Abstract
Pombe Cdc15 homology proteins, characterized by Fer/CIP4 homology Bin-Amphiphysin-Rvs/extended Fer/CIP4 homology (F-BAR/EFC) domains with membrane invaginating property, play critical roles in a variety of membrane reorganization processes. Among them, Rapostlin/formin-binding protein 17 (FBP17) has attracted increasing attention as a critical coordinator of endocytosis. Here we found that Rapostlin was expressed in the developing rat brain, including the hippocampus, in late developmental stages when accelerated dendritic spine formation and maturation occur. In primary cultured rat hippocampal neurons, knockdown of Rapostlin by shRNA or overexpression of Rapostlin-QQ, an F-BAR domain mutant of Rapostlin that has no ability to induce membrane invagination, led to a significant decrease in spine density. Expression of shRNA-resistant wild-type Rapostlin effectively restored spine density in Rapostlin knockdown neurons, whereas expression of Rapostlin deletion mutants lacking the protein kinase C-related kinase homology region 1 (HR1) or Src homology 3 (SH3) domain did not. In addition, knockdown of Rapostlin or overexpression of Rapostlin-QQ reduced the uptake of transferrin in hippocampal neurons. Knockdown of Rnd2, which binds to the HR1 domain of Rapostlin, also reduced spine density and the transferrin uptake. These results suggest that Rapostlin and Rnd2 cooperatively regulate spine density. Indeed, Rnd2 enhanced the Rapostlin-induced tubular membrane invagination. We conclude that the F-BAR protein Rapostlin, whose activity is regulated by Rnd2, plays a key role in spine formation through the regulation of membrane dynamics.
Keywords: Endocytosis, G Proteins, Neurons, Rho, Signal Transduction
Introduction
Regulation of cell morphology is one of the most important cellular processes in the development of various tissues, including brain. In studies about the mechanisms underlying the regulation of cell morphology, emphasis has been placed on proteins that are known to regulate the reorganization of actin cytoskeleton, such as Rho family small GTPases and their downstream effectors (1–4). In contrast, less is known about the regulatory mechanisms of membrane dynamics involved in the regulation of cell morphology. Therefore, the interplay between membrane dynamics and actin reorganization has been receiving considerable attention in recent years.
PCH (pombe Cdc15 homology)2 family adaptor proteins with evolutionally conserved F-BAR/EFC domains (Fer/CIP4 homology (FCH) domain and BAR (Bin-Amphiphysin-Rvs)/extended Fer/CIP4 homology) have emerged as important coordinators of membrane dynamics (5–9). Rapostlin (also known as forming-binding protein 17, FBP17) is a PCH family member that accumulates at an endocytic site at the invagination step and plays an important role in endocytosis (10–14). The N-terminal F-BAR domain of Rapostlin binds directly to the membrane through the interaction with phosphatidylinositol 4,5-bisphosphate and phosphatidylserine and thereby induces tubular membrane invagination with its concave surface, which is functionally linked to clathrin-mediated endocytosis. Indeed, perturbation of Rapostlin function by shRNA-mediated knockdown shows defects in endocytosis in A431 and HeLa cells (11, 12). On the other hand, Rapostlin binds both N-WASP and dynamin through the C-terminal SH3 domain and recruits them to endocytic sites. N-WASP stimulates actin polymerization through the activation of the Arp2/3 complex (15, 16), whereas dynamin has an established role in driving scission of the endocytic vesicle (17). Thus, Rapostlin mediates the efficient formation of coated vesicles by coordinating membrane invagination and local actin polymerization during endocytosis.
In addition to the N-terminal F-BAR domain and the C-terminal SH3 domain, both of which PCH family members have in common, Rapostlin has an HR1 (protein kinase C-related kinase homology region 1, also referred to as acidic region) domain in the central region that mediates the interaction with a Rho family small GTPase Rnd2 (18), although the functional importance of the interaction remains unsolved. Rnd2 belongs to the Rnd subfamily of Rho family small GTPases that consists of Rnd1, Rnd2, and Rnd3/RhoE (19, 20). Rnd3 antagonizes RhoA-mediated signaling pathways by inhibiting a RhoA effector, ROCK I (21), and by activating a RhoA inactivator, p190 RhoGAP (22). Rnd1 and Rnd2 are predominantly expressed in brain, whereas Rnd3 is expressed ubiquitously (19). Rnd1 is involved in dendritic growth and spine development (23, 24) and also mediates semaphorin-induced repulsive responses through direct interaction with semaphorin receptor plexins (25–29). On the other hand, Rnd2 regulates neuronal migration in the cerebral cortex (30, 31) and is also involved in the regulation of axon growth by semaphorin (32).
Dendritic spines are specialized protrusions from neuronal dendrites where most of the excitatory synapses are formed in the central nervous system (33–38). Spine morphogenesis is a critical step in the establishment of functional synaptic connections. Indeed, abnormal spine morphology is observed in several neurological diseases, such as mental retardation. In addition to extensively dissected cytoskeletal regulation of spine morphogenesis, growing evidence indicates that the endocytic and recycling pathways are also required for spine formation, although the underlying molecular mechanisms are far from being clear (37–40).
Rapostlin is predominantly expressed in brain (18), although its physiological neural functions are largely unknown so far. In this study, we found that Rapostlin was strongly expressed in neurons in late developmental stages when accelerated spine formation occurs. Knockdown experiments in primary cultured hippocampal neurons showed that Rapostlin regulates spine development. We also found that Rnd2 promotes the membrane invaginating activity of Rapostlin and regulates spine formation. These results strongly suggest that Rapostlin and Rnd2 cooperatively regulate spine morphogenesis through the reorganization of the plasma membrane.
EXPERIMENTAL PROCEDURES
Plasmid Constructions and Antibodies
The cDNA encoding wild-type rat Rapostlin (amino acids 2–620 of RapostlinL, the most abundant splicing variant of Rapostlin in brain, Ref. 41) was subcloned into the expression vector pcDNA3 (Invitrogen) or pCMV (Clontech, Mountain View, CA) containing a GFP or Myc epitope tag sequence at the amino terminus. The expression plasmid pCAG encoding YFP was a generous gift from Drs. J. Miyazaki (Osaka University, Osaka, Japan) and T. Saito (Chiba University, Chiba, Japan). The F-BAR domain mutant Rapostlin-QQ (K51Q, K52Q) was created by PCR-mediated mutagenesis. The cDNAs encoding Toca-1 and Rnd2 were obtained as described previously (42–44). The target sequences used for the shRNAs are as follows: Rapostlin shRNA-A, nucleotides 132–150, 5′-ACAGCTCAGGAATCTTTCA-3′ (45); Rapostlin shRNA-B, nucleotides 419–437, 5′-GGTTTGAGCGGGACTGTAA-3′; and Rapostlin shRNA-C, nucleotides 945–963, 5′-GTCCAGAGGCAAGCTCTGG-3′. These were expressed using an shRNA expression vector pSilencer (Ambion, Inc., Austin, TX). The Rnd2 (Rnd2 shRNA-665) and firefly luciferase shRNA expression vectors were obtained as described previously (32, 46, 47). The shRNA-resistant wild-type Rapostlin and two deletion mutants of Rapostlin, ΔHR1 (amino acids 2–398 and 553–620) and ΔSH3 (amino acids 2–552), were subcloned into pEF-BOS with the Myc epitope tag sequence at the amino terminus. The shRNA-resistant mutants were created by PCR-mediated mutagenesis at six nucleotides in the Rapostlin shRNA-B targeting region, which did not change the amino acid sequence of Rapostlin.
A rabbit polyclonal antibody for Rapostlin was raised against a bacterially expressed glutathione S-transferase-fused peptide corresponding to the amino acid residues 339–369 of rat Rapostlin (PPPPPPASASPSAVPNGPQSPKQQKEPLSHR). The specific antibody against Rapostlin was purified with a peptide-conjugated affinity column corresponding to the amino acid residues 354–369 of Rapostlin (CNGPQSPKQQKEPLSHR) or the amino acid residues 339–353 of Rapostlin (CPPPPPPASASPSAVP). A rabbit polyclonal antibody against Rnd2 was obtained as described previously (30). The following antibodies were purchased from commercial sources: a rabbit polyclonal antibody against GFP (Molecular Probes, Inc., Eugene, OR), a mouse monoclonal antibody against Myc (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), a mouse monoclonal antibody against PSD-95 (Abcam, Inc., Cambridge, UK), a mouse monoclonal antibody against Tau-1 (Chemicon, Temecula, CA), a mouse monoclonal antibody against MAP2 (2a+2b) (Sigma-Aldrich), mouse monoclonal antibodies against β-actin and α-tubulin (Sigma-Aldrich), horseradish peroxidase-conjugated secondary antibodies (DAKO, Denmark), Alexa Fluor 488- and 594-conjugated secondary antibodies (Molecular Probes, Inc.), and biotinylated donkey antibody against rabbit IgG (Chemicon).
Immunoblotting
Immunoblotting was performed as described previously (41). Briefly, proteins were separated by SDS-PAGE and were electrophoretically transferred onto a polyvinylidene difluoride membrane (Millipore Corp., Bedford, MA). The membrane was blocked with 3% low-fat milk in Tris-buffered saline and then incubated with primary antibodies. The primary antibodies were detected with horseradish peroxidase-conjugated secondary antibodies and a chemiluminescence detection kit (Chemi-Lumi One, Nacalai Tesque, Japan).
Preparation of Rat Tissue Homogenates
Brains of Wistar rats were homogenized with a Teflon homogenizer in homogenizing buffer (20 mm Tris-HCl (pH 7.4), 0.32 m sucrose, 10 mm MgCl2, 1 mm EDTA, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 0.1 mm benzamidine, 0.2 mm phenylmethylsulfonyl fluoride) as described previously (23, 43). The homogenates were lysed with Laemmli sample buffer and analyzed by immunoblotting.
Immunohistochemistry
Immunohistochemistry was performed as described previously (48). Briefly, postnatal day 14 (P14) Wistar rats were anesthetized with chloral hydrate and perfused transcardially with a fixative containing 4% paraformaldehyde in 0.1 m phosphate buffer (pH 7.4). The brains were removed and postfixed in the fixative at 4 °C overnight. The brains were then saturated with 0.1 m phosphate buffer containing 30% sucrose at 4 °C. The brains were cut into 40-μm-thick coronal sections on a cryostat (CM3050 S, Leica, Nussloch, Germany) at −18 °C. Free-floating sections were treated with 1 mg/ml pepsin (DAKO) in 0.2 N HCl and then incubated at 4 °C overnight with the rabbit antibody against Rapostlin in PBS, containing 0.3% Triton X-100, 0.25% λ-carrageenan, and 0.5% normal donkey serum. After a rinse in PBS containing 0.3% Triton X-100, the sections were incubated for 1 h with a biotinylated donkey antibody against rabbit IgG. The sections were rinsed again and reacted for 1 h with avidin-biotin-peroxidase complex (Elite ABC kit, Vector Laboratories, Burlingame, CA). After a wash of the sections in PBS, bound peroxidase was visualized by incubation with 0.02% 3,3′-diaminobenzidine tetrahydrochloride and 0.001% hydrogen peroxide in 50 mm Tris-HCl (pH 7.6).
Cell Culture and Transfection
HEK293T and HeLa cells were grown in DMEM containing 10% FBS, 4 mm glutamine, 100 units/ml of penicillin, and 0.1 mg/ml of streptomycin under humidified air containing 5% CO2 at 37 °C. HEK293T cells (1 × 106 cells) cultured on 60-mm culture dishes and HeLa cells (3 × 104 cells) cultured in 24-well plates on glass coverslips (circular, 13 mm in diameter) were transfected with test plasmids using Lipofectamine plus and Lipofectamine 2000 (Invitrogen), respectively, according to the manufacturer's instructions.
Cultured hippocampal neurons were prepared from the hippocampi of embryonic day 19 (E19) rats as described previously (23, 24). Briefly, hippocampi of Wistar rat embryos were dissected in ice-cold calcium- and magnesium-free Hanks' balanced salt solution. The hippocampi were washed in Hanks' balanced salt solution and incubated in Hanks' balanced salt solution with 0.25% trypsin and 0.1% DNase for 10 min at 37 °C. After the incubation, the hippocampi were washed in Hanks' balanced salt solution, followed by trituration with Pasteur pipettes. The cells were seeded onto poly-l-lysine-coated glass coverslips (circular, 13 mm in diameter) at a density of 3 × 104 cells in Dulbecco's modified Eagle's medium containing 10% FBS, 4 mm glutamine, 100 units/ml of penicillin, and 0.1 mg/ml of streptomycin and cultured under humidified air containing 5% CO2 at 37 °C. After 5 h, the medium was replaced with neurobasal medium (Invitrogen) with 2% B27 supplement (Invitrogen), 0.5 mm glutamine, 50 units/ml of penicillin, and 0.05 mg/ml of streptomycin and cultured under humidified air containing 5% CO2 at 37 °C. For immunofluorescence analysis, hippocampal neurons were transfected with the indicated plasmids using Lipofectamine 2000. For immunoblot analyses, hippocampal neurons were cultured on poly-l-lysine-coated 35-mm culture dishes. Neurons were transfected with test plasmids using the rat neuron nucleofector kit (Amaxa Biosystems, Cologne, Germany) and lysed with Laemmli sample buffer at 8 DIV.
Immunofluorescence Microscopy
Immunocytochemistry was performed as described previously (41, 43). Primary cultured rat hippocampal neurons on coverslips were fixed with 4% paraformaldehyde in PBS. HeLa cells on coverslips were fixed at 9 h after transfection. After residual paraformaldehyde had been quenched with 50 mm NH4Cl in PBS, cells were permeabilized with 0.2% Triton X-100 in PBS and incubated with 10% FBS in PBS. Cells were incubated with the antibody against Myc or PSD-95 in PBS followed by incubation with an Alexa Fluor 594-conjugated goat antibody against mouse IgG in PBS. We used immunoreaction enhancer solutions (Can Get Signal Immunostain, Toyobo, Osaka, Japan) instead of PBS for dilution of Rapostlin antibody. Cells on coverslips were mounted on 90% glycerol containing 0.1% p-phenylenediamine dihydrochloride in PBS. Images were captured using a Nikon C1 laser scanning confocal imaging system equipped with a Nikon Eclipse TE2000-U microscope or a Leica DC350F digital camera system equipped with a Nikon Eclipse E800 microscope.
Transferrin Uptake Assay and TUNEL Assay
For the transferrin uptake assay, hippocampal neurons were transfected with the indicated plasmids at 7 DIV using Lipofectamine 2000. At 10 DIV, neurons were starved for 6 h in B27 supplement-free neurobasal medium and incubated with 10 μg/ml transferrin conjugated to Alexa Fluor 594 (Molecular Probes, Inc.) for 10 min at 37 °C. After washing once with PBS, neurons were fixed with 4% paraformaldehyde in PBS and mounted for analysis. For the TUNEL assay, hippocampal neurons were transfected with the indicated plasmids at 11 DIV using Lipofectamine 2000 and fixed at 15 DIV. The TUNEL assay was performed as described previously (49).
Data Analysis
Quantification of dendritic spines, axon morphology, and dendrite morphology in hippocampal neurons was performed as described previously (23, 32, 50). The number of dendritic spines was measured from confocal z-series YFP image stacks of the proximal segments of dendrites using ImageJ image analysis software (National Institutes of Health). A PSD-95-positive protrusion with a bulbous head wider than the base was scored as a spine. At least 15 neurons were collected per construct from three experiments, and spines were analyzed within dendritic segments of 100–200 μm in each neuron. Statistical significance of the difference between means was determined by t test. Quantification of fluorescence intensities of Alexa Fluor 594-transferrin was performed as described previously (39, 51, 52). Relative fluorescence intensity was determined from the average fluorescence intensity of Alexa Fluor 594-transferrin in dendrites and the soma of YFP-positive neurons normalized to that of YFP-negative untransfected neurons in the same field. HeLa cells with membrane tubulation were counted as described previously (11, 12, 43).
RESULTS
The Expression of Rapostlin Is Increased during Neuronal Development
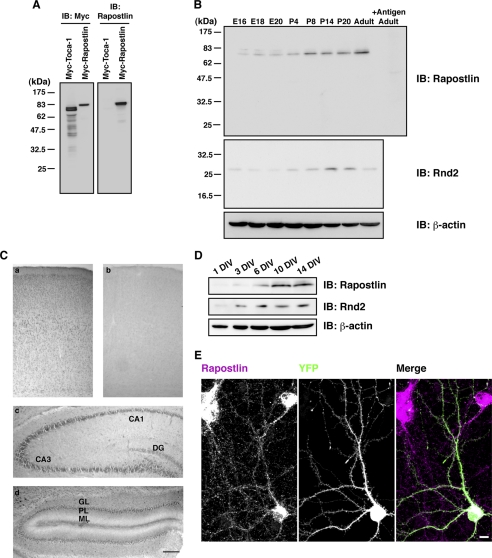
We found previously that Rapostlin is predominantly expressed in brain (18). To characterize the expression of Rapostlin protein in rat brains, we raised an antibody against Rapostlin that recognized Rapostlin overexpressed in HEK293T cells but did not cross-react with the close paralog Toca-1 (Fig. 1A) (43). Immunoblot analysis of rat brain lysates from E16 to adult using this antibody showed that the expression of endogenous Rapostlin protein in the brain was low during embryonic stages and gradually increased in postnatal days (Fig. 1B). The immunoreactivity was abolished by preabsorption of the antibody with antigenic peptide. The expression level of endogenous Rnd2 in brain also increased gradually in postnatal days (Fig. 1B).
FIGURE 1.
Expression and distribution of Rapostlin in rat brain. A, specificity of the antibody against Rapostlin. Cell lysates from HEK293T cells transfected with Myc-tagged Toca-1 or Rapostlin were immunoblotted (IB) with an antibody against Myc or Rapostlin. B, immunoblot analysis of Rapostlin and Rnd2 from rat brain lysates at various ages. Lysates (50 μg of total protein) of rat brain at the indicated ages were subjected to immunoblotting with the antibody against Rapostlin or Rnd2. We also used the antibody against Rapostlin preincubated with excess antigenic peptide to confirm the specificity of the immunoreactivity observed. The level of β-actin was also analyzed as a loading control. C, immunohistochemistry was performed with the antibody against Rapostlin for coronally sectioned P14 rat cerebral cortex (a and b), hippocampus (c), and cerebellum (d). No immunoreactivity was observed in the section adjacent to a when the antibody was preincubated with an excess amount of antigenic peptide (b). DG, dentate gyrus; GL, granular layer; PL, Purkinje layer; ML, molecular layer. Scale bar = 200 μm. D, expression of Rapostlin and Rnd2 in developing rat hippocampal cell cultures. Lysates of hippocampal cell cultures at various stages were subjected to immunoblotting with the antibody against Rapostlin, Rnd2, or β-actin. E, staining with Rapostlin antibody in primary cultured rat hippocampal neurons at 15 DIV. YFP transfection was used to visualize cell morphology. Scale bar = 10 μm.
To further examine the distribution of Rapostlin in the developing brain, we performed immunohistochemistry using the Rapostlin antibody in the brain section prepared at P14. Expression of Rapostlin was observed in cortical neurons, hippocampal neurons, and cerebellar Purkinje cells with high intensity (Fig. 1C). No signal was detected after absorption of the primary antibody with an excess amount of antigen as a control (Fig. 1C and data not shown). This expression pattern was similar to that of Rnd2 mRNA (53). We next prepared primary cultured rat hippocampal neurons and examined whether the expression levels of Rapostlin and Rnd2 were also increased in cultured neurons during development. Immunoblot analysis showed that expression of Rapostlin and Rnd2 was also increased during development in cultured hippocampal neurons (Fig. 1D). Immunocytochemical analysis in 15 DIV cultured hippocampal neurons with the Rapostlin antibody showed that Rapostlin was localized to both cell bodies and dendrites (Fig. 1E).
Knockdown of Rapostlin by shRNA in Rat Hippocampal Neurons Impairs Spine Formation
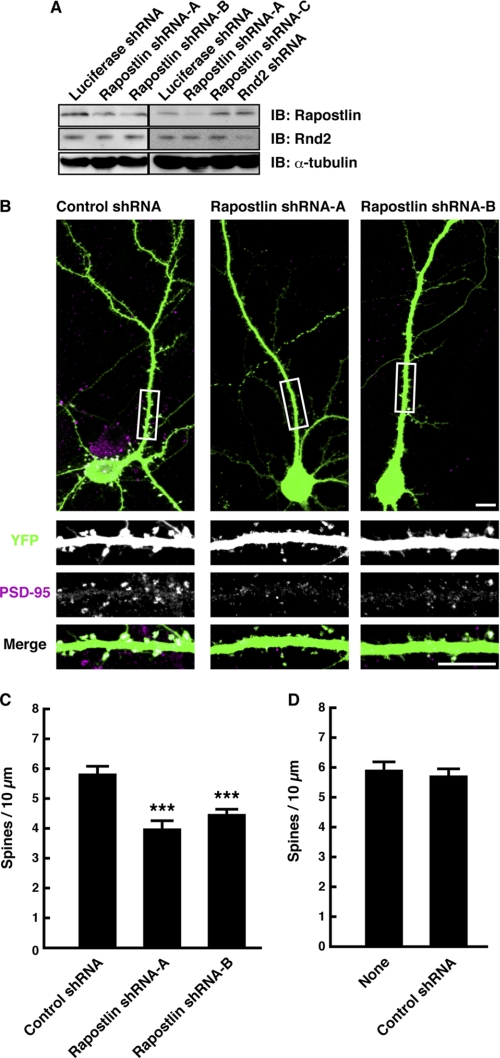
To investigate the physiological function of Rapostlin, we generated shRNA expression vectors targeted to Rapostlin. Rapostlin shRNA-A or -B effectively reduced the level of endogenous Rapostlin protein in cultured hippocampal neurons, whereas luciferase shRNA, Rapostlin shRNA-C, or Rnd2 shRNA had no effect on the amount of Rapostlin protein (Fig. 2A). Therefore, the shRNA-C was used as a control (hereafter called control shRNA). This result also confirmed the specificity of the Rapostlin antibody. Because the expression level of Rapostlin was increased in the period of accelerated spine formation in hippocampal neurons, we examined the effect of shRNA-mediated knockdown of Rapostlin on spine development. Cultured hippocampal neurons at 11 DIV were transiently cotransfected with shRNA and YFP expression vectors to visualize dendrite morphology and were then observed at 15 DIV by staining with an antibody against PSD-95, a postsynaptic marker. We found that expression of Rapostlin shRNA-A or -B significantly decreased spine density compared with that of control shRNA (Fig. 2, B and C). Expression of control shRNA had no obvious effect on neuronal morphology, with little change in spine density compared with untransfected neurons (Fig. 2D). These results suggest that Rapostlin is involved in spine formation in hippocampal neurons.
FIGURE 2.
Knockdown of Rapostlin decreases spine density in primary cultured rat hippocampal neurons. A, cell lysates from neurons transfected with the luciferase shRNA, Rapostlin shRNA, or Rnd2 shRNA expression vector were subjected to immunoblotting (IB) with the antibody against Rapostlin, Rnd2, or α-tubulin to detect endogenous proteins. B, neurons were transiently transfected with control shRNA (Rapostlin shRNA-C), Rapostlin shRNA-A, or Rapostlin shRNA-B and YFP at 11 DIV, and then they were fixed at 15 DIV and stained with the antibody against PSD-95, a postsynaptic marker protein. The transfected cells are shown by the fluorescence of YFP. The three lower panels show enlargements of the boxed regions in the top panel. The top and bottom panels show the merge of the two images with YFP (green) and PSD-95 (magenta). Scale bars = 10 μm. C and D, quantification of the effect of Rapostlin knockdown on spine density in neurons. The number of spines within dendritic segments of 100–200 μm in YFP-positive neurons was counted, and the spine density was calculated (spines/10 μm dendritic shaft). At least 15 neurons were collected per construct from three experiments, and data represent mean ± S.E. ***, p < 0.01 (versus control shRNA, n = 45, t test).
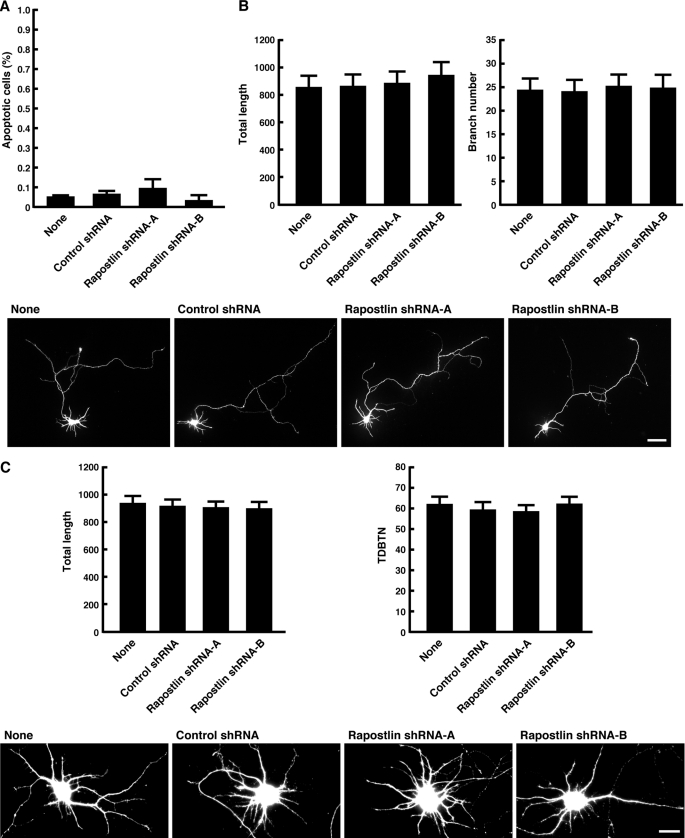
To examine whether the effect of Rapostlin knockdown on the spine formation was not due to a decrease in cell viability, cultured hippocampal neurons at 11 DIV were transiently cotransfected with Rapostlin shRNA and YFP and observed at 15 DIV by TUNEL staining to visualize apoptotic cells. We found that TUNEL-positive cells were low in control neurons transfected with YFP alone and that cotransfection of Rapostlin shRNAs had no effect on the number of TUNEL-positive neurons (Fig. 3A). We also examined the effects of Rapostlin knockdown on axon and dendrite morphology. However, expression of Rapostlin shRNA had no obvious effects on the total length and branching of axons and dendrites (Fig. 3, B and C).
FIGURE 3.
Knockdown of Rapostlin had no effects on axon and dendrite morphology. A, knockdown of Rapostlin does not induce apoptosis. Neurons were transiently transfected with shRNA and YFP at 11 DIV, fixed at 15 DIV, and then TUNEL assays were performed. TUNEL and YFP double-positive cells were scored for TUNEL-positive nuclei, and data represent mean ± S.E. from three independent experiments. B, quantification of axon morphology. Neurons were transiently transfected with shRNA and YFP at 2 DIV, and then they were fixed at 4 DIV. The axons were identified by staining with the axonal marker Tau-1 (data not shown). Scale bar = 50 μm. The axon length and branch number of transfected neurons were measured. Processes longer than 5 μm were considered branches. At least 15 neurons were collected per construct from three experiments, and data represent mean ± S.E. C, quantification of dendrite morphology. Neurons were transiently transfected with shRNA and YFP at 5 DIV, and then they were fixed at 7 DIV. The dendrites were identified by staining with the dendritic marker MAP2 (data not shown). The total length of dendrites and total dendritic branch tip number (TDBTN) of transfected neurons were measured. Dendritic tips were scored when they were longer than 5 μm. At least 15 neurons were collected per construct from three experiments, and data represent mean ± S.E. Scale bar = 20 μm.
Overexpression of Rapostlin-QQ Inhibits Spine Formation
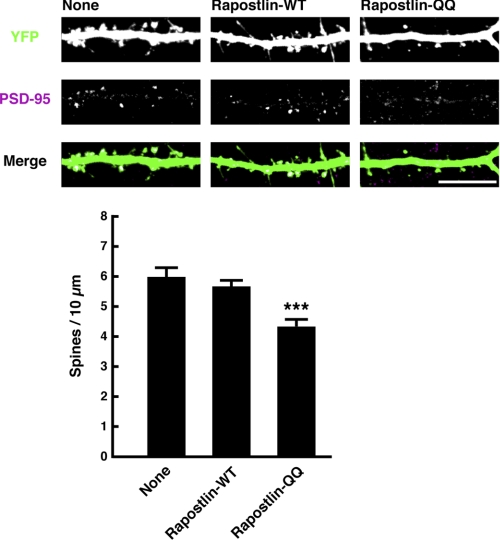
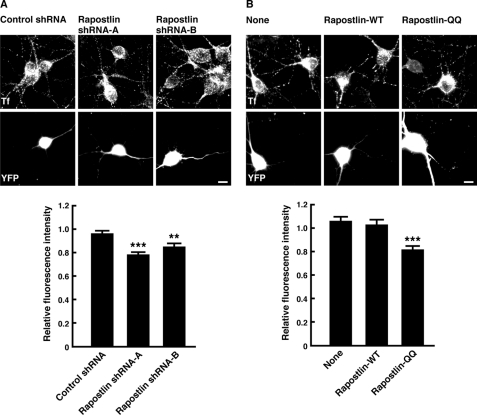
To figure out further functions of Rapostlin, we examined the effect of Rapostlin or its mutant overexpression on spine formation. Overexpression of Myc-tagged wild-type Rapostlin with YFP in cultured hippocampal neurons had no significant effect on spine density compared with overexpression of YFP alone. However, overexpression of Rapostlin-QQ, an F-BAR domain mutant of Rapostlin that has lost the ability to associate with the membrane and to induce plasma membrane invagination (12, 13), significantly decreased spine density (Fig. 4). These results suggest that the F-BAR domain of Rapostlin is important for spine formation in hippocampal neurons and that Rapostlin-QQ acts in a dominant negative manner.
FIGURE 4.
Overexpression of Rapostlin-QQ decreases spine density in primary cultured rat hippocampal neurons. Neurons were transiently transfected with the indicated plasmids at 11 DIV, and then they were fixed at 15 DIV and stained with the antibody against PSD-95. Scale bar = 10 μm. For quantification, the number of spines within dendritic segments of 100–200 μm in YFP-positive neurons was counted, and the spine density was calculated (spines/10 μm dendritic shaft). At least 15 neurons were collected per construct from three experiments, and data represent mean ± S.E. ***, p < 0.01 (versus none, n = 45, t test).
Knockdown of Rapostlin or Overexpression of Rapostlin-QQ Suppresses Transferrin Uptake
Rapostlin is recruited to clathrin-coated pits and plays a role in clathrin-mediated endocytosis through the F-BAR domain-dependent tubular membrane invagination (10–14). Therefore, we next examined whether knockdown of Rapostlin or Rapostlin-QQ overexpression also affected clathrin-mediated endocytosis in hippocampal neurons by measuring the uptake of fluorescently labeled transferrin (Alexa Fluor 594-transferrin) because constitutive clathrin-mediated endocytosis of transferrin occurs in dendrites and the soma of cultured hippocampal neurons (39, 54). Coexpression of YFP with the control shRNA had little effect on transferrin uptake compared with YFP-negative untransfected neurons. However, knockdown of Rapostlin caused a significant reduction in transferrin uptake (Fig. 5A). In addition, overexpression of Rapostlin-QQ also reduced the uptake of transferrin, whereas it was unaffected by overexpression of wild-type Rapostlin (Fig. 5B). These results suggest that Rapostlin mediates constitutive endocytosis of transferrin in hippocampal neurons.
FIGURE 5.
Knockdown of Rapostlin and overexpression of Rapostlin-QQ suppress the uptake of transferrin in primary cultured rat hippocampal neurons. A, neurons were transiently transfected with shRNA and YFP at 7 DIV. After 3 days, they were incubated with Alexa Fluor 594-transferrin for 10 min at 37 °C. The images of Alexa Fluor 594-transferrin (Tf, upper panels) and YFP (lower panels) are shown. Relative fluorescence intensity was determined from the average fluorescence intensity of Alexa Fluor 594-transferrin in dendrites and the soma of YFP-positive neurons normalized to that of YFP-negative untransfected neurons in the same field. At least 20 neurons were collected per construct from three experiments, and data represent mean ± S.E. **, p < 0.01; ***, p < 0.001 (versus control shRNA, n = 60, t test). B, neurons were transiently transfected with Rapostlin-WT or Rapostlin-QQ and YFP at 7 DIV. After 3 days, they were incubated with Alexa Fluor 594-transferrin for 10 min at 37 °C. ***, p < 0.001 (versus none, n = 45, t test). Scale bars = 10 μm.
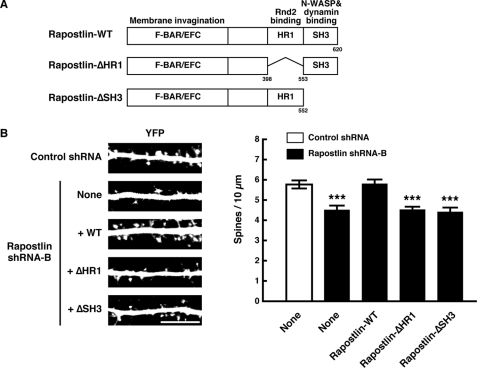
The HR1 and SH3 Domains Are Involved in the Rapostlin-mediated Spine Formation
In addition to the N-terminal F-BAR domain, Rapostlin has the HR1 domain in the central region, which mediates the interaction with Rnd2 (18), and the C-terminal SH3 domain, which mediates the interaction with N-WASP or dynamin (10–12, 55) (Fig. 6A). To examine whether the HR1 and SH3 domains of Rapostlin were also important for Rapostlin-mediated spine formation, we attempted a rescue experiment with deletion mutants of Rapostlin (Rapostlin-ΔHR1 and -ΔSH3). Expression of the shRNA-resistant wild-type Rapostlin completely rescued the impaired spine phenotype caused by Rapostlin shRNA-B, whereas expression of the shRNA-resistant Rapostlin-ΔHR1 and -ΔSH3 failed to rescue the defect caused by Rapostlin knockdown (Fig. 6B). These results suggest that the HR1 and SH3 domains are indispensable for Rapostlin-mediated spine formation.
FIGURE 6.
The HR1 and SH3 domains are involved in the Rapostlin-mediated spine formation. A, the domain structure of Rapostlin. B, primary cultured rat hippocampal neurons were transiently cotransfected with the indicated plasmids at 11 DIV and fixed at 15 DIV. Scale bar = 10 μm. For quantification, the number of spines within dendritic segments of 100–200 μm in YFP-positive neurons was counted, and the spine density was calculated (spines/10 μm dendritic shaft). At least 15 neurons were collected per construct from three experiments, and data represent mean ± S.E. ***, p < 0.001 (versus control shRNA, n = 45, t test).
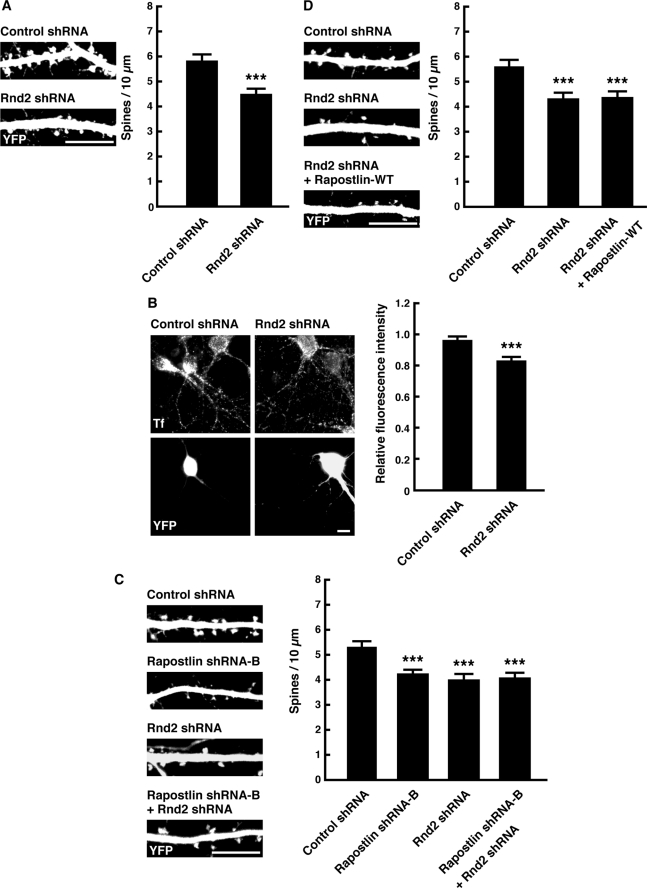
Knockdown of Rnd2 Decreases Both Spine Density and Transferrin Uptake
The involvement of the HR1 domain in the regulation of spine density by Rapostlin prompted us to examine whether Rnd2 also regulates spine formation and transferrin uptake in hippocampal neurons. Expression of Rnd2 shRNA, which effectively reduced the level of endogenous Rnd2 protein in cultured hippocampal neurons (Fig. 2A), significantly decreased spine density and transferrin uptake compared with control shRNA (Fig. 7, A and B). These results suggest that Rnd2 regulates constitutive endocytosis of transferrin and spine formation in hippocampal neurons.
FIGURE 7.
Knockdown of Rnd2 suppresses spine formation and transferrin uptake in primary cultured rat hippocampal neurons. A, neurons were transiently transfected with control or Rnd2 shRNA and YFP at 11 DIV, and then they were fixed at 15 DIV and stained with the antibody against PSD-95. Scale bar = 10 μm. For quantification, the number of spines within dendritic segments of 100–200 μm in YFP-positive neurons was counted, and the spine density was calculated (spines/10 μm dendritic shaft). At least 15 neurons were collected per construct from three experiments, and data represent mean ± S.E. ***, p < 0.001 (versus control shRNA, n = 45, t test). B, neurons were transiently transfected with control or Rnd2 shRNA and YFP at 7 DIV. After 3 days, they were incubated with Alexa Fluor 594-transferrin for 10 min at 37 °C. The images of Alexa Fluor 594-transferrin (Tf, upper panels) and YFP (lower panels) are shown. Scale bar = 10 μm. Relative fluorescence intensity was determined from the average fluorescence intensity of Alexa Fluor 594-transferrin in dendrites and the soma of YFP-positive neurons normalized to that of YFP-negative untransfected neurons in the same field. At least 20 neurons were collected per construct from three experiments, and data represent mean ± S.E. ***, p < 0.001 (versus control shRNA, n = 60, t test). C and D, neurons were transiently transfected with the indicated plasmids at 11 DIV, and then they were fixed at 15 DIV. Scale bar = 10 μm. For quantification, the number of spines within dendritic segments of 100–200 μm in YFP-positive neurons was counted, and the spine density was calculated (spines/10 μm dendritic shaft). At least 15 neurons were collected per construct from three experiments, and data represent mean ± S.E. ***, p < 0.001 (versus control shRNA, n = 45, t test).
To investigate a further relationship between Rapostlin and Rnd2, we performed double knockdown of Rapostlin and Rnd2, and it had no additive inhibitory effect on spine formation (Fig. 7C). This result suggests that Rapostlin and Rnd2 act in the same pathway. We next examined whether Rapostlin overexpression could rescue the impaired spine phenotype caused by Rnd2 knockdown (Fig. 7D). However, overexpression of Rapostlin failed to rescue the defect caused by Rnd2 knockdown. One possible reason for this may be that other binding partners of Rnd2 are also required for the spine formation downstream of Rnd2.
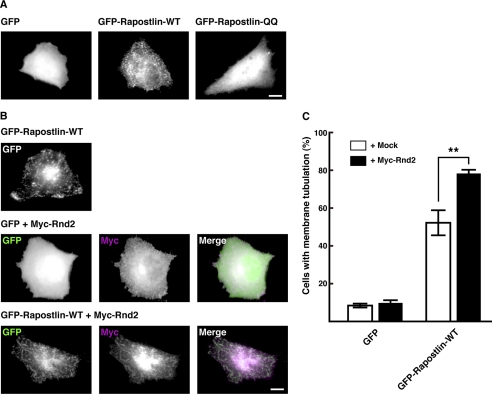
Rnd2 Enhances the Rapostlin-mediated Tubular Plasma Membrane Invagination
We next examined whether Rnd2 affects the Rapostlin-induced membrane tubulating activity in living cells. Rapostlin induces tubular plasma membrane invagination through the N-terminal F-BAR domain when it was overexpressed in COS cells (10–12). These features are widely used to assess the membrane invaginating ability of the overexpressed F-BAR protein. We found that overexpression of GFP-tagged Rapostlin in HeLa cells also induced plasma membrane invagination, which was similar to that observed in COS cells in previous reports (10–12). On the other hand, overexpression of GFP-Rapostlin-QQ failed to induce membrane invagination and showed a diffuse distribution (Fig. 8A). Interestingly, coexpression of Myc-tagged Rnd2 with GFP-Rapostlin enhanced tubular plasma membrane invagination compared with expression of GFP-Rapostlin alone (Fig. 8, B and C). In the cells, Myc-Rnd2 was colocalized with GFP-Rapostlin in tubular structures (Fig. 8B). These results suggest that Rnd2 regulates the Rapostlin-mediated membrane tubulating activity.
FIGURE 8.
Effect of coexpression of Rnd2 on Rapostlin-induced plasma membrane tubulation in HeLa cells. A, HeLa cells were transfected with GFP, GFP-Rapostlin, or GFP-Rapostlin-QQ, and the images of GFP are shown. B, HeLa cells were cotransfected with GFP or GFP-tagged Rapostlin with Myc-tagged Rnd2 and stained with the antibody against Myc. The right panels show the merge of the two images with GFP (green) and Myc (magenta). Scale bars = 20 μm. C, quantification of the effect of Rnd2 expression on Rapostlin-induced membrane tubulation in HeLa cells. Cells with membrane tubulation were quantified, and results were scored as a percentage of the number of the transfected cells. Results are the means ± S.E. of at least three independent experiments in which more than 50 cells were counted. **, p < 0.01 (versus + mock, t test).
DISCUSSION
The F-BAR domains are potent inducers of membrane curvature, and there is growing evidence that Rapostlin, a member of the F-BAR proteins, is recruited to clathrin-coated pits and regulates clathrin-mediated endocytosis through the F-BAR domain-dependent membrane tubulation (11–14). However, its physiological function has been largely unknown. In this study, we found that Rapostlin is expressed in neurons, including hippocampal neurons, in late developmental stages when spine formation occurs. In primary cultured hippocampal neurons, Rapostlin positively regulates constitutive endocytosis and spine formation through the F-BAR domain. The HR1 and SH3 domains also play a role in the regulation of spine formation by Rapostlin. On the other hand, the Rho family small GTPase Rnd2, which directly interacts with Rapostlin through the HR1 domain (18), is involved in the constitutive endocytosis and spine formation. In addition, Rnd2 enhances the tubular membrane invaginating activity of Rapostlin. Collectively, our findings suggest that Rapostlin-mediated membrane dynamics, which are regulated by Rnd2, play a key role in spine morphogenesis, providing a novel molecular mechanism for dendritic development.
Rapostlin belongs to a subfamily consisting of Rapostlin, CIP4, and Toca-1, and recent studies suggest their involvement in the regulation of cell motility and adhesion. CIP4 promotes breast cancer cell motility and invasion (56), and a study with CIP4 knockout mice shows a defect in T-cell trafficking (57). On the other hand, Toca-1 promotes EGF-induced cell motility and invasiveness in epidermoid carcinoma cells (58), and Rapostlin regulates podosome formation in macrophages (59). These results strongly support a role for the F-BAR domain protein family in the regulation of cell morphology. We previously reported that Toca-1 is expressed in brain during early developmental stages and regulates axon branching in hippocampal neurons (43). Here we found that Rapostlin is expressed in late developmental stages, which is in clear contrast to the expression of Toca-1, and regulates dendritic spine development in hippocampal neurons. These results imply that despite their shared homology, Rapostlin and Toca-1 have distinct roles in neuronal development.
Dendritic spines are tiny protrusions that receive most excitatory synaptic inputs and show structural plasticity during development. Recent studies indicate that the endocytic, sorting, and recycling pathways, including recycling endosomes, bring AMPA receptors into spines as well as additional membrane necessary for spine growth and remodeling (38–40). Preventing recycling endosomal transport abolishes activity-dependent new spine formation and maintenance of preexisting spines, and endosomal trafficking occurs concurrently with spine enlargement (38), suggesting that membrane trafficking is important for the regulation of spine morphology. However, the detailed molecular mechanisms underlying membrane trafficking required for spine formation is poorly understood. Our results suggest that Rapostlin may be required for the maintenance of recycling endosomes in dendrites by promoting endocytosis to regulate spine formation. Thus, the present study provides new insight into the molecular mechanism that regulates membrane dynamics for spine formation.
Rapostlin binds both N-WASP and dynamin through the SH3 domain and recruits them to endocytic sites (10–12, 55). N-WASP stimulates actin polymerization through the activation of the Arp2/3 complex (15, 16), and the N-WASP-mediated actin polymerization plays an important role in endocytosis in part by generating forces for scission of endocytic vesicles (60–62). Dynamin also plays a key role in driving scission of endocytic vesicles (17). Thus, Rapostlin plays a pivotal role in endocytosis by coordinating membrane invagination and reorganization of actin cytoskeleton. Our experiments using a knockdown/rescue approach suggest that the SH3 domain is essential for the Rapostlin-mediated spine formation. Interestingly, N-WASP and dynamin also regulate spine formation in hippocampal neurons (63–66). Therefore, Rapostlin may cooperate with N-WASP or dynamin to promote spine formation in hippocampal neurons.
Our results also show that the HR1 domain is important for the Rapostlin-mediated spine formation. The HR1 domain of Rapostlin directly binds to Rnd2 among Rho family small GTPases (18). This study indicates that Rnd2 is involved in the regulation of endocytosis and spine formation in cultured hippocampal neurons. In addition, Rnd2 enhances the membrane-tubulating activity of Rapostlin. Therefore, we hypothesize that Rnd2 might act as a key upstream regulator for Rapostlin in the regulation of endocytosis and spine formation in hippocampal neurons. However, we cannot rule out the possibility that the HR1 domain has additional functions in the regulation of spine formation by Rapostlin because the HR1 domain has been reported to mediate the actin polymerization induced by Rapostlin and N-WASP (55). On the other hand, it is currently unknown how Rnd2 promotes the membrane-tubulating activity of Rapostlin. It has been reported that oligomerization of the F-BAR domain is required for the Rapostlin-induced membrane tubulation (13). Therefore, Rnd2 may promote the oligomerization of Rapostlin. Alternatively, Rnd2 may promote the efficient translocation of Rapostlin to a plasma membrane subdomain because many Rho family members recruit their downstream effectors to the plasma membrane (16, 44), although the F-BAR domain itself has the ability to interact directly with the plasma membrane. Indeed, a Rho family GTPase, TC10, recruits a Rapostlin paralog, CIP4, from intracellular compartments to the plasma membrane upon insulin stimulation (67, 68).
This study has shed light on a novel physiological function of the F-BAR protein Rapostlin in neural development. We also identified Rnd2 as an upstream regulator for the membrane deformation activity of Rapostlin, although it is unclear how the activity of Rnd2 is regulated during dendritic development. In the embryonic cerebral cortex, Rnd2 is regulated by the transcription factor neurogenin2 at the level of its expression to promote migration of neurons (31). Other studies also reported the regulation of the expression level of Rnd2 by a transcription factor or drug treatment (69–71). It will be interesting in future studies to investigate whether the expression of Rnd2 in dendrites is regulated by synaptic activity in hippocampal neurons.
Acknowledgments
We thank Dr. J. Miyazaki (Osaka University) and Dr. T. Saito (Chiba University) for supplying the pCAG-YFP expression plasmid.
This work was supported in part by grants-in-aid for the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
- PCH
- pombe Cdc15 homology
- F-BAR/EFC
- Fer/CIP4 homology (FCH) and BAR (Bin-Amphiphysin-Rvs)/extended Fer/CIP4 homology
- HR1
- protein kinase C-related kinase homology region 1
- SH3
- Src homology 3
- N-WASP
- neural Wiskott-Aldrich syndrome protein
- P14
- postnatal day 14
- E19
- embryonic day 19
- DIV
- day(s) in vitro.
REFERENCES
- 1. Hall A. (1998) Science 279, 509–514 [DOI] [PubMed] [Google Scholar]
- 2. Luo L. (2000) Nat. Rev. Neurosci. 1, 173–180 [DOI] [PubMed] [Google Scholar]
- 3. Negishi M., Katoh H. (2002) J. Biochem. 132, 157–166 [DOI] [PubMed] [Google Scholar]
- 4. Burridge K., Wennerberg K. (2004) Cell 116, 167–179 [DOI] [PubMed] [Google Scholar]
- 5. Aspenström P., Fransson A., Richnau N. (2006) Trends Biochem. Sci. 31, 670–679 [DOI] [PubMed] [Google Scholar]
- 6. Dawson J. C., Legg J. A., Machesky L. M. (2006) Trends Cell Biol. 16, 493–498 [DOI] [PubMed] [Google Scholar]
- 7. Itoh T., De Camilli P. (2006) Biochim. Biophys. Acta 1761, 897–912 [DOI] [PubMed] [Google Scholar]
- 8. Chitu V., Stanley E. R. (2007) Trends Cell Biol. 17, 145–156 [DOI] [PubMed] [Google Scholar]
- 9. Suetsugu S. (2010) J. Biochem. 148, 1–12 [DOI] [PubMed] [Google Scholar]
- 10. Kamioka Y., Fukuhara S., Sawa H., Nagashima K., Masuda M., Matsuda M., Mochizuki N. (2004) J. Biol. Chem. 279, 40091–40099 [DOI] [PubMed] [Google Scholar]
- 11. Itoh T., Erdmann K. S., Roux A., Habermann B., Werner H., De Camilli P. (2005) Dev. Cell 9, 791–804 [DOI] [PubMed] [Google Scholar]
- 12. Tsujita K., Suetsugu S., Sasaki N., Furutani M., Oikawa T., Takenawa T. (2006) J. Cell Biol. 172, 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shimada A., Niwa H., Tsujita K., Suetsugu S., Nitta K., Hanawa-Suetsugu K., Akasaka R., Nishino Y., Toyama M., Chen L., Liu Z. J., Wang B. C., Yamamoto M., Terada T., Miyazawa A., Tanaka A., Sugano S., Shirouzu M., Nagayama K., Takenawa T., Yokoyama S. (2007) Cell 129, 761–772 [DOI] [PubMed] [Google Scholar]
- 14. Wu M., Huang B., Graham M., Raimondi A., Heuser J. E., Zhuang X., Camilli P. (2010) Nat. Cell Biol. 12, 902–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miki H., Takenawa T. (2003) J. Biochem. 134, 309–313 [DOI] [PubMed] [Google Scholar]
- 16. Takenawa T., Suetsugu S. (2007) Nat. Rev. Mol. Cell Biol. 8, 37–48 [DOI] [PubMed] [Google Scholar]
- 17. Hinshaw J. E. (2000) Annu. Rev. Cell Dev. Biol. 16, 483–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fujita H., Katoh H., Ishikawa Y., Mori K., Negishi M. (2002) J. Biol. Chem. 277, 45428–45434 [DOI] [PubMed] [Google Scholar]
- 19. Nobes C. D., Lauritzen I., Mattei M. G., Paris S., Hall A., Chardin P. (1998) J. Cell Biol. 141, 187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chardin P. (2006) Nat. Rev. Mol. Cell Biol. 7, 54–62 [DOI] [PubMed] [Google Scholar]
- 21. Riento K., Guasch R. M., Garg R., Jin B., Ridley A. J. (2003) Mol. Cell Biol. 23, 4219–4229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wennerberg K., Forget M. A., Ellerbroek S. M., Arthur W. T., Burridge K., Settleman J., Der C. J., Hansen S. H. (2003) Curr. Biol. 13, 1106–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ishikawa Y., Katoh H., Negishi M. (2003) J. Neurosci. 23, 11065–11072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ishikawa Y., Katoh H., Negishi M. (2006) Neurosci. Lett. 400, 218–223 [DOI] [PubMed] [Google Scholar]
- 25. Oinuma I., Katoh H., Harada A., Negishi M. (2003) J. Biol. Chem. 278, 25671–25677 [DOI] [PubMed] [Google Scholar]
- 26. Oinuma I., Ishikawa Y., Katoh H., Negishi M. (2004) Science 305, 862–865 [DOI] [PubMed] [Google Scholar]
- 27. Oinuma I., Katoh H., Negishi M. (2004) J. Neurosci. 24, 11473–11480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ito Y., Oinuma I., Katoh H., Kaibuchi K., Negishi M. (2006) EMBO Rep. 7, 704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oinuma I., Katoh H., Negishi M. (2006) J. Cell Biol. 173, 601–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakamura K., Yamashita Y., Tamamaki N., Katoh H., Kaneko T., Negishi M. (2006) Neurosci. Res. 54, 149–153 [DOI] [PubMed] [Google Scholar]
- 31. Heng J. I., Nguyen L., Castro D. S., Zimmer C., Wildner H., Armant O., Skowronska-Krawczyk D., Bedogni F., Matter J. M., Hevner R., Guillemot F. (2008) Nature 455, 114–118 [DOI] [PubMed] [Google Scholar]
- 32. Uesugi K., Oinuma I., Katoh H., Negishi M. (2009) J. Biol. Chem. 284, 6743–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hering H., Sheng M. (2001) Nat. Rev. Neurosci. 2, 880–888 [DOI] [PubMed] [Google Scholar]
- 34. Nimchinsky E. A., Sabatini B. L., Svoboda K. (2002) Annu. Rev. Physiol. 64, 313–353 [DOI] [PubMed] [Google Scholar]
- 35. Yuste R., Bonhoeffer T. (2004) Nat. Rev. Neurosci. 5, 24–34 [DOI] [PubMed] [Google Scholar]
- 36. Tada T., Sheng M. (2006) Curr. Opin. Neurobiol. 16, 95–101 [DOI] [PubMed] [Google Scholar]
- 37. Cooney J. R., Hurlburt J. L., Selig D. K., Harris K. M., Fiala J. C. (2002) J. Neurosci. 22, 2215–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Park M., Salgado J. M., Ostroff L., Helton T. D., Robinson C. G., Harris K. M., Ehlers M. D. (2006) Neuron 52, 817–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thomas S., Ritter B., Verbich D., Sanson C., Bourbonnière L., McKinney R. A., McPherson P. S. (2009) J. Biol. Chem. 284, 12410–12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saneyoshi T., Fortin D. A., Soderling T. R. (2010) Curr. Opin. Neurobiol. 20, 108–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kakimoto T., Katoh H., Negishi M. (2004) J. Biol. Chem. 279, 14104–14110 [DOI] [PubMed] [Google Scholar]
- 42. Katoh H., Harada A., Mori K., Negishi M. (2002) Mol. Cell Biol. 22, 2952–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kakimoto T., Katoh H., Negishi M. (2006) J. Biol. Chem. 281, 29042–29053 [DOI] [PubMed] [Google Scholar]
- 44. Katoh H., Negishi M. (2003) Nature 424, 461–464 [DOI] [PubMed] [Google Scholar]
- 45. Tanaka H., Katoh H., Negishi M. (2006) J. Biol. Chem. 281, 10355–10364 [DOI] [PubMed] [Google Scholar]
- 46. Yamaki N., Negishi M., Katoh H. (2007) Exp. Cell Res. 313, 2821–2832 [DOI] [PubMed] [Google Scholar]
- 47. Fujito T., Ikeda W., Kakunaga S., Minami Y., Kajita M., Sakamoto Y., Monden M., Takai Y. (2005) J. Cell Biol. 171, 165–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Katoh H., Fujimoto S., Ishida C., Ishikawa Y., Negishi M. (2006) Brain Res. 1073, 103–108 [DOI] [PubMed] [Google Scholar]
- 49. Fujimoto S., Negishi M., Katoh H. (2009) Mol. Biol. Cell 20, 4941–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ueda S., Fujimoto S., Hiramoto K., Negishi M., Katoh H. (2008) J. Neurosci. Res. 86, 3052–3061 [DOI] [PubMed] [Google Scholar]
- 51. Zhang H., Webb D. J., Asmussen H., Niu S., Horwitz A. F. (2005) J. Neurosci. 25, 3379–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim S. H., Choi H. J., Lee K. W., Hong N. H., Sung B. H., Choi K. Y., Kim S. M., Chang S., Eom S. H., Song W. K. (2006) Genes Cells 11, 1197–1211 [DOI] [PubMed] [Google Scholar]
- 53. Nishi M., Takeshima H., Houtani T., Nakagawara K., Noda T., Sugimoto T. (1999) Mol. Brain Res. 67, 74–81 [DOI] [PubMed] [Google Scholar]
- 54. Mundigl O., Matteoli M., Daniell L., Thomas-Reetz A., Metcalf A., Jahn R., De Camilli P. (1993) J. Cell Biol. 122, 1207–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takano K., Takano K., Toyooka K., Suetsugu S. (2008) EMBO J. 27, 2817–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pichot C. S., Arvanitis C., Hartig S. M., Jensen S. A., Bechill J., Marzouk S., Yu J., Frost J. A., Corey S. J. (2010) Cancer Res. 70, 8347–8356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Koduru S., Kumar L., Massaad M. J., Ramesh N., Le Bras S., Ozcan E., Oyoshi M. K., Kaku M., Fujiwara Y., Kremer L., King S., Fuhlbrigge R., Rodig S., Sage P., Carman C., Alcaide P., Luscinskas F. W., Geha R. S. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 16252–16256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hu J., Mukhopadhyay A., Craig A. W. B. (2011) J. Biol. Chem. 286, 2261–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tsuboi S., Takada H., Hara T., Mochizuki N., Funyu T., Saitoh H., Terayama Y., Yamaya K., Ohyama C., Nonoyama S., Ochs H. D. (2009) J. Biol. Chem. 284, 8548–8556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Merrifield C. J., Qualmann B., Kessels M. M., Almers W. (2004) Eur. J. Cell Biol. 83, 13–18 [DOI] [PubMed] [Google Scholar]
- 61. Benesch S., Polo S., Lai F. P., Anderson K. I., Stradal T. E., Wehland J., Rottner K. (2005) J. Cell Sci. 118, 3103–3115 [DOI] [PubMed] [Google Scholar]
- 62. Kaksonen M., Toret C. P., Drubin D. G. (2006) Nat. Rev. Mol. Cell Biol. 7, 404–414 [DOI] [PubMed] [Google Scholar]
- 63. Irie F., Yamaguchi Y. (2002) Nat. Neurosci. 5, 1117–1118 [DOI] [PubMed] [Google Scholar]
- 64. Gray N. W., Kruchten A. E., Chen J., McNiven M. A. (2005) J. Cell Sci. 118, 1279–1290 [DOI] [PubMed] [Google Scholar]
- 65. Wegner A. M., Nebhan C. A., Hu L., Majumdar D., Meier K. M., Weaver A. M., Webb D. J. (2008) J. Biol. Chem. 283, 15912–15920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jaskolski F., Mayo-Martin B., Jane D., Henley J. M. (2009) J. Biol. Chem. 284, 12491–12503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chang L., Adams R. D., Saltiel A. R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 12835–12840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chang L., Chiang S. H., Saltiel A. R. (2007) Endocrinology 148, 27–33 [DOI] [PubMed] [Google Scholar]
- 69. Armentano M., Filosa A., Andolfi G., Studer M. (2006) Development 133, 4151–4162 [DOI] [PubMed] [Google Scholar]
- 70. Marie-Claire C., Salzmann J., David A., Courtin C., Canestrelli C., Noble F. (2007) Brain Res. 1134, 12–17 [DOI] [PubMed] [Google Scholar]
- 71. Shimomura A., Ohama T., Hori M., Ozaki H. (2009) J. Vet. Med. Sci. 71, 1591–1597 [DOI] [PubMed] [Google Scholar]