Abstract
The human equilibrative nucleoside transporters hENT1 and hENT2 (each with 456 residues) are 40% identical in amino acid sequence and contain 11 putative transmembrane helices. Both transport purine and pyrimidine nucleosides and are distinguished functionally by a difference in sensitivity to inhibition by nanomolar concentrations of nitrobenzylmercaptopurine ribonucleoside (NBMPR), hENT1 being NBMPR-sensitive. Previously, we used heterologous expression in Xenopus oocytes to demonstrate that recombinant hENT2 and its rat ortholog rENT2 also transport purine and pyrimidine bases, h/rENT2 representing the first identified mammalian nucleobase transporter proteins (Yao, S. Y., Ng, A. M., Vickers, M. F., Sundaram, M., Cass, C. E., Baldwin, S. A., and Young, J. D. (2002) J. Biol. Chem. 277, 24938–24948). The same study also revealed lower, but significant, transport of hypoxanthine by h/rENT1. In the present investigation, we have used the enhanced Xenopus oocyte expression vector pGEMHE to demonstrate that hENT1 additionally transports thymine and adenine and, to a lesser extent, uracil and guanine. Fluxes of hypoxanthine, thymine, and adenine by hENT1 were saturable and inhibited by NBMPR. Ratios of Vmax (pmol/oocyte·min−1):Km (mm), a measure of transport efficiency, were 86, 177, and 120 for hypoxantine, thymine, and adenine, respectively, compared with 265 for uridine. Hypoxanthine influx was competitively inhibited by uridine, indicating common or overlapping nucleobase and nucleoside permeant binding pockets, and the anticancer nucleobase drugs 5-fluorouracil and 6-mercaptopurine were also transported. Nucleobase transport activity was absent from an engineered cysteine-less version hENT1 (hENT1C−) in which all 10 endogenous cysteine residues were mutated to serine. Site-directed mutagenesis identified Cys-414 in transmembrane helix 10 of hENT1 as the residue conferring nucleobase transport activity to the wild-type transporter.
Keywords: Membrane Proteins, Nucleoside, Nucleoside Nucleotide Analogs, Nucleoside Nucleotide Transport, Site-directed Mutagenesis, Xenopus Oocytes, hENT1, hENT2, Nucleobase Transport, Nucleoside Transport
Introduction
Physiologic nucleosides and the majority of synthetic analogs with antineoplastic and/or antiviral activities are hydrophilic molecules that require specialized plasma membrane nucleoside transporter (NT)2 proteins for passage into and out of cells (1–6). Necessary for nucleoside metabolism by salvage pathways, NTs are also critical determinants of pharmacologic actions of nucleoside drugs. By regulating availability of adenosine to purinoreceptors, NTs also modulate a diverse array of physiological processes, including neurotransmission, immune responses, platelet aggregation, renal function and coronary vasodilation. Two structurally unrelated NT families of integral membrane proteins exist in human and other mammalian cells and tissues: the SLC28 concentrative nucleoside transporter (CNT) family and the SLC29 equilibrative nucleoside transporter (ENT) family (7–10). CNTs mediate inwardly directed Na+- or Na+/H+-dependent accumulation of nucleosides within cells and are found predominantly in intestinal and renal epithelia and other specialized cells. ENTs, in contrast, mediate bidirectional Na+-independent movement of nucleosides across plasma and intracellular membranes and are normally present in most, possibly all, cell types. Both protein families are evolutionarily old: ENTs are widely distributed in mammalian, lower vertebrate, and other eukaryote species; CNTs are present in both eukaryotes and prokaryotes.
In humans, the CNT protein family is represented by three members: hCNT1, hCNT2, and hCNT3. All three transport uridine but exhibit different preferences for other permeants, with hCNT1 being selective for pyrimidine nucleosides (and, with lower fluxes, adenosine), hCNT2 for purine nucleosides, and hCNT3 for both purine and pyrimidine nucleosides. The human ENT protein family has four members, hENT1, hENT2, hENT3, and hENT4. hENT1 and hENT2, the two major NTs of plasma membranes, are broadly selective for both purine and pyrimidine nucleosides and are distinguished functionally by a difference in sensitivity to inhibition by nitrobenzylmercaptopurine ribonucleoside (NBMPR), hENT1 being NBMPR-sensitive and ENT2 being NBMPR-insensitive (11–13). hENT3 also transports purine and pyrimidine nucleosides but functions predominantly in intracellular membranes (14, 15). hENT4 transports adenosine and monoamines in the brain and heart (16, 17). Known previously by their functional designations of es, ei, cit, cif, and cib,3 hENT1, hENT2, hCNT1, hCNT2, and hCNT3, respectively, account for the five major nucleoside transport processes of human cells and tissues (2, 9, 10).
Purine nucleobases are also important salvage metabolites in humans and other mammals, and independent transport processes specific for nucleobases as well as shared mechanisms for nucleoside and nucleobase transport have been described (5, 6). Some ENT family members, for example, are also capable of transporting nucleobases (9). Human (h) and rat (r) ENT2 transport purine and pyrimidine nucleosides and nucleobases with equivalent kinetic efficiencies (18), whereas human and mouse (m) ENT3 and mENT4 also transport adenine (16, 17). Similarly, ENTs from parasitic protozoa such as Leishmania major, Trypanosoma brucei, and Plasmodium falciparum accept nucleosides, nucleobases, or both nucleosides and nucleobases as permeants (19, 20). ENTs have a common 11-transmembrane (TM) helical architecture (21), and chimeric studies utilizing rENT1 and rENT2 have identified TMs 5 and 6 of rENT2 as major determinants of hypoxanthine transport activity (18).
In our studies of nucleobase transport by h/rENT2, we found that h/rENT1 produced in Xenopus oocytes mediated smaller, but still significant, fluxes of hypoxanthine (18). There is also evidence of hypoxanthine transport by native mENT1 in mouse S49 lymphoma cells (22). In the present investigation, we have taken advantage of the enhanced Xenopus oocyte expression vector pGEMHE (23) to kinetically evaluate and characterize nucleobase transport by recombinant hENT1 and, for comparison, hCNT1–3. The results establish hENT1 as a dual nucleoside/nucleobase transporter that accepts not only hypoxanthine but also adenine and thymine as permeants. Lesser fluxes of uracil and guanine were also demonstrated, whereas hCNT1–3 did not transport nucleobases and thus were nucleoside-specific. Nucleobase transport activity was blocked by NBMPR and competitively inhibited by uridine. The clinically used anticancer nucleobase drugs 5-fluorouracil and 6-mercaptopurine were also transported. In contrast to wild-type hENT1, an engineered cysteine-less version of hENT1 (hENT1C−) lacked nucleobase transport activity, and we established that mutation of a single cysteine residue (Cys-414) at the cytoplasmic interface of TM 10 was responsible for the different transport selectivities of the native and engineered transporters.
EXPERIMENTAL PROCEDURES
Production of hENT1 and hENT2 in Xenopus Oocytes
cDNAs encoding hENT1 (GenBankTM accession number AAC51103) (11) or hENT2 (GenBankTM accession number NP_001523) (12) in pBluescript II KS(+) (Stratagene) were subcloned into the enhanced Xenopus laevis oocyte expression vector pGEMHE (23) for production of the recombinant transporters in Xenopus oocytes by standard procedures (24). Briefly, plasmids were linearized with NheI and transcribed with T7 RNA polymerase in the presence of m7GpppG cap using the mMessage mMachineTM (Ambion) transcription system. Defolliculated stage VI Xenopus oocytes were microinjected with 20 nl of water alone or 20 nl of water containing capped RNA transcript (20 ng) and incubated in modified Barth's medium (changed daily) at 18 °C for 72 h prior to the assay of transport activity.
pGEMHE provides additional 5′- and 3′-untranslated regions from the Xenopus β-globin gene flanking the multiple cloning sites and gives enhanced production and functional activity of recombinant proteins in Xenopus oocytes (23). To illustrate this, we compared in the same experiment initial rates of uridine uptake (20 μm) by hENT1-producting oocytes using both pGEMHE and the standard plasmid vector pBluescript II KS(+). pGEMHE generated mediated fluxes of uridine (uptake in RNA transcript-injected oocytes minus uptake in control water-injected oocytes) of 1.04 ± 0.07 pmol/oocyte·min−1 compared with 0.13 ± 0.01 pmol/oocyte·min−1 for pBluescript II KS(+), a difference of 8-fold.
Construction of Cysteine-less hENT1C− and hENT1C− Mutants S1 to S10
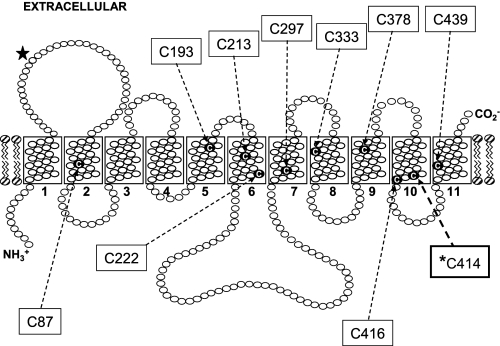
hENT1/pGEMHE was used as template for the construction of a cysteine-less version of hENT1 (hENT1C−) in which all 10 endogenous cysteine residues of hENT1 were converted to serine by PCR-based mutagenesis using the QuikChangeTM multisite-directed mutagenesis kit (Stratagene). In turn, hENT1C−/pGEMHE provided the template to individually reconvert serine back into cysteine at residue positions 87 (designated as hENT1C− mutant S1), 193 (S2), 213 (S3), 222 (S4), 297 (S5), 333 (S6), 378 (S7), 414 (S8), 416 (S9), and 439 (S10). All of the constructs were sequenced in both directions by Taq DyeDeoxy terminator cycle sequencing to ensure that the correct mutations had been introduced.
Radioisotope Flux Assays
Transport assays were performed as previously described (11–14) on groups of 12 oocytes at room temperature (20 °C) using 3H-labeled uridine (GE Healthcare) and 3H- or 14C-labeled nucleobases (Moravek Biochemicals) (2 μCi/ml for 3H permeants and 1 μCi/ml for 14C permeants) in 200 μl of transport medium containing 100 mm NaCl, 2 mm KCl, 1 mm CaCl2, 1 mm MgCl2, and 10 mm HEPES, pH 7.5. Unless otherwise indicated, uptake was determined at concentrations of 20 μm. Incubation periods of 2 min were used to measure initial rates of transport (influx) (11–13), and values are reported in units of pmol/oocytes·min−1. For NBMPR inhibition studies, the oocytes were pretreated with varying concentrations of NBMPR at room temperature for 1 h before addition of permeant. At the end of incubation periods, extracellular label was removed by six rapid washes in ice-cold transport medium. Individual oocytes were then dissolved in 1% (w/v) sodium dodecyl sulfate for quantitation of radioactivity by liquid scintillation counting.
The results for transport experiments are given as the means ± S.E. for 10–12 oocytes. Kinetic parameters (Km and Vmax) (± S.E.) for mediated transport corrected for basal uptake in control water-injected oocytes were determined using SigmaPlot software (Jandel Scientific Software). Inhibitor IC50 values (± S.E.) were also determined using SigmaPlot software. Each experiment was performed at least twice on oocytes from different frogs, yielding closely similar results. Statistical significance was evaluated using Student's t test.
RESULTS AND DISCUSSION
In contrast to nucleosides, there is little information on the molecular basis of nucleobase transport in mammals, although both equilibrative and concentrative (Na+-dependent) nucleobase-specific transport activities have been found in a variety of cell types and tissues (5, 6). Equilibrative nucleobase transport has been described in human erythrocytes, human T-lymphoblastoid cells, pig renal epithelial cells, S49 mouse-derived lymphoma cells, human cardiac microvascular endothelial cells, and pig kidney PK15NTD cells (6, 27–31), and Na+-dependent nucleobase transport has been described in kidney, intestine, placenta, choroid plexus, and rat primary cultured Sertoli cells and mouse Sertoli-like TM 4 cells (27–38). However, the corresponding transporter proteins (with the exceptions noted below) have yet to be discovered.
In bacteria, fungi, and plants, three protein families contribute to nucleobase transport: the nucleobase-ascorbate transporter family, the microbial purine-related transporter family, and the plant purine-related transporter family (39). Only the nucleobase-ascorbate transporter family has known orthologs in mammals. Of these, the Na+-dependent vitamin C transporters SVCT1 and SVCT2 mediate uptake of l-ascorbic acid, whereas SVCT3 is an orphan protein of unknown function. Recently, rat intestinal SVCT4 was identified as a Na+-dependent transporter specific for the nucleobases uracil, thymine, guanine, hypoxanthine and xanthine and was renamed SNBT1 (sodium-dependent nucleobase transporter 1) (40). The corresponding snbt gene in humans lacks three key TM-encoding exons and does not produce functional nucleobase transporter protein (40). Although urate, the end product of human purine degradation, is a permeant of the human glucose transporter family member hGLUT9 (41); salvage nucleobases are not transported.4
Shared mechanisms for nucleoside and nucleobase transport have also been described (5, 6), and hENT2 (and intracellular hENT3) are the first discovered, and so far only identified, transporter proteins for salvage nucleobases in human cells and tissues. The present investigation adds hENT1 to this select group of transporters and identifies a key amino acid residue that contributes to hENT1 nucleobase transport activity.
Uptake of Nucleobases by Wild-type hENT1
In our previous studies of nucleobase transport by h/rENT2 produced in Xenopus oocytes, we noted modest transport of hypoxanthine by h/rENT1 (18). Fluxes, however, were relatively small and insufficient for further study. In the present investigation, we have used the enhanced Xenopus oocyte expression vector pGEMHE (23) to undertake in depth functional and molecular characterization of nucleobase transport by hENT1.
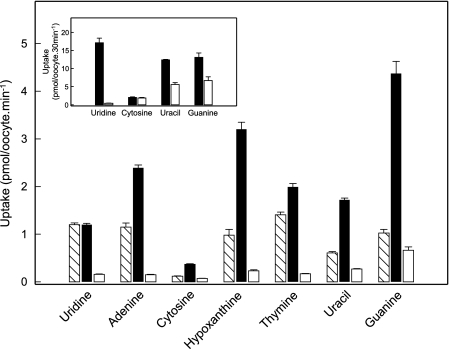
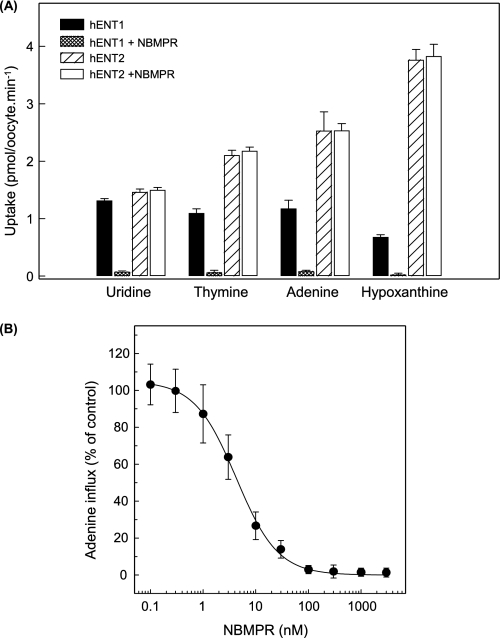
Using pGEMHE, Fig. 1 shows uridine influx in hENT1-producing and water-injected oocytes compared with corresponding 2-min fluxes for hypoxanthine and a panel of other purine (adenine and guanine) and pyrimidine (cytosine, thymine, and uracil) nucleobases measured in the same batch of cells. The results show clear evidence of hENT1-mediated transport of thymine, adenine, and hypoxanthine, with fluxes of 1.15 ± 0.05, 1.00 ± 0.17, and 0.87 ± 0.11 pmol/oocyte·min−1, respectively, compared with 1.04 ± 0.04 pmol/oocyte·min−1 for uridine. Smaller differences in uptake between transcript-injected and water-injected oocytes were apparent for uracil and guanine (0.34 ± 0.03 and 0.36 ± 0.10 pmol/oocyte·min−1, respectively), and there was no measurable transport of cytosine, a nucleobase not likely salvaged in humans and other mammals. Extending the incubation period to 30 min (Fig. 1, inset) confirmed significant hENT1-mediated transport of uracil and adenine, but not cytosine.
FIGURE 1.
Uptake of radiolabeled uridine and a panel of purine and pyrimidine nucleobases by recombinant wild-type hENT1 and hENT2 produced in Xenopus oocytes. Uptake of uridine and nucleobases (20 μm) by oocytes microinjected with recombinant RNA transcripts for hENT1 or hENT2 (hatched and solid columns, respectively) or water alone (open columns) was measured at room temperature (20 °C) using 2-min uptake intervals. The inset shows uptake of uridine, cytosine, uracil, or guanine by hENT1-injected oocytes (solid columns) or water-injected oocytes (open columns) using extended uptake intervals of 30-min. Error bars (S.E. of 10–12 oocytes) are not shown where values were smaller than the thickness of lines.
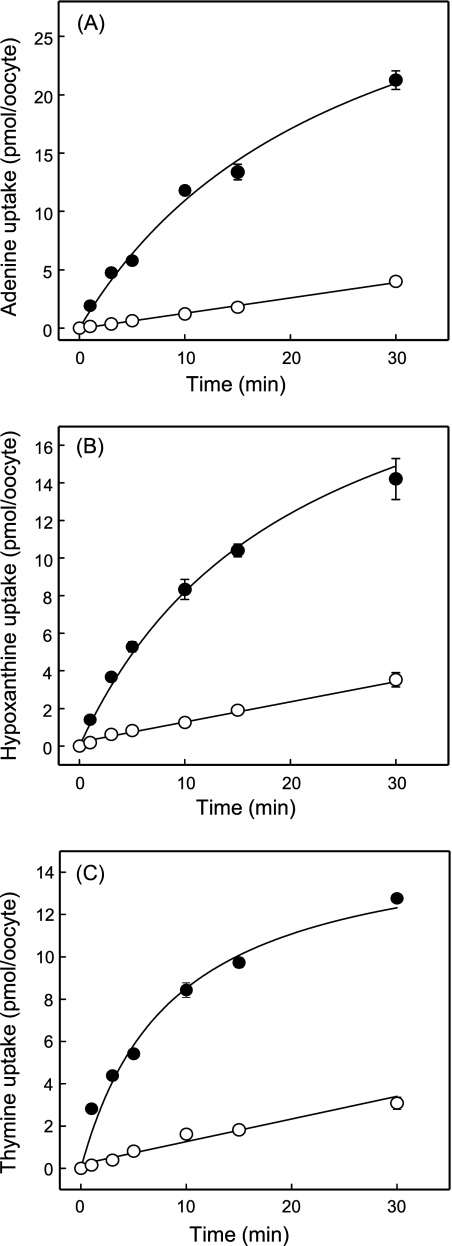
Time courses of thymine, adenine, and hypoxanthine uptake in hENT1-producing and control water-injected oocytes are compared in Fig. 2. The results confirmed the large differences in thymine, adenine, and hypoxanthine uptake between hENT1-producing and control water-injected oocytes uptake shown in Fig. 1 and established that a 2-min flux interval is appropriate to measure initial rates of hENT1-mediated nucleobase transport.
FIGURE 2.
Time courses of uptake of radiolabeled adenine (A), hypoxanthine (B), or thymine (C) by recombinant wild-type hENT1 produced in Xenopus oocytes. Uptake of nucleobases (20 μm) by oocytes microinjected with recombinant RNA transcripts for hENT1 (solid circles) or water alone (open circles) was measured at room temperature (20 °C) using uptake intervals of 1–30 min. Error bars (S.E. of 10–12 oocytes) are not shown where values were smaller than the size of data points. In subsequent kinetic experiments, an uptake interval intermediate between the 1- and 3-min time points (2 min) was used to measure initial rates of transport.
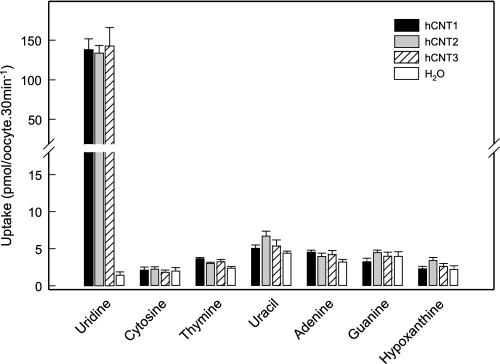
Uptake of Nucleobases by Wild-type hCNT1–3
Corresponding studies of nucleobase transport by hCNT1–3 have been restricted to uracil and hypoxanthine (42–44). As a more rigorous test of their nucleobase transport capabilities, Fig. 3 illustrates a comparison using pGEMHE of uptake of uridine by hCNT1-, hCNT2-, and hCNT3-producing and control water-injected oocytes against that of the same panel of radiolabeled nucleobases shown in Fig. 1 for hENT1. In the experiments with hCNTs, extended incubation periods of 30 min were used to maximize possible detection of nucleobase transport activities. In contrast to large mediated fluxes of uridine of 130–150 pmol/oocyte·30 min−1, none of the transporters tested showed significantly greater uptake of cytosine, thymine, uracil, adenine, guanine, or hypoxanthine than water-injected oocytes. Unlike hENTs, therefore, hCNTs were demonstrated to lack nucleobase transport activities and to be nucleoside-specific.
FIGURE 3.
Uptake of radiolabeled uridine and a panel of purine and pyrimidine nucleobases by recombinant wild-type hCNT1, hCNT2, or hCNT3 produced in Xenopus oocytes. Uptake of uridine or various nucleobases (20 μm) by oocytes microinjected with recombinant RNA transcripts for hCNT1, hCNT2, or hCNT3 (solid, gray, and hatched columns, respectively) or water alone (open columns) was measured at room temperature (20 °C) using 30-min uptake intervals. Error bars (S.E. of 10–12 oocytes) are not shown where values were smaller than the thickness of lines.
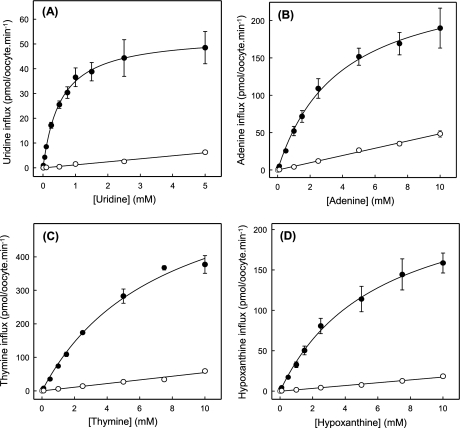
Kinetics of Nucleobase Transport by Wild-type hENT1
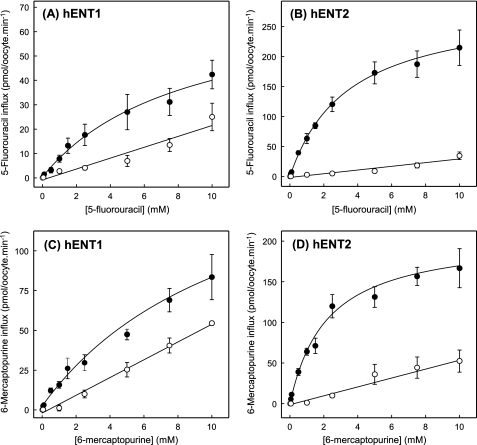
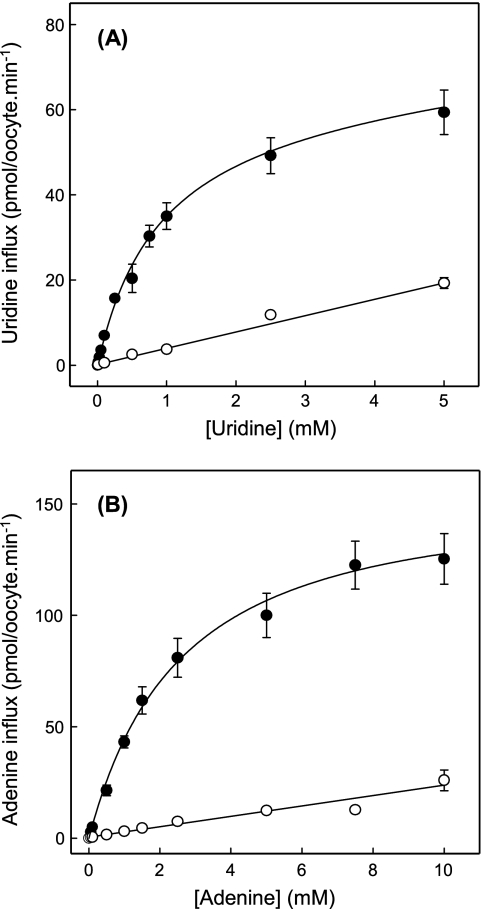
To further characterize nucleobase transport by recombinant wild-type hENT1, Fig. 4 shows the concentration dependence of uptake of uridine, thymine, adenine, or hypoxanthine in hENT1-producing and control water-injected oocytes. Although previous studies indicated that Xenopus oocytes possess endogenous nucleobase transport activities (36), uptake of thymine, adenine, or hypoxanthine by control water-injected oocytes was, like that of uridine, nonsaturable. Rates of uptake at 1 mm extracellular concentrations were: 37 ± 4 pmol/oocyte·min−1 for uridine (Fig. 4A), 52 ± 6 pmol/oocyte·min−1 for adenine (Fig. 4B), 74 ± 4 pmol/oocyte·min−1 for thymine (Fig. 4C), and 33 ± 4 pmol/oocyte·min−1 for hypoxanthine (Fig. 4D). Fluxes in hENT1-producing oocytes corrected for basal uptake rates in water-injected oocytes, in contrast, were saturable and conformed to simple Michaelis-Menten kinetics. Calculated kinetic parameters (apparent Km and Vmax values) derived from the data shown in Fig. 4 are summarized in Table 1. To enable a direct comparison of the nucleobase transport capabilities of hENT1 and hENT2, Table 1 includes corresponding pGEMHE kinetic data for hENT2 measured under identical conditions. Also included in Table 1 are the calculated Vmax/Km ratios for the two transporters for uridine, thymine, adenine, and hypoxanthine.
FIGURE 4.
Concentration dependence of uptake of radiolabeled uridine (A), adenine (B), thymine (C), or hypoxanthine (D) by recombinant wild-type hENT1 produced in Xenopus oocytes. Uptake of uridine or nucleobases by oocytes microinjected with recombinant RNA transcripts for hENT1 (solid circles) or water alone (open circles) was measured at room temperature (20 °C). Calculated kinetic constants (Km and Vmax) for the mediated components of transport (uptake in RNA transcript-injected oocytes minus uptake in water-injected oocytes) are presented in Table 1. Error bars (S.E. of 10–12 oocytes) are not shown where values were smaller than the size of data points.
TABLE 1.
Kinetic properties of recombinant hENT1, S414C/hENT1C−, and hENT2 produced in Xenopus oocytes
Kinetic constants for mediated transport are derived from the influx data presented in Figs. 4 and 7.
| Permeant | Apparent Km | Vmax | Vmax/Km |
|---|---|---|---|
| mm | pmol/oocyte·min−1 | ||
| hENT1 | |||
| Uridine | 0.4 ± 0.1 | 53 ± 2.0 | 133 |
| Adenine | 3.2 ± 0.4 | 192 ± 10 | 60 |
| Thymine | 6.3 ± 1.1 | 558 ± 51 | 89 |
| Hypoxanthine | 6.0 ± 0.5 | 257 ± 12 | 43 |
| 5-Fluorouracil | 2.3 ± 0.6 | 24 ± 2.2 | 10 |
| 6-Meracaptopurine | 1.2 ± 0.4 | 31 ± 2.8 | 26 |
| S414C/hENT1C− | |||
| Uridine | 0.5 ± 0.1 | 41 ± 2.0 | 82 |
| Adenine | 2.0 ± 0.2 | 128 ± 5.0 | 64 |
| hENT2 | |||
| Uridine | 0.5 ± 0.1 | 70 ± 4.2 | 140 |
| Adenine | 1.8 ± 0.3 | 259 ± 14 | 144 |
| Thymine | 6.0 ± 0.5 | 707 ± 31 | 118 |
| Hypoxanthine | 1.5 ± 0.2 | 269 ± 9.0 | 179 |
| 5-Fluorouracil | 2.6 ± 0.2 | 232 ± 6.1 | 89 |
| 6-Mercaptopurine | 1.1 ± 0.2 | 129 ± 7.5 | 117 |
hENT1- and hENT2-mediated transport of uridine gave similar apparent Km values of 0.4 and 0.5 mm and Vmax values of 53 and 70 pmol/oocyte·min−1, respectively, resulting in very similar Vmax/Km ratios (a measure of transport efficiencies) of 133 and 140, respectively. Therefore, with the caveat that relative levels of hENT1 and hENT2 protein expression at the oocyte cell surface are not known, both transporters handled uridine similarly, providing an internal reference for comparison with corresponding kinetic data for transport of nucleobases. Confirming previous findings with pBluescript II KS(+) (18), hENT2 transported thymine, adenine, and hypoxanthine with lower apparent affinities (apparent Km values of 6.0, 1.8, and 1.5 mm, respectively) than that of uridine (apparent Km value of 0.5 mm). This, however, was more than compensated by the higher Vmax values (707, 259, and 269 pmol/oocyte·min−1, respectively), such that the Vmax/Km ratios (118, 144, and 179, respectively) were similar to the corresponding reference value of 140 for uridine. Apparent affinities for hENT1 transport of thymine, adenine, and hypoxanthine were somewhat lower (apparent Km values of 6.3, 3.2, and 6.0 mm, respectively) than those for hENT2 and, compared with uridine, were only partly compensated by higher Vmax values (558, 192, and 257 pmol/oocyte·min−1, respectively). As a result, calculated Vmax/Km ratios (89, 60, and 43, respectively) were lower than the corresponding ratios for nucleobase transport by hENT2 and also somewhat lower than the ratio for hENT1 transport of uridine (133). Setting the Vmax/Km ratios of uridine transport by hENT1 and hENT2 arbitrarily at 1.0, the relative transport efficiencies for thymine, adenine, and hypoxanthine influx via hENT1 were 0.7, 0.5, and 0.3, respectively, and via hENT2 were 0.8, 1.0, and 1.3, respectively. Consistent with the findings of Fig. 1, therefore, these data revealed that hENT1 mediated influx of thymine, adenine, and hypoxanthine with transport efficiencies only slightly lower than that of uridine.
For comparison with the present data, it has been reported that hypoxanthine is transported by system es in S49 mouse lymphoma cells with an apparent Km value of 1 mm (5, 22). However, the apparent Km value for uridine transport in S49 cells is also lower than that reported here for recombinant hENT1 (46). Thus, there is good agreement for the relative affinities for uridine versus hypoxanthine transport by ENT1 in the two studies.
Inhibition of hENT1-mediated Nucleobase Transport by NBMPR and Uridine
Nucleoside transport by hENT1 is inhibited by nanomolar concentrations of NBMPR, whereas hENT2 is NBMPR-insensitive (11, 12, 47, 48). As shown in Fig. 5A, hENT1-mediated transport of thymine, adenine, or hypoxanthine was also inhibited by NBMPR (1 μm), whereas the same concentration of NBMPR had no effect on nucleobase transport by hENT2. In Fig. 5B, the concentration dependence of NBMPR inhibition of hENT1-mediated transport of adenine gave an IC50 value of 4.1 ± 0.3 nm. Assuming competitive inhibition (49) and an adenine apparent Km of 3.2 mm (Table 1), the calculated apparent Ki from these data was also 4.1 nm, a value similar to that determined previously (2.0 nm) for NBMPR inhibition of hENT1-mediated transport of uridine in oocytes (11). In S49 cells, the concentration of NBMPR required to inhibit es-mediated adenine transport by 50%, was ∼10 nm (22).
FIGURE 5.
Effects of NBMPR on nucleobase transport by recombinant wild-type hENT1 and hENT2 produced in Xenopus oocytes. A, mediated uptake of uridine, thymine, adenine, or hypoxanthine (20 μm, 20 °C) in oocytes producing recombinant hENT1 (solid and stippled columns) or hENT2 (hatched and open columns) was measured in the presence or absence of NBMPR (1 μm) (stippled/open and solid/hatched columns, respectively). B, concentration dependence of NBMPR inhibition of adenine uptake (20 μm, 20 °C) in oocytes producing recombinant hENT1. The values in A and B are corrected for endogenous uptake by water-injected oocytes. The calculated IC50 and Ki values for NBMPR inhibition of adenine transport in B are presented in the text. Error bars (S.E. of 10–12 oocytes) are not shown where values were smaller than the size of data points.
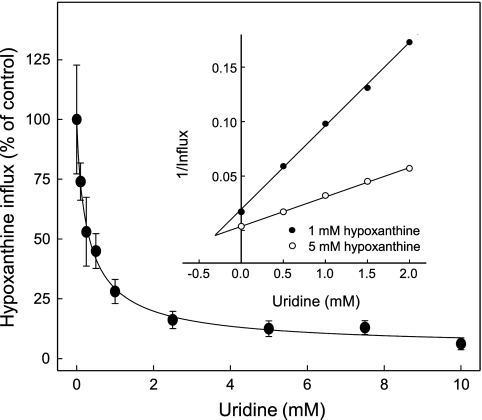
NBMPR binds to the outward-facing conformation of the nucleoside-binding pocket (9, 10). NBMPR inhibition of hENT1-mediated nucleobase transport therefore suggests that nucleosides and nucleobases share common or overlapping permeant binding pockets within the transporter. Similar to experiments undertaken previously for hENT2 (see Fig. 8B in Ref. 18), this was verified kinetically by showing that uridine inhibited hENT1-mediated hypoxanthine transport (IC50 value 0.3 ± 0.1 mm at 20 μm hypoxanthine) (Fig. 6) and by demonstrating that this inhibition was competitive (apparent Ki value also 0.3 mm) (Fig. 6, inset). Numerically consistent with these inhibition constants, the apparent Km value for hENT1-mediated uridine transport was 0.4 mm (Table 1).
FIGURE 8.
Concentration dependence of uptake of radiolabeled 5-fluorouracil (A and B) and 6-mercaptopurine (C and D) by recombinant wild-type hENT1 and hENT2 produced in Xenopus oocytes. Uptake of 5-fluorouracil or 6-mercaptopurine by oocytes microinjected with recombinant RNA transcripts for hENT1 or hENT2 (solid circles) or water alone (open circles) was measured at room temperature (20 °C). Calculated kinetic constants (Km and Vmax) for the mediated components of transport (uptake in RNA transcript-injected oocytes minus uptake in water-injected oocytes) are presented in Table 1. Error bars (S.E. of 10–12 oocytes) are not shown where values were smaller than the size of data points.
FIGURE 6.
Effect of uridine on nucleobase transport by recombinant wild-type hENT1 produced in Xenopus oocytes. Mediated uptake of hypoxanthine (20 μm, 20 °C) in oocytes producing recombinant hENT1 was measured in the presence of graded concentrations of uridine. The inset is a corresponding Dixon plot of uridine inhibition (0.5–2.0 mm) of hENT1-mediated hypoxanthine transport (pmol/oocyte·min−1) measured at two different substrate concentrations (1.0 and 5.0 mm). The values are corrected for endogenous uptake by water-injected oocytes. The calculated IC50 value for uridine inhibition of 20 μm hypoxanthine uptake and the uridine Ki value determined by linear regression analysis of the Dixon plot are presented in the text.
Transport of Anticancer Nucleobase Analogs by hENT1 and hENT2
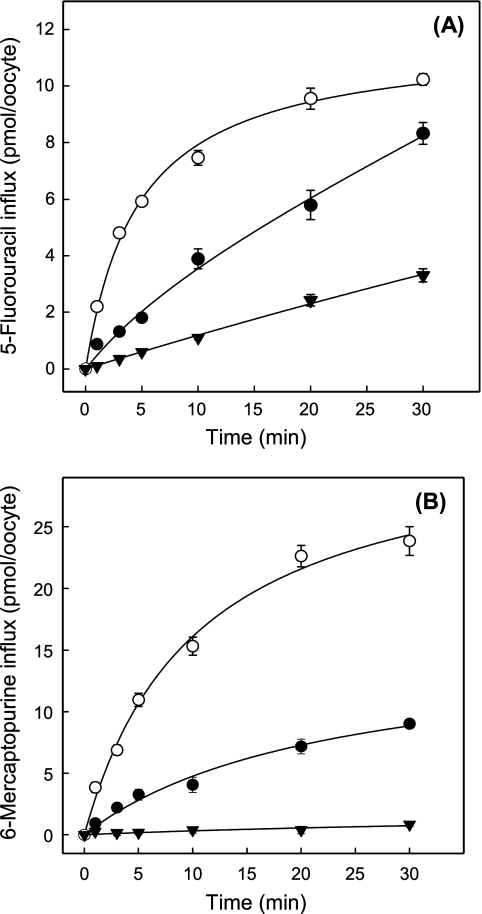
Because 5-fluorouracil and 6-mercaptopurine are analogs of uracil and hypoxanthine, respectively, and are used clinically for the treatment of various cancers, their uptake was studied in hENT1- and hENT2-producing Xenopus oocytes. Fig. 7 shows time courses of 5-fluorouracil or 6-mercaptopurine uptake in hENT1-producing, hENT2-producing, and control water-injected oocytes. The results established that both drugs were transported by hENT1 and hENT2. Fluxes in hENT1- and hENT2-producing oocytes were substantially greater than those in control water-injected oocytes, with hENT2-mediated fluxes of both drugs being greater than those with hENT1. The results further established that 2-min flux intervals were appropriate to measure initial rates of uptake of 5-fluorouracil and 6-mercaptopurine for both hENT1 and hENT2.
FIGURE 7.
Time courses of uptake of radiolabeled 5-fluorouracil (A) and 6-mercaptopurine (B) by recombinant wild-type hENT1 and hENT2 produced in Xenopus oocytes. Uptake of 5-fluorouracil and 6-mercaptopurine (20 μm) by oocytes microinjected with recombinant RNA transcripts of hENT1 (solid circles) or hENT2 (open circles) or water alone (solid inverted triangles) was measured at room temperature (20 °C) using uptake intervals of 1–30 min. Error bars (S.E. of 10–12 oocytes) are not shown where values were smaller than the size of data points. In subsequent kinetic experiments, an uptake interval intermediate between the 1- and 3-min time points (2 min) was used to measure initial rates of transport.
Fig. 8 shows the concentration dependence of 5-fluorouracil and 6-mercaptopurine uptake in hENT1-producing, hENT2-producing and control water-injected oocytes. Mediated fluxes of both drugs by both transporters were saturable and conformed to simple Michaelis-Menten kinetics. Kinetic parameters (apparent Km and Vmax) derived from the data shown in Fig. 8 are summarized in Table 1. hENT1 and hENT2 exhibited similar apparent affinities for 5-fluorouracil (apparent Km values of 2.3 and 2.6 mm, respectively) but very different activities (Vmax values of 24 and 232 pmol/oocyte·min−1, respectively), resulting in Vmax/Km ratios of 10 and 89, respectively. hENT1 and hENT2 also handled 6-mercaptopurine with similar apparent affinities (1.2 and 1.1 mm, respectively), although the difference in Vmax values was less marked than for 5-fluorouracil (31 and 129 pmol/oocyte·min−1, respectively), giving Vmax/Km ratios of 26 and 117, respectively. The Efficiencies for hENT1 transport of 5-fluorouracil and 6-mercaptopurine, therefore, were lower than for the corresponding transport of physiologic nucleobases, whereas the efficiencies for hENT2 transport of fluorouracil, 6-mercaptopurine, and physiologic nucleobases were similar. In contrast to the present findings, previous studies of recombinant mENT2 produced in COS-7 cells suggested that 6-mercaptopurine, but not 5-fluorouracil, is a permeant of mENT2, the latter having an anomalously low reported apparent Km value of 14 μm (50).
Nucleobase Transport by hENT1C− and hENT1C− Mutants S1 to S10
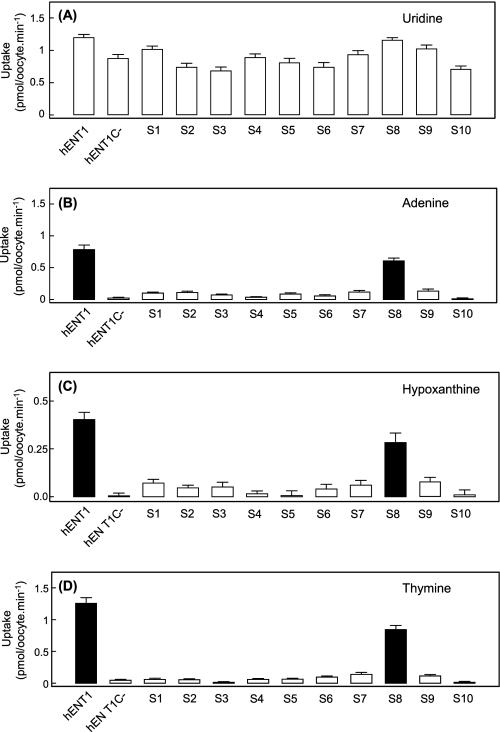
hENT1 is a 456-amino acid residue protein with 11 predicted TM helices, a membrane architecture confirmed experimentally by introduction of engineered N-glycosylation sites and immunological approaches (51). All 10 endogenous cysteine residues are located in TM regions (Fig. 9). An engineered cysteine-less version of hENT1 (hENT1C−) in which each cysteine residues is replaced by serine is fully functional with respect to nucleoside transport and is sensitive to inhibition by NBMPR and the vasoactive drugs dipyridamole and dilazep.4 Unlike wild-type hENT1, however, hENT1C− was unable to transport either purine or pyrimidine nucleobases (mediated fluxes were <1% that of uridine) (Fig. 10). To identify the cysteine-to-serine conversion(s) responsible for this phenotypic change, hENT1C− was used as a template to prepare 10 revertant mutants in which each endogenous cysteine residue was individually reintroduced back into its original position (Table 2). In Fig. 10, the uridine, thymine, adenine, and hypoxanthine transport capabilities of each of these 10 mutants were compared with both wild-type hENT1 and hENT1C−. All 10 S-series mutants transported uridine when produced in Xenopus oocytes (Fig. 10A), but only mutant S8 (S414C/hENT1C−) also transported thymine, adenine, and hypoxanthine (Fig. 10, B–D) with mediated fluxes relative to that of uridine of 71, 46, and 25%, respectively, which were broadly similar to those of wild-type hENT1 (104, 63, and 36%, respectively).
FIGURE 9.
Membrane topology of hENT1 showing locations of endogenous cysteine residues. The positions of putative TM helices are indicated as open rectangles and are numbered. Cysteine residues are indicated by the letter C. The single N-glycosylation site of hENT1 is highlighted by a solid star. *C414 is the single cysteine residue implicated in hENT1 nucleobase transportability.
FIGURE 10.
Uptake of radiolabeled uridine (A), adenine (B), hypoxanthine (C), or thymine (D) by recombinant wild-type hENT1, cysteine-less hENT1C−, or S-series hENT1C− mutants produced in Xenopus oocytes. Mediated uptake of uridine, adenine, hypoxanthine, or thymine (20 μm) in oocytes producing recombinant wild-type hENT1, hENT1C−, or S-series hENT1C− mutants was measured at room temperature (20 °C). The values are corrected for endogenous uridine and nucleobase uptake by water-injected oocytes.
TABLE 2.
Nomenclature of hENT1C− mutants
| Name | Region | Mutation |
|---|---|---|
| S1 | TM 2 | S87C/hENT1C− |
| S2 | TM 5 | S193C/hENT1C− |
| S3 | TM 6 | S213C/hENT1C− |
| S4 | TM 6 | S222C/hENT1C− |
| S5 | TM 7 | S297C/hENT1C− |
| S6 | TM 8 | S333C/hENT1C− |
| S7 | TM 9 | S378C/hENT1C− |
| S8 | TM 10 | S414C/hENT1C− |
| S9 | TM 10 | S416C/hENT1C− |
| S10 | TM 11 | S439C/hENT1C− |
The relatively small variability in uridine fluxes shown in Fig. 10A suggests that all S-series mutants were expressed at the oocyte cell surface in amounts similar to hENT1 and hENT1C−. For mutant S8 (S414C/hENT1C−), the kinetics of uridine and adenine transport were also investigated (Fig. 11). Mediated fluxes of uridine (Fig. 11A) and adenine (Fig. 11B) were saturable and conformed to simple Michaelis-Menten kinetics, with apparent Km and Vmax values of 0.5 mm and 41 pmol/oocyte·min−1, respectively, for uridine (Vmax/Km ratio, 82), and 2.0 mm and 128 pmol/oocyte·min−1, respectively, for adenine (Vmax/Km ratio, 64) (Table 1). The close correspondence between these values and those for hENT1 (see also Table 1) established that reintroduction of Cys-414 into hENT1C− functionally restored wild-type nucleobase transport capability. Cys-414 in TM 10 is therefore a major determinant of nucleobase transport by hENT1.
FIGURE 11.
Concentration dependence of uptake of radiolabeled uridine (A) or adenine (B) by recombinant S-series mutant S414C/hENT1C− (S8) produced in Xenopus oocytes. Uptake of uridine or adenine by oocytes microinjected with recombinant RNA transcripts for S414C/hENT1C− (solid circles) or water alone (open circles) was measured at room temperature (20 °C). Calculated kinetic constants (Km and Vmax) for the mediated components of transport (uptake in RNA transcript-injected oocytes minus uptake in water-injected oocytes) are presented in the text. Error bars (S.E. of 10–12 oocytes) are not shown where values were smaller than the size of data points.
Consistent with a similar role in other species, hENT1 Cys-414 is conserved in other mammalian ENT1s (supplemental Fig. S1). Present as threonine in h/m/r ENT2, chimeric studies suggest that the major determinant(s) of nucleobase transportability in ENT2 family members instead reside in the TM 5–6 region of the transporter. For hENT1, mutation of Cys-414 of hENT1C− to theronine or serine did not restore nucleobase transport by hENT1C− (data not shown). The molecular determinants of nucleobase transport activity in ENT1 and ENT2 proteins are therefore different.
Currently, no ENT crystal structure is known, and there is no statistically significant sequence similarity between ENT family members and any transporters for which a crystallographic structure has been determined. Despite this, we have hypothesized that ENTs may share a common evolutionary origin with the major facilitator superfamily of transporters (52). The crystal structures of two Escherichia coli members of the latter family, the lactose transporter LacY and the glycerol-3-phosphate/phosphate exchanger GlpT, have been determined (53, 54). Support for structural commonality between ENTs and major facilitator superfamily transporters is provided by site-directed mutagenesis studies of mammalian and protozoan ENTs, which suggest similar packing of TMs helices around a solvent-accessible permeant binding site (10). This has allowed construction of putative tertiary structures of several protozoan ENTs, including Leishmania donovani LdNT2, P. falciparum PfENT1, and T. brucei TbNBT1 (25, 55, 56). Supplemental Fig. S2 shows a model of hENT1 modeled using TMs 1–11 of the 12-TM GlpT protein as template (10). Recently, an ab initio 3-D model of L. donovani LdNT1.1 was generated with structural features common to those obtained by threading approaches (57).
In supplemental Fig. S2, TM 10 is one of the cluster of helices forming the translocation pore, with residue Cys-414 located toward the cytoplasmic end of the TM and adjacent to another cysteine residue (Cys-416) at the helix boundary. The latter residue has been recently been identified as the cysteine residue in hENT1 responsible for N-ethylmaleimide inhibition of the transporter by a mechanism constraining the protein in an inward facing conformation.4 Rather than directly participate in nucleobase binding, therefore, Cys-414 most likely has other roles. In the major facilitator superfamily peptide transporter PepTSo, for example, residues in a similar position in the TM 10–11 hairpin are known to contribute, together with residues in the TM 4–5 hairpin, to the intracellular gate of the protein (45). In the Arabidopsis thaliana protein CHL1, a homolog of PepTSo, phosphorylation of the conserved residue Thr-101 switches the former from being a low affinity to a high affinity nitrate transporter (26). The residue in PepTSo corresponding to CHL1 Thr-101 (Thr-87) is near the cytoplasmic end of TM 3 close to the cytoplasmic loop connecting this helix to TM 2 (45), and, like hENT1 Cys-414, it is also distant from the likely permeant-binding site at the center of the protein. Nucleobases lack the sugar moiety present in nucleosides and may require the additional structural stability provided by hENT1 Cys-414 to enable binding and therefore translocation of nucleobases.
CONCLUSIONS
Known previously from flux studies by their functional designations as the es and ei equilibrative nucleoside transport processes (2, 3), hENT1 and hENT2 have been distinguished functionally by their sensitivity and insensitivity, respectively, to inhibition by NBMPR, and by the ability of hENT2, but not hENT1, to transport hypoxanthine and other nucleobases. The apparent affinities of hENT2 for nucleobases are lower than for nucleosides (as indicated by higher Km values) but are compensated by higher Vmax values (18). At physiological concentrations, therefore, the efficiencies of the nucleoside and nucleobase transport processes, defined kinetically as the ratio Vmax:Km, are similar. To date, hENT2 (and intracellular hENT3) are the first discovered, and so far only identified transporter proteins for nucleobases in human cells and tissues. Here, we have used heterologous expression in Xenopus oocytes to demonstrate that recombinant hENT1 is also capable of transporting nucleobases and the therapeutic anticancer nucleobase drugs 5-fluorouracil and 6-mercaptopurine. Both purine (hypoxanthine, adenine > guanine) and pyrimidine nucleobases (thymine > uracil) were accepted by hENT1 as permeants, albeit with lesser kinetic efficiencies than hENT2. Human concentrative nucleoside transporters hCNT1, hCNT2, and hCNT3, in contrast, did not transport nucleobases and thus should be considered nucleoside-specific. Counterbalancing the lower kinetic efficiencies of hENT1 for nucleobase transport, most human cells and tissues have substantially greater plasma membrane abundance of hENT1 than hENT2 (8–11). Representing the larger of the two transport activities, therefore, hENT1 likely plays a significant role in human nucleobase and nucleobase drug transport and homeostasis. In structure-function studies, Cys-414 in TM 10 of hENT1 was identified as a molecular determinant of hENT1 nucleobase transportability.
Supplementary Material
This work was supported in part by the Canadian Institutes of Health Research and the Alberta Cancer Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Nucleoside transport system nomenclature is as follows: es, equilibrative, sensitive to inhibition by NBMPR; ei, equilibrative, insensitive to inhibition by NBMPR; cit, concentrative, NBMPR-insensitive, transports thymidine; cif, concentrative, NBMPR-insensitive, transports formycin B; cib, concentrative, NBMPR-insensitive, broadly selective for both purine and pyrimidine nucleosides.
S. Y. M. Yao and J. D. Young, unpublished observations.
- NT
- nucleoside transporter
- CNT
- concentrative nucleoside transporter
- ENT
- equilibrative nucleoside transporter
- TM
- putative transmembrane helix
- NBMPR
- nitrobenzylmercaptopurine ribonucleoside
- h
- human
- r
- rat
- m
- mouse.
REFERENCES
- 1. Zhang J., Visser F., King K. M., Baldwin S. A., Young J. D., Cass C. E. (2007) Metastasis Rev. 26, 85–110 [DOI] [PubMed] [Google Scholar]
- 2. King A. E., Ackley M. A., Cass C. E., Young J. D., Baldwin S. A. (2006) Trends Pharmacol. Sci. 27, 416–425 [DOI] [PubMed] [Google Scholar]
- 3. Cabrita M. A., Baldwin S. A., Young J. D., Cass C. E. (2002) Biochem. Cell. Biol. 80, 623–638 [DOI] [PubMed] [Google Scholar]
- 4. Young J. D., Cheeseman C. I., Mackey J. R., Cass C. E., Baldwin S. A. (2000) in Current Topics in Membranes (Barrett K. E., Donowitz M. eds.) pp. 329–378, Academic Press, San Diego, CA [Google Scholar]
- 5. Griffith D. A., Jarvis S. M. (1996) Biochim. Biophys. Acta 1286, 153–181 [DOI] [PubMed] [Google Scholar]
- 6. de Koning H., Diallinas G. (2000) Mol. Membr. Biol. 17, 75–94 [DOI] [PubMed] [Google Scholar]
- 7. Gray J. H., Owen R. P., Giacomini K. M. (2004) Pflugers Arch. 447, 728–734 [DOI] [PubMed] [Google Scholar]
- 8. Baldwin S. A., Beal P. R., Yao S. Y., King A. E., Cass C. E., Young J. D. (2004) Pflugers Arch. 447, 735–743 [DOI] [PubMed] [Google Scholar]
- 9. Young J. D., Yao S. Y., Sun L., Cass C. E., Baldwin S. A. (2008) Xenobiotica. 38, 995–1021 [DOI] [PubMed] [Google Scholar]
- 10. Parkinson F. E., Damaraju V. L., Graham K., Yao S. Y., Baldwin S. A., Cass C. E., Young J. D. (2011) Curr. Top. Med. Chem. 11, 948–972 [DOI] [PubMed] [Google Scholar]
- 11. Griffiths M., Beaumont N., Yao S. Y., Sundaram M., Boumah C. E., Davies A., Kwong F. Y., Coe I., Cass C. E., Young J. D., Baldwin S. A. (1997) Nat. Med. 3, 89–93 [DOI] [PubMed] [Google Scholar]
- 12. Griffiths M., Yao S. Y., Abidi F., Phillips S. E., Cass C. E., Young J. D., Baldwin S. A. (1997) Biochem. J. 328, 739–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yao S. Y., Ng A. M., Muzyka W. R., Griffiths M., Cass C. E., Baldwin S. A., Young J. D. (1997) J. Biol. Chem. 272, 28423–28430 [DOI] [PubMed] [Google Scholar]
- 14. Baldwin S. A., Yao S. Y., Hyde R. J., Ng A. M., Foppolo S., Barnes K., Ritzel M. W., Cass C. E., Young J. D. (2005) J. Biol. Chem. 280, 15880–15887 [DOI] [PubMed] [Google Scholar]
- 15. Govindarajan R., Leung G. P., Zhou M., Tse C. M., Wang J., Unadkat J. D. (2009) Am. J. Physiol. Gastrointest. Liver Physiol. 296, G910–G922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Engel K., Zhou M., Wang J. (2004) J. Biol. Chem. 279, 50042–50049 [DOI] [PubMed] [Google Scholar]
- 17. Barnes K., Dobrzynski H., Foppolo S., Beal P. R., Ismat F., Scullion E. R., Sun L., Tellez J., Ritzel M. W., Claycomb W. C., Cass C. E., Young J. D., Billeter-Clark R., Boyett M. R., Baldwin S. A. (2006) Circ. Res. 99, 510–519 [DOI] [PubMed] [Google Scholar]
- 18. Yao S. Y., Ng A. M., Vickers M. F., Sundaram M., Cass C. E., Baldwin S. A., Young J. D. (2002) J. Biol. Chem. 277, 24938–24948 [DOI] [PubMed] [Google Scholar]
- 19. Landfear S. M., Ullman B., Carter N. S., Sanchez M. A. (2004) Eukaryot. Cell 3, 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parker M. D., Hyde R. J., Yao S. Y., McRobert L., Cass C. E., Young J. D., McConkey G. A., Baldwin S. A. (2000) Biochem. J. 349, 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sundaram M., Yao S. Y., Ingram J. C., Berry Z. A., Abidi F., Cass C. E., Baldwin S. A., Young J. D. (2001) J. Biol. Chem. 276, 45270–45275 [DOI] [PubMed] [Google Scholar]
- 22. Aronow B., Ullman B. (1986) J. Biol. Chem. 261, 2014–2019 [PubMed] [Google Scholar]
- 23. Liman E. R., Tytgat J., Hess P. (1992) Neuron 9, 861–871 [DOI] [PubMed] [Google Scholar]
- 24. Yao S. Y., Cass C. E., Young J. D. (2000) in Membrane Transport: A Practical Approach (Baldwin S. A. ed.) pp. 47–78, Oxford University Press, New York [Google Scholar]
- 25. Baldwin S. A., McConkey G. A., Cass C. E., Young J. D. (2007) Curr. Pharm. Des. 13, 569–580 [DOI] [PubMed] [Google Scholar]
- 26. Liu K. H., Tsay Y. F. (2003) EMBO J. 22, 1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Plagemann P. G., Wohlhueter R. M., Woffendin C. (1988) Biochim. Biophys. Acta 947, 405–443 [DOI] [PubMed] [Google Scholar]
- 28. Gati W. P., Paterson A. R., Tyrrell D. L., Cass C. E., Moravek J., Robins M. J. (1992) J. Biol. Chem. 267, 22272–22276 [PubMed] [Google Scholar]
- 29. Griffith D. A., Doherty A. J., Jarvis S. M. (1992) Biochim. Biophys. Acta 1106, 303–310 [DOI] [PubMed] [Google Scholar]
- 30. Ullman B., Patrick J., McCartan K. (1987) Mol. Cell. Biol. 7, 97–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hoque K. M., Chen L., Leung G. P., Tse C. M. (2008) Am. J. Physiol. Regul. Integr. Comp. Physiol. 294, R1988–R1995 [DOI] [PubMed] [Google Scholar]
- 32. Griffith D. A., Jarvis S. M. (1993) J. Biol. Chem. 268, 20085–20090 [PubMed] [Google Scholar]
- 33. Griffith D. A., Jarvis S. M. (1994) Biochem. J. 303, 901–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barros L. F. (1994) Am. J. Obstet. Gynecol. 171, 111–117 [DOI] [PubMed] [Google Scholar]
- 35. Washington C. B., Giacomini K. M. (1995) J. Biol. Chem. 270, 22816–22819 [DOI] [PubMed] [Google Scholar]
- 36. Shayeghi M., Akerman R., Jarvis S. M. (1999) Biochim. Biophys. Acta 1416, 109–118 [DOI] [PubMed] [Google Scholar]
- 37. Kato R., Maeda T., Akaike T., Tamai I. (2006) Am. J. Physiol. Endocrinol. Metab. 290, E968–E975 [DOI] [PubMed] [Google Scholar]
- 38. Kato R., Maeda T., Akaike T., Tamai I. (2009) Biol. Pharm. Bull. 32, 450–455 [DOI] [PubMed] [Google Scholar]
- 39. Pantazopoulou A., Diallinas G. (2007) FEMS Microbiol. Rev. 31, 657–675 [DOI] [PubMed] [Google Scholar]
- 40. Yamamoto S., Inoue K., Murata T., Kamigaso S., Yasujima T., Maeda J. Y., Yoshida Y., Ohta K. Y., Yuasa H. (2010) J. Biol. Chem. 285, 6522–6531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Caulfield M. J., Munroe P. B., O'Neill D., Witkowska K., Charchar F. J., Doblado M., Evans S., Eyheramendy S., Onipinla A., Howard P., Shaw-Hawkins S., Dobson R. J., Wallace C., Newhouse S. J., Brown M., Connell J. M., Dominiczak A., Farrall M., Lathrop GM., Samani N. J., Kumari M., Marmot M., Brunner E., Chambers J., Elliott P., Kooner J., Laan M., Org E., Veldre G., Viigimaa M., Cappuccio F. P., Ji C., Iacone R., Strazzullo P., Moley K. H., Cheeseman C. (2008) PLoS Med. 5, e197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smith K. M., Ng A. M., Yao S. Y., Labedz K. A., Knaus E. E., Wiebe L. I., Cass C. E., Baldwin S. A., Chen X. Z., Karpinski E., Young J. D. (2004) J. Physiol. 558, 807–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ritzel M. W., Yao S. Y., Ng A. M., Mackey J. R., Cass C. E., Young J. D. (1998) Mol. Membr. Biol. 15, 203–211 [DOI] [PubMed] [Google Scholar]
- 44. Ritzel M. W., Ng A. M., Yao S. Y., Graham K., Loewen S. K., Smith K. M., Ritzel R. G., Mowles D. A., Carpenter P., Chen X. Z., Karpinski E., Hyde R. J., Baldwin S. A., Cass C. E., Young J. D. (2001) J. Biol. Chem. 276, 2914–2927 [DOI] [PubMed] [Google Scholar]
- 45. Newstead S., Drew D., Cameron A. D., Postis V. L., Xia X., Fowler P. W., Ingram J. C., Carpenter E. P., Sansom M. S., McPherson M. J., Baldwin S. A., Iwata S. (2011) EMBO J. 30, 417–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Belt J. A., Noel L. D. (1985) Biochem. J. 232, 681–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Crawford C. R., Patel D. H., Naeve C., Belt J. A. (1998) J. Biol. Chem. 273, 5288–5293 [DOI] [PubMed] [Google Scholar]
- 48. Ward J. L., Sherali A., Mo Z. P., Tse C. M. (2000) J. Biol. Chem. 275, 8375–8381 [DOI] [PubMed] [Google Scholar]
- 49. Stein W. D. (1986) in Transport and Diffusion across Cell Membrane, pp. 231–337, Academic Press, Orlando, FL [Google Scholar]
- 50. Nagai K., Nagasawa K., Kihara Y., Okuda H., Fujimoto S. (2007) Int. J. Pharm. 333, 56–61 [DOI] [PubMed] [Google Scholar]
- 51. Yao S. Y., Sundaram M., Chomey E. G., Cass C. E., Baldwin S. A., Young J. D. (2001) Biochem. J. 353, 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Young J. D., Yao S. Y., Cass C. E., Baldwin S. A. (2003) Equilibrative Nucleoside Transport Proteins: Red Cell Transport in Health and Disease, pp. 321–337, Springer, Berlin [Google Scholar]
- 53. Abramson J., Smirnova I., Kasho V., Verner G., Kaback H. R., Iwata S. (2003) Science 301, 610–615 [DOI] [PubMed] [Google Scholar]
- 54. Huang Y., Lemieux M. J., Song J., Auer M., Wang D. N. (2003) Science 301, 616–620 [DOI] [PubMed] [Google Scholar]
- 55. Arastu-Kapur S., Arendt C. S., Purnat T., Carter N. S., Ullman B. (2005) J. Biol. Chem. 280, 2213–2219 [DOI] [PubMed] [Google Scholar]
- 56. Papageorgiou I., De Koning H. P., Soteriadou K., Diallinas G. (2008) Int. J. Parasitol. 38, 641–653 [DOI] [PubMed] [Google Scholar]
- 57. Valdés R., Arastu-Kapur S., Landfear S. M., Shinde U. (2009) J. Biol. Chem. 284, 19067–19076 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.