Abstract
Aberrant microglial activation has been proposed to contribute to the cognitive decline in Alzheimer disease (AD), but the underlying molecular mechanisms remain enigmatic. Fractalkine signaling, a pathway mediating the communication between microglia and neurons, is deficient in AD brains and down-regulated by amyloid-β. Although fractalkine receptor (CX3CR1) on microglia was found to regulate plaque load, no functional effects have been reported. Our study demonstrates that CX3CR1 deficiency worsens the AD-related neuronal and behavioral deficits. The effects were associated with cytokine production but not with plaque deposition. Ablation of CX3CR1 in mice overexpressing human amyloid precursor protein enhanced Tau pathology and exacerbated the depletion of calbindin in the dentate gyrus. The levels of calbindin in the dentate gyrus correlated negatively with those of tumor necrosis factor α and interleukin 6, suggesting neurotoxic effects of inflammatory factors. Functionally, removing CX3CR1 in human amyloid precursor protein mice worsened the memory retention in passive avoidance and novel object recognition tests, and their memory loss in the novel object recognition test is associated with high levels of interleukin 6. Our findings identify CX3CR1 as a key microglial pathway in protecting against AD-related cognitive deficits that are associated with aberrant microglial activation and elevated inflammatory cytokines.
Keywords: Alzheimers Disease, Amyloid, Cytokine, Inflammation, Neurodegeneration, Cognitive Deficits, Microglia
Introduction
One pathogenic hallmark of neurodegenerative diseases, including Alzheimer disease (AD),3 is aberrant microglial activation (1–4). A recent genome-wide association study, which linked CR1 and CD33 to AD risk, further supported the involvement of inflammatory pathway in AD etiology (5, 6). In AD, besides causing direct toxic effects on neuronal and synaptic functions, amyloid-β (Aβ) and/or Tau serve as inflammatory stimuli to provoke microglial activation and proinflammatory responses (7–14). However, how aberrantly activated microglia lead to the neuronal dysfunction and cognitive decline in AD remains to be determined.
The fractalkine receptor CX3CR1 is expressed by microglia in the central nervous system (CNS) (15–17). The exclusive ligand for CX3CR1 is fractalkine (CX3CL1), which is expressed in neurons as a full-length transmembrane protein and binds CX3CR1 on microglia with high affinity. CX3CL1 can be proteolytically cleaved by tumor necrosis factor-α (TNF-α)-converting enzyme (TACE/ADAM17, ADAM10) or cathepsin S (18–20) and released in a soluble form that is a potent chemoattractant for monocytes, T cells, and natural killer cells.
From the complementary expression of CX3CL1 on neurons and CX3CR1 on microglia, it has been proposed that neuron signaling to microglia is mediated through this receptor (17). Indeed, deleting CX3CR1 by replacing CX3CR1 with a green fluorescent protein (GFP) reporter gene (21) exacerbated microglial neurotoxicity induced by systemic inflammation in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine model of Parkinson disease and in the SOD1G93A model of amyotrophic lateral sclerosis (22). Notably, CX3CR1−/− microglia appeared to exacerbate cell-autonomous microglial neurotoxicity induced by lipopolysaccharide (LPS), suggesting that fractalkine signaling is important for limiting microglia toxicity (22). In agreement with these in vivo findings, in vitro studies showed that CX3CL1 suppressed neuronal cell death induced by microglia activated with LPS and interferon-γ in a dose-dependent manner (23). The production of NO, IL-6, and TNF-α was also inhibited by CX3CL1 (24, 25). In addition, CX3CL1 protects against excitotoxicity through the activation of the ERK1/2 and PI3K/Akt pathways (26, 27).
The role of fractalkine signaling in AD pathogenesis is complex and poorly understood. The effects of CX3CR1 deficiency have been somewhat discordant due in part to the use of differing models and analytical approaches. Deleting CX3CR1 in Tau transgenic mice exacerbated Tau phosphorylation and aggregation as well as behavioral impairments (11). In contrast, deleting CX3CR1 prevented the neuronal loss and microglial migration without affecting amyloid deposition in 3×Tg AD mice (PS1M146V knock-in, transgenic APPswe, and TauP301L) (28). More recent studies focusing on amyloidosis showed that CX3CR1 deficiency attenuated amyloid deposition in AD mouse models with extensive plaque deposition (29, 30). However, no functional effects have been demonstrated in these studies. Because plaque load correlates poorly with the synaptic and functional changes in AD patients (31, 32) and in AD mouse models (33), the current study addressed the functional outcome of CX3CR1 deletion in hAPP-J20 mice by examining its effects on neuronal and cognitive dysfunction and inflammatory responses.
EXPERIMENTAL PROCEDURES
Mice
To remove CX3CR1 genetically in hAPP mice, hAPP-J20 mice (C57BL/6) were crossed with CX3CR1 GFP knock-in mice in which the CX3CR1 gene was replaced with a cDNA encoding GFP (referred to as CX3CR1−/− here) (21). The first cross resulted in hAPP/CX3CR1+/− mice, which were crossed with CX3CR1+/− mice to generate six genotypes: hAPP/CX3CR1−/−, hAPP/CX3CR1+/+, hAPP/CX3CR1+/−, CX3CR1−/−, CX3CR1+/+, and CX3CR1+/−. PCR-based genotyping for CX3CR1 allele or hAPP transgene was performed as described (21, 33). All animal procedures were carried out under University of California, San Francisco, Institutional Animal Care and Use Committee-approved guidelines.
Human Samples
Human brain samples were obtained from the New York Brain Bank at Columbia University, the Alzheimer's Disease Research Center at University of California at San Diego (UCSD), and the Laboratory of Cellular and Molecular Neurobiology at Sun Health Research Institute (Sun City, AZ). See Table 1 for details.
TABLE 1.
Summary of patient information used in the study
| Source | Case number | Gender | Age | Braak stage | Clinical/neuropathological characterization | Brain region |
|---|---|---|---|---|---|---|
| UCSDa | 4870 | F | 63 | 1 | Control | HPb |
| UCSD | 5302 | F | 83 | 1.1 | Control | HP |
| UCSD | 5356 | M | 79 | 1.2 | Alzheimer changes | HP |
| UCSD | 5372 | F | 84 | 1.2 | Alzheimer changes | HP |
| UCSD | 5381 | M | 78 | 2.1 | Alzheimer changes | HP |
| UCSD | 5300 | F | 84 | 5.2 | Alzheimer disease | HP |
| UCSD | 5308 | M | 80 | 5.2 | Alzheimer disease | HP |
| UCSD | 5309 | M | ? | 5.2 | Alzheimer disease | HP |
| UCSD | 5337 | M | 61 | 6.2 | Alzheimer disease | HP |
| UCSD | 5366 | F | 85 | 6.2 | Alzheimer disease | HP |
| UCSD | 5367 | F | 94 | 6.2 | Alzheimer disease | HP |
| UCSD | 5373 | F | 77 | 6.2 | Alzheimer disease | HP |
| UCSD | 5315 | M | 67 | 5.2 | Alzheimer disease | HP |
| UCSD | 5321 | M | 84 | 5.2 | Alzheimer disease | HP |
| UCSD | 5332 | M | 75 | 5.2 | Alzheimer disease | HP |
| UCSD | 5338 | M | 79 | 5.2 | Alzheimer disease | HP |
| UCSD | 5380 | M | 85 | 5.2 | Alzheimer disease | HP |
| UCSD | 5316 | F | 86 | 6.2 | Alzheimer disease | HP |
| UCSD | 5376 | M | 85 | 6.2 | Alzheimer disease | HP |
| UCSD | 5387 | M | 77 | 6.2 | Alzheimer disease | HP |
| UCSD | 5390 | M | 74 | 6.2 | Alzheimer disease | HP |
| UCSD | 5325 | F | 45 | 6.3 | Alzheimer disease | HP |
| NYBBc | 4096 | M | 87 | Severe AD | HP | |
| NYBB | 4134 | F | 65 | Severe AD | HP | |
| NYBB | 4135 | M | 75 | Severe AD | HP | |
| NYBB | 4160 | F | 88 | Severe AD | HP | |
| NYBB | 3833 | M | 85 | Control | HP | |
| NYBB | 3962 | M | 89 | Control | HP | |
| Sun Healthd | 99-44 | M | 69 | 1 | Non-diagnostic; control | Frontal cortex |
| Sun Health | 97-17 | M | 78 | 2 | Non-demented control | Frontal cortex |
| Sun Health | 96-35 | M | 84 | 3 | Control; Cerebral amyloid angiopathy | Frontal cortex |
| Sun Health | 00-49 | F | 86 | 3 | Non-demented control | Frontal cortex |
| Sun Health | 01-13 | F | 91 | 5 | AD, argyrophilic grains, cingulate gyrus, and temporal lobe | Frontal cortex |
| Sun Health | 01-09 | F | 83 | 6 | AD; cerebral white matter rarefaction | Frontal cortex |
| Sun Health | 99-15 | M | 83 | 6 | AD; small old infarct, cerebral white matter rarefaction (CWMR) | Frontal cortex |
| Sun Health | 01-34 | F | 89 | 6 | AD; CWMR; Alzheimer type II astrocytosis | Frontal cortex |
| Sun Health | 01-48 | F | 97 | 6 | AD; CWMR | Frontal cortex |
a UCSD, University of California, San Diego.
b HP, hippocampus.
c NYBB, New York Brain Bank at Columbia University.
d Sun Health, Sun Health Research Institute; now Banner Health.
Western Blot Analysis
Protein extracts were prepared from brain tissue samples with lysis buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 0.5% sodium deoxycholate, 1% Nonidet P-40, 0.1% SDS, protease inhibitors), and concentrations were determined by bicinchoninic acid assay (Pierce). Fifty μg of proteins were resolved in SDS-PAGE and transferred to nitrocellulose membranes. After blocking, membranes were probed with a rabbit polyclonal anti-CX3CR1 (1:500, Abcam, Cambridge, MA), anti-CT15 (1:1000, a kind gift of E. H. Koo, UCSD), a goat polyclonal anti-CX3CL1 (1:2000, R&D Systems, Minneapolis, MN), a mouse monoclonal anti-GAPDH (1:1000, Millipore, Billerica, MA), or anti-AT8 (1:500, Thermo Fisher Scientific) overnight at 4 °C. Horseradish peroxidase-conjugated goat anti-mouse secondary antibody (1:2000, EMD Chemicals, Gibbstown, NJ), rabbit anti-goat secondary antibody (1:2000, EMD Chemicals), or goat anti-rabbit secondary antibody (1:2000, EMD Chemicals) was used to detect the primary antibodies. After several washes, peroxidase activity on the membrane was detected by enhanced chemiluminescence (GE Healthcare) and quantitated by the Quantity One 4.0 software (Bio-Rad).
Aβ ELISA and Immunoblotting
Two-to-four-month-old mice were used to quantitate Aβ levels before plaque formation. Snap-frozen hippocampi were homogenized in 5 m guanidine buffer and quantified as described (49). Low molecular mass Aβ oligomers was detected as described (50). Hippocampal samples were homogenized in buffer containing 0.5 mm EDTA, 1 mm DTT, 100 mm PMSF, phosphatase inhibitor cocktails I and II (Sigma-Aldrich), and protease inhibitors (Roche Applied Science) in PBS. Fifty μg of proteins were immunoprecipitated with anti-Aβ1–40 (5C3, 5 μl, EMD Chemicals) and anti-Aβ1–42 (8G7, 5 μl, EMD Chemicals) antibodies overnight at 4 °C. The amounts of dimers and monomers of Aβ were then determined by Western blotting with the additional step of microwaving the nitrocellulose membrane for 2.5 min before blocking in 3% BSA/Tris-buffered saline with 0.5% Tween and incubating with the primary anti-Aβ antibodies.
Quantitative Reverse Transcription-PCR
Total RNA in primary mixed cultures and primary microglia was isolated with TRIzol (Invitrogen) method, and the total RNA in brain cortex was isolated with the RNeasy mini kit (Qiagen, Valencia, CA). The remaining DNA in the RNA samples was removed by incubation with RNase-free DNase (Ambion, Austin, TX) at 37 °C for 30 min, and the RNA was converted into cDNA by the TaqMan reverse transcription (RT) kit (Applied Biosystems, Foster City, CA). Real-time RT-PCR was carried out on an ABI 7900 HT sequence detector (Applied Biosystems) with SYBR Green PCR master mix (Applied Biosystems). The relative gene expression was quantified as 2−ΔCt. The mean value of replicates of each sample was expressed as the threshold cycle (Ct) at which the amount of fluorescence begins to increase rapidly. The amount of gene expression was calculated as the difference (ΔCT) between the CT value of the sample for target gene and the CT value of that sample for the endogenous control GAPDH (ΔCt = Ct(target gene) − Ct(GAPDH)). The relative amount of gene expression for each target gene was determined by 2−ΔCt and expressed as the -fold change as compared with a control. The following primers were used for real-time RT-PCR: mouse TNF-α (forward, 5′-CATCAGTTCTATGGCCCAGA-3′; reverse, 5′-TGCTCCTCCACTTGGTGGTT-3′), mouse IL-6 (forward, 5′-ACCACTTCACAAGTCGGAGGCT-3′; reverse, 5′-TCTGCAAGTGCATCATCGTTGT-3′), mouse GAPDH (forward, 5′-GGGAAGCCCATCACCATCTT-3′; reverse, 5′-GCCTTCTCCATGGTGGTGAA-3′), rat GAPDH (forward, 5′-CTCAAGATTGTCAGCAATGC-3′; reverse, 5′-TTCCACGATGCCAAAGTTGT-3′), rat CX3CR1 (forward, 5′-CCATGTGCAAGCTCACGACT-3′; reverse, 5′-ACTGTCCGGTTGTTCATGGA-3′), and rat CD11b (forward, 5′-GTGTGACTACAGCACAA-3′; reverse, 5′-CCCAAGGACATATTCAC-3′).
Primary Cortical Cultures and Treatments
To generate mixed cortical culture, cortices from rat pups were isolated on postnatal day 0 or 1 and dissociated. Cells were plated at 160,000 cells/ml in plating medium containing Dulbecco's modified Eagle's medium, 10% fetal bovine serum, 0.5 mm GlutaMAX, 100 units/ml penicillin, and 100 μg/ml streptomycin. For primary microglial culture, cortical cultures were maintained in DMEM medium mentioned above for 2 weeks with changing the medium at day 5. After 2 weeks of culture, flasks were gently shaken, and the microglia containing supernatants were harvested. Cells were plated at 4 × 105 cells/well into 24-well dishes. Purity of primary culture was characterized by immunocytochemistry with the microglia marker Iba-I (Wako). Aβ1–42 peptide lyophilized in hydroxyfluroisopropanol was obtained from rPeptide (Athens, GA). Lyophilized Aβ powder was reconstituted before use in dry dimethyl sulfoxide and further diluted in Dulbecco's modified Eagle's medium at 100 μm. Primary mixed cortical cultures and microglia cultures are treated with Aβ1–42 peptide for 48 or 24 h, respectively. To activate CX3CR1, primary microglia cultures were pretreated with CX3CL1 peptide (R&D Systems) for 16 h before being treated with Aβ1–42 for an additional 24 h. All the supplies for the cell culture were from Invitrogen.
Immunohistochemistry and Quantitation of Immunoreactive Structures
Mice were transcardially perfused with 0.9% saline solution, and then the brains were fixed in 4% paraformaldehyde in phosphate-buffered saline buffer for 48 h followed by an incubation with 30% sucrose. After quenching endogenous peroxidase activity, microtome sections (30 μm) were incubated with primary antibody solution for mouse anti-AT8 (1:200; Thermo Fisher Scientific), rat anti-Mac-2 (1:200; Cedarlane Laboratories), anti-calbindin (1:15000; Swant, Bellinzona, Switzerland), or biotinylated mouse monoclonal antibody 3D6 (5 μg/ml, Elan Pharmaceuticals, South San Francisco, CA). Expression of specific proteins was visualized with the avidin-biotin-peroxidase complex from the VECTASTAIN ABC Elite kit (Vector Laboratories, Burlingame, CA). Calbindin expression in the dentate gyrus (DG) was quantified as described (37). Images were viewed using DEI-470 digital camera (Optronics, Goleta, CA). The BIOQUANT image analysis system (BIOQUANT Image Analysis Corp.) was employed to perform integrated optical density measurement from three coronal sections. Integrated optical density was averaged in two areas of the molecular layer of the DG and of the stratum radiatum of the CA-1 region, in which calbindin levels are unaffected by AD. Relative calbindin levels were expressed as the ratio of integrated optical density in the molecular layer and in the stratum. The plaque load and the Mac-2 immunoreactive microglia were measured as the percentage of the area of the hippocampus covered by 3D6 or Mac-2 staining, respectively, using ImageJ (National Institutes of Health). Three coronal sections were analyzed per mouse, and the average of the individual measurements was used to do the statistics.
Confocal Staining
Brain tissue sections (30 μm) were co-incubated with polyclonal rabbit anti-GFAP at 4 °C for overnight. The antigens were visualized with goat anti-rabbit Alexa Fluor 546 used in 1:200 for 2 h at room temperature. Confocal images were captured using Nikon ECLIPSE Ti 2000 spinning disk confocal microscopy. The percentage of the area of immunoreactive structures from three coronal sections per mouse were measured using ImageJ, and the average value was used to compare group means. For immunostaining of CX3CR1 and CD68 in rat primary cultures, cells were fixed in 4% paraformaldehyde in phosphate-buffered saline for 15 min at room temperature. After 1 h of blocking, cells were incubated with primary antibodies to CD68 (1:200; Serotec, Oxford, UK) and CX3CR1 (1:100; Abcam) overnight at 4 °C and visualized with anti-rat or anti-rabbit conjugated with fluorescein isothiocyanate or Texas Red (Vector Laboratories).
Passive Avoidance
The apparatus consisted of two chambers, one illuminated and the other darkened, separated by a vertical sliding door. Mice were initially placed in the light chamber. After 15 s, a sliding door to the dark chamber was opened, and the latency to enter the dark chamber was recorded. The door was closed as soon as all four paws of the mouse were inside the dark chamber, and an electric shock of 0.3 mA (duration, 2 s) was delivered. Retention was tested 24 or 120 h later, and the latency to enter the dark chamber was recorded up to 500-s cutoff.
Novel Object Recognition
Mice were transferred to the testing room and acclimated for at least 1 h. Mice were tested in a white round plastic chamber 35 cm in diameter. On the first day, mice were habituated to the chamber for 15 min, and then two identical objects were placed into the same chamber. Mice were allowed to explore the objects and the chamber for 10 min as training. Twenty-four h later, one of the original objects was replaced with a novel object of different shape. Mice were then allowed to explore the objects and the chamber for 10 min. Time spent exploring each object was recorded for data analysis. The recognition index was obtained as the ratio of time spent with the novel object to that spent with the familiar object.
Statistical Analysis
Statistical analyses were calculated with GraphPad Prism version 5 (GraphPad, San Diego, CA). All values in figures were expressed as means ± S.E. Differences between means were assessed with paired or unpaired Student's t test or one-way analysis of variance followed by Tukey-Kramer or Newman-Keuls multiple comparison post hoc test. The linear relationship between two measurement variables was made by Pearson's correlation analysis. A p value less than 0.05 was considered significant.
RESULTS
Fractalkine Signaling Was Down-regulated by Aβ in Cultured Microglia and in AD Brains
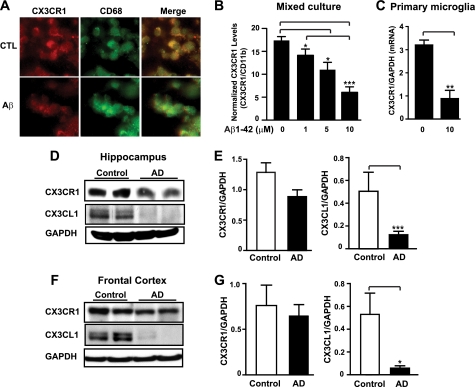
We first examined the expression of CX3CR1 in rat mixed cortical cultures, which contain neurons, astroglia, and microglia (34). Consistent with previous findings that CX3CR1 is exclusively expressed in microglia (22), immunoreactivity of CX3CR1 completely colocalized with that of CD68, a microglial marker. Treatment with Aβ oligomers decreased the immunoreactivity of CX3CR1, but not that of CD68 (Fig. 1A). We then used real-time RT-PCR to measure the effects of Aβ oligomers on CX3CR1 expression. Because Aβ treatment was found to increase microglial numbers in mixed cortical cultures (34), levels of CX3CR1 mRNA were normalized with those of CD11b to account for possible differences in the numbers of microglia (Fig. 1B). Treatment with Aβ oligomers significantly reduced levels of normalized CX3CR1 mRNA in a dose-dependent manner, suggesting that Aβ impairs fractalkine signaling in microglia cultured with neurons and astroglia. Aβ oligomers also reduced levels of CX3CR1 mRNA in highly enriched primary microglial cultures, suggesting regulation on the transcriptional level (Fig. 1C). To assess how fractalkine signaling might be affected in AD brains, we measured the protein levels of CX3CR1 and the ligand CX3CL1. In the hippocampus, levels of CX3CR1 were modestly lower in AD brains than in age-matched non-demented controls (Fig. 1, D and E). Notably, levels of the CX3CL1 were markedly lower in AD patients than controls (Fig. 1, D and E). A similar reduction was observed in the frontal cortices of AD brains (Fig. 1, F and G). Because CX3CL1 is the exclusive ligand of CX3CR1, these results indicate that fractalkine signaling is deficient in the vulnerable regions of AD brains.
FIGURE 1.
Fractalkine signaling is down-regulated by Aβ in cultured microglia and in AD brains. A, representative images of CX3CR1 and CD68 immunostaining in rat primary mixed cortical cultures treated with or without 10 μm oligomeric Aβ for 48 h. B and C, quantification of CX3CR1 mRNA in rat primary mixed cortical cultures (n = 6 from triplicate wells) (B) or microglia cultures (n = 3) (C), treated with designated concentrations of oligomeric Aβ1–42 or vehicle for 48 or 24 h, respectively. Because CX3CL1 is exclusively expressed in neurons, primary microglia cultures were pretreated with 3.3 nm CX3CL1 peptide for 16 h before being treated with Aβ1–42 for an additional 24 h. The levels of CX3CR1 were normalized with microglial marker (CD11b) in mixed cortical cultures (B) or with GAPDH in primary microglial cultures (C). D–G, deficiency in fractalkine signaling in AD brains. D and F, representative Western blots of CX3CR1 and CX3CL1 in the hippocampus (D) or frontal cortex (F) of AD patients and age-matched controls. E and G, quantification of CX3CR1 or CX3CL1 protein in the hippocampus (E) or frontal cortex (G) of AD (hippocampus, n = 24; frontal cortex, n = 5) patients and non-demented controls (hippocampus and frontal cortex, n = 4). ***, p < 0.001; **, p < 0.01; *, p < 0.05, unpaired Student's t test (C, E, and G). ***, p < 0.001; *, p < 0.05, one-way analysis of variance Tukey-Kramer post hoc analysis (B). Values are means ± S.E. (B, C, E, and G).
Deletion of CX3CR1 Did Not Affect APP Processing or Aβ Levels in hAPP Mice
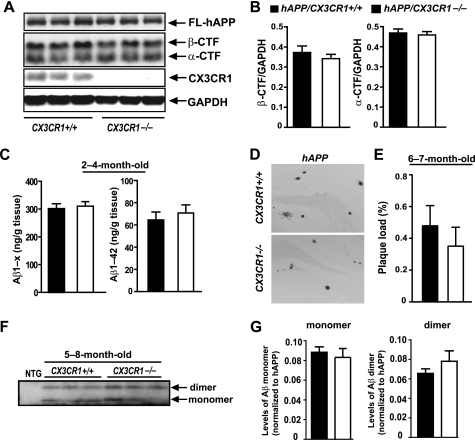
To examine the effects of fractalkine signaling on AD-related neurodegeneration, we crossed CX3CR1GFP/GFP knock-in mice (referred to as CX3CR1−/− for simplicity) with hAPP-J20 mice. We first determined whether CX3CR1 deletion affected hAPP processing by comparing levels of metabolic fragments of hAPP in hAPP/CX3CR1+/+ and hAPP/CX3CR1−/− mice. Consistent with previous findings (29, 30), removal of CX3CR1 did not affect the levels of β-CTF or α-CTF by Western blot analysis (Fig. 2, A and B). In agreement with the lack of effects on Aβ processing, levels of soluble total Aβ (Aβ1-x) or Aβ1–42 in 2–4-month-old mice before plaque deposition remained similar in hAPP mice with or without CX3CR1 (Fig. 2C). This result suggests that fractalkine signaling exerts no discernible effects on Aβ metabolism. Plaque deposition starts at 5–6 months of age in hAPP-J20 mice. We examined total plaque load, which includes diffuse plaques without a dense core and thioflavin S-positive plaques with β-sheet structure. In 5–8-month-old hAPP mice, early plaque deposition was not altered significantly by CX3CR1 deletion (Fig. 2, D and E). We also measured the levels of toxic species of Aβ dimers and presumably non-toxic monomers in 5–8-month-old hAPP mice by immunoprecipitation and Western blot analysis and found that levels of Aβ dimers and Aβ monomers did not differ with CX3CR1 ablation (Fig. 2, F and G).
FIGURE 2.
Deletion of CX3CR1 does not affect Aβ levels or APP processing. A, representative Western blot of the metabolic fragments of hAPP. FL-hAPP, full-length hAPP. B, quantification of hAPP fragments (β-CTF and α-CTF) in the cortex of 5–8-month-old hAPP mice with or without CX3CR1. n = 6–11/genotype. C, ELISA measurements of soluble Aβ1-x or Aβ1–42 in the hippocampus of 2–4-month-old hAPP mice with or without CX3CR1. n = 5/genotype. D, representative picture of 3D6-immunoreactive plaques in the hippocampus of 6–7-month-old hAPP mice with or without CX3CR1. E, quantification of plaque load in the hippocampus of 6–7-month-old hAPP mice with or without CX3CR1. n = 8–11/genotype. F, representative Western blot of Aβ dimers and monomers in the hippocampus of 5–8-month-old hAPP/CX3CR1+/+ or hAPP/CX3CR1−/− mice. NTG, non-transgenic. G, quantification of levels of Aβ monomers or dimers in the hippocampus of 5–8-month-old hAPP/CX3CR1+/+ or hAPP/CX3CR1−/− mice. n = 6–7/genotype. Values are means ± S.E. (B, C, E, and G).
Deletion of CX3CR1 Exacerbated Microglial Activation and Inflammatory Responses in hAPP Mice
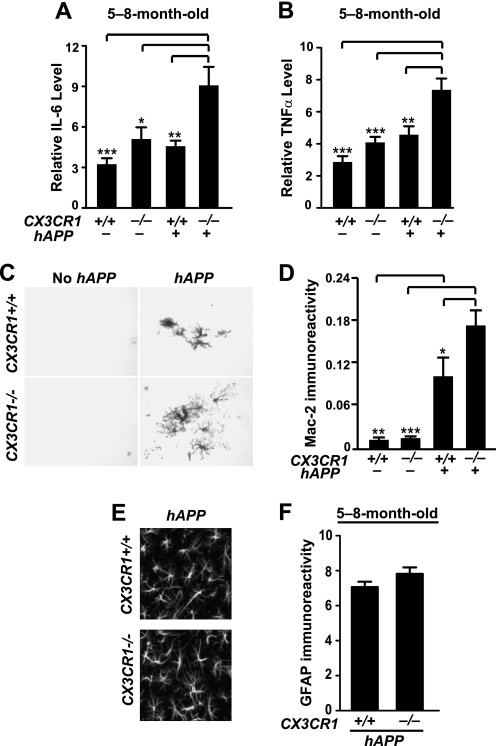
Fractalkine signaling limits microglial toxicity induced by LPS or in models of amyotrophic lateral sclerosis, Parkinson disease (22), or Tau pathology (11). Using quantitative RT-PCR, we examined the levels of inflammatory factors. Deleting CX3CR1 in 5–8-month-old hAPP mice significantly increased levels of IL-6 and TNF-α (Fig. 3, A and B). The higher levels of IL-6 or TNF-α could not be attributed to more Aβ because 3D6-positive plaque load in hAPP/CX3CR1−/− mice was not significantly elevated, and levels of TNF-α or IL-6 were not correlated with the plaque load in this age group (data not shown). Deleting CX3CR1 alone did not induce a significant increase in the expression of TNF-α or IL-6, suggesting that CX3CR1 limits Aβ-associated proinflammatory responses.
FIGURE 3.
Deletion of CX3CR1 exacerbates microglial activation. A and B, quantification of levels of IL-6 (A) or TNF-α (B) mRNA in the cortex of 5–8-month-old CX3CR1+/+, hAPP/CX3CR1+/+, CX3CR1−/−, or hAPP/CX3CR1−/− mice. n = 13–19 mice/genotype. C, representative picture of Mac-2 immunostaining in the hippocampus of 5–8-month-old wild type (CX3CR1+/+), hAPP/CX3CR1+/+, CX3CR1−/−, or hAPP/CX3CR1−/− mice. D, quantification of Mac-2 immunoreactive protein in the hippocampus. n = 8–9/genotype. E, representative images of GFAP immunostaining in the hippocampus of 5–8-month-old hAPP/CX3CR1+/+ or hAPP/CX3CR1−/− mice. F, quantification of immunoreactive GFAP in the hippocampus. n = 8–10/genotype. *, p < 0.05; **, p < 0.01; ***, p < 0.001, one-way analysis of variance Tukey-Kramer post hoc analysis (A, B, and D). Values are means ± S.E.
To further assess the effects of CX3CR1 on Aβ-associated microglial activation, we examined Mac-2 immunoreactivity, which was detected in activated microglia associated with chronic or acute neuronal injury but absent in quiescent microglia (35, 36). The number of Mac-2-positive microglia was significantly higher in 5–8-month-old hAPP mice than in those that do not express hAPP, indicating the presence of microglia activated by Aβ regardless of the CX3CR1 genotype (Fig. 3, C and D). Consistent with elevated levels of TNF-α or IL-6, the Mac-2 immunoreactivity in hAPP/CX3CR1−/− mice was also significantly higher than that in hAPP/CX3CR1+/+ mice (Fig. 3, C and D). In contrast, deleting CX3CR1 did not affect astrogliosis significantly, as measured with GFAP staining (Fig. 3, E and F). Our findings suggest that fractalkine signaling suppresses inflammatory responses in hAPP, likely by modulating microglial activation.
Deletion of CX3CR1 Exacerbated Microglial Neurotoxicity in hAPP Mice
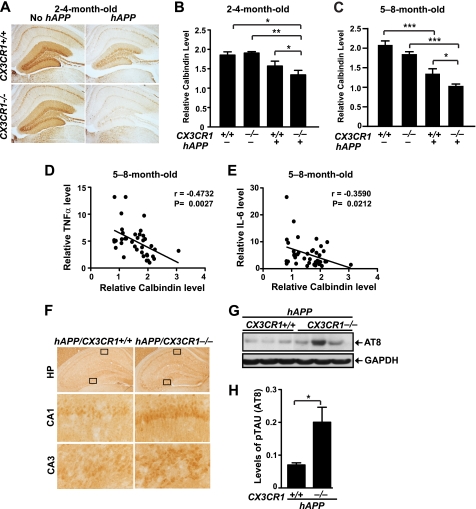
To investigate the effects of fractalkine signaling on Aβ-associated neuronal deficits, we examined the depletion of calbindin in the DG, which correlated tightly with the cognitive deficits in hAPP-J20 mice (37). A modest reduction in calbindin levels in the DG was observed in 2–4-month-old hAPP mice with CX3CR1 (Fig. 4A). Removing CX3CR1 in hAPP mice led to a significant reduction in calbindin in the DG, whereas removing CX3CR1 in mice that do not express hAPP did not have an effect at this age (Fig. 4, A and B). In 5–8-month-old hAPP mice, levels of calbindin were significantly reduced in the DG, consistent with previous findings (37). Ablation of CX3CR1 in hAPP mice exacerbated calbindin loss in the DG (Fig. 4C). Moreover, levels of calbindin correlated negatively with levels of TNF-α (Fig. 4D) and IL-6 (Fig. 4E), supporting the notion that the worsened neuronal deficits are associated with higher levels of inflammatory cytokines.
FIGURE 4.
Deletion of CX3CR1 exacerbates microglial toxicity in hAPP mice. A, representative images of calbindin immunostaining in the DG of 2–4-month-old CX3CR1+/+, hAPP/CX3CR1+/+, CX3CR1−/−, or hAPP/CX3CR1−/− mice. B and C, quantification of calbindin in the DG of 2–4-month-old (B) or 5–8-month-old (C) mice. n = 5–14/genotype *, p < 0.05; **, p < 0.01; ***, p < 0.001 (one-way analysis of variance with Newman-Keuls multiple comparison test). D and E, levels of calbindin in the DG correlated negatively with TNF-α or IL-6 in 5–8-month-old mice. n = 8–13 mice/genotype, Pearson's correlation. F, representative picture of phospho-Tau (AT-8) immunostaining in the hippocampus (HP) of 5–8-month-old hAPP mice with or without CX3CR1. G, representative Western blot of the phospho-Tau protein in the cortex of hAPP mice (hAPP/CX3CR1+/+ or hAPP/CX3CR1−/−). H, quantification of levels of phospho-Tau (p-Tau) (AT8; Ser-202/Thr-205) in the cortex of 5–8-month-old hAPP mice with or without CX3CR1. n = 5–6/genotype. *, p < 0.05, unpaired Student's t test. Values are means ± S.E. (B, C, and H).
Deficiency of fractalkine signaling was found to exacerbate microglia-mediated Tau phosphorylation in a mouse model of systematic inflammation and a humanized Tau transgenic mouse model (11). To determine the effects of fractalkine signaling on Tau phosphorylation in a hAPP mouse model, we examined phosphorylated Tau (phospho-Tau) (AT8; Ser-202/Thr-205) immunoreactivity in 5–8-month-old hAPP mice with or without CX3CR1. Increased AT8 immunoreactivity was observed in the hippocampus of hAPP mice without CX3CR1 (Fig. 4F). In the cortex, Western blot analysis revealed that the levels of phospho-Tau were significantly higher in hAPP/CX3CR1−/− mice than in hAPP/CX3CR1+/+ mice. Thus, fractalkine signaling attenuates pathogenic forms of phospho-Tau in hAPP mice (Fig. 4, G and H).
Deletion of CX3CR1 Exacerbated Cognitive Deficits in hAPP Mice
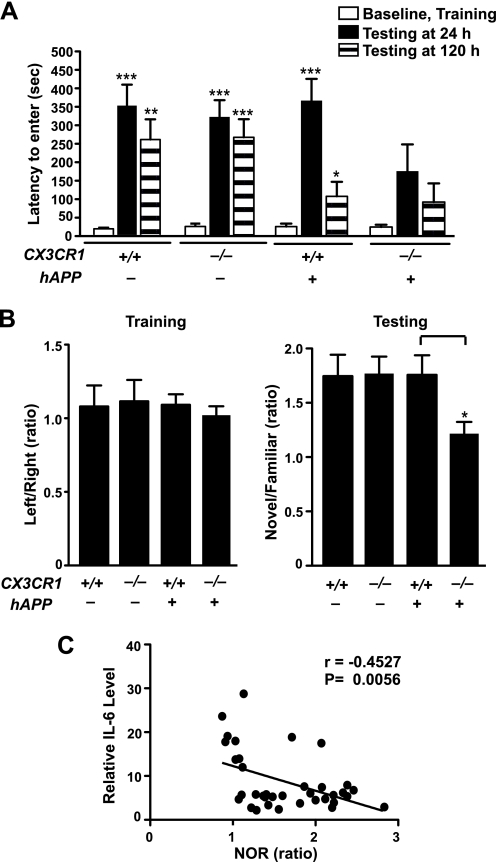
To determine the effects of fractalkine signaling on cognitive deficits, we first performed passive avoidance tests in 5–8-month-old hAPP mice. During the training session, mice readily entered the dark chamber due to their innate preference for darkness. No difference was detected among the genotypes (Fig. 5A). Twenty-four h after being shocked in the dark chamber, control mice exhibited a significant delay in entering the dark chamber again, a reflection of normal memory retention associated with an aversive signal (Fig. 5A) (38, 39). hAPP/CX3CR1+/+ mice exhibited normal delay 24 h after the training but were significantly impaired 120 h after. In contrast, hAPP/CX3CR1−/− mice exhibited no significant delay even at 24 h, suggesting much worse memory retention than hAPP/CX3CR1+/+ mice. Thus, CX3CR1 removal exacerbated Aβ-associated memory deficits.
FIGURE 5.
CX3CR1 ablation exacerbates cognitive deficits in hAPP mice. A, in the passive avoidance task, the increased escape latency to enter the dark chamber during the test indicates memory retention. No difference was detected during the training (Baseline). During the test performed 24 or 120 h after the shock, latency was significantly increased in CX3CR1+/+, hAPP/CX3CR1+/+, and CX3CR1−/− mice, but not in hAPP/CX3CR1−/− mice. hAPP/CX3CR1+/+ mice exhibited reduced latency 120 h after the training. n = 8–10/genotype, 3–8 months old. *, p < 0.05; **, p < 0.01; ***, p < 0.001, paired Student's t test as compared with the correspondent training latency. Latency was recorded up to 500-s cutoff. B, in the novel object recognition task, the preference to the novel object, as measured by the ratios of the time spent on the novel versus familiar object during the test session, reflects memory retention. All genotypes exhibited no preference during the training session when left and right objects were the same. During the test session, hAPP/CX3CR1−/− mice exhibited less preference to the novel object than hAPP/CX3CR1+/+ mice. n = 8–10/genotype. *, p < 0.05, unpaired Student's t test. C, levels of IL-6 negatively correlated with the preference of the novel object recognition (NOR) test. n = 8–10 mice/genotype, Pearson's correlation. Values are means ± S.E. (A and B).
Next, novel object recognition was performed to measure non-spatial memory that is not associated with aversive signal. Normal mice spent more time with the novel object than with the familiar one; mice inherently prefer novel objects more than familiar objects (40, 41). During the training session, mice spent an equal amount of time with two identical objects, right and left, regardless of their genotypes (Fig. 5B). At 24 h after the training (testing period), control mice (CX3CR1+/+) preferred the novel object to the familiar one as expected (Fig. 5B), reflecting memory retention. CX3CR1−/− mice and hAPP mice exhibited similar preference (Fig. 5B). In contrast, hAPP/CX3CR1−/− mice exhibited less preference for the novel object than hAPP/CX3CR1+/+ mice, suggesting less memory retention (Fig. 5B). Notably, the lack of preference (lower ratios) is associated with higher levels of IL-6 in these mice (Fig. 5C). Our results indicate that CX3CR1 deficiency worsens Aβ-associated memory deficits, possibly by exacerbating inflammatory responses.
DISCUSSION
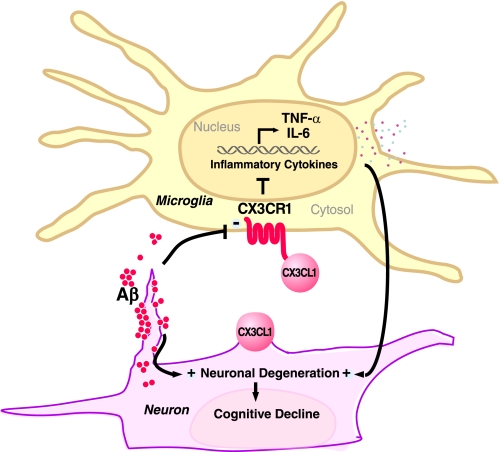
Our study demonstrates that CX3CR1 deficiency worsens the functional outcome in the hAPP-J20 model of AD. Removing CX3CR1 in hAPP-J20 mice enhanced Tau pathology, exacerbated calbindin depletion, and worsened memory deficits in two cognitive tests. These effects were associated with elevated cytokine production but not with altered hAPP metabolism or plaque deposition. Our findings identify CX3CR1 deficiency as a key pathway leading to the aberrant activation of microglia, which promotes inflammatory responses and exacerbates AD-related neuronal and cognitive deficits (Fig. 6).
FIGURE 6.
Diagram depicts potential mechanisms by which fractalkine signaling limits microglial toxicity in AD. Down-regulation of CX3CR1 by Aβ treatment leads to increased expression of inflammatory factors, including IL-6 and TNF-α, and exacerbates cognitive decline in AD. Reduced expression of CX3CL1 by injured neurons could further activate microglia and feed into the vicious cycle. − indicates down-regulation; + indicates up-regulation.
We found that fractalkine signaling is down-regulated by Aβ and in AD brains. Treatment with Aβ oligomers lowered levels of its receptor CX3CR1 in primary cortical and microglial cultures. Although only a modest reduction of CX3CR1 was observed in AD brains, the per cell reduction of CX3CR1 is likely to be biologically meaningful given the marked microgliosis in AD brains. Further, because CX3CL1 is the only ligand of CX3CR1, reduction of CX3CL1 would drastically diminish fractalkine signaling in AD. As CX3CL1 is produced in neurons, loss of neurons in AD brains likely accounts for diminished CX3CL1 levels.
Using multiple markers, our study showed that deleting CX3CR1 exacerbated microglial activation in hAPP mice independent of plaque deposition. CX3CR1 deletion elevated levels of the inflammatory cytokines TNF-α and IL-6. As indicated by Mac-2 immunoreactivity, more active microglia were identified in the brains of hAPP mice lacking CX3CR1. These findings are consistent with previous studies that CX3CR1 deficiency exacerbates microglial activation induced by LPS, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine injection, or expression of mutant SOD1 (22), as well as the production of NO, IL-6, and TNF-α in cultured cells (24, 25). On the other hand, deleting CX3CR1 elevates phagocytosis and reduces fibrillar Aβ plaques (29, 30). Because Aβ deposits, especially senile plaques, strongly activate microglia, the effects on plaque load could confound the effects on inflammatory factors. For example, CX3CR1 deletion could lower the levels of inflammatory factors induced by fibrillar Aβ due to the reduction in plaque load. Our data showed instead that inflammatory cytokine production was elevated in hAPP/CX3CR1−/− mice (Fig. 3) despite a trend toward reduced plaque burden.
Our studies show that CX3CR1 deficiency exacerbates functional deficits in hAPP mice, associated with elevated cytokine production and Tau pathology. CX3CR1 removal exacerbated calbindin depletion in the DG in hAPP mice before and after plaque deposition. Deleting CX3CR1 also exacerbated levels of phospho-Tau in hAPP mice, confirming and extending previous observations in transgenic mice overexpressing human Tau (11). The negative correlation of calbindin levels in the DG with the levels of TNF-α and IL-6 suggests that the toxic effects of CX3CR1 deficiency could be partially attributed to exacerbated inflammatory responses. Functionally, removing CX3CR1 in hAPP mice worsened the memory retention in passive avoidance and novel object recognition tests. The poor performance in the novel object recognition test is associated with high levels of IL-6, supporting a causal role of inflammatory responses in neuronal dysfunction and cognitive deficits. Consistent with this interpretation, high levels of IL-6 via infusion or overexpression increased levels of Tau phosphorylation and impaired long-term potentiation in the DG (42, 43). In contrast, administration of IL-6-neutralizing antibodies prolonged long-term potentiation and improved spatial alternation behavior (44). In a similar vein, ablation of IL-6 protected against LPS-induced disruption of working memory (45). Future studies will be needed to determine whether the worsened cognitive deficits in hAPP/CX3CR1−/− mice are mediated by elevated levels of IL-6.
Our study provides the first evidence for protective effects of fractalkine signaling on Aβ-associated cognitive deficits that are plaque-independent. These results differ from a previous report that removing CX3CR1 protected against neuronal loss in 5×Tg mice, which express mutant Tau, hAPP on a PS1 knock-in background, and Thy1-YFP (28). By multiphoton imaging, these mice showed loss of small numbers of layer III cortical neurons at 4–6 months of age (28). It is uncertain whether the lack of neuronal loss in 5×Tg mice deficient in CX3CR1 would lead to functional benefits. Nevertheless, our results are consistent with previous findings that deletion of CX3CR1 elevates the state of microglial activation in various models of acute and chronic neuronal injury (11, 29, 30, 46). Both beneficial and detrimental effects are associated with microglia activation (4, 47, 48). On the one hand, our study showed that deleting CX3CR1 in hAPP mice increased expression of inflammatory factors, such as TNF-α and IL-6, and exacerbated plaque-independent neuronal dysfunction and cognitive deficits. On the other hand, in mice with extensive senile plaque load, the heightened microglial activation induced by CX3CR1 deficiency elevated Aβ clearance, possibly by phagocytosis, exhibiting a protective role. These findings highlight the complexity of enhanced microglial activation as a “double-edged sword,” leading to either toxic or protective outcomes. Understanding the underlying signaling pathways and associated functional outcomes will have important implications in AD and other neurodegenerative diseases.
Acknowledgments
We thank Drs. Lennart Mucke and Israel Charo for insightful discussions, Dr. Nino Devidze and the Gladstone/University of California, San Francisco (UCSF) Behavioral Core to help with behavioral analyses, Dr. Dan Littman from New York University for CX3CR1 null mice, Kaitlyn Ho for advice on measuring Aβ dimers, Dr. Katerina Mancevska from New York Brain Bank at Columbia University, Dr. Yong Shen from Sun Health Institute (now Banner Health), Dr. Eliezer Masliah from UCSD for human brain samples, Gary Howard for editorial review, and Kelley Nelson for administrative assistance. The J. David Gladstone Institutes were supported by National Institutes of Health Grant NCRR CO6 RRO18928.
This work was supported, in whole or in part, by grants from National Institute on Aging 1R01AG030207, Consortium for Frontotemporal Dementia Research, and S. D. Bechtel Jr. Foundation (to L. G.).
- AD
- Alzheimer disease
- Aβ
- amyloid-β
- APP
- amyloid precursor protein
- hAPP
- human APP
- DG
- dentate gyrus
- GFAP
- glial fibrillary acidic protein
- CTF
- C-terminal fragment.
REFERENCES
- 1. Ilieva H., Polymenidou M., Cleveland D. W. (2009) J. Cell Biol. 187, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wyss-Coray T. (2006) Nat. Med. 12, 1005–1015 [DOI] [PubMed] [Google Scholar]
- 3. McGeer P. L., Rogers J., McGeer E. G. (2006) J. Alzheimers Dis. 9, 271–276 [DOI] [PubMed] [Google Scholar]
- 4. Frank-Cannon T. C., Alto L. T., McAlpine F. E., Tansey M. G. (2009) Mol. Neurodegener. 4, 47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Seshadri S., Fitzpatrick A. L., Ikram M. A., DeStefano A. L., Gudnason V., Boada M., Bis J. C., Smith A. V., Carassquillo M. M., Lambert J. C., Harold D., Schrijvers E. M., Ramirez-Lorca R., Debette S., Longstreth W. T., Jr., Janssens A. C., Pankratz V. S., Dartigues J. F., Hollingworth P., Aspelund T., Hernandez I., Beiser A., Kuller L. H., Koudstaal P. J., Dickson D. W., Tzourio C., Abraham R., Antunez C., Du Y., Rotter J. I., Aulchenko Y. S., Harris T. B., Petersen R. C., Berr C., Owen M. J., Lopez-Arrieta J., Varadarajan B. N., Becker J. T., Rivadeneira F., Nalls M. A., Graff-Radford N. R., Campion D., Auerbach S., Rice K., Hofman A., Jonsson P. V., Schmidt H., Lathrop M., Mosley T. H., Au R., Psaty B. M., Uitterlinden A. G., Farrer L. A., Lumley T., Ruiz A., Williams J., Amouyel P., Younkin S. G., Wolf P. A., Launer L. J., Lopez O. L., van Duijn C. M., Breteler M. M. (2010) JAMA 303, 1832–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hollingworth P., Harold D., Sims R., Gerrish A., Lambert J. C., Carrasquillo M. M., Abraham R., Hamshere M. L., Pahwa J. S., Moskvina V., Dowzell K., Jones N., Stretton A., Thomas C., Richards A., Ivanov D., Widdowson C., Chapman J., Lovestone S., Powell J., Proitsi P., Lupton M. K., Brayne C., Rubinsztein D. C., Gill M., Lawlor B., Lynch A., Brown K. S., Passmore P. A., Craig D., McGuinness B., Todd S., Holmes C., Mann D., Smith A. D., Beaumont H., Warden D., Wilcock G., Love S., Kehoe P. G., Hooper N. M., Vardy E. R., Hardy J., Mead S., Fox N. C., Rossor M., Collinge J., Maier W., Jessen F., Rüther E., Schürmann B., Heun R., Kölsch H., van den Bussche H., Heuser I., Kornhuber J., Wiltfang J., Dichgans M., Frölich L., Hampel H., Gallacher J., Hüll M., Rujescu D., Giegling I., Goate A. M., Kauwe J. S., Cruchaga C., Nowotny P., Morris J. C., Mayo K., Sleegers K., Bettens K., Engelborghs S., De Deyn P. P., Van Broeckhoven C., Livingston G., Bass N. J., Gurling H., McQuillin A., Gwilliam R., Deloukas P., Al-Chalabi A., Shaw C. E., Tsolaki M., Singleton A. B., Guerreiro R., Mühleisen T. W., Nöthen M. M., Moebus S., Jöckel K. H., Klopp N., Wichmann H. E., Pankratz V. S., Sando S. B., Aasly J. O., Barcikowska M., Wszolek Z. K., Dickson D. W., Graff-Radford N. R., Petersen R. C., van Duijn C. M., Breteler M. M., Ikram M. A., DeStefano A. L., Fitzpatrick A. L., Lopez O., Launer L. J., Seshadri S., Berr C., Campion D., Epelbaum J., Dartigues J. F., Tzourio C., Alpérovitch A., Lathrop M., Feulner T. M., Friedrich P., Riehle C., Krawczak M., Schreiber S., Mayhaus M., Nicolhaus S., Wagenpfeil S., Steinberg S., Stefansson H., Stefansson K., Snaedal J., Björnsson S., Jonsson P. V., Chouraki V., Genier-Boley B., Hiltunen M., Soininen H., Combarros O., Zelenika D., Delepine M., Bullido M. J., Pasquier F., Mateo I., Frank-Garcia A., Porcellini E., Hanon O., Coto E., Alvarez V., Bosco P., Siciliano G., Mancuso M., Panza F., Solfrizzi V., Nacmias B., Sorbi S., Bossù P., Piccardi P., Arosio B., Annoni G., Seripa D., Pilotto A., Scarpini E., Galimberti D., Brice A., Hannequin D., Licastro F., Jones L., Holmans P. A., Jonsson T., Riemenschneider M., Morgan K., Younkin S. G., Owen M. J., O'Donovan M., Amouyel P., Williams J. (2011) Nat. Genet. 43, 429–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rogers J., Webster S., Lue L. F., Brachova L., Civin W. H., Emmerling M., Shivers B., Walker D., McGeer P. (1996) Neurobiol. Aging 17, 681–686 [DOI] [PubMed] [Google Scholar]
- 8. Hickman S. E., Allison E. K., El Khoury J. (2008) J. Neurosci. 28, 8354–8360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kitazawa M., Yamasaki T. R., LaFerla F. M. (2004) Ann. N.Y. Acad. Sci. 1035, 85–103 [DOI] [PubMed] [Google Scholar]
- 10. Yoshiyama Y., Higuchi M., Zhang B., Huang S. M., Iwata N., Saido T. C., Maeda J., Suhara T., Trojanowski J. Q., Lee V. M. (2007) Neuron 53, 337–351 [DOI] [PubMed] [Google Scholar]
- 11. Bhaskar K., Konerth M., Kokiko-Cochran O. N., Cardona A., Ransohoff R. M., Lamb B. T. (2010) Neuron 68, 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Colton C. A. (2009) J. Neuroimmune Pharmacol. 4, 399–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ransohoff R. M., Cardona A. E. (2010) Nature 468, 253–262 [DOI] [PubMed] [Google Scholar]
- 14. Ransohoff R. M., Perry V. H. (2009) Annu. Rev. Immunol 27, 119–145 [DOI] [PubMed] [Google Scholar]
- 15. Imai T., Hieshima K., Haskell C., Baba M., Nagira M., Nishimura M., Kakizaki M., Takagi S., Nomiyama H., Schall T. J., Yoshie O. (1997) Cell 91, 521–530 [DOI] [PubMed] [Google Scholar]
- 16. Combadiere C., Salzwedel K., Smith E. D., Tiffany H. L., Berger E. A., Murphy P. M. (1998) J. Biol. Chem. 273, 23799–23804 [DOI] [PubMed] [Google Scholar]
- 17. Harrison J. K., Jiang Y., Chen S., Xia Y., Maciejewski D., McNamara R. K., Streit W. J., Salafranca M. N., Adhikari S., Thompson D. A., Botti P., Bacon K. B., Feng L. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10896–10901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garton K. J., Gough P. J., Blobel C. P., Murphy G., Greaves D. R., Dempsey P. J., Raines E. W. (2001) J. Biol. Chem. 276, 37993–38001 [DOI] [PubMed] [Google Scholar]
- 19. Tsou C. L., Haskell C. A., Charo I. F. (2001) J. Biol. Chem. 276, 44622–44626 [DOI] [PubMed] [Google Scholar]
- 20. Clark A. K., Yip P. K., Malcangio M. (2009) J. Neurosci. 29, 6945–6954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jung S., Aliberti J., Graemmel P., Sunshine M. J., Kreutzberg G. W., Sher A., Littman D. R. (2000) Mol. Cell. Biol. 20, 4106–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cardona A. E., Pioro E. P., Sasse M. E., Kostenko V., Cardona S. M., Dijkstra I. M., Huang D., Kidd G., Dombrowski S., Dutta R., Lee J. C., Cook D. N., Jung S., Lira S. A., Littman D. R., Ransohoff R. M. (2006) Nat. Neurosci. 9, 917–924 [DOI] [PubMed] [Google Scholar]
- 23. Mizuno T., Kawanokuchi J., Numata K., Suzumura A. (2003) Brain Res 979, 65–70 [DOI] [PubMed] [Google Scholar]
- 24. Zujovic V., Benavides J., Vigé X., Carter C., Taupin V. (2000) Glia 29, 305–315 [PubMed] [Google Scholar]
- 25. Kim H. C., Yamada K., Nitta A., Olariu A., Tran M. H., Mizuno M., Nakajima A., Nagai T., Kamei H., Jhoo W. K., Im D. H., Shin E. J., Hjelle O. P., Ottersen O. P., Park S. C., Kato K., Mirault M. E., Nabeshima T. (2003) Neuroscience 119, 399–419 [DOI] [PubMed] [Google Scholar]
- 26. Limatola C., Lauro C., Catalano M., Ciotti M. T., Bertollini C., Di Angelantonio S., Ragozzino D., Eusebi F. (2005) J. Neuroimmunol. 166, 19–28 [DOI] [PubMed] [Google Scholar]
- 27. Meucci O., Fatatis A., Simen A. A., Miller R. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8075–8080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fuhrmann M., Bittner T., Jung C. K., Burgold S., Page R. M., Mitteregger G., Haass C., LaFerla F. M., Kretzschmar H., Herms J. (2010) Nat. Neurosci. 13, 411–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee S., Varvel N. H., Konerth M. E., Xu G., Cardona A. E., Ransohoff R. M., Lamb B. T. (2010) Am. J. Pathol. 177, 2549–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Z., Condello C., Schain A., Harb R., Grutzendler J. (2010) J. Neurosci. 30, 17091–17101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lue L. F., Kuo Y. M., Roher A. E., Brachova L., Shen Y., Sue L., Beach T., Kurth J. H., Rydel R. E., Rogers J. (1999) Am. J. Pathol. 155, 853–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Terry R. D., Masliah E., Salmon D. P., Butters N., DeTeresa R., Hill R., Hansen L. A., Katzman R. (1991) Ann. Neurol. 30, 572–580 [DOI] [PubMed] [Google Scholar]
- 33. Mucke L., Masliah E., Yu G. Q., Mallory M., Rockenstein E. M., Tatsuno G., Hu K., Kholodenko D., Johnson-Wood K., McConlogue L. (2000) J. Neurosci. 20, 4050–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen J., Zhou Y., Mueller-Steiner S., Chen L. F., Kwon H., Yi S., Mucke L., Gan L. (2005) J. Biol. Chem. 280, 40364–40374 [DOI] [PubMed] [Google Scholar]
- 35. Dong S., Hughes R. C. (1997) Glycoconj. J. 14, 267–274 [DOI] [PubMed] [Google Scholar]
- 36. Lalancette-Hébert M., Gowing G., Simard A., Weng Y. C., Kriz J. (2007) J. Neurosci. 27, 2596–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Palop J. J., Jones B., Kekonius L., Chin J., Yu G. Q., Raber J., Masliah E., Mucke L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9572–9577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pittenger C., Fasano S., Mazzocchi-Jones D., Dunnett S. B., Kandel E. R., Brambilla R. (2006) J. Neurosci. 26, 2808–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Potter M. C., Elmer G. I., Bergeron R., Albuquerque E. X., Guidetti P., Wu H. Q., Schwarcz R. (2010) Neuropsychopharmacology 35, 1734–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Squire L. R., Wixted J. T., Clark R. E. (2007) Nat. Rev. Neurosci. 8, 872–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brown M. W., Aggleton J. P. (2001) Nat. Rev. Neurosci. 2, 51–61 [DOI] [PubMed] [Google Scholar]
- 42. Quintanilla R. A., Orellana D. I., González-Billault C., Maccioni R. B. (2004) Exp. Cell Res. 295, 245–257 [DOI] [PubMed] [Google Scholar]
- 43. Bellinger F. P., Madamba S. G., Campbell I. L., Siggins G. R. (1995) Neurosci. Lett. 198, 95–98 [DOI] [PubMed] [Google Scholar]
- 44. Balschun D., Wetzel W., Del Rey A., Pitossi F., Schneider H., Zuschratter W., Besedovsky H. O. (2004) FASEB J. 18, 1788–1790 [DOI] [PubMed] [Google Scholar]
- 45. Sparkman N. L., Buchanan J. B., Heyen J. R., Chen J., Beverly J. L., Johnson R. W. (2006) J. Neurosci. 26, 10709–10716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cardona A. E., Sasse M. E., Liu L., Cardona S. M., Mizutani M., Savarin C., Hu T., Ransohoff R. M. (2008) Blood 112, 256–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wyss-Coray T., Mucke L. (2002) Neuron 35, 419–432 [DOI] [PubMed] [Google Scholar]
- 48. Cameron B., Landreth G. E. (2010) Neurobiol. Dis. 37, 503–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Johnson-Wood K., Lee M., Motter R., Hu K., Gordon G., Barbour R., Khan K., Gordon M., Tan H., Games D., Lieberburg I., Schenk D., Seubert P., McConlogue L. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 1550–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Harris J. A., Devidze N., Verret L., Ho K., Halabisky B., Thwin M. T., Kim D., Hamto P., Lo I., Yu G. Q., Palop J. J., Masliah E., Mucke L. (2010) Neuron 68, 428–441 [DOI] [PMC free article] [PubMed] [Google Scholar]