scholar]
- 25. Sanchez J. C., Schaller D., Ravier F., Golaz O., Jaccoud S., Belet M., Wilkins M. R., James R., Deshusses J., Hochstrasser D. (1997) Electrophoresis 18, 150–155 [DOI] [PubMed] [Google Scholar]
- 26. Kim M., Jung Y., Lee K., Kim C. (2000) Arch. Pharm. Res. 23, 633–636 [DOI] [PubMed] [Google Scholar]
- 27. Rao K. V., Chen L., Gnanasekar M., Ramaswamy K. (2002) J. Biol. Chem. 277, 31207–31213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Feng Y., Liu D., Yao H., Wang J. (2007) Arch. Biochem. Biophys. 467, 48–57 [DOI] [PubMed] [Google Scholar]
- 29. Graidist P., Yazawa M., Tonganunt M., Nakatomi A., Lin C. C., Chang J. Y., Phongdara A., Fujise K. (2007) Biochem. J. 408, 181–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee J. H., Rho S. B., Park S. Y., Chun T. (2008) FEBS Lett. 582, 1210–1218 [DOI] [PubMed] [Google Scholar]
- 31. Fritsche M., Haessler C., Brandner G. (1993) Oncogene 8, 307–318 [PubMed] [Google Scholar]
- 32. Miyashita T., Reed J. C. (1995) Cell 80, 293–299 [DOI] [PubMed] [Google Scholar]
- 33. Gong L., Kamitani T., Fujise K., Caskey L. S., Yeh E. T. (1997) J. Biol. Chem. 272, 28198–28201 [DOI] [PubMed] [Google Scholar]
- 34. Fujise K., Zhang D., Liu J., Yeh E. T. (2000) J. Biol. Chem. 275, 39458–39465 [DOI] [PubMed] [Google Scholar]
- 35. Pellegrini M., Corda M., Manca L., Olianas A., Sanna M. T., Fais A., De Rosa M. C., Bertonati C., Masala B., Giardina B. (2001) Eur. J. Biochem. 268, 3313–3320 [DOI] [PubMed] [Google Scholar]
- 36. Mnjoyan Z. H., Dutta R., Zhang D., Teng B. B., Fujise K. (2003) Circulation 108, 464–471 [DOI] [PubMed] [Google Scholar]
- 37. Monia B. P., Johnston J. F., Geiger T., Muller M., Fabbro D. (1996) Nat. Med. 2, 668–675 [DOI] [PubMed] [Google Scholar]
- 38. Flørenes V. A., Maelandsmo G. M., Forus A., Andreassen A., Myklebost O., Fodstad O. (1994) J. Natl. Cancer Inst. 86, 1297–1302 [DOI] [PubMed] [Google Scholar]
- 39. Levine A. J. (1997) Cell 88, 323–331 [DOI] [PubMed] [Google Scholar]
- 40. Hansen S., Lane D. P., Midgley C. A. (1998) J. Mol. Biol. 275, 575–588 [DOI] [PubMed] [Google Scholar]
- 41. Lu B. Y., Chang J. Y. (2002) Biochem. J. 364, 81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Salamanca S., Chang J. Y. (2005) Biochemistry 44, 744–750 [DOI] [PubMed] [Google Scholar]
- 43. Lu X., Lane D. P. (1993) Cell 75, 765–778 [DOI] [PubMed] [Google Scholar]
- 44. Masuda H., Miller C., Koeffler H. P., Battifora H., Cline M. J. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 7716–7719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rho S. B., Lee J. H., Park M. S., Byun H. J., Kang S., Seo S. S., Kim J. Y., Park S. Y. (2011) FEBS Lett. 585, 29–35 [DOI] [PubMed] [Google Scholar]
- 46. Lane D. P. (1992) Nature 358, 15–16 [DOI] [PubMed] [Google Scholar]
- 47. Kruse J. P., Gu W. (2009) Cell 137, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Iwabuchi K., Bartel P. L., Li B., Marraccino R., Fields S. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 6098–6102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Iwabuchi K., Li B., Massa H. F., Trask B. J., Date T., Fields S. (1998) J. Biol. Chem. 273, 26061–26068 [DOI] [PubMed] [Google Scholar]
- 50. Wang B., Matsuoka S., Carpenter P. B., Elledge S. J. (2002) Science 298, 1435–1438 [DOI] [PubMed] [Google Scholar]
- 51. Das S., Raj L., Zhao B., Kimura Y., Bernstein A., Aaronson S. A., Lee S. W. (2007) Cell 130, 624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Budhram-Mahadeo V., Morris P. J., Smith M. D., Midgley C. A., Boxer L. M., Latchman D. S. (1999) J. Biol. Chem. 274, 15237–15244 [DOI] [PubMed] [Google Scholar]
- 53. Budram-Mahadeo V., Morris P. J., Latchman D. S. (2002) Oncogene 21, 6123–6131 [DOI] [PubMed] [Google Scholar]
- 54. Samuels-Lev Y., O'Connor D. J., Bergamaschi D., Trigiante G., Hsieh J. K., Zhong S., Campargue I., Naumovski L., Crook T., Lu X. (2001) Mol. Cell 8, 781–794 [DOI] [PubMed] [Google Scholar]
- 55. Kussie P. H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A. J., Pavletich N. P. (1996) Science 274, 948–953 [DOI] [PubMed] [Google Scholar]
- 56. Böttger V., Böttger A., Garcia-Echeverria C., Ramos Y. F., van der Eb A. J., Jochemsen A. G., Lane D. P. (1999) Oncogene 18, 189–199 [DOI] [PubMed] [Google Scholar]
- 57. Okamoto T., Izumi H., Imamura T., Takano H., Ise T., Uchiumi T., Kuwano M., Kohno K. (2000) Oncogene 19, 6194–6202 [DOI] [PubMed] [Google Scholar]
- 58. Tian C., Xing G., Xie P., Lu K., Nie J., Wang J., Li L., Gao M., Zhang L., He F. (2009) Nat. Cell Biol. 11, 580–591 [DOI] [PubMed] [Google Scholar]
- 59. Vousden K. H., Lane D. P. (2007) Nat. Rev. Mol. Cell Biol. 8, 275–283 [DOI] [PubMed] [Google Scholar]
- 60. Bensaad K., Vousden K. H. (2005) Nat. Med. 11, 1278–1279 [DOI] [PubMed] [Google Scholar]
- 61. Maltzman W., Czyzyk L. (1984) Mol. Cell. Biol. 4, 1689–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Haupt Y., Maya R., Kazaz A., Oren M. (1997) Nature 387, 296–299 [DOI] [PubMed] [Google Scholar]
- 63. Kubbutat M. H., Jones S. N., Vousden K. H. (1997) Nature 387, 299–303 [DOI] [PubMed] [Google Scholar]
- 64. Buschmann T., Fuchs S. Y., Lee C. G., Pan Z. Q., Ronai Z. (2000) Cell 101, 753–762 [DOI] [PubMed] [Google Scholar]
- 65. Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E. A. (2004) Science 303, 844–848 [DOI] [PubMed] [Google Scholar]
- 66. Shangary S., Qin D., McEachern D., Liu M., Miller R. S., Qiu S., Nikolovska-Coleska Z., Ding K., Wang G., Chen J., Bernard D., Zhang J., Lu Y., Gu Q., Shah R. B., Pienta K. J., Ling X., Kang S., Guo M., Sun Y., Yang D., Wang S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3933–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Merched A. J., Williams E., Chan L. (2003) Arterioscler. Thromb. Vasc. Biol. 23, 1608–1614 [DOI] [PubMed] [Google Scholar]
- 68. van Vlijmen B. J., Gerritsen G., Franken A. L., Boesten L. S., Kockx M. M., Gijbels M. J., Vierboom M. P., van Eck M., van De Water B., van Berkel T. J., Havekes L. M. (2001) Circ. Res. 88, 780–786 [DOI] [PubMed] [Google Scholar]
- 69. Guevara N. V., Kim H. S., Antonova E. I., Chan L. (1999) Nat. Med. 5, 335–339 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.