Abstract
Transforming growth factor-β (TGF-β) signaling is controlled by a variety of regulators, of which Smad7, c-Ski, and SnoN play a pivotal role in its negative regulation. Arkadia is a RING-type E3 ubiquitin ligase that targets these negative regulators for degradation to enhance TGF-β signaling. In the present study we identified a candidate human tumor suppressor gene product RB1CC1/FIP200 as a novel positive regulator of TGF-β signaling that functions as a substrate-selective cofactor of Arkadia. Overexpression of RB1CC1 enhanced TGF-β signaling, and knockdown of endogenous RB1CC1 attenuated TGF-β-induced expression of target genes as well as TGF-β-induced cytostasis. RB1CC1 down-regulated the protein levels of c-Ski but not SnoN by enhancing the activity of Arkadia E3 ligase toward c-Ski. Substrate selectivity is primarily attributable to the physical interaction of RB1CC1 with substrates, suggesting its role as a scaffold protein. RB1CC1 thus appears to play a unique role as a modulator of TGF-β signaling by restricting substrate specificity of Arkadia.
Keywords: Signal Transduction, SMAD Transcription Factor, Transcription Repressor, Transforming Growth Factor β (TGFβ), Ubiquitylation
Introduction
Transforming growth factor-β (TGF-β) is a multifunctional cytokine that regulates various cellular responses, including growth, motility, differentiation, and apoptosis in a wide variety of target cells. TGF-β binds to two different types of serine/threonine kinase receptors, type I and II, and induces the formation of heterotetrameric complexes consisting of two type II receptors and two type I receptors on the cell surface (1–3). Type II receptor kinase, which is constitutively active, phosphorylates type I receptor kinase in the glycine/serine-rich domain (GS domain) and activates it. Type I receptor kinase then predominantly transmits signals through a group of cytoplasmic signaling mediators termed Smads. Smad2 and Smad3, which are TGF-β-specific receptor-regulated Smads (R-Smads), are phosphorylated by type I receptor and form a complex with Smad4, a common Smad. The Smad complexes then translocate into the nucleus (4) where they regulate transcription of target genes in cooperation with transcriptional activators and/or repressors (5).
TGF-β signaling is regulated by various cytoplasmic as well as nuclear proteins through physical interactions and posttranslational modifications, including phosphorylation, ubiquitylation, SUMOylation, and acetylation. Smad7, an inhibitory Smad, competitively inhibits R-Smad phosphorylation by binding to activated type I receptor (6–8). c-Ski and SnoN, which are nuclear co-repressor proteins of the Ski family, inactivate the activated Smad complexes through physical interaction (9, 10). Several HECT-type E3 ubiquitin ligases are reported to be involved in the regulation of TGF-β signaling (11). Active type I receptor is down-regulated by the cooperative action of Smurf1 or Smurf2 with Smad7 through ubiquitylation and subsequent proteasomal degradation (12, 13). WWP1/Tiul1 and NEDD4–2 also have similar functions in down-regulation of active type I receptor (14, 15). In addition, Smurfs and WWP1/Tiul1 down-regulate Smad proteins in a signaling- or cofactor-dependent fashion (16–20). Other types of E3 ligases, including an SCF ubiquitin E3 ligase complex ROC1-SCFFbw1a (21), a U-box ubiquitin E3 ligase CHIP (22), and a RING-type E3 ligase Ectodermin (23), are also involved in the degradation or inactivation of Smad proteins. All these E3 ubiquitin ligases down-regulate TGF-β signaling. Arkadia, which up-regulates TGF-β signaling, is the sole exception (24, 25).
Arkadia was originally identified as a protein essential for the induction of the mammalian node (26) and was subsequently shown to enhance Nodal signaling during embryonic development (27). We and others previously found that Arkadia is a RING-type ubiquitin ligase that targets negative regulators of TGF-β signaling, Smad7, c-Ski, and SnoN to enhance TGF-β signaling (24, 25, 28–30). Recently we found that Arkadia ubiquitylates adaptor protein-2μ to enhance epidermal growth factor receptor signaling, indicating possible involvement of Arkadia in the regulation of other signal transduction pathways (31, 32). However, unlike the Smurf family E3 ligases (33–35), regulation of Arkadia function is not well understood, except for a study reporting that Axin1 functions as a scaffold protein to promote Smad7 degradation by Arkadia (36).
In this study we employed a yeast two-hybrid system to identify proteins that interact with Arkadia and identified RB1-inducible coiled-coil 1 (RB1CC1),2 also termed focal adhesion kinase interacting protein of 200 kDa (FIP200), as a novel Arkadia-binding protein. We found that RB1CC1 enhances TGF-β signaling by acting as a cofactor of Arkadia. Notably, RB1CC1 preferentially enhances ubiquitylation of c-Ski by Arkadia, whereas Axin1 exclusively enhances Arkadia-dependent ubiquitylation of Smad7. Our findings indicate that the E3 ligase activity of Arkadia is regulated by cofactors with distinct substrate preferences.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Screening
The yeast two-hybrid screening was performed as described previously (31).
Cell Culture
HEK293, HEK293T, COS7, and HaCaT cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 100 units/ml of penicillin G, and 100 μg/ml of streptomycin. Cells were grown in a 5% CO2 atmosphere at 37 °C.
cDNA Constructs and Chemicals
Mouse Arkadia cDNA and C937A mutant (Arkadia-CA) were as described (24). Other Arkadia mutants have been described previously (28). Human RB1CC1 cDNA was described previously (37). RB1CC1 deletion mutants were constructed using a PCR-based method. cDNA for a constitutively active form of TGF-β type I receptor (ALK-5-TD), mouse Smad7, and human ubiquitin (Ub) were as previously described (24). cDNA for c-Ski, mutants of c-Ski, and SnoN were prepared as described (28). Rat Axin1 cDNA was a kind gift from Dr. M. Kato. TGF-β3 was from R&D Systems (Minneapolis, MN). A44-03, an ALK-4/5/7 inhibitor, is a dihydrochloride salt form of A-77-01 (38).
Cell Fractionation
Subcellular fractionation of HEK293 and HaCaT cells was performed using NE-PER nuclear and cytoplasmic extraction reagent (Pierce/Thermo Fisher Scientific, Waltham, MA) according to the manufacturer's protocol.
Transfection of cDNAs, Immunoprecipitation, and Immunoblotting
Transfection of DNA was performed using FuGENE 6 (Roche Diagnostics) according to the manufacturer's recommendations. Cell lysates were prepared in Igepal CA630 lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Igepal CA630, 1 mm phenylmethylsulfonyl fluoride, and 100 units/ml aprotinin). Radioimmunoprecipitation assay buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton-X100, and protease inhibitors (P8340, Sigma) was also used in some experiments to detect endogenous protein expression. After centrifugation, protein concentrations of cell lysates were quantified using a DC protein assay kit (Bio-Rad). For some experiments, cells were treated with 10 μm lactacystin (Kyowa Medex, Tokyo, Japan) for 6 h or 50 μm MG132 (Peptide Institute, Osaka, Japan) for 4 h before harvesting to inhibit proteasomal degradation. For immunoprecipitation, cell lysates and antibodies were incubated between 2 h and overnight. Immune complexes were then precipitated with protein A/G-Sepharose beads or Dynabeads-M280 sheep anti-mouse IgG (Dynal/Invitrogen) and washed four times with lysis buffer. SDS-PAGE and immunoblotting were performed as described (39). Stripping and re-probing of blotted membranes were performed using stripping buffer (62.5 mm Tris-HCl, pH 6.7, 2% SDS, and 0.8% 2-mercaptoethanol; for transfection experiments) or Restore Western blot stripping buffer (Pierce; for endogenous proteins). We used ImageJ software to quantify immunoblot data. All quantification data were obtained from the image files acquired using an LAS-3000 mini luminoimage analyzer (Fuji Film, Tokyo, Japan).
Antibodies
Anti-RB1CC1 antibody was prepared by immunizing a rabbit with human RB1CC1 (amino acid residues 1247–1396) expressed as a fusion protein with glutathione S-transferase (GST). Anti-Arkadia antibody (#62) was raised against mouse Arkadia (amino acid residues 1–203) as described previously (24) and used for immunoblotting. Commercially available antibodies used were as follows: mouse anti-FLAG (M2) (Sigma), anti-α-tubulin (DM1A) (Sigma), goat anti-Smad6/7 (N19) (Santa Cruz Biotechnology, Santa Cruz, CA), anti-Arkadia (H16) (Santa Cruz Biotechnology; for immunoprecipitation), rabbit anti-phospho-Smad2 (138D4, Cell Signaling Technology, Danvers, MA), anti-Axin1 (Cell Signaling Technology), anti-histone deacetylase 1 (Upstate Biotechnology/Millipore Corp., Billerica, MA), anti-SnoN (H-317) (Santa Cruz Biotechnology), mouse anti-HA (Invivogen, San Diego, CA), anti-c-Ski (G8) (Cascade Bioscience, Winchester, MA), anti-RNF111 (Arkadia) (Abnova, Taipei, Taiwan; for immunoblotting of human Arkadia), and anti-myc (9E10) (BD Pharmingen). Normal mouse IgG1 (MB002) (R&D Systems), normal goat IgG (500-G00) (Peprotech, London, UK), and normal rabbit serum (S-5000) (Vector Laboratories, Burlingame, CA) were used as controls.
GST Pulldown Assay
The C-terminal region of RB1CC1 (amino acid residues 1397–1594) was expressed as a fusion protein with GST. Arkadia or c-Ski was expressed using TNT Quick Coupled Transcription/Translation System (Promega, Madison, WI). The pulldown assay was performed as described previously (40).
Luciferase Assay
A luciferase reporter assay was performed as described previously with only minor changes (24). For normalization, pGL4.75-SV40-hRluc was used. (CAGA)9-MLP-Luc2 was derived from (CAGA)9-MLP-Luc (41) by replacing the plasmid backbone from pGL3 to pGL4.
RNA Interference and Oligonucleotides
Stealth small interfering RNAs (siRNAs) were purchased from Invitrogen as follows: human Arkadia (sense, 5′-GCAGAGGAAGAAACGAGAAGUGUUA-3′), RB1CC1 #1 (sense, 5′-CCUGGACCAGAUGAUUGCUAGCUGU-3′), RB1CC1 #2 (5′-CCGAGUUAAUUAGUAGACAUGAAGA-3′), Axin1 (sense, 5′-GGCAGCUACAGAUACUACUUCAAGA-3′), Smad7 (sense, 5′-AGAAGAAGUUGGGAAUCUGAAAGCC-3′), and control oligos (12935–112, sequence not available). siRNAs were introduced into HEK293, HEK293T, or COS7 cells using Lipofectamine2000 reagent (Invitrogen) according to the manufacturer's instructions with 100–120 pmol of siRNA/well of 6-well tissue culture plates. For transfection of siRNAs into HaCaT cells, Lipofectamine RNAiMAX (Invitrogen) was used.
Quantitative Real-time PCR Analysis
Total RNAs were extracted using TRIzol (Invitrogen). First-strand cDNAs were synthesized using PrimeScript reverse transcriptase (Takara Bio, Shiga, Japan) and oligo(dT)12–18 primers (Invitrogen). Quantitative real-time PCR analysis was performed using Platinum SYBR Green qPCR SuperMix-UDG with ROX (Invitrogen) and the ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Foster City, CA). Detected signals were confirmed as specific by a dissociation protocol. Primers used for quantitative real-time PCR were as follows: human p21WAF (sense, 5′-CGGCAGACCAGCATGACAGA-3′; antisense, 5′-GAAGATCAGCCGGCGTTTG-3′); p15INK4b (sense, 5′-GGTAATGAAGCTGAGCCCAGGT-3′; antisense, 5′-GATAATCCACCGTTGGCCGTA-3′); PAI-1 (sense, 5′-GGCTGACTTCACGAGTCTTTCA-3′; antisense, 5′-ATGCGGGCTGAGACTATGACA-3′); Smad7 (sense, 5′-GATACCCGATGGATTTTCTC-3′; antisense 5′-CTCCAGAAGAAGTTGGGAAT-3′); SnoN (sense, 5′-TACTGGGCATACTTCCATTC-3′; antisense, 5′-GGAAGTACGCTACCATTTTG-3′); c-Ski (sense, 5′-CGACTATGGCAACAAGTACA-3′; antisense, 5′-CTGTTTTGGGTCTTATGGAG-3′); GAPDH (sense, 5′-GAAGGTGAAGGTCGGAGTC-3′; antisense, 5′-GAAGATGGTGATGGGATTTC-3′).
Thymidine Incorporation Assay
Cells were seeded at a density of 2.0 × 104 cells/well in 24-well plates and transfected with siRNAs. Two days after transfection cells were stimulated by the addition of TGF-β into the medium containing 0.1% FBS. Thirty-six hours later cells were labeled with 0.6 μCi/ml [3H]thymidine (PerkinElmer Life Sciences) for 2 h. Radioactive DNA in the cells was precipitated using 5% trichloroacetic acid, solubilized with 1 n NaOH and quantified using liquid scintillation counting.
Pulse-chase Assay
Twenty-four hours after transfection, COS7 cells were starved in methionine/cysteine-free DMEM for 3.5 h, labeled with [35S]methionine/cysteine for 10 min, and chased in DMEM supplemented with 10% FBS, 2 mm methionine, and 0.5 mm cysteine.
Statistical Analysis
For the quantitative real-time-PCR experiments, luciferase reporter assays, and the thymidine incorporation assay, results are expressed as the means. Error bars represent the S.D. The Tukey-Kramer test of R program was used for multiple comparisons of data. p values of less than 0.01 were considered to indicate significance of the experiment.
RESULTS
Identification of RB1CC1 as an Arkadia-binding Protein
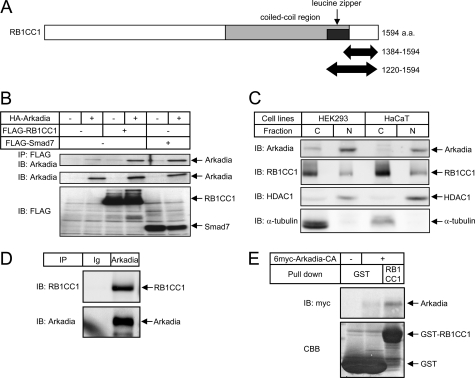
Arkadia is an E3 ubiquitin ligase that enhances TGF-β family signaling by targeting Smad7, c-Ski, and SnoN (24, 25, 28, 29). To identify novel substrates or regulators of Arkadia, we performed yeast two-hybrid screening of a human lung cDNA library using full-length Arkadia as bait. We isolated 342 positive clones from 1.6 × 106 transformants. Among the positive clones, we identified four encoding partial sequences of RB1CC1, a protein with 1594 amino acid residues (37, 42) that is widely expressed among various tissues and cell lines (supplemental Fig. S1). The clones encoded amino acid residues 1384–1594 or 1220–1594 of RB1CC1 (Fig. 1A). Physical interaction of full-length RB1CC1 with Arkadia was confirmed in HEK293T cells exogenously expressing differently tagged proteins (Fig. 1B). Co-precipitation of Arkadia with RB1CC1 was comparable to that with Smad7.
FIGURE 1.
Identification of RB1CC1 as a novel binding protein of Arkadia. A, shown is a schematic representation of RB1CC1. RB1CC1 is characterized by a large coiled-coil region (amino acids (a.a.) 860-1391, light gray) containing a leucine zipper motif (1371–1391, dark gray). Bold arrows represent two identified yeast clones that encode amino acid residues 1384–1594 or 1220–1594 of RB1CC1. B, shown is the physical interaction of full-length RB1CC1 with Arkadia in transfected HEK293T cells. Smad7 was used as a positive control to compare the intensity of interaction. IP, immunoprecipitation; IB, immunoblotting. C, subcellular localization of Arkadia and RB1CC1 is shown. HEK293 cells and HaCaT cells were lysed and fractionated into cytoplasmic (C) and nuclear (N) fractions. Endogenous Arkadia as well as RB1CC1 in each fraction was examined by immunoblotting. Anti-histone deacetylase 1 (HDAC1) and anti-α-tubulin were used to verify the fractionation integrity. D, interaction of endogenous RB1CC1 with Arkadia in HaCaT cells is shown. Cells were treated with 50 μm MG132 for 4 h before harvesting. Arkadia was then immunoprecipitated with anti-Arkadia antibody (H16), and co-precipitated RB1CC1 was detected by immunoblotting with anti-RB1CC1 antibody (upper panel). Immunoprecipitated Arkadia is also shown (lower panel). Ig, normal goat IgG. E, shown is the direct interaction between GST-RB1CC1 (amino acid residues 1397–1594) and Arkadia. A GST pulldown assay of Arkadia was performed. Input of the GST fusion protein was visualized by Coomassie Brilliant Blue (CBB) staining.
Immunocytochemical analyses have previously revealed that RB1CC1 is located in the cytoplasm (42) or in the nucleus (43). We conducted subcellular fractionation of HEK293 cells as well as HaCaT cells and examined the distribution of endogenous Arkadia and RB1CC1. Arkadia was found to be located primarily in the nucleus (31). In contrast, RB1CC1 was predominantly observed in the cytoplasm as well as in the nucleus (Fig. 1C). We then examined the interaction of endogenous Arkadia and RB1CC1 and detected RB1CC1 in the immunoprecipitates obtained using an anti-Arkadia antibody (Fig. 1D). We thus identified RB1CC1 as a novel binding protein of Arkadia.
We confirmed their direct interaction using bacterially expressed RB1CC1 (amino acid residues 1397–1594) and in vitro translated Arkadia (Fig. 1E). We also observed that the localization as well as the interaction of RB1CC1 and Arkadia was not affected by TGF-β signaling (data not shown).
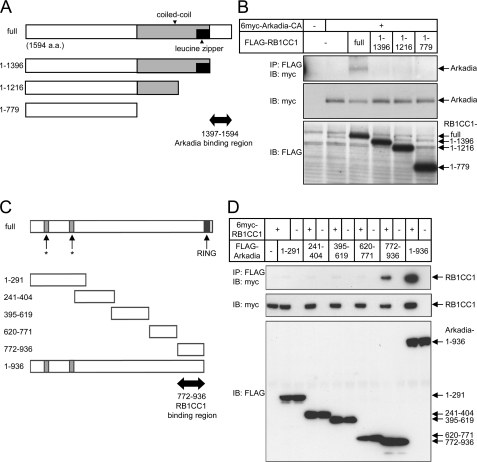
To determine the Arkadia binding region in RB1CC1, we constructed a series of C-terminal-truncated mutants of RB1CC1 (Fig. 2A) and examined their interaction with Arkadia (Fig. 2B). An RB1CC1 mutant lacking residues 1397–1594 failed to interact with Arkadia, suggesting that this region is essential for the binding. This finding together with the observation that a C-terminal fragment of RB1CC1 (residues 1384–1594) interacted with Arkadia in the yeast two-hybrid system indicates that the C-terminal region of RB1CC1 is necessary and sufficient for interaction with Arkadia.
FIGURE 2.
Mapping of regions responsible for interaction between Arkadia and RB1CC1. A, shown is a schematic representation of RB1CC1 deletion mutants. The putative Arkadia-binding region is indicated by a bold arrow. a.a., amino acids. B, shown is mapping of the Arkadia binding region on RB1CC1. HEK293T cells were transfected with the deletion mutants of RB1CC1 and the Arkadia C937A mutant lacking E3 ligase activity (Arkadia-CA). RB1CC1 was immunoprecipitated, and co-precipitated Arkadia was detected. Expression of Arkadia and RB1CC1 mutants is shown in the two bottom panels. C, Arkadia fragments used for binding assay are schematically presented. Asterisks represent putative nuclear localization signals. A putative RB1CC1 binding region is indicated by a bold arrow. D, mapping of the RB1CC1 binding region on Arkadia is shown. HEK293T cells were transfected with RB1CC1 and various Arkadia fragments. Arkadia fragments were immunoprecipitated, and co-precipitated RB1CC1 was visualized. Expression of each protein is presented in the two bottom panels.
We next mapped the RB1CC1 binding region in Arkadia. For this, we divided Arkadia into small units (Fig. 2C) and examined their interactions with RB1CC1. The C-terminal region (residues 772–936) of Arkadia was found to interact with RB1CC1 (Fig. 2D). Because Arkadia interacts with its substrates through this region (28), we examined whether RB1CC1 is a substrate of Arkadia E3 ubiquitin ligase. We did not detect significant ubiquitylation of RB1CC1 by Arkadia (data not shown), suggesting that RB1CC1 is not a substrate but may be a cofactor of Arkadia. We, therefore, examined the possibility that RB1CC1 affects TGF-β signaling.
RB1CC1 Is a Novel Positive Regulator of TGF-β Signaling
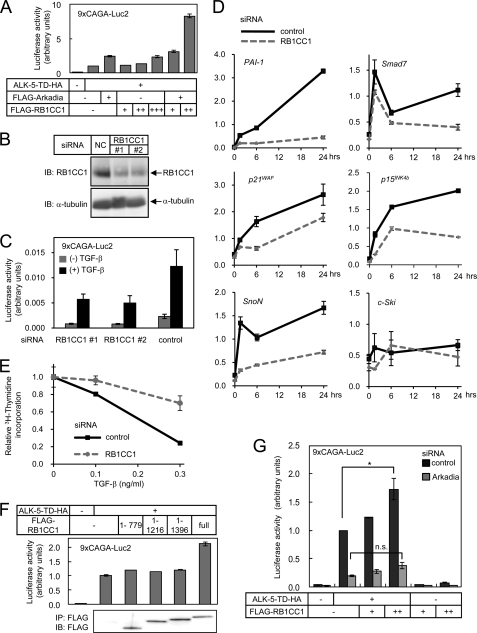
The effect of RB1CC1 on TGF-β signaling was examined using luciferase reporter assays. Overexpression of RB1CC1 enhanced transactivation of a TGF-β-responsive luciferase reporter (CAGA)9-MLP-Luc2 induced by a constitutively active TGF-β type I receptor (ALK-5-TD) (Fig. 3A). Moreover, when RB1CC1 was co-expressed with Arkadia, luciferase reporter activity was synergistically increased (Fig. 3A).
FIGURE 3.
RB1CC1 is a novel positive regulator of TGF-β signaling. A, a luciferase reporter assay was performed in HEK293 cells using (CAGA)9-MLP-Luc2. Cells were stimulated by co-expression of a constitutively active TGF-β type I receptor (ALK-5-TD). Error bars represent S.D. B, knockdown of RB1CC1 using siRNA oligos is shown. HaCaT cells were transfected with siRNA oligos and harvested 48 h later. Expression of RB1CC1 protein in cell lysates was examined by immunoblotting. 80 μg of protein was applied on each lane. NC, control siRNA. C, knockdown of RB1CC1 down-regulates Smad signaling. siRNA oligos were transfected into HaCaT cells 1 day before transfection of luciferase reporter plasmids. Cells were then stimulated with TGF-β, and luciferase activities were measured. Error bars represent S.D. D, shown is the effect of RB1CC1 knockdown on TGF-β-responsive gene expression in HaCaT cells. Cells were transfected with control or RB1CC1 siRNA #2. Forty-eight hours later cells were stimulated with TGF-β3 (1 ng/ml) and were harvested at 1.5, 6, or 24 h after stimulation. Induction of target genes was examined by quantitative real-time-PCR analysis and normalized by GAPDH expression levels. Expression of c-Ski was also examined as a control. Error bars represent S.D. E, RB1CC1 siRNAs inhibit the cytostatic effect of TGF-β. HaCaT cells were transfected with siRNAs and stimulated with TGF-β 36 h later. [3H]Thymidine incorporation was measured the next day. The obtained data were normalized to the incorporation without TGF-β. Error bars represent S.D. F, RB1CC1 enhances Smad signaling via its C-terminal region. HEK293 cells were transfected with deletion mutants of RB1CC1, (CAGA)9-MLP-Luc2 reporter, and ALK-5-TD. Error bars represent S.D. The lower panel shows expression of RB1CC1 deletion mutants. G, RB1CC1 fails to enhance TGF-β signaling upon knockdown of Arkadia. Luciferase reporter assay was performed using HEK293 cells transfected with control siRNA or Arkadia siRNA. Error bars represent S.D. *, p < 0.01; n.s., not significant.
To examine whether endogenous RB1CC1 exhibits positive effects on TGF-β signaling, we knocked down RB1CC1 in HEK293 cells by transfecting siRNA duplex (Fig. 3B). Knockdown of endogenous RB1CC1 resulted in decreased transactivation of (CAGA)9-MLP-Luc2 induced by TGF-β (Fig. 3C). We also examined the effects of RB1CC1 knockdown in HaCaT cells on cellular responses induced by TGF-β. Induction of genes, including plasminogen activator inhibitor-1 (PAI-1), Smad7, p21WAF, p15INK4b, and SnoN, by TGF-β was reduced, whereas the basal expression levels of these genes were minimally affected (Fig. 3D). TGF-β-induced inhibition of DNA synthesis was also attenuated by knockdown of RB1CC1 (Fig. 3E). These findings suggest that endogenous RB1CC1 functions as a positive regulator of TGF-β signaling.
We next determined the region of RB1CC1 required for the enhancement of TGF-β signaling using the C-terminal-truncated mutants. We found that a mutant lacking residues 1397–1594 failed to enhance TGF-β-induced reporter activity of (CAGA)9-MLP-Luc2 (Fig. 3F), indicating that the C-terminal region is required for the interaction with Arkadia and also for enhancing TGF-β signaling. Because the C-terminal region of RB1CC1 appears to contain no conserved functional motifs, it was anticipated that the interaction with Arkadia is important for enhancing TGF-β signaling by RB1CC1. Consistently, the enhancement of TGF-β signaling by RB1CC1 was considerably attenuated when Arkadia was knocked down (Fig. 3G).
RB1CC1 Down-regulates c-Ski and Smad7
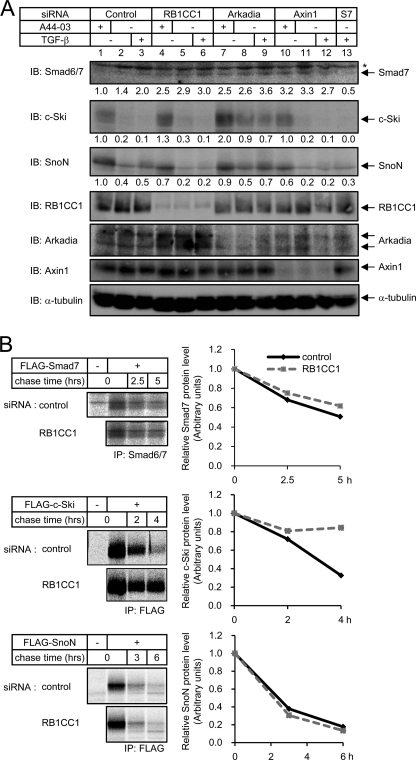
To examine the role of RB1CC1 in regulation of Arkadia function, we knocked down endogenous RB1CC1 and measured the resulting protein expression of Arkadia substrates i.e. Smad7, c-Ski, and SnoN in HaCaT cells (Fig. 4A). As references, Arkadia and Axin1, another cofactor of Arkadia (36), were also knocked down. Smad7 expression was weakly induced in response to TGF-β stimulation (Fig. 4A, lanes 2 and 3, and Ref. 7). Although knockdown of RB1CC1 attenuated the induction of Smad7 mRNA (Fig. 3D), Smad7 protein expression was enhanced by RB1CC1 knockdown (Fig. 4A, lanes 4–6). Endogenous RB1CC1 thus appears to down-regulate Smad7 protein. Up-regulation of Smad7 protein was also observed after knockdown of Arkadia (lanes 7–9) or Axin1 (lanes 10 and 11).
FIGURE 4.
RB1CC1 down-regulates Smad7 and c-Ski. A, shown are changes in protein expression of Smad7, c-Ski, and SnoN by knockdown of RB1CC1, Arkadia, or Axin1 in HaCaT cells. Cells were transfected with the indicated siRNA oligos. Forty-four hours later cells were treated with 1 ng/ml of TGF-β3 4 h before harvest or left untreated. Alternatively, cells were treated with 3 μm A44-03 for 5 h. Cell lysates (100 μg of protein) were analyzed for substrate expression by immunoblotting (upper three panels). Protein expression was quantified using Image J and normalized to that of lane 1 and is indicated below each panel. Lower panels show expression of knocked-down genes and α-tubulin as loading control. *, nonspecific band. We reproducibly observed reduced expression of Arkadia protein by knock-down of Axin1. B, turnover of Smad7, c-Ski, and SnoN protein after knockdown of RB1CC1 is shown. For pulse-chase analysis, COS7 cells were sequentially transfected with the indicated siRNA oligos and an expression plasmid for Smad7, c-Ski, or SnoN. Cells were then pulse-labeled with [35S]Met/Cys, washed, and incubated for indicated time. Total cell lysates were subjected to immunoprecipitation using anti-Smad6/7 or anti-FLAG antibody. Precipitated proteins were separated by SDS-PAGE and analyzed by autoradiography.
When HaCaT cells were cultured in the presence of A44-03, an inhibitor of TGF-β type I receptor kinase, protein expression of c-Ski and SnoN was significantly up-regulated (Fig. 4A, lanes 1 and 2). Endogenous TGF-β signaling thus appears to down-regulate c-Ski and SnoN under the culture condition used in this study, which is consistent with a previous report stating that c-Ski and SnoN proteins are down-regulated in response to TGF-β (44). In the presence of A44-03, c-Ski accumulated after the knockdown of RB1CC1, whereas SnoN did not (Fig. 4A, lanes 1 and 4). Similarly, knockdown of Arkadia caused accumulation of c-Ski but not SnoN (Fig. 4A, lane 7), whereas knockdown of Axin1 resulted in accumulation of neither of these proteins (lane 10). These findings indicate that RB1CC1 and Arkadia are involved in the down-regulation of c-Ski in resting states. In the presence of TGF-β signaling, however, c-Ski was down-regulated even when RB1CC1 was knocked down (Fig. 4A, lane 2 and 3 and lanes 5 and 6), suggesting that c-Ski is down-regulated through an RB1CC1-independent mechanism after TGF-β stimulation. SnoN protein expression was decreased after RB1CC1 knockdown in the presence of TGF-β (Fig. 4A, lane 2 and 3 and lanes 5 and 6) probably because SnoN mRNA was down-regulated (Fig. 3D). In contrast, knockdown of Arkadia up-regulated the expression of SnoN protein (Fig. 4A, lane 2 and 3 and lanes 8 and 9), indicating that Arkadia is involved in the down-regulation of SnoN following TGF-β stimulation, as reported previously (25). Thus, RB1CC1 does not control SnoN protein levels in the absence as well as in the presence of TGF-β signaling.
We also examined the effect of RB1CC1 on turnover of Smad7, c-Ski, and SnoN. A pulse-chase assay was performed in the cells transfected with siRNA targeting RB1CC1 (Fig. 4B). Knockdown of RB1CC1 resulted in substantial turnover delay of c-Ski and modest turnover delay of Smad7 but not of SnoN. Endogenous RB1CC1 thus promotes intracellular degradation of c-Ski and Smad7. We concluded that RB1CC1 is involved in down-regulation of c-Ski and Smad7 but not SnoN, among Arkadia substrates that affect TGF-β signaling under physiological conditions.
RB1CC1 Physically Interacts with c-Ski and Smad7
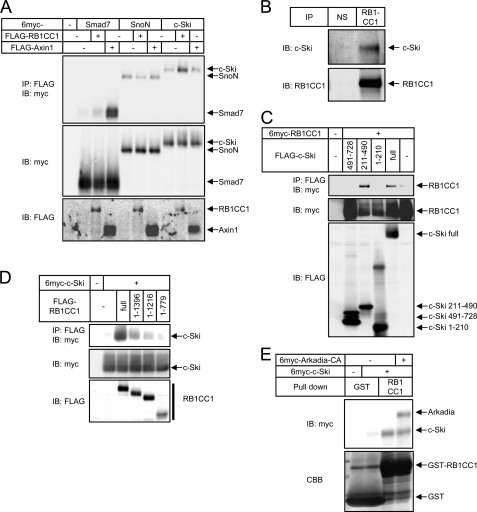
To elucidate the mechanism underlying the selective down-regulation of c-Ski and Smad7 by RB1CC1, we next examined the physical interaction of RB1CC1 with Arkadia substrates (Fig. 5A). RB1CC1 interacted with c-Ski and weakly with Smad7, but the interaction with SnoN was at near background level. Target selectivity of RB1CC1 is thus principally attributed to its physical interaction with target proteins. These findings indicate that RB1CC1 plays a role as a cofactor of Arkadia in the down-regulation of its substrates through physical interaction both with Arkadia and its substrates. Interestingly, Axin1 exclusively interacted with Smad7, which is consistent with its target specificity. Thereafter we focused on c-Ski to examine how RB1CC1 contributes to the down-regulation of Arkadia substrates as RB1CC1 interacted more strongly with c-Ski than with Smad7.
FIGURE 5.
Physical interaction of RB1CC1 with substrates of Arkadia. A, interaction of RB1CC1 or Axin1 with Arkadia substrates is shown. COS7 cells were transfected with the indicated plasmids and lysed. RB1CC1 or Axin1 was immunoprecipitated, and co-precipitated proteins were visualized. The top panel shows the interaction, and the lower two panels show expression of each protein. B, interaction between endogenous RB1CC1 and c-Ski in HaCaT cells is shown. A lysate of HaCaT cells was subjected to immunoprecipitation using anti-RB1CC1 serum or normal serum (NS) as a negative control followed by immunoblotting with anti-c-Ski (upper panel) or anti-RB1CC1 (lower panel). C, RB1CC1-interacting region in c-Ski is shown. HEK293T cells were transfected with FLAG-tagged c-Ski fragments and 6myc-tagged RB1CC1 as indicated. Cell lysates were subjected to immunoprecipitation using anti FLAG-antibody followed by immunoblotting with anti-myc antibody. D, c-Ski interacting region in RB1CC1 is shown. HEK293T cells were transfected with FLAG-tagged RB1CC1 fragments and 6myc-tagged c-Ski as indicated. Cell lysates were subjected to immunoprecipitation using anti FLAG-antibody followed by immunoblotting with anti-myc antibody. E, shown is direct interaction between GST-RB1CC1 (amino acid residues 1397–1594) and c-Ski. A GST pulldown assay of c-Ski was performed in the presence or absence of Arkadia. Input of GST-fusion protein was visualized by CBB staining.
Endogenous c-Ski was precipitated by anti-RB1CC1 antibody in HaCaT cells (Fig. 5B), indicating their physical interaction under physiological conditions. We then determined the region responsible for the RB1CC1-c-Ski interaction in transfected HEK293T cells. RB1CC1 interacted with the middle region of c-Ski (211–490) containing a SAND domain (Fig. 5C) through which c-Ski interacts with Arkadia (28). Although SnoN also contains a SAND domain, it fails to interact with RB1CC1. The amino acid identity of the SAND domains from c-Ski and SnoN is as low as 50%, which may result in the differential affinity to RB1CC1. Ski principally interacted with the C-terminal region (1397–1594) of RB1CC1 (Fig. 5D), through which RB1CC1 interacts with Arkadia. These multiple protein-protein interactions appear to occur in close proximity to each other. Moreover, we found that bacterially expressed RB1CC1 (amino acid residues 1397–1594) pulled down in vitro translated c-Ski, which was not affected by the co-presence of Arkadia (Fig. 5E). These findings indicate that RB1CC1 interacts directly with c-Ski.
RB1CC1 Plays a Role as Cofactor of Arkadia in Ubiquitylation of c-Ski and Smad7
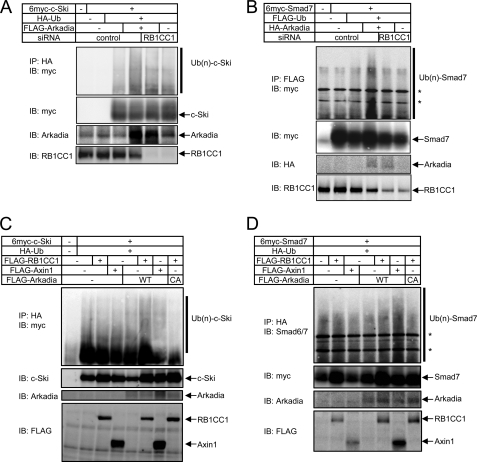
Because RB1CC1 interacts with c-Ski (Fig. 5, A and B) and accelerates its turnover (Fig. 4B), we next examined the effect of RB1CC1 on Arkadia-induced ubiquitylation of c-Ski (Fig. 6A). Ubiquitylation of c-Ski induced by Arkadia was attenuated by knockdown of RB1CC1. Similar results were obtained for Smad7 ubiquitylation induced by Arkadia (Fig. 6B). These findings indicate that RB1CC1 acts as a cofactor of Arkadia in the ubiquitylation of c-Ski and Smad7.
FIGURE 6.
RB1CC1 enhances Arkadia-induced ubiquitylation of c-Ski/Smad7. A and B, Arkadia-induced ubiquitylation of c-Ski (A) and Smad7 (B) was repressed by knockdown of RB1CC1. HEK293T cells were sequentially transfected with siRNAs and expression plasmids as indicated. Ubiquitylation of c-Ski and Smad7 was determined by immunoprecipitation using antibodies for tagged ubiquitin followed by immunoblotting with anti-myc antibody. Ub(n)-Ski and Ub(n)-/Smad7 denote ubiquitylated c-Ski/Smad7. The lower three panels show expression of each protein. C and D, the effects of RB1CC1 and Axin1 on ubiquitylation of c-Ski (C) and Smad7 (D) by Arkadia are shown. HEK293T cells were transfected with 6myc-c-Ski or 6myc-Smad7, HA-Ub), and FLAG-RB1CC1 or FLAG-Axin1 expression vectors. Ubiquitylation of c-Ski and Smad7 was determined by immunoprecipitation using anti-HA antibody followed by immunoblotting with anti-myc or anti-Smad 6/7 antibody. Ub(n)-Ski and Ub(n)-/Smad7 denote ubiquitylated c-Ski/Smad7. *, nonspecific band.
We further examined the possible cooperativity between Arkadia and RB1CC1 in the ubiquitylation of c-Ski (Fig. 6C) and Smad7 (Fig. 6D). The effects of Axin1, another cofactor of Arkadia, were also examined for comparison. Arkadia, RB1CC1, and Axin1 were ectopically expressed at a low level such that they did not apparently enhance the ubiquitylation of c-Ski or Smad7. Co-expression of Arkadia and RB1CC1 cooperatively enhanced the ubiquitylation of c-Ski, but that of Arkadia and Axin1 did it only marginally (Fig. 6C). In contrast, co-expression of Arkadia and Axin1 enhanced the ubiquitylation of Smad7, whereas that of Arkadia and RB1CC1 did it only modestly (Fig. 6D). Cooperativity between Arkadia and RB1CC1 was not observed when an inactive mutant of Arkadia (Fig. 6C) was used, suggesting that RB1CC1-mediated enhancement of the ubiquitylation of c-Ski and Smad7 is dependent on the E3 ligase activity of Arkadia. These findings indicate that RB1CC1 is an Arkadia cofactor that principally assists the E3 ubiquitin ligase activity of Arkadia on c-Ski, which is distinct from the effects of Axin1 on Arkadia activity.
DISCUSSION
TGF-β regulates various processes in a wide variety of target cells to support embryonic development as well as to maintain adult tissue homeostasis. Accumulating evidence suggests that aberrant TGF-β signal transduction leads to the progression of various diseases (45, 46). Elucidation of TGF-β signaling should thus provide findings valuable for understanding the pathogenic mechanisms of such diseases. Although the principal pathway of TGF-β signaling from the cell membrane to the nucleus has been elucidated (3, 4), how positive and negative regulators affect TGF-β signaling, either singly or cooperatively, in context-dependent fashion is still not completely understood.
Since the identification of the Smad signaling pathway, various negative and positive regulators of this process, which act in the cytoplasm or in the nucleus, have been identified. Interestingly, some positive regulators exhibit effects through modulating the function of negative regulators. For example, TIEG and Foxp3 enhance TGF-β signaling through the repression of Smad7 gene expression (47, 48), AMSH and AMSH2 interact with inhibitory Smads and suppress their effects (49, 50), and Jab1/CSN5 induce Smad7 degradation (51). Arkadia induces ubiquitylation and proteasome-dependent degradation of three important negative regulators of TGF-β signaling, Smad7, c-Ski, and SnoN (24, 25, 28, 29). Among these positive regulators, an important function of Arkadia in TGF-β family signaling has been genetically demonstrated; that is, Arkadia-deficient mice fail to maintain anterior embryonic structures and lack the node as a result of perturbation of signaling of Nodal, a member of TGF-β family (27). In the present study we found that a cell regulatory protein RB1CC1 is a binding partner of Arkadia and positively controls TGF-β signaling through enhancement of the ubiquitin ligase activity of Arkadia.
RB1CC1 Regulates Cellular Responses through Multiple Mechanisms
RB1CC1/FIP200 was first identified as a binding partner of focal adhesion kinase family members including focal adhesion kinase and Pyk2, which inhibits their kinase activities through binding to their kinase domain and suppresses cell spreading and migration (42). Subsequently, RB1CC1/FIP200 was independently identified as a positive regulator of RB1 gene expression (37). RB1CC1 is a protein with 1594 amino acid residues, which is characterized by a large coiled-coil region containing a leucine zipper motif, and interacts with various cellular proteins through the N-terminal, coiled-coil, or C-terminal region to modulate cellular functions. Previous studies have also shown that RB1CC1 plays important roles in controlling G1-S cell cycle progression through up-regulation of p21WAF together with down-regulation of cyclin D1 (52, 53), cell size control through the inhibition of the TSC1-TSC2 complex (54, 55), regulation of TNF-α-induced apoptosis through the modulation of TRAF2-ASK1 signal transduction (56), and autophagosome formation together with ULK kinases (57). RB1CC1 is thus regarded as a multifunctional protein that affects distinct cell functions depending on its binding partner as well as cellular contexts (58).
RB1CC1 as a Cofactor of Arkadia
Interestingly, RB1CC1 enhances the ubiquitin ligase activity of Arkadia in a substrate-dependent fashion. RB1CC1 facilitates Arkadia-induced ubiquitylation of c-Ski and Smad7 but not SnoN. Because the selectivity is attributed to physical interactions between RB1CC1 and substrates of Arkadia, it is possible that RB1CC1 may function as a scaffold protein. RB1CC1 interacts with both Arkadia and its substrates at its C-terminal region, which may facilitate the enzymatic reaction through proximity effects or arranging each molecule in a favorable orientation for a reaction to occur. RB1CC1 may also function in the recruitment of another molecule(s) that accelerates the ubiquitylation reaction or elimination of a molecule(s) that inhibits the ubiquitylation reaction. The detailed mechanism by which RB1CC1 promotes Arkadia-induced ubiquitylation remains to be elucidated.
Arkadia also induces ubiquitylation of Smad2 or Smad3 in some cellular situations (59, 60). Arkadia down-regulates Smad2 when Smad2 is C-terminal-phosphorylated (59). Arkadia weakly interacts with Smad3 in a signaling-dependent fashion (24) and ubiquitylates Smad3, which is enhanced in the presence of c-Ski (60). RB1CC1 may not enhance ubiquitylation of phospho-Smad2/3 by Arkadia because RB1CC1 fails to interact with phospho-Smad2/3 (data not shown). We did not observe up-regulation of protein level of C-terminal-phosphorylated Smad2/3 after knockdown of RB1CC1 (data not shown). However, the effects of RB1CC1 on protein levels of phosphorylated Smad2/3 remain unclear because knockdown of RB1CC1 down-regulates TGF-β signaling, which may result in decreased Smad phosphorylation, thus attenuating the direct effects on the protein levels of phosphorylated Smad2/3.
RB1CC1 and Axin1 Have Distinct Roles in Regulation of TGF-β Signaling
Axin1, another tumor suppressor gene product, has been reported to function as a scaffold protein in promoting the degradation of Smad7 by Arkadia (36). Axin1 was originally identified as a scaffold protein involved in negative regulation of Wnt signaling (61). Axin1 interacts with Arkadia and Smad7 to form a ternary complex and appears to facilitate the ubiquitin ligase activity of Arkadia on Smad7 by ensuring that Smad7 is located in the proximity to Arkadia.
Importantly, Axin1 exclusively affects Smad7, whereas RB1CC1 principally affects c-Ski but not SnoN. These selective effects on Arkadia substrates may account for the differential function of Axin1 and RB1CC1. Axin1 is down-regulated by Wnt signaling through the action of Dvl (62), suggesting that Axin1 plays a role in cross-talk between Wnt and TGF-β signaling (36). Therefore, RB1CC1 and Axin1 have similar but distinct roles in the regulation of TGF-β signaling and in cross-talk with other signaling pathways. The contribution of RB1CC1 or Axin1 to Arkadia-induced enhancement of TGF-β signaling could thus differ depending on the type of cell as well as cellular contexts.
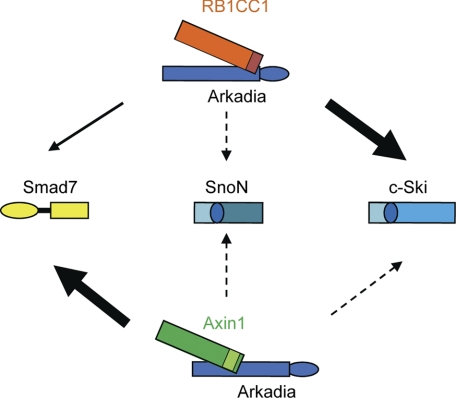
Although Arkadia has the ability to ubiquitylate Smad7, c-Ski, and SnoN, its effects on the substrates are modified by cofactors. Arkadia is widely expressed at a nearly equivalent level in various cell lines (30). Cofactors, including RB1CC1 and Axin1, appear to play important roles in the regulation of Arkadia function. Fig. 7 shows a schematic model of how RB1CC1 and Axin1 affect TGF-β signaling. At present, the cofactor(s) for ubiquitylation of SnoN remains to be determined. One candidate is phosphorylated Smad3, which has been reported to be required for Arkadia-induced degradation of SnoN (25).
FIGURE 7.
Model of modulation of Arkadia E3 ubiquitin ligase activity by RB1CC1 and Axin1. The Arkadia-RB1CC1 complex preferentially targets c-Ski, whereas the Arkadia-Axin1 complex targets Smad7. The RB1CC1 C-terminal region (red boxed) interacts with Arkadia at the region preceding the RING domain (ellipse) and c-Ski at the SAND domain (circled). The Axin1 (PP2A/MEKK4 binding region, yellow-green box) interacts with Arkadia at the middle region and Smad7 at the MH2 domain (yellow box).
Role of RB1CC1 in Regulation of Smad Signaling in Health and Disease
Phenotypes of RB1CC1 knock-out mice have previously been reported (56). Homozygous deletion of RB1CC1 leads to embryonic lethality in mid/late gestation and is associated with heart failure and liver degeneration. Arkadia-null mice lack the node and node-derived mesoderm and die in mid-gestation (26). The earlier embryonic lethality associated with Arkadia deficiency suggests that RB1CC1 is not necessary for Arkadia function in the early stages of embryonic development. It is possible that node formation requires degradation of a substrate(s) of Arkadia other than c-Ski and Smad7. Alternatively RB1CC1 may selectively affect certain subset of TGF-β target genes because its effect on expression of Smad7 was less than that on expression of other genes including PAI-1 (Fig. 3D).
Although RB1CC1 appears to be dispensable for the regulation of Smad signaling during early embryonic development, it may play a role in adults. The RB1CC1 gene is located in chromosome 8q11, which contains several loci of putative tumor suppressor genes (43, 63). RB1CC1 potently inhibits proliferation of breast cancer cells through up-regulation of p21WAF (53, 64), p16INK4a, and RB1 (64) as well as down-regulation of cyclin D (53). It is thus considered a candidate human tumor suppressor gene, although a recent report using mice with conditional RB1CC1 deletion in skin has challenged this idea (65). The TGF-β-Smad pathway is also thought to have tumor suppressor functions during the early stages of tumorigenesis as TGF-β exhibits potent antiproliferative activity in a wide variety of cells. Additionally, mutations of signaling components downstream of TGF-β have been identified in tumor tissues (66). In this study we demonstrated that RB1CC1 is a positive regulator of TGF-β signaling under physiological conditions; knockdown of RB1CC1 attenuated expression of target genes as well as cytostasis induced by TGF-β. Thus, enhancement of TGF-β signaling may be one of the mechanisms by which RB1CC1 exerts tumor suppressor functions in vivo.
In conclusion, we have shown that RB1CC1 is a novel positive regulator of TGF-β signaling. RB1CC1 modifies the substrate selectivity of Arkadia E3 ubiquitin ligase. Control of the effects of RB1CC1 as well as its relevance to diseases is the next issue to be addressed.
Supplementary Material
Acknowledgments
We thank Takao Yamori for mRNAs from 39 tumor cell lines, Aki Hanyu, Ayako Nakano, Mami Takahata, Yasushi Yuki, Etsuko Kobayashi, Noriko Kaneniwa, Keiko Yuki, So-ichi Yaguchi, and Etsuko Ohara for technical assistance, and Yasumichi Inoue for critical reading of the manuscript.
This work was supported by KAKENHI (grants-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- RB1CC1
- RB1-inducible coiled-coil 1
- FIP200
- focal adhesion kinase interacting protein of 200 kDa
- Ub
- ubiquitin
- IP
- immunoprecipitation
- IB
- immunoblotting.
REFERENCES
- 1. Attisano L., Wrana J. L. (2000) Curr. Opin. Cell Biol. 12, 235–243 [DOI] [PubMed] [Google Scholar]
- 2. Derynck R., Zhang Y. E. (2003) Nature 425, 577–584 [DOI] [PubMed] [Google Scholar]
- 3. Shi Y., Massagué J. (2003) Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 4. Heldin C. H., Miyazono K., ten Dijke P. (1997) Nature 390, 465–471 [DOI] [PubMed] [Google Scholar]
- 5. Miyazawa K., Shinozaki M., Hara T., Furuya T., Miyazono K. (2002) Genes Cells 7, 1191–1204 [DOI] [PubMed] [Google Scholar]
- 6. Hayashi H., Abdollah S., Qiu Y., Cai J., Xu Y. Y., Grinnell B. W., Richardson M. A., Topper J. N., Gimbrone M. A., Jr., Wrana J. L., Falb D. (1997) Cell 89, 1165–1173 [DOI] [PubMed] [Google Scholar]
- 7. Nakao A., Afrakhte M., Morén A., Nakayama T., Christian J. L., Heuchel R., Itoh S., Kawabata M., Heldin N. E., Heldin C. H., ten Dijke P. (1997) Nature 389, 631–635 [DOI] [PubMed] [Google Scholar]
- 8. Kamiya Y., Miyazono K., Miyazawa K. (2010) J. Biol. Chem. 285, 30804–30813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu J. W., Krawitz A. R., Chai J., Li W., Zhang F., Luo K., Shi Y. (2002) Cell 111, 357–367 [DOI] [PubMed] [Google Scholar]
- 10. Suzuki H., Yagi K., Kondo M., Kato M., Miyazono K., Miyazawa K. (2004) Oncogene 23, 5068–5076 [DOI] [PubMed] [Google Scholar]
- 11. Inoue Y., Imamura T. (2008) Cancer Sci. 99, 2107–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kavsak P., Rasmussen R. K., Causing C. G., Bonni S., Zhu H., Thomsen G. H., Wrana J. L. (2000) Mol. Cell 6, 1365–1375 [DOI] [PubMed] [Google Scholar]
- 13. Ebisawa T., Fukuchi M., Murakami G., Chiba T., Tanaka K., Imamura T., Miyazono K. (2001) J. Biol. Chem. 276, 12477–12480 [DOI] [PubMed] [Google Scholar]
- 14. Komuro A., Imamura T., Saitoh M., Yoshida Y., Yamori T., Miyazono K., Miyazawa K. (2004) Oncogene 23, 6914–6923 [DOI] [PubMed] [Google Scholar]
- 15. Kuratomi G., Komuro A., Goto K., Shinozaki M., Miyazawa K., Miyazono K., Imamura T. (2005) Biochem. J. 386, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin X., Liang M., Feng X. H. (2000) J. Biol. Chem. 275, 36818–36822 [DOI] [PubMed] [Google Scholar]
- 17. Zhang Y., Chang C., Gehling D. J., Hemmati-Brivanlou A., Derynck R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 974–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Seo S. R., Lallemand F., Ferrand N., Pessah M., L'Hoste S., Camonis J., Atfi A. (2004) EMBO J. 23, 3780–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morén A., Imamura T., Miyazono K., Heldin C. H., Moustakas A. (2005) J. Biol. Chem. 280, 22115–22123 [DOI] [PubMed] [Google Scholar]
- 20. Gao S., Alarcón C., Sapkota G., Rahman S., Chen P. Y., Goerner N., Macias M. J., Erdjument-Bromage H., Tempst P., Massagué J. (2009) Mol. Cell 36, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fukuchi M., Imamura T., Chiba T., Ebisawa T., Kawabata M., Tanaka K., Miyazono K. (2001) Mol. Biol. Cell 12, 1431–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xin H., Xu X., Li L., Ning H., Rong Y., Shang Y., Wang Y., Fu X. Y., Chang Z. (2005) J. Biol. Chem. 280, 20842–20850 [DOI] [PubMed] [Google Scholar]
- 23. Dupont S., Zacchigna L., Cordenonsi M., Soligo S., Adorno M., Rugge M., Piccolo S. (2005) Cell 121, 87–99 [DOI] [PubMed] [Google Scholar]
- 24. Koinuma D., Shinozaki M., Komuro A., Goto K., Saitoh M., Hanyu A., Ebina M., Nukiwa T., Miyazawa K., Imamura T., Miyazono K. (2003) EMBO J. 22, 6458–6470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Levy L., Howell M., Das D., Harkin S., Episkopou V., Hill C. S. (2007) Mol. Cell. Biol. 27, 6068–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Episkopou V., Arkell R., Timmons P. M., Walsh J. J., Andrew R. L., Swan D. (2001) Nature 410, 825–830 [DOI] [PubMed] [Google Scholar]
- 27. Niederländer C., Walsh J. J., Episkopou V., Jones C. M. (2001) Nature 410, 830–834 [DOI] [PubMed] [Google Scholar]
- 28. Nagano Y., Mavrakis K. J., Lee K. L., Fujii T., Koinuma D., Sase H., Yuki K., Isogaya K., Saitoh M., Imamura T., Episkopou V., Miyazono K., Miyazawa K. (2007) J. Biol. Chem. 282, 20492–20501 [DOI] [PubMed] [Google Scholar]
- 29. Le Scolan E., Zhu Q., Wang L., Bandyopadhyay A., Javelaud D., Mauviel A., Sun L., Luo K. (2008) Cancer Res. 68, 3277–3285 [DOI] [PubMed] [Google Scholar]
- 30. Nagano Y., Koinuma D., Miyazawa K., Miyazono K. (2010) J. Biochem. 147, 545–554 [DOI] [PubMed] [Google Scholar]
- 31. Mizutani A., Saitoh M., Imamura T., Miyazawa K., Miyazono K. (2010) J. Biochem. 148, 733–741 [DOI] [PubMed] [Google Scholar]
- 32. Miyazono K., Koinuma D. (2011) J. Biochem. 149, 1–3 [DOI] [PubMed] [Google Scholar]
- 33. Ogunjimi A. A., Briant D. J., Pece-Barbara N., Le Roy C., Di Guglielmo G. M., Kavsak P., Rasmussen R. K., Seet B. T., Sicheri F., Wrana J. L. (2005) Mol. Cell 19, 297–308 [DOI] [PubMed] [Google Scholar]
- 34. Chong P. A., Lin H., Wrana J. L., Forman-Kay J. D. (2006) J. Biol. Chem. 281, 17069–17075 [DOI] [PubMed] [Google Scholar]
- 35. Wiesner S., Ogunjimi A. A., Wang H. R., Rotin D., Sicheri F., Wrana J. L., Forman-Kay J. D. (2007) Cell 130, 651–662 [DOI] [PubMed] [Google Scholar]
- 36. Liu W., Rui H., Wang J., Lin S., He Y., Chen M., Li Q., Ye Z., Zhang S., Chan S. C., Chen Y. G., Han J., Lin S. C. (2006) EMBO J. 25, 1646–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chano T., Ikegawa S., Kontani K., Okabe H., Baldini N., Saeki Y. (2002) Oncogene 21, 1295–1298 [DOI] [PubMed] [Google Scholar]
- 38. Tojo M., Hamashima Y., Hanyu A., Kajimoto T., Saitoh M., Miyazono K., Node M., Imamura T. (2005) Cancer Sci. 96, 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koinuma D., Tsutsumi S., Kamimura N., Taniguchi H., Miyazawa K., Sunamura M., Imamura T., Miyazono K., Aburatani H. (2009) Mol. Cell. Biol. 29, 172–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ikushima H., Komuro A., Isogaya K., Shinozaki M., Hellman U., Miyazawa K., Miyazono K. (2008) EMBO J. 27, 2955–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dennler S., Itoh S., Vivien D., ten Dijke P., Huet S., Gauthier J. M. (1998) EMBO J. 17, 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ueda H., Abbi S., Zheng C., Guan J. L. (2000) J. Cell Biol. 149, 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chano T., Kontani K., Teramoto K., Okabe H., Ikegawa S. (2002) Nat. Genet. 31, 285–288 [DOI] [PubMed] [Google Scholar]
- 44. Sun Y., Liu X., Ng-Eaton E., Lodish H. F., Weinberg R. A. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 12442–12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Massagué J. (2008) Cell 134, 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yoshimura A., Wakabayashi Y., Mori T. (2010) J. Biochem. 147, 781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Johnsen S. A., Subramaniam M., Janknecht R., Spelsberg T. C. (2002) Oncogene 21, 5783–5790 [DOI] [PubMed] [Google Scholar]
- 48. Fantini M. C., Becker C., Monteleone G., Pallone F., Galle P. R., Neurath M. F. (2004) J. Immunol. 172, 5149–5153 [DOI] [PubMed] [Google Scholar]
- 49. Itoh F., Asao H., Sugamura K., Heldin C. H., ten Dijke P., Itoh S. (2001) EMBO J. 20, 4132–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ibarrola N., Kratchmarova I., Nakajima D., Schiemann W. P., Moustakas A., Pandey A., Mann M. (2004) BMC Cell Biol. 5, 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim B. C., Lee H. J., Park S. H., Lee S. R., Karpova T. S., McNally J. G., Felici A., Lee D. K., Kim S. J. (2004) Mol. Cell. Biol. 24, 2251–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Abbi S., Ueda H., Zheng C., Cooper L. A., Zhao J., Christopher R., Guan J. L. (2002) Mol. Biol. Cell 13, 3178–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Melkoumian Z. K., Peng X., Gan B., Wu X., Guan J. L. (2005) Cancer Res. 65, 6676–6684 [DOI] [PubMed] [Google Scholar]
- 54. Gan B., Melkoumian Z. K., Wu X., Guan K. L., Guan J. L. (2005) J. Cell Biol. 170, 379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chano T., Saji M., Inoue H., Minami K., Kobayashi T., Hino O., Okabe H. (2006) Int. J. Mol. Med. 18, 425–432 [PubMed] [Google Scholar]
- 56. Gan B., Peng X., Nagy T., Alcaraz A., Gu H., Guan J. L. (2006) J. Cell Biol. 175, 121–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J. L., Mizushima N. (2008) J. Cell Biol. 181, 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gan B., Guan J. L. (2008) Cell. Signal. 20, 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mavrakis K. J., Andrew R. L., Lee K. L., Petropoulou C., Dixon J. E., Navaratnam N., Norris D. P., Episkopou V. (2007) PLoS Biol. 5, e67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yuzawa H., Koinuma D., Maeda S., Yamamoto K., Miyazawa K., Imamura T. (2009) Bone 44, 53–60 [DOI] [PubMed] [Google Scholar]
- 61. Ikeda S., Kishida S., Yamamoto H., Murai H., Koyama S., Kikuchi A. (1998) EMBO J. 17, 1371–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yamamoto H., Kishida S., Kishida M., Ikeda S., Takada S., Kikuchi A. (1999) J. Biol. Chem. 274, 10681–10684 [DOI] [PubMed] [Google Scholar]
- 63. Chano T., Ikegawa S., Saito-Ohara F., Inazawa J., Mabuchi A., Saeki Y., Okabe H. (2002) Gene 291, 29–34 [DOI] [PubMed] [Google Scholar]
- 64. Chano T., Ikebuchi K., Ochi Y., Tameno H., Tomita Y., Jin Y., Inaji H., Ishitobi M., Teramoto K., Nishimura I., Minami K., Inoue H., Isono T., Saitoh M., Shimada T., Hisa Y., Okabe H. (2010) PLoS One 5, e11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wei H., Gan B., Wu X., Guan J. L. (2009) J. Biol. Chem. 284, 6004–6013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Piek E., Roberts A. B. (2001) Adv. Cancer Res. 83, 1–54 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.