Abstract
GIV (Gα-interacting vesicle-associated protein, also known as Girdin) is a bona fide enhancer of PI3K-Akt signals during a diverse set of biological processes, e.g. wound healing, macrophage chemotaxis, tumor angiogenesis, and cancer invasion/metastasis. We recently demonstrated that tyrosine phosphorylation of GIV by receptor and non-receptor-tyrosine kinases is a key step that is required for GIV to directly bind and enhance PI3K activity. Here we report the discovery that Src homology 2-containing phosphatase-1 (SHP-1) is the major protein-tyrosine phosphatase that targets two critical phosphotyrosines within GIV and antagonizes phospho-GIV-dependent PI3K enhancement in mammalian cells. Using phosphorylation-dephosphorylation assays, we demonstrate that SHP-1 is the major and specific protein-tyrosine phosphatase that catalyzes the dephosphorylation of tyrosine-phosphorylated GIV in vitro and inhibits ligand-dependent tyrosine phosphorylation of GIV downstream of both growth factor receptors and GPCRs in cells. In vitro binding and co-immunoprecipitation assays demonstrate that SHP-1 and GIV interact directly and constitutively and that this interaction occurs between the SH2 domain of SHP-1 and the C terminus of GIV. Overexpression of SHP-1 inhibits tyrosine phosphorylation of GIV and formation of phospho-GIV-PI3K complexes, and specifically suppresses GIV-dependent activation of Akt. Consistently, depletion of SHP-1 enhances peak tyrosine phosphorylation of GIV, which coincides with an increase in peak Akt activity. We conclude that SHP-1 antagonizes the action of receptor and non-receptor-tyrosine kinases on GIV and down-regulates the phospho-GIV-PI3K-Akt axis of signaling.
Keywords: Akt PKB, Cancer Biology, Cell Migration, Cell Signaling, Cytoskeleton, Phosphotyrosine Signaling, PI 3-Kinase (PI3K), Receptor-tyrosine Kinase, Protein-tyrosine Kinase (Tyrosine Kinase), Protein-tyrosine Phosphatase (Tyrosine Phosphatase)
Introduction
GIV4 (Gα-interacting, vesicle-associated protein, also known as Girdin), is a multidomain protein that is required for growth factors (EGF (1, 2), insulin-like growth factor (3), VEGF (4), and insulin (5–8)) to enhance Akt activation in a PI3K-dependent manner, directly link Akt to the actin cytoskeleton, remodel actin, and trigger cell migration. GIV is also required for Akt enhancement downstream of G-protein coupled receptors (GPCRs) (7–9). Working downstream of both growth factor receptor-tyrosine kinases (RTKs) and GPCRs, GIV serves as a common enhancer of Akt signals for two large and distinct classes of receptors during a diverse set of biological processes, e.g. epithelial wound healing, macrophage chemotaxis, development, neuronal migration, autophagy, tumor angiogenesis, tumor cell migration, and cancer invasion/metastasis (1–4, 6, 8, 10–13).
Recent work has provided mechanistic insights into these biological functions of GIV. We previously demonstrated that GIV is a non-receptor guanine-nucleotide exchange factor (GEF) for Gαi (7) and subsequently demonstrated that GIV directly binds ligand-activated epidermal growth factor receptor (EGFR) (2, 13). By linking G protein signaling to EGFR and assembling a Gαi-GIV-EGFR signaling complex, GIV enhances EGFR autophosphorylation, prolongs receptor association with the plasma membrane (PM) and selectively enhances PI3K-Akt signals initiated by ligand-activated receptors at the PM.
In depth understanding of how exactly GIV relays receptor-initiated signals selectively through the PI3K-Akt pathway remained poorly understood until recently when we defined (14) GIV as a central molecule in the tyrosine signaling pathway. We found that GIV is a common substrate for multiple receptor and non-receptor-tyrosine kinases and identified the sites of phosphorylation as tyrosines 1764 and 1798 in its C terminus. We demonstrated that these phosphotyrosines serve as docking sites for the two Src homology 2 (SH2) domains of the p85α regulatory subunit of PI3K, thereby allowing tyrosine-phosphorylated GIV to directly bind and activate Class 1 PI3K downstream of both RTKs and GPCRs. We found that these molecular mechanisms orchestrate a distinct cascade of events when cells expressing wild-type GIV are stimulated with growth factors (2, 14); GIV interacts with ligand-activated RTKs and enhances receptor autophosphorylation; subsequently, RTKs and non-RTKs phosphorylate GIV on two key tyrosines; phospho-GIV-PI3K complexes are assembled and stabilized at the receptor tail; receptor-initiated activation of PI3K is further enhanced by direct interaction of PI3K with phosphotyrosines within the C terminus of GIV, and enhanced PI3K activity generates PIP3 at the PM, which in turn triggers recruitment and activation of Akt. By contrast, in cells expressing a mutant GIV in which both tyrosines are replaced by phenylalanines, GIV interacts with ligand-activated RTKs, but the subsequent steps within the cascade are aborted. Furthermore, this “phospho-GIV-PI3K-Akt” cascade is progressively hyperactivated in breast cancer cells with advancing clinical stage and increasing degrees of tumor invasiveness (14), leading to the speculation that inhibition of tyrosine phosphorylation of GIV or disruption of the GIV-p85α interface are feasible therapeutic strategies to halt cancer progression. Despite the insights gained into which tyrosine kinases trigger GIV phosphorylation and its biological relevance during cancer invasion, the molecular mechanism(s) that down-regulates or attenuates this tyrosine-based signaling pathway remains unknown.
A major superfamily of proteins that down-regulate tyrosine-based signaling pathways by coordinately antagonizing the actions of protein-tyrosine kinases is protein-tyrosine phosphatases (PTPs) (15). Because of their antagonistic catalytic functions, PTPs and protein-tyrosine kinases act together to control a wide range of phosphotyrosine-mediated signaling pathways in mammalian cells. Of the PTPs known to date, we noted that the cytosolic PTP, Src homology 2 (SH2)-containing phosphatase-1 (SHP-1; also known as PTP1C, HCP, and SH-PTP1) stands out as the phosphatase that carries out a set of functions that antagonize almost all known functions of GIV during signal transduction. Most important of them all, whereas GIV has been shown to function as an enhancer of PI3K-Akt signals (2, 5, 7, 14), SHP-1 is a major PTP that inhibits the PI3K-Akt pathway (16–18). Although GIV binds the EGFR, enhances its autophosphorylation, and prolongs its signaling from the PM (2), SHP-1 binds to the activated receptor, dephosphorylates it, and promotes its endocytosis and degradation (19, 20). Given these striking and diametrically opposite sets of functions, we wondered if SHP-1 may serve as the specific phosphatase that targets the phosphotyrosines in GIV and thereby down-regulates the phospho-GIV-PI3K-Akt axis.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Unless otherwise indicated, all reagents were of analytical grade and obtained from Sigma. Cell culture media were purchased from Invitrogen. All restriction endonucleases and Escherichia coli strain DH5α were purchased from New England Biolabs (Beverly, MA). E. coli strain BL21(DE3) was purchased from Invitrogen. Pfu ultra DNA polymerase was purchased from Stratagene (La Jolla, CA). Rabbit antisera against the coiled-coil region of GIV was raised as described (21). Mouse mAbs against hexahistidine (His6), FLAG (M2), and α-tubulin were obtained from Sigma and against HA was purchased from Covance Inc. (Princeton, NJ). Rabbit anti-Gαi3, anti-SHP-1, anti-SHP2, anti-total EGFR (cytoplasmic tail), and anti-GIV/Girdin(T-13) IgGs were from Santa Cruz Biotechnology (Santa Cruz, CA), anti-p85α was from Millipore Inc. (Billerica, MA), anti-phosphotyrosine was from BD Biosciences, and anti-phospho-Akt (Ser-473), anti-phospho-EGFR(Tyr-1173), and anti-phospho-ERK1/2 IgGs were from Cell Signaling (Beverly, MA). Goat anti-rabbit and goat anti-mouse Alexa Fluor 680 or IRDye 800 F(ab′)2 used for Odyssey Infrared Imaging were from Li-Cor Biosciences (Lincoln, NE).
Plasmid Constructs and Mutagenesis
Hexahistidine (His6)- and GST-tagged human SHP-1 PTP (GenBankTM accession number BC002523) cloned into pET28 and pET42 (Novagen) expression vectors, respectively, were generous gifts from Richard Anderson (University of Wisconsin, Madison, WI) (22). For mammalian expression, human SHP-1 was cloned from pET28 and inserted between EcoR1 and Xho1 restriction sites of pcDNA 3.1. HA-tagged mouse SHP-1 (NM_013545.2) was cloned from pBS(Bluescript), a generous gift from Ulrike Lorenz (University of Virginia, Charlottesville, VA) (23), and inserted between two EcoR1 restriction sites of pcDNA 3.1. To generate the GST-tagged ΔNSH2 (amino acids 166–end) and ΔN+CSH2 (amino acids 276–end) truncations of SHP-1 for use in in vitro binding assays, corresponding fragments of human SHP-1 (NM_002831) were cloned from pcDNA3.1 and inserted between the Bam H1 and EcoR1 restriction sites of the pGEX 4T-1 vector. Cloning of His-GIV-CT (amino acids 1660–1870) into pET28b and GIV-FLAG into 3×FLAG-pCMV-14 were described previously (9). C-terminal HA-tagged c-Src (accession number NM_001025395) for mammalian expression was generated by cloning the entire coding sequence into pcDNA 3.1 between Xho1 and EcoR1 restriction site. GST-SHP-1 C453S, HA-SHP-1 C453S, His-GIV-CT Y1764F and Y1798F, and c-Src-HA K295R (inactive) and Y527F (active) mutants were generated by site-directed mutagenesis (sequences of primers available upon request) using the QuikChange kit (Stratagene, San Diego, CA) as per the manufacturer's protocols. All constructs were checked by DNA sequencing.
Protein Expression and Purification
GST, GST-SHP-1 (full-length, wild type, and C453S mutant and ΔNSH2 and ΔN+CSH2), His-SHP-1, and His-GIV-CT fusion constructs were expressed in E. coli strain BL21(DE3) (Invitrogen) and purified as described previously (7, 9). Briefly, bacterial cultures were induced overnight at 25 °C with 1 mm isopropyl β-d-1-thiogalactopyranoside. Pelleted bacteria from 1 liter of culture were resuspended in 10 ml of GST lysis buffer (25 mm Tris-HCl, pH 7.5, 20 mm NaCl, 1 mm EDTA, 20% (v:v) glycerol, 1% (v:v) Triton X-100, 2× protease inhibitor mixture (Complete EDTA-free, Roche Diagnostics)) or His lysis buffer (50 mm NaH2PO4, pH 7.4, 300 mm NaCl, 10 mm imidazole, 1% (v:v) Triton X-100, 2× protease inhibitor mixture (Complete EDTA-free, Roche Diagnostics)) for GST or His-fused proteins, respectively. After sonication (4 × 20s, 1 min between cycles), lysates were centrifuged at 12,000 × g at 4 °C for 20 min. Solubilized proteins were affinity-purified on glutathione-Sepharose 4B beads (GE Healthcare) or HisPur Cobalt Resin (Pierce). Proteins were eluted, dialyzed overnight against PBS, and stored at −80 °C.
In Vitro Phosphorylation and Dephosphorylation Assays
In vitro kinase-phosphatase assays were carried out in tandem using the following protocol. First, the kinase assays were performed using purified His-GIV-CT (∼3–5 μg/reaction) and commercially obtained recombinant tyrosine kinases (EGFR, Millipore; c-Src, Cell Signaling). Reactions were started by the addition of 1 mm ATP and carried out at 25 °C for 60 min in tyrosine kinase buffer (60 mm HEPES, pH 7.5, 5 mm MgCl2, 5 mm MnCl2). Phosphorylated His-GIV-CT was subsequently used as the substrate in in vitro dephosphorylation assays using either purified His-SHP-1 (Fig. 1A), GST-SHP-1 WT, GST-SHP-1 C453S (Fig. 1C), HA-SHP-1 immuno-isolated from COS7 cells (Fig. 1D), or lysates of COS7 cells immuno-depleted of endogenous SHP-1 (Fig. 1F) as the source for phosphatase. Phosphatase reactions were carried out at 30 °C for 60 min in phosphatase buffer (25 mm HEPES, 2.5 mm EDTA, 5 mm DTT, 50 mm NaCl, 65 ng/μl BSA at pH 7.4) and stopped by adding Laemmli sample buffer and boiling at 100 °C.
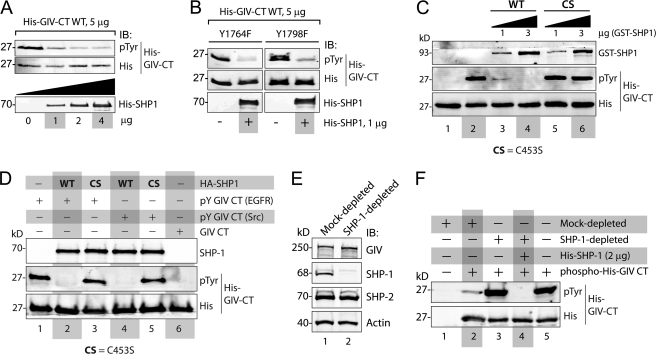
FIGURE 1.
SHP-1 specifically dephosphorylates tyrosine-phosphorylated GIV in vitro. A, His-SHP-1 dephosphorylated tyrosine-phosphorylated His-GIV-CT. His-GIV-CT (1660–1870) was phosphorylated in vitro using recombinant EGFR kinase and was subsequently used as the substrate in phosphatase assays using increasing (1, 2, and 4 μg) amounts of recombinant His-SHP-1. Residual tyrosine phosphorylation on His-GIV-CT was assessed by immunoblotting (IB) using phosphotyrosine (pTyr) and His mAbs. His-GIV-CT was efficiently dephosphorylated in the presence of 1 μg and maximally in the presence of 2 μg of His-SHP-1. B, SHP-1 dephosphorylated both phosphotyrosines (Tyr-1764 and -1798) in the C terminus of GIV. His-GIV-CT mutants with one intact tyrosine (Y1764F mutant, in which Tyr-1798 is intact, and Y1798F mutant, in which Tyr-1764 is intact) were phosphorylated in vitro using recombinant EGFR kinase as in A. Equal aliquots (5 μg) of these phosphorylated His-GIV-CT mutants were subsequently used in phosphatase assays in the presence (+) or absence (−) of 1 μg His-SHP-1 and analyzed for residual tyrosine phosphorylation by immunoblotting as in A. C, WT, but not the catalytically inactive CS mutant of SHP-1 dephosphorylates tyrosine-phosphorylated His-GIV-CT in vitro. Equal aliquots of tyrosine-phosphorylated His-GIV-CT (phosphorylated using recombinant EGFR kinase as in A) were used as substrates in phosphatase assays with the indicated amounts of bacterially expressed, purified WT, or C453S mutant of GST-SHP-1. Samples were subsequently analyzed for residual tyrosine phosphorylation by immunoblotting as in A. GST-SHP-1 WT efficiently dephosphorylated His-GIV-CT at 1 μg (lane 3) and maximally at 3 μg (lane 4), whereas GST-SHP-1 CS failed to dephosphorylate His-GIV-CT at either dose (lanes 5 and 6). D, WT, but not the catalytically inactive CS mutant of SHP-1, immuno-isolated from COS7 cells dephosphorylated tyrosine-phosphorylated His-GIV-CT in vitro. Tyrosine-phosphorylated His-GIV-CT was generated by carrying out in vitro phosphorylation assays using recombinant EGFR (pY-GIV-CT (EGFR); lanes 1–3) or Src (pY-GIV-CT (Src); lanes 4–6) kinases as in A. Lysates of COS7 cells transiently expressing WT or the CS mutant of HA-SHP-1 were incubated sequentially with anti-HA mAb and protein G-agarose beads to immuno-isolate active (WT) and catalytically inactive CS phosphatase, respectively. In vitro phosphatase assays were subsequently carried out by incubating equal aliquots of Tyr(P)-GIV-CT with the bead-bound SHP-1 WT (lanes 2 and 4), SHP-1 CS (lanes 3 and 5), or control beads (lane 1). Both EGFR and Src-phosphorylated GIV-CT were efficiently dephosphorylated in the presence of HA-SHP-1 WT (lanes 2 and 4) but not HA-SHP-1 CS (lanes 3 and 5) or control beads (lane 1). E and F, COS7 cell lysates immuno-depleted of endogenous SHP-1 fail to dephosphorylate tyrosine-phosphorylated His-GIV-CT in vitro. E, lysates of COS7 cells were immuno-depleted of SHP-1 (lane 2) or mock-depleted (lane 1) using anti-SHP-1 or preimmune control IgGs, respectively. Equal aliquots of lysates were analyzed for GIV, SHP-1, SHP-2, and actin by immunoblotting. The efficacy of SHP-1 immuno-depletion was confirmed as ∼>99% by optical densitometry. F, equal aliquots of tyrosine-phosphorylated His-GIV-CT (∼5 μg; prepared as in A) were used as substrates in in vitro dephosphorylation assays with either buffer alone (lane 5) or equal aliquots (∼75 μg) of mock-depleted (lanes 1 and 2) or SHP-1-depleted (lanes 3 and 4) lysates. Residual phosphorylation was assessed by immunoblotting for Tyr(P) mAb. Tyrosine-phosphorylated His-GIV-CT was efficiently dephosphorylated in the presence of mock-depleted (lane 2) but not SHP-1-depleted lysate (lane 3). The phosphatase activity of SHP-1-depleted lysate was restored upon the addition of bacterially expressed His-SHP-1 (lane 4).
Cell Culture, Transfection, and Lysis
COS7 and HeLa cells were grown at 37 °C in DMEM supplemented with 10% FBS, 100 units/ml penicillin, 100 μg/ml streptomycin, 1% l-glutamine, and 5% CO2. siRNA transfection of HeLa cells was carried out using Oligofectamine (Invitrogen) following the manufacturer's protocol. Oligos against human SHP-1 were from Santa Cruz Biotechnology. Briefly, HeLa cells at 70% confluence were transfected with 0.83 nm (final) of SHP-1 siRNA in Opti-MEM. Transfection of COS7 cells with HA-SHP-1, GIV-3×FLAG, or c-Src-HA were carried out using GeneJuice (Novagen). Lysates used as a source for SHP-1 for in vitro dephosphorylation assays or as a source for GIV in immunoprecipitation assays were prepared by resuspending cells in lysis buffer (20 mm HEPES, pH 7.2, 5 mm Mg(CH3COO)2, 125 mm K(CH3COO), 0.4% Triton X-100, 1 mm DTT supplemented with sodium orthovanadate (500 μm), phosphatase (Sigma), and protease (Roche Diagnostics) inhibitor cocktails) and subsequently passing them through a 30-gauge needle at 4 °C and cleared (centrifuged at 14,000 × g for 10 min) before use in subsequent experiments. Whole cell lysates used to study the extent of Akt and ERK phosphorylation were prepared by resuspending the entire cell pellet in Laemmli sample buffer and boiling at 100 °C.
In Vivo Phosphorylation Assays
For in vivo phosphorylation assays on endogenous GIV, HeLa cells were serum-starved for 12–16 h before stimulation with 50 nm EGF (Invitrogen). For in vivo phosphorylation assays using overexpressed GIV, FLAG-tagged GIV was co-expressed with either EGFR, Src, or SHP-1 plasmids in COS7 cells in various assays. ∼30 h after transfection, cells were serum-starved (0% FBS) for an additional 16–18 h followed by stimulation with 50 nm EGF or 10 μm LPA. Reactions were stopped using PBS chilled at 4 °C supplemented with 200 μm sodium orthovanadate and immediately scraped and lysed for immunoprecipitation.
Immunoprecipitation and GST Pulldown Assays
These assays were carried out exactly as described previously (2, 9). Briefly, cell lysates (∼1–2 mg of protein) were incubated for 4 h at 4 °C with 2 μg of anti-HA mAb (Covance), anti-FLAG mAb (Sigma), 1.6 μg of anti-SHP-1 PTP pAb, or anti SHP-2 PTP pAb, or 1 μg of their respective preimmune IgGs followed by incubation with protein G (for all mAbs) or A (for all pAbs)-Sepharose beads (GE Healthcare) at 4 °C for an additional 60 min. Beads were washed in PBS-T wash buffer (4.3 mm Na2HPO4, 1.4 mm KH2PO4, pH 7.4, 137 mm NaCl, 2.7 mm KCl, 0.1% (v:v) Tween 20, 10 mm MgCl2, 5 mm EDTA, 2 mm DTT, and 0.5 mm sodium orthovanadate), and bound proteins were eluted by boiling in Laemmli sample buffer. In vitro binding assays with GST-fused proteins were carried out exactly as previously described (7, 9). Buffers were supplemented with 0.5 mm sodium orthovanadate for all steps of the assay.
Immunoblotting
Protein samples were separated on 10% SDS-PAGE and transferred to PVDF membranes (Millipore). Membranes were blocked with PBS supplemented with 5% nonfat milk (or with 5% BSA whenever probing for phosphorylated proteins) before incubation with primary Abs. Infrared imaging with two-color detection was performed using an Odyssey imaging system (Li-Cor Biosciences, Lincoln, NE). Primary antibodies were diluted as follows: anti-Tyr(P) 1:500, anti-HA 1:1000; anti-SHP-1 1:500, anti-SHP-2 1:500, anti-GIV/Girdin(CTAb) 1:500, anti-GIV ccAb (sera) 1:500, anti-EGFR 1:500, anti-pAkt(Ser-473) 1:250; anti-pERK1/2 1:500, anti-Tyr(P)-1173(EGFR) 1:250, anti-Gαi3 1:300, and anti-FLAG 1:1000. All Odyssey images were processed using Image J software (National Institutes of Health) and assembled for presentation using Photoshop and Illustrator software (both Adobe).
Statistical Analysis
Each experiment presented in the figures is representative of at least three independent experiments. Statistical significance between various conditions was assessed with the Wilcoxon Signed-Rank test. Graphical data presented was prepared using GraphPad Software, Inc. (San Diego, CA).
RESULTS
Protein-tyrosine Phosphatase SHP-1 Dephosphorylates GIV in Vitro
To determine whether GIV is a substrate of SHP-1, we carried out in vitro phosphatase assays using bacterially expressed, recombinant His-SHP-1 and tyrosine-phosphorylated His-GIV-CT (amino acids 1660–1870) as substrate. We examined the C terminus of GIV because the only two sites of tyrosine phosphorylation in the entire protein are located within that region (14). To generate the phosphorylated GIV-CT substrate, we first carried out in vitro kinase assays using His-GIV-CT and recombinant EGFR kinase (Millipore) as described previously (14). When phosphorylated, His-GIV-CT was subsequently incubated with increasing quantities of His-SHP-1, SHP-1 efficiently dephosphorylated GIV in a dose-dependent manner (Fig. 1A) indicating that GIV is a substrate of SHP-1 in vitro.
Because GIV is tyrosine-phosphorylated at two sites, Tyr-1764 and -1798 (14), next we asked if SHP-1 selectively dephosphorylates one or both of those phosphotyrosines. When phospho-mutants of His-GIV-CT with either Tyr-1764 or -1798 mutated to phenylalanine were phosphorylated in vitro by EGFR kinase and subsequently incubated with His-SHP-1, SHP-1 efficiently dephosphorylated both His-GIV-CT mutants (Fig. 1B). We conclude that SHP-1 is capable of dephosphorylating both phosphotyrosines Tyr(P)-1764 and -1798 in the C terminus of GIV.
To investigate if the catalytic activity of SHP-1 is required to dephosphorylate GIV, in vitro phosphatase assays were carried out using phosphorylated His-GIV-CT and GST-tagged SHP-1 WT or the constitutively inactive mutant in which cysteine 453 in the catalytic center is replaced with serine (SHP-1 C453S, hereby referred to as CS). The CS mutation selectively abolishes PTP activity while retaining the ability of SHP-1 to bind substrate proteins (24). SHP-1 WT, but not SHP-1 CS dephosphorylated tyrosine-phosphorylated His-GIV-CT in a dose-dependent manner (Fig. 1C). Identical results were obtained when SHP-1-HA immuno-isolated from COS7 cells was used to dephosphorylate EGFR- or Src-phosphorylated His-GIV-CT in vitro (Fig. 1D). These results demonstrate that an intact catalytic domain is indeed required for the protein-tyrosine phosphatase SHP-1 to dephosphorylate GIV-CT in vitro.
Next we asked whether other tyrosine phosphatases besides SHP-1 can dephosphorylate GIV. To this end we prepared mock-depleted and SHP-1-depleted COS7 lysates (Fig. 1E) and used them as sources of phosphatases in in vitro phosphatase assays with tyrosine-phosphorylated His-GIV-CT. Although mock-depleted lysates efficiently dephosphorylated tyrosine-phosphorylated GIV-CT, lysates immuno-depleted of SHP-1 (by >99%) failed to do so (Fig. 1F). Furthermore, the addition of recombinant His-SHP-1 to SHP-1-depleted lysates restored the ability of this lysate to dephosphorylate tyrosine-phosphorylated His-GIV-CT (Fig. 1F). Thus, the key catalytic activity that is required for removal of the phosphotyrosines in GIV was lost with depletion of SHP-1 and restored by adding back recombinant His-SHP-1. These results indicate that SHP-1 is the major cellular phosphatase that dephosphorylates the tyrosines in GIV in vitro. Of note, SHP-2, the closest relative of SHP-1, was abundant in both mock- and SHP-1-depleted cytosols (Fig. 1E) but failed to dephosphorylate His-GIV-CT (Fig. 1F), highlighting the specificity of SHP-1 toward GIV-CT as its substrate.
SHP-1 Inhibits Tyrosine Phosphorylation of GIV after Activation of EGF Receptor or Src Kinases
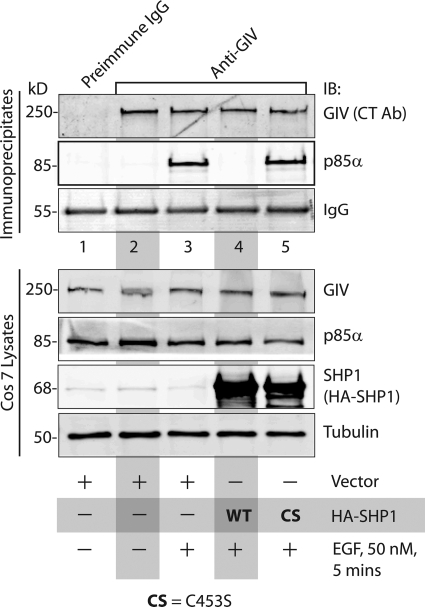
We recently demonstrated that both receptor (EGFR and InsulinR) and non-receptor (c-Src) tyrosine kinases can phosphorylate the GIV C-terminal tyrosines in vivo (14). To determine whether SHP-1 can inhibit EGFR- or Src-induced tyrosine phosphorylation of GIV in cells, we carried out in vivo phosphorylation assays using an approach identical to what we used previously (14). When we immunoprecipitated GIV from COS7 cells co-transfected with GIV-FLAG and HA-SHP-1 and assessed the extent of tyrosine phosphorylation on GIV, we found that the co-expression of SHP-1 virtually abolished the EGF-triggered tyrosine phosphorylation (supplemental Fig. S1). When we performed the in vivo phosphorylation assay in cells co-transfected with either HA-SHP-1-WT or HA-SHP-1-CS mutant, we found that WT, but not the CS, mutant virtually abolished the EGF-triggered tyrosine-phosphorylation (Fig. 2A), thus confirming that the catalytic activity of SHP-1 is required for down-regulation of tyrosine-phosphorylated GIV after EGF stimulation. We conclude that SHP-1 dephosphorylates tyrosine-phosphorylated GIV downstream of EGFR.
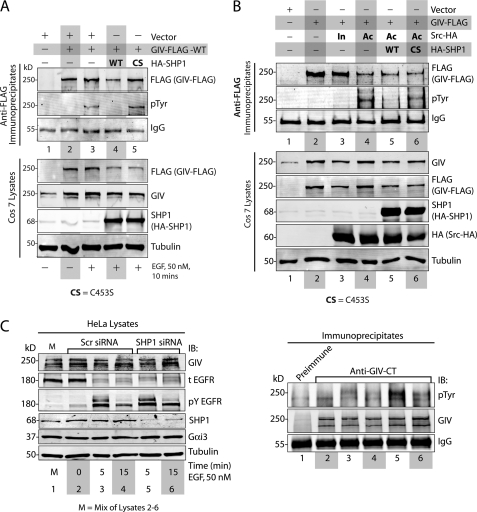
FIGURE 2.
SHP-1 inhibits tyrosine phosphorylation of GIV by EGFR and Src. A, WT, but not the catalytically inactive CS mutant of SHP-1, dephosphorylates GIV after EGF stimulation. COS7 cells transfected with vector alone (lane 1), GIV-FLAG alone (lanes 2 and 3), GIV-FLAG and HA-SHP-1 WT (lane 4), or GIV-FLAG and HA-SHP-1 CS mutant (lane 5) were serum-starved (−) and subsequently stimulated with EGF (50 nm, +) for 10 min. Equal aliquots of lysates (bottom) were incubated sequentially with anti-FLAG mAb and protein G-agarose beads. Immune complexes (top) were analyzed by immunoblotting (IB) for GIV and Tyr(P) (pTyr) using the Li-COR Odyssey Infrared Western blot Imaging System. Single channel images for GIV and Tyr(P) are displayed in grayscale, which shows that immunoprecipitated GIV is phosphorylated on tyrosine(s) exclusively after EGFR stimulation (compare lanes 2 and 3). This EGF-dependent tyrosine phosphorylation of GIV was undetectable in cells co-transfected with HA-SHP-1 WT (lane 4) but robust in those expressing HA-SHP-1 CS (lane 5). Expression of GIV and SHP-1 in all lysates was analyzed by immunoblotting for FLAG, GIV, SHP-1, and tubulin (bottom). B, WT, but not the catalytically inactive CS mutant of SHP-1, dephosphorylates Src-phosphorylated GIV. COS7 cells were transfected with vector alone (lane 1) or GIV-FLAG alone (lane 2) or GIV-FLAG and Src-HA K295R inactive mutant (In, lane 3), or GIV-FLAG and Src-HA Y527F active mutant (Ac, lanes 4–6). HA-SHP-1 WT and HA-SHP-1 CS were co-transfected with active Src Y527F in lanes 5 and 6, respectively. Equal aliquots of lysates (bottom) were incubated with anti-FLAG mAb and protein G-agarose beads. Immune complexes (top) were analyzed by immunoblotting for GIV and Tyr(P) as in A. Tyrosine phosphorylation was detectable in GIV-FLAG immunoprecipitated from cells co-expressing the active Src-HA mutant (lane 4, pTyr panel) but not from cells expressing vector (lane 1) or GIV-FLAG alone (lane 2) or those expressing the inactive Src-HA mutant (lane 3). Src-induced tyrosine phosphorylation of GIV-FLAG was undetectable in cells co-expressing HA-SHP-1 WT (lane 5) but restored in cells co-expressing HA-SHP-1 CS (lane 6). Expression of GIV, Src, and SHP-1 in all lysates was analyzed by immunoblotting for FLAG, GIV, SHP-1, HA (Src), and tubulin (bottom). C, EGF-dependent tyrosine-phosphorylation of GIV is enhanced in cells depleted of endogenous SHP-1. HeLa cells treated with scrambled (Scr) or SHP-1 siRNA were serum-starved (0 min) followed by stimulation with 50 nm EGF for 5 and 15 min. Equal aliquots of lysates (left) were incubated sequentially with anti-GIV-CT and protein A-agarose beads. Immune complexes (right) were analyzed for GIV and Tyr(P) by immunoblotting. Tyrosine-phosphorylated GIV (pTyr) was undetectable in starved cells (lane 2), peaked at 5 min after EGF stimulation in both scrambled and SHP-1 siRNA-treated cells (lanes 3 and 5), and decreased at 15 min (lanes 4 and 6). Phosphorylation of GIV in the immunoprecipitates (right panel) is increased in SHP-1-depleted cells as compared with control by ∼4.2-fold at 5 min (compare lanes 3 and 5) and by ∼1.5-fold at 15 min (compare lanes 4 and 6). Activation of EGFR and depletion of SHP-1 (by ∼70%) were confirmed by analyzing equal aliquots of lysates for GIV, total EGFR (t EGFR), phosphotyrosine 1173 EGFR (pY EGFR), SHP-1, Gαi3, and tubulin by immunoblotting.
To discern if SHP-1 can also dephosphorylate GIV after activation of non-RTKs like Src, we immunoprecipitated GIV from COS7 cells co-expressing GIV-FLAG with constitutively active (Y527F (25)) or inactive (K295R (26)) mutants of Src-HA and the WT or the inactive CS mutant of HA-SHP-1. As shown previously by us (14), GIV was tyrosine-phosphorylated in a Src activity-dependent manner; co-transfection of GIV with active, but not inactive Src mutant resulted in increased tyrosine phosphorylation of GIV (Fig. 2B). Co-expression of SHP-1 WT, but not SHP-1 CS abolished Src-dependent tyrosine phosphorylation of GIV (Fig. 2B), demonstrating that the catalytic activity of SHP-1 is required for down-regulation of Src-phosphorylated GIV. We conclude that SHP-1 antagonizes the action of Src kinase by dephosphorylating GIV once it is phosphorylated by Src.
To investigate how SHP-1 affects the timing and extent of tyrosine phosphorylation of endogenous GIV, we depleted HeLa cells of endogenous SHP-1 using target-specific or control siRNA. We subsequently immunoprecipitated GIV and assessed the extent of tyrosine phosphorylation at 5 or 15 min after EGF stimulation. Consistent with our recent work (14), in control siRNA-treated cells tyrosine phosphorylation of GIV occurred exclusively after ligand stimulation, reached peak levels at 5 min, and was down-regulated at 15 min (Fig. 2C). In cells depleted of SHP-1 (by ∼70%), although the timing of phospho-dephosphorylation of GIV remained same, the extent of peak phosphorylation was enhanced by ∼4.2-fold at 5 min and ∼1.5-fold at 15 min compared with controls. Of note, levels of total EGFR, the peak extent of receptor autophosphorylation (Tyr(P) EGFR), and the rate of receptor degradation were almost similar in both control and SHP-1-depleted cells at 5 min and only slightly increased in SHP-1-depleted cells at 15 min, indicating that the tyrosine kinase activity of EGFR was similar under these conditions. This indicates that the observed increase in the abundance of tyrosine-phosphorylated GIV in cells depleted of SHP-1 at 5 min is unlikely to be due to hyperphosphorylation by EGFR and instead is a consequence of impaired dephosphorylation of GIV by SHP-1. Taken together, these results demonstrate that SHP-1 can dephosphorylate GIV in vivo after activation of both RTKs and non-RTKs.
SHP-1 Inhibits Tyrosine Phosphorylation of GIV Downstream of G Protein-coupled Receptors
Because activation of GPCRs is known to trigger tyrosine phosphorylation of GIV (via Src kinase) (14) and to activate SHP-1 (via Gαq) (27), next we investigated if SHP-1 can down-regulate tyrosine phosphorylation of GIV after activation of lysophosphatidic acid (LPA) receptor, a member of the GPCR family. Consistent with our recent work (14), activation of LPA receptor triggered tyrosine phosphorylation of GIV (supplemental Fig. S2, Fig. 3). This LPA-triggered tyrosine phosphorylation was inhibited in cells co-expressing SHP-1 WT but maintained in those cells co-expressing the inactive SHP-1 CS mutant (Fig. 3), indicating that the catalytic activity of SHP-1 is required for down-regulation of tyrosine-phosphorylated GIV after LPA stimulation. We conclude that SHP-1 also dephosphorylates GIV downstream of GPCRs.
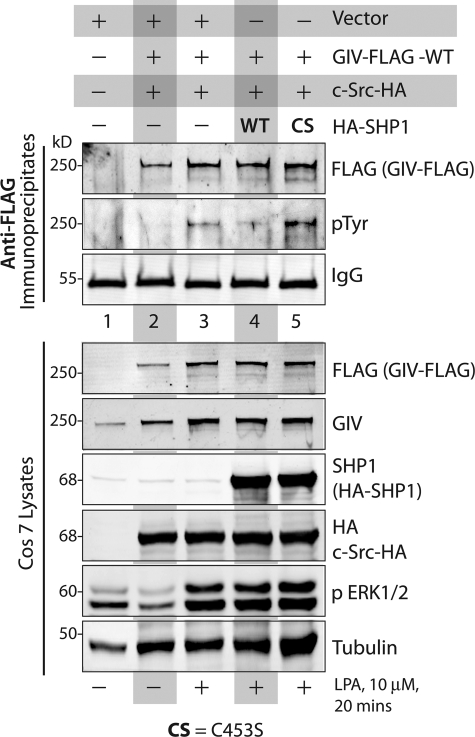
FIGURE 3.
WT, but not the catalytically inactive SHP-1C453S (CS) mutant, inhibits tyrosine phosphorylation of GIV upon activation of LPA receptor. COS7 cells transfected with vector alone (lane 1), GIV-FLAG alone (lanes 2 and 3), GIV-FLAG and HA-SHP-1 WT (lane 4), or GIV-FLAG and HA-SHP-1 CS mutant (lane 5) were serum-starved (−) and subsequently stimulated with LPA (10 μm, +) for 20 min. Equal aliquots of lysates (bottom) were incubated with anti-FLAG mAb and protein G-agarose beads. Immune complexes (top) were analyzed for GIV and Tyr(P) (pTyr) by immunoblotting. Immunoprecipitated GIV was phosphorylated on tyrosine(s) exclusively after LPA receptor stimulation (compare lanes 2 and 3). LPA-dependent tyrosine phosphorylation of GIV was markedly reduced in cells co-transfected with HA-SHP-1 WT (lane 4) but robust in those expressing HA-SHP-1 CS (lane 5). Adequate stimulation of cells by LPA and expression of GIV, Src, and SHP-1 in all lysates was analyzed by immunoblotting for FLAG, GIV, SHP-1, HA (Src), phospho-ERK1/2, and tubulin (bottom).
SHP-1 Interacts Directly and Constitutively with GIV
Most PTPs interact with their substrates before catalyzing the removal of phosphates on target tyrosines (28, 29). To investigate if endogenous SHP-1 and GIV interact in vivo, we immunoprecipitated SHP-1 from COS7 (Fig. 4A) or HEK (supplemental Fig. S3) cells and immunoblotted for GIV. GIV co-immunoprecipitated with SHP-1 in both cell lines, indicating that they interact at steady state in vivo. Of note, this GIV-SHP-1 interaction is specific because GIV did not co-immunoprecipitate with the closely related family member SHP-2 (Fig. 4A). To assess if the GIV-SHP-1 interaction is ligand-dependent, we immunoprecipitated SHP-1 from serum-starved or EGF-stimulated COS7 cells and analyzed the immune complexes for GIV. We found that the ratio of GIV:SHP-1 in immune complexes remained unchanged before and after EGF stimulation, indicating that the interaction occurs constitutively, i.e. independent of receptor activation. The abundance of GIV-SHP-1 interaction also remained unchanged regardless of the activation status of Src (data not shown). In contrast to GIV, Gαi3 did not interact with SHP-1 in either COS7 or HEK cell lines (Fig. 4B, supplemental Fig. S3). To determine whether the GEF motif of GIV, via which GIV activates Gαi subunits (1–3, 7), is required for the GIV-SHP-1 interaction, we immunoprecipitated SHP-1 from COS7 cells expressing WT or the GEF-deficient F1685A (FA) mutant of GIV-FLAG and analyzed the immune complexes for GIV-FLAG. We found that the relative abundance of GIV:SHP-1 complexes was similar between GIV-FA and GIV-WT (Fig. 4C), demonstrating that GIV GEF motif and GIV-dependent activation of Gαi are not required for GIV to interact with SHP-1 in cells. Taken together, we conclude that GIV, but not Gαi3, interacts specifically with SHP-1 (and not SHP-2) in mammalian cells, and this interaction occurs constitutively, i.e. independent of both receptor and G protein activation.
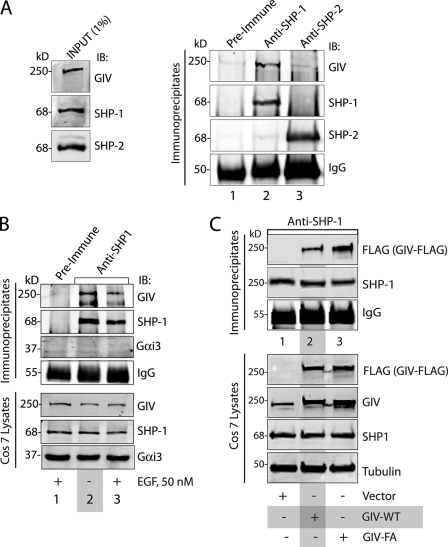
FIGURE 4.
SHP-1 interacts with GIV in vivo. A, GIV co-immunoprecipitates with SHP-1, but not SHP-2. Equal aliquots of COS7 lysates (left) were incubated with either rabbit preimmune (first lane) or anti-SHP-1 (second lane) or anti-SHP-2 (third lane) IgGs and protein A-agarose beads. Immune complexes (25) were analyzed for GIV, SHP-1, and SHP-2 by immunoblotting (IB). SHP-1 and SHP-2 were immunoprecipitated efficiently and specifically. GIV co-immunoprecipitated with SHP-1 (second lane) but not SHP-2 (third lane) or control IgG (first lane). B, GIV, but not Gαi3, interacts constitutively with SHP-1. COS7 cells were serum-starved (−) and subsequently stimulated with 50 nm EGF for 10 min (+). Equal aliquots of lysates (bottom) were incubated with either rabbit preimmune (lane 1) or anti-SHP-1 (lanes 2 and 3) IgGs and protein A-agarose beads. Immune complexes (top) were analyzed for GIV, SHP-1, and Gαi3 by immunoblotting. SHP-1 was immunoprecipitated efficiently and specifically. GIV, but not Gαi3, co-immunoprecipitated with SHP-1 from both starved and EGF-stimulated cells (lanes 2 and 3). C, GIV GEF motif is not required for the GIV-SHP-1 interaction. COS7 cells were transfected with vector alone (lane 1) or FLAG-tagged wild-type GIV (GIV-WT; lane 2), or the GEF-deficient F1685A (FA) mutant (GIV-FA; lane 3). Equal aliquots of lysates (bottom) were incubated with anti-SHP-1 IgGs and protein A-agarose beads. Immune complexes were analyzed for FLAG (GIV-FLAG) and SHP-1 by immunoblotting.
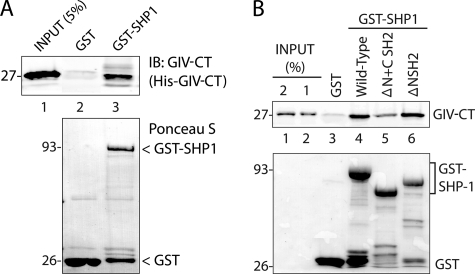
To determine whether the GIV-SHP-1 interaction we observe in vivo is direct, we carried out in vitro binding assays using recombinant His-GIV-CT and GST or GST-SHP-1 immobilized on glutathione beads. GST-SHP-1, but not GST alone, bound His-GIV-CT (Fig. 5A), indicating that SHP-1 directly binds GIV and that the C terminus of GIV is sufficient to mediate such an interaction. Because SHP-1 is recruited to its substrates by one or both of its in-tandem SH2 domains (30), next we asked if the SH2 domains (if so, which one (NSH2 or CSH2)) is responsible for mediating the direct interaction of SHP-1 with the C terminus of GIV. For this we generated GST-tagged truncated versions of SHP-1 lacking either both SH2 domains (ΔN+C SH2) or just the NSH2 domain (ΔNSH2, in which the CSH2 is intact) and used them alongside GST-SHP-1 (full-length) in binding assays. When these GST-SHP-1 proteins were immobilized on glutathione beads and incubated with His-GIV-CT, GIV-CT bound equally to full-length SHP-1 and to SHP-1 ΔNSH2, whereas binding to SHP-1 ΔN+CSH2 was dramatically reduced by ∼80% (Fig. 5B), indicating that the NSH2 domain is not required for SHP-1 to bind GIV, but the CSH2 domain is, and that the CSH2 domain can account for the observed interaction between GIV-CT and SHP-1 in vitro. Thus, the second (CSH2) but not the first (NSH2) of the two in-tandem SH2 domains of SHP-1 directly binds the C terminus of GIV. Noteworthy, the observed interaction between GST-SHP-1 and His-GIV-CT was not affected by tyrosine phosphorylation of GIV-CT (data not shown). Taken together with our findings by co-immunoprecipitation assays on cell lysates (Fig. 4), we conclude that the GIV-SHP-1 interaction is direct and constitutive.
FIGURE 5.
The C terminus of GIV directly binds to SHP-1. A, His-GIV-CT (amino acids 1660–1870) directly binds GST-SHP-1. Equal aliquots (∼3 μg) of His-GIV-CT were incubated with ∼30 μg of GST and ∼25 μg of GST-SHP-1 immobilized on glutathione beads. Bound His-GIV-CT was analyzed by immunoblotting (IB) with GIV-CT pAb. Relative amounts of bead-bound GST and GST-SHP-1 were confirmed by Ponceau S staining. B, His-GIV-CT preferentially binds the C-terminal SH2 domain of SHP-1. Equal aliquots (∼3 μg) of His-GIV-CT were incubated with ∼30 μg of GST, wild-type GST-SHP-1, GST-SHP-1 lacking both SH2 domains (ΔN+CSH2), and GST-SHP-1 lacking only the N-terminal SH2 domain (ΔNSH2). Bound His-GIV-CT was analyzed by immunoblotting with GIV-CT pAb. His-GIV-CT specifically bound wild-type GST-SHP-1 but not GST alone (lanes 3 and 4); binding was reduced in the absence of both SH2 domains (lane 5) but restored when the C-terminal SH2 domain was present (lane 6).
SHP-1 Attenuates GIV-dependent Enhancement of PI3K-Akt Signals by Dephosphorylating Tyrosine-phosphorylated GIV and Inhibiting the Assembly of Phospho-GIV-p85α(PI3K) Complexes
We recently demonstrated (14) that tyrosine-phosphorylated GIV directly binds the p85α-regulatory subunit of PI3K, activates Class 1 PI3K, enhances the generation of second messenger PIP3 at the PM, and triggers the subsequent recruitment and activation of Akt kinase. Because SHP-1 dephosphorylates tyrosine-phosphorylated GIV, we predicted that SHP-1 might also inhibit the ligand-dependent assembly of GIV-PI3K complexes in cells. We found that is indeed the case because EGF-induced formation of complexes between p85α(PI3K) and endogenous GIV (Fig. 6) or FLAG-tagged GIV (supplemental Fig. S4) in COS7 cells was inhibited in the presence of WT but not the inactive CS mutant of HA-SHP-1. Of note, the failure to assemble GIV-p85α(PI3K) complexes in cells expressing SHP-1 WT coincided with a dramatic suppression of tyrosine-phosphorylated GIV to virtually undetectable levels. These findings indicate that the catalytic activity is required for SHP-1 to simultaneously dephosphorylate tyrosine-phosphorylated GIV and to inhibit the formation of phospho-GIV-p85α complexes. These results indicate that the catalytic activity of SHP-1 is required to dephosphorylate the phosphotyrosines in GIV and thereby inhibit the formation of phospho-GIV-p85α complexes.
FIGURE 6.
WT, but not the catalytically inactive CS mutant of SHP-1, inhibits the formation of GIV-p85α(PI3K) complexes after EGF stimulation. COS7 cells transfected with vector alone (lanes 1–3), HA-SHP-1 WT (lane 4), or HA-SHP-1 CS mutant (lane 5) were serum-starved (−) and subsequently treated with 50 nm EGF before lysis. Equal aliquots of lysates (bottom) were incubated with preimmune (lane 1) or anti-GIV-CT (lanes 2–5) IgGs and protein A-agarose beads. Immune complexes (top) were analyzed for endogenous GIV and p85α by immunoblotting (IB). p85α was detectable in GIV-bound complexes after EGF stimulation but not in starved cells (compare lanes 2 and 3), undetectable in cells expressing SHP-1 WT but restored in cells expressing SHP-1 C453S. Ab, antibody.
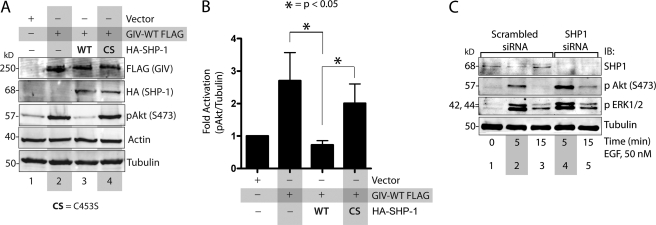
To determine whether SHP-1 affects GIV function as an enhancer of Akt signaling, we co-transfected COS7 cells with GIV-FLAG and either SHP-1-WT or SHP-1-CS mutant and maintained the cells in 2% FBS before lysis. Consistent with the previously established role of GIV as an enhancer of Akt phosphorylation (5, 7, 14), we found that overexpression of GIV-FLAG alone enhanced Akt activity by ∼2.5-fold, as determined by the extent of Akt phosphorylation at Ser-473 (Fig. 7, A and B). Co-expression of WT, but not the inactive CS mutant of SHP-1, returned Akt activity to levels seen in control cells without GIV-FLAG (Fig. 7, A and B), indicating that a catalytically active SHP-1 is required to inhibit GIV from enhancing the phosphorylation and subsequent activation of Akt kinase. Furthermore, depletion of endogenous SHP-1 from HeLa cells by siRNA was associated with enhanced Akt phosphorylation at both 5 and 15 min after EGF stimulation (Fig. 7C), temporally coinciding with the enhanced phosphorylation of GIV observed in these cells at those time points (Fig. 2C). Of note, the peak phosphorylation of ERK1/2 in SHP-1-depleted cells was similar to controls, indicating that the observed effect of SHP-1 on Akt signaling is pathway-specific. Based on these findings, we conclude that SHP-1 antagonizes/attenuates GIV-dependent enhancement of Akt signaling and that enhancement of Akt activity in the absence of SHP-1 is mediated at least in part due to a failure to down-regulate tyrosine-phosphorylated GIV and the phospho-GIV-PI3K-AKt signaling cascade.
FIGURE 7.
SHP-1 inhibits the GIV ability to enhance Akt phosphorylation. A and B, WT, but not the catalytically inactive CS mutant of SHP-1 inhibits GIV-dependent Akt phosphorylation. A, COS7 cells were transfected with empty vector (lane 1), GIV-FLAG alone (lane 2), GIV-FLAG and HA-SHP-1 WT (lane 3), or GIV-FLAG and HA-SHP-1 CS (lane 4). Cells were maintained in the presence of 2% FBS for 20 h before lysis. Equal aliquots of whole cell lysates were analyzed for FLAG, HA (HA-SHP-1), phospho-Akt (pAkt), actin, and tubulin by immunoblotting. Akt phosphorylation at Ser-473 (pAkt) was increased in cells expressing GIV-FLAG (compare lanes 1 and 2), decreased in cells co-expressing GIV-FLAG and SHP-1 WT (lane 3), and increased in cells co-expressing GIV-FLAG and SHP-1 CS (lane 4). B, bar graphs showing quantification of pAkt:actin ratios in A expressed as -fold increase compared with vector control. Quantifications were performed by band densitometry using LiCOR Odyssey Infrared Imager. Results are shown as the mean ± S.E. (n = 5). C, depletion of SHP-1 enhances Akt phosphorylation at 5 min after EGF stimulation. HeLa cells treated with scrambled (lanes 1–3) or SHP-1 (lanes 4 and 5) siRNA were serum-starved and subsequently stimulated with EGF for 5 and 15 min as in Fig. 2C. Equal aliquots of whole cell lysates were analyzed for phosphorylated Akt, phosphorylated ERK (pERK1/2), SHP-1, and tubulin by immunoblotting (IB). pAkt, but not pERK1/2, was specifically and significantly enhanced at both 5 and 15 min after EGF stimulation in SHP-1-depleted cells as compared with control cells (compare lanes 4 and 5 with 2 and 3), temporally coinciding with the enhancement of tyrosine phosphorylation of GIV observed in Fig. 2C.
In summary (Fig. 8), here we demonstrate that SHP-1 is the major cellular phosphatase that down-regulates tyrosine-phosphorylated GIV downstream of both growth factor RTKs and GPCRs and attenuates the previously characterized (14) prometastatic phospho-GIV-PI3K-Akt axis of signaling.
FIGURE 8.
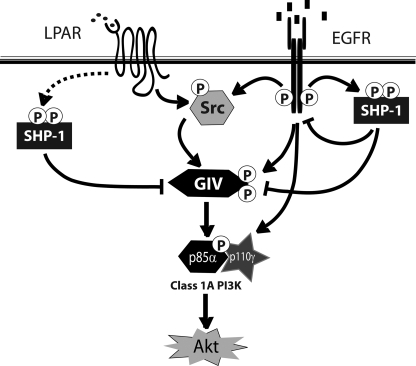
Summary and working model. Tyrosine phosphorylation of GIV is triggered by ligands for either RTKs (i.e. EGFR) or GPCRs (i.e. LPA receptor). Activation of non-RTKs (i.e. Src) downstream of both classes of receptors can also phosphorylate GIV on identical tyrosines (14). Tyrosine-phosphorylated GIV directly binds p85α and activates Class 1 PI3Ks, which in turn activate Akt. SHP-1 protein-tyrosine phosphatase is activated downstream of both RTKs and GPCRs. Activated SHP-1 is known to bind and dephosphorylate the autophosphorylation site(s) on the cytoplasmic tail of EGFR (19, 20) and is known to effectively dephosphorylate substrates of Src kinase (56). Here we demonstrate that SHP-1 binds and dephosphorylates GIV, prevents the formation of GIV-p85α(PI3K) complexes, and thereby inhibits activation of Akt via the GIV-PI3K axis.
DISCUSSION
The key finding in this work is the identification of SHP-1 as the major and specific PTP that dephosphorylates GIV and attenuates GIV-dependent enhancement of the PI3K-Akt signaling pathway. We previously demonstrated (14) that ligand stimulation of either RTKs or GPCRs leads to tyrosine phosphorylation of GIV by cooperative action of activated RTKs and non-RTKs, triggers the formation of phospho-GIV-PI3K complexes via two key phosphotyrosines within the C terminus of GIV that serve as docking sites for p85α(PI3K), and thereby enhances the activity of PI3K and Akt kinases. Here we show that ligand stimulation of either RTKs or GPCRs also simultaneously activates SHP-1, which binds and dephosphorylates tyrosine-phosphorylated GIV, dissociates GIV-PI3K complexes, and prevents GIV from enhancing the activity of PI3K, thereby serving as a negative feedback for down-regulating the receptor-GIV-PI3K axis of signal transduction. Our results also demonstrate that SHP-1 is the specific PTP that dephosphorylates both tyrosines in the GIV C terminus and that SHP-1 dephosphorylates these tyrosines irrespective of whether they are phosphorylated by receptor (EGFR) or non-receptor (Src) tyrosine kinase. We conclude that regardless of the type of receptor activated, SHP-1 attenuates the GIV-PI3K-Akt axis of signaling by antagonizing the action of protein-tyrosine kinases on GIV.
We also demonstrate that SHP-1 directly and constitutively binds the C terminus of GIV. By domain mapping, we further established that this interaction is mediated maximally via the second of the two in-tandem SH2 domains (CSH2) in SHP-1. Although SH2 domains commonly recognize and bind phosphorylated tyrosines, thereby triggering the formation of protein complexes in a ligand-dependent manner (31), in the case of the SHP-1 SH2 domains, a preference for phosphorylated GIV in vitro or ligand dependence in cells was not observed. However, this is not unusual because the SH2 domains of SHP-1 are known to often bind interacting partners/substrates in a phosphotyrosine-independent manner (32–37), including its own C terminus (38). Our immunoprecipitation assays demonstrate that only a portion of GIV and SHP-1 molecules is constitutively associated (Fig. 4). Given that both the C terminus of GIV (2) and the SH2 domains of SHP-1 (19, 20) can independently and directly bind EGFR via phosphotyrosines in the cytoplasmic tails of the ligand-activated receptor, our findings are compatible with the possibility that in unstimulated cells the SHP-1-GIV interaction is direct and is independent of receptor, whereas ligand stimulation may trigger the formation of ternary complexes between autophosphorylated EGFR, the C terminus of GIV, and SHP-1 via three-way direct or indirect interactions; (i) GIV and Tyr(P)-1148 EGFR (2),5 (ii) SHP-1(NSH2) and Tyr(P)-1173 EGFR (19, 20), and (iii) GIV and SHP-1(CSH2) (this work). Although these receptor-dependent interactions are acute and are mostly implicated in fine-tuning receptor autophosphorylation and down-regulation of receptor signaling (2, 19, 20), the receptor-independent, direct and constitutive association between GIV and SHP-1 we report here may allow a more flexible and diverse regulation of both GIV functions and SHP-1 activity.
With regard to the biological significance of such partial constitutive interaction, there are several possibilities. We identify one major consequence of such an interaction as dephosphorylation and down-regulation of tyrosine-phosphorylated GIV by SHP-1. Besides ensuring a rapid down-regulation of tyrosine-based signaling by GIV, another possible consequence of the constitutive proximity of SHP-1 to its substrates could be that such a setup allows the substrate to feedback regulate SHP-1 catalytic activity, as seen in other instances (32, 35, 37, 39). For example, GIV interaction with the CSH2 domain of SHP-1 could “release” the conformation-dependent autoinhibition of SHP-1 by its in-tandem NSH2 domain (38), as shown for RTKs (38). Noteworthy, we did not find any homology between GIV sequences flanking its two C-terminal phosphotyrosines and the conserved immunoreceptor tyrosine-based inhibition motifs that are found in the cytoplasmic tails of many receptors of the immune system that bind to the CSH2 domain of SHP-1 and activate the phosphatase (40). Thus, it is unlikely that binding of GIV to the CSH2 domain directly activates the catalytic activity of SHP-1. However, it is tempting to speculate that the ability of GIV to bind the CSH2 domain of SHP-1 might affect the ability of other immunoreceptor tyrosine-based inhibition motifs to activate the phosphatase in cells, and this might be another mechanism via which GIV may interfere with activation of SHP-1 after receptor stimulation and further enhance phosphotyrosine-based signaling.
We also define the molecular mechanism by which SHP-1 antagonizes the phospho-GIV-PI3K-Akt axis of signaling. Here we demonstrate that by dephosphorylating phosphotyrosines 1764 and 1798 within the C terminus of GIV, the two critical phosphotyrosines of GIV that serve as docking sites for the SH2 domains of p85α-regulatory subunit of Class 1A PI3Ks, SHP-1 inhibits the formation of GIV-PI3K complexes and abrogates phospho-GIV-dependent enhancement of PI3K activity and inhibits the subsequent phosphorylation and activation of Akt in cells. We also demonstrate that SHP-1, but not its closest structurally related family member, SHP-2, is the specific phosphatase for GIV. This is in keeping with the fact that whereas SHP-1 antagonizes Akt signaling (18, 41–44) and inhibits chemotaxis (27, 45–48), SHP-2 is frequently associated with enhancement of the PI3K-Akt pathway (49–53) during growth factor-initiated cell migration (54). Of note, all of the experimental evidence supporting SHP-1 ability to antagonize the Akt pathway was obtained using mesenchymal and hematopoietic cells and the biological role of the epithelium-specific isoform of SHP-1 and how it might inhibit chemotaxis/cell migration remained elusive (55). We show here that in epithelial cells depleted of SHP-1, both peak tyrosine phosphorylation of GIV and Akt activity are coincidently enhanced. By contrast, in cells expressing a constitutively dephosphorylated mutant GIV in which both tyrosines 1764 and 1798 are mutated to phenylalanines, PI3K-Akt signals are abrogated, actin remodeling fails to generate stress fibers, and consequently, cells do not migrate efficiently in scratch-wound assays (14). Noteworthy, these phenotypes (Akt enhancement, actin remodeling, and cell migration) have been previously implicated in contributing to the prometastatic functions of GIV expressed in tumor cells (1–4, 7, 11, 12). These findings together with the data presented in this work define the biological importance of the epithelial isoform of SHP-1 (55).
Our findings also underscore the clinical significance of SHP-1 in antagonizing the previously defined prometastatic functions of GIV that are expressed in the tumor epithelium (1–4, 12). GIV and SHP-1 display a striking contrast with regard to their level of expression and role in tumor cell invasion and angiogenesis during cancer progression (expression of GIV mRNA and protein increase during cancer progression (2, 3, 12)), and GIV is essential for tumor cell invasion and neoangiogenesis (3, 4), whereas expression of SHP-1 is suppressed during cancer progression, and SHP-1 is known to inhibit angiogenesis (57–62). Because the role of GIV-dependent PI3K-Akt enhancement and cell migration extends beyond tumor cell invasion/metastasis into other diverse biological processes, e.g. epithelial wound healing (1, 8), neuronal migration (10, 63, 64), cellular autophagy (6), maintenance of vascular integrity after injury (65), and neoangiogenesis (4), it is plausible that SHP-1 antagonizes GIV functions during all these processes via a common underlying mechanism outlined here.
In conclusion, we not only identify SHP-1 as the phosphatase that dephosphorylates GIV and elucidate the molecular mechanism(s) by which it attenuates a major tyrosine-based signaling pathway, but we also provide insights into the hitherto elusive role of epithelial SHP-1.
Supplementary Material
Acknowledgments
We thank Marilyn G. Farquhar (University of California, San Diego) for scientific advice and thoughtful comments during preparation of this manuscript and Jason Ear for assistance with protein expression and purification.
This work was funded by the Burroughs Wellcome Fund, Research Scholar Award (American Gastroenterology Association), and a Clinical Scientist Development Award (Doris Duke Charitable Foundation) (to P. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- GIV
- Gα-interacting, vesicle-associated protein
- GEF
- guanine nucleotide exchange factor
- RTK
- receptor-tyrosine kinase
- GPCR
- G protein-coupled receptor
- PTP
- protein-tyrosine phosphatase
- EGFR
- EGF receptor
- LPA
- lysophosphatidic acid
- SH2
- c-Src homology 2
- SHP-1
- SH2-containing protein-tyrosine phosphatase-1
- PM
- plasma membrane
- CS
- C453S
- FA
- F1685A
- pAb
- polyclonal antibody
- CT
- C terminus.
REFERENCES
- 1. Enomoto A., Murakami H., Asai N., Morone N., Watanabe T., Kawai K., Murakumo Y., Usukura J., Kaibuchi K., Takahashi M. (2005) Dev. Cell 9, 389–402 [DOI] [PubMed] [Google Scholar]
- 2. Ghosh P., Beas A. O., Bornheimer S. J., Garcia-Marcos M., Forry E. P., Johannson C., Ear J., Jung B. H., Cabrera B., Carethers J. M., Farquhar M. G. (2010) Mol. Biol. Cell 21, 2338–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jiang P., Enomoto A., Jijiwa M., Kato T., Hasegawa T., Ishida M., Sato T., Asai N., Murakumo Y., Takahashi M. (2008) Cancer Res. 68, 1310–1318 [DOI] [PubMed] [Google Scholar]
- 4. Kitamura T., Asai N., Enomoto A., Maeda K., Kato T., Ishida M., Jiang P., Watanabe T., Usukura J., Kondo T., Costantini F., Murohara T., Takahashi M. (2008) Nat. Cell Biol. 10, 329–337 [DOI] [PubMed] [Google Scholar]
- 5. Anai M., Shojima N., Katagiri H., Ogihara T., Sakoda H., Onishi Y., Ono H., Fujishiro M., Fukushima Y., Horike N., Viana A., Kikuchi M., Noguchi N., Takahashi S., Takata K., Oka Y., Uchijima Y., Kurihara H., Asano T. (2005) J. Biol. Chem. 280, 18525–18535 [DOI] [PubMed] [Google Scholar]
- 6. Garcia-Marcos M., Ear J., Farquhar M. G., Ghosh P. (2011) Mol. Biol. Cell 22, 673–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Garcia-Marcos M., Ghosh P., Farquhar M. G. (2009) Proc. Natl. Acad. Sci. U. S. A. 106, 3178–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ghosh P., Garcia-Marcos M., Bornheimer S. J., Farquhar M. G. (2008) J. Cell Biol. 182, 381–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garcia-Marcos M., Ghosh P., Ear J., Farquhar M. G. (2010) J. Biol. Chem. 285, 12765–12777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Enomoto A., Asai N., Namba T., Wang Y., Kato T., Tanaka M., Tatsumi H., Taya S., Tsuboi D., Kuroda K., Kaneko N., Sawamoto K., Miyamoto R., Jijiwa M., Murakumo Y., Sokabe M., Seki T., Kaibuchi K., Takahashi M. (2009) Neuron 63, 774–787 [DOI] [PubMed] [Google Scholar]
- 11. Enomoto A., Ping J., Takahashi M. (2006) Ann. N.Y. Acad. Sci. 1086, 169–184 [DOI] [PubMed] [Google Scholar]
- 12. Garcia-Marcos M., Jung B. H., Ear J., Cabrera B., Carethers J. M., Ghosh P. (2011) FASEB J. 25, 590–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ghosh P., Garcia-Marcos M., Farquhar M. G. (2011) Cell Adh. Migr. 5, 237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin C., Ear J., Pavlova Y., Mittal Y., Kufareva I.., Ghassemian M., Abagyan R., Garcia-Marcos M., Ghosh P. (2011) Science's STKE, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Larsen M., Tremblay M. L., Yamada K. M. (2003) Nat. Rev. Mol. Cell Biol. 4, 700–711 [DOI] [PubMed] [Google Scholar]
- 16. Cuevas B., Lu Y., Watt S., Kumar R., Zhang J., Siminovitch K. A., Mills G. B. (1999) J. Biol. Chem. 274, 27583–27589 [DOI] [PubMed] [Google Scholar]
- 17. Yu Z., Su L., Hoglinger O., Jaramillo M. L., Banville D., Shen S. H. (1998) J. Biol. Chem. 273, 3687–3694 [DOI] [PubMed] [Google Scholar]
- 18. Lu Y., Yu Q., Liu J. H., Zhang J., Wang H., Koul D., McMurray J. S., Fang X., Yung W. K., Siminovitch K. A., Mills G. B. (2003) J. Biol. Chem. 278, 40057–40066 [DOI] [PubMed] [Google Scholar]
- 19. Keilhack H., Tenev T., Nyakatura E., Godovac-Zimmermann J., Nielsen L., Seedorf K., Böhmer F. D. (1998) J. Biol. Chem. 273, 24839–24846 [DOI] [PubMed] [Google Scholar]
- 20. Tenev T., Keilhack H., Tomic S., Stoyanov B., Stein-Gerlach M., Lammers R., Krivtsov A. V., Ullrich A., Böhmer F. D. (1997) J. Biol. Chem. 272, 5966–5973 [DOI] [PubMed] [Google Scholar]
- 21. Le-Niculescu H., Niesman I., Fischer T., DeVries L., Farquhar M. G. (2005) J. Biol. Chem. 280, 22012–22020 [DOI] [PubMed] [Google Scholar]
- 22. Bairstow S. F., Ling K., Anderson R. A. (2005) J. Biol. Chem. 280, 23884–23891 [DOI] [PubMed] [Google Scholar]
- 23. Fawcett V. C., Lorenz U. (2005) J. Immunol. 174, 2849–2859 [DOI] [PubMed] [Google Scholar]
- 24. Mizuno K., Tagawa Y., Mitomo K., Arimura Y., Hatano N., Katagiri T., Ogimoto M., Yakura H. (2000) J. Immunol. 165, 1344–1351 [DOI] [PubMed] [Google Scholar]
- 25. Cooper J. A., Gould K. L., Cartwright C. A., Hunter T. (1986) Science 231, 1431–1434 [DOI] [PubMed] [Google Scholar]
- 26. Kamps M. P., Sefton B. M. (1986) Mol. Cell. Biol. 6, 751–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ngai J., Inngjerdingen M., Berge T., Taskén K. (2009) BMC Immunol. 10, 27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blanchetot C., Chagnon M., Dubé N., Hallé M., Tremblay M. L. (2005) Methods 35, 44–53 [DOI] [PubMed] [Google Scholar]
- 29. Côté J. F., Charest A., Wagner J., Tremblay M. L. (1998) Biochemistry 37, 13128–13137 [DOI] [PubMed] [Google Scholar]
- 30. Pei D., Wang J., Walsh C. T. (1996) Proc. Natl. Acad. Sci. U. S. A. 93, 1141–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yaffe M. B. (2002) Nat. Rev. Mol. Cell Biol. 3, 177–186 [DOI] [PubMed] [Google Scholar]
- 32. Jones M. L., Craik J. D., Gibbins J. M., Poole A. W. (2004) J. Biol. Chem. 279, 40475–40483 [DOI] [PubMed] [Google Scholar]
- 33. Feng Y. H., Sun Y., Douglas J. G. (2002) Proc. Natl. Acad. Sci. U. S. A. 99, 12049–12054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Berg K. L., Siminovitch K. A., Stanley E. R. (1999) J. Biol. Chem. 274, 35855–35865 [DOI] [PubMed] [Google Scholar]
- 35. Jiao H., Berrada K., Yang W., Tabrizi M., Platanias L. C., Yi T. (1996) Mol. Cell. Biol. 16, 6985–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mizuno K., Katagiri T., Hasegawa K., Ogimoto M., Yakura H. (1996) J. Exp. Med. 184, 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yoshida K., Kharbanda S., Kufe D. (1999) J. Biol. Chem. 274, 34663–34668 [DOI] [PubMed] [Google Scholar]
- 38. Pei D., Lorenz U., Klingmüller U., Neel B. G., Walsh C. T. (1994) Biochemistry 33, 15483–15493 [DOI] [PubMed] [Google Scholar]
- 39. Kharbanda S., Bharti A., Pei D., Wang J., Pandey P., Ren R., Weichselbaum R., Walsh C. T., Kufe D. (1996) Proc. Natl. Acad. Sci. U. S. A. 93, 6898–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Burshtyn D. N., Lam A. S., Weston M., Gupta N., Warmerdam P. A., Long E. O. (1999) J. Immunol. 162, 897–902 [PubMed] [Google Scholar]
- 41. Dubois M. J., Bergeron S., Kim H. J., Dombrowski L., Perreault M., Fournès B., Faure R., Olivier M., Beauchemin N., Shulman G. I., Siminovitch K. A., Kim J. K., Marette A. (2006) Nat. Med. 12, 549–556 [DOI] [PubMed] [Google Scholar]
- 42. Sugano M., Tsuchida K., Hata T., Makino N. (2005) FASEB J. 19, 2054–2056 [DOI] [PubMed] [Google Scholar]
- 43. Cui T. X., Nakagami H., Nahmias C., Shiuchi T., Takeda-Matsubara Y., Li J. M., Wu L., Iwai M., Horiuchi M. (2002) Mol. Endocrinol. 16, 2113–2123 [DOI] [PubMed] [Google Scholar]
- 44. Krötz F., Engelbrecht B., Buerkle M. A., Bassermann F., Bridell H., Gloe T., Duyster J., Pohl U., Sohn H. Y. (2005) J. Am. Coll. Cardiol. 45, 1700–1706 [DOI] [PubMed] [Google Scholar]
- 45. Christophi G. P., Massa P. T. (2009) Viral Immunol. 22, 371–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fortin C. F., Larbi A., Lesur O., Douziech N., Fulop T., Jr. (2006) J. Leukoc. Biol. 79, 1061–1072 [DOI] [PubMed] [Google Scholar]
- 47. Kruger J., Butler J. R., Cherapanov V., Dong Q., Ginzberg H., Govindarajan A., Grinstein S., Siminovitch K. A., Downey G. P. (2000) J. Immunol. 165, 5847–5859 [DOI] [PubMed] [Google Scholar]
- 48. Kim C. H., Qu C. K., Hangoc G., Cooper S., Anzai N., Feng G. S., Broxmeyer H. E. (1999) J. Exp. Med. 190, 681–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hakak Y., Hsu Y. S., Martin G. S. (2000) Oncogene 19, 3164–3171 [DOI] [PubMed] [Google Scholar]
- 50. Ivins Zito C., Kontaridis M. I., Fornaro M., Feng G. S., Bennett A. M. (2004) J. Cell. Physiol. 199, 227–236 [DOI] [PubMed] [Google Scholar]
- 51. Wadley G. D., Konstantopoulos N., Macaulay L., Howlett K. F., Garnham A., Hargreaves M., Cameron-Smith D. (2007) J. Appl. Physiol. 102, 1624–1631 [DOI] [PubMed] [Google Scholar]
- 52. Bard-Chapeau E. A., Yuan J., Droin N., Long S., Zhang E. E., Nguyen T. V., Feng G. S. (2006) Mol. Cell. Biol. 26, 4664–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Burks J., Agazie Y. M. (2006) Oncogene 25, 7166–7179 [DOI] [PubMed] [Google Scholar]
- 54. Laramée M., Chabot C., Cloutier M., Stenne R., Holgado-Madruga M., Wong A. J., Royal I. (2007) J. Biol. Chem. 282, 7758–7769 [DOI] [PubMed] [Google Scholar]
- 55. Banville D., Stocco R., Shen S. H. (1995) Genomics 27, 165–173 [DOI] [PubMed] [Google Scholar]
- 56. Frank C., Burkhardt C., Imhof D., Ringel J., Zschörnig O., Wieligmann K., Zacharias M., Böhmer F. D. (2004) J. Biol. Chem. 279, 11375–11383 [DOI] [PubMed] [Google Scholar]
- 57. Witkiewicz A., Raghunath P., Wasik A., Junkins-Hopkins J. M., Jones D., Zhang Q., Odum N., Wasik M. A. (2007) Hum. Pathol. 38, 462–467 [DOI] [PubMed] [Google Scholar]
- 58. Benali N., Cordelier P., Calise D., Pages P., Rochaix P., Nagy A., Esteve J. P., Pour P. M., Schally A. V., Vaysse N., Susini C., Buscail L. (2000) Proc. Natl. Acad. Sci. U. S. A. 97, 9180–9185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Seo D. W., Li H., Qu C. K., Oh J., Kim Y. S., Diaz T., Wei B., Han J. W., Stetler-Stevenson W. G. (2006) J. Biol. Chem. 281, 3711–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Koyama M., Oka T., Ouchida M., Nakatani Y., Nishiuchi R., Yoshino T., Hayashi K., Akagi T., Seino Y. (2003) Lab. Invest. 83, 1849–1858 [DOI] [PubMed] [Google Scholar]
- 61. Tassidis H., Brokken L. J., Jirström K., Ehrnström R., Pontén F., Ulmert D., Bjartell A., Härkönen P., Wingren A. G. (2010) Int. J. Cancer 126, 2296–2307 [DOI] [PubMed] [Google Scholar]
- 62. Tassidis H., Culig Z., Wingren A. G., Härkönen P. (2010) Prostate 70, 1491–1500 [DOI] [PubMed] [Google Scholar]
- 63. Bradshaw N. J., Porteous D. J. (2011) Neuropharmacology, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Porteous D., Millar K. (2009) Neuron 63, 711–713 [DOI] [PubMed] [Google Scholar]
- 65. Miyake H., Maeda K., Asai N., Shibata R., Ichimiya H., Isotani-Sakakibara M., Yamamura Y., Kato K., Enomoto A., Takahashi M., Murohara T. (2011) Circ. Res. 108, 1170–1179 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.