Abstract
Chronic myelogenous leukemia (CML) is a myeloproliferative disorder characterized at the molecular level by the expression of Bcr-Abl, a chimeric protein with deregulated tyrosine kinase activity. The protein-tyrosine phosphatase 1B (PTP1B) is up-regulated in Bcr-Abl-expressing cells, suggesting a regulatory link between the two proteins. To investigate the interplay between these two proteins, we inhibited the activity of PTP1B in Bcr-Abl-expressing TonB.210 cells by either pharmacological or siRNA means and examined the effects of such inhibition on Bcr-Abl expression and function. Herein we describe a novel mechanism by which the phosphatase activity of PTP1B is required for Bcr-Abl protein stability. Inhibition of PTP1B elicits tyrosine phosphorylation of Bcr-Abl that triggers the degradation of Bcr-Abl through ubiquitination via the lysosomal pathway. The degradation of Bcr-Abl consequently inhibits tyrosine phosphorylation of Bcr-Abl substrates and the downstream production of intracellular reactive oxygen species. Furthermore, PTP1B inhibition reduces cell viability and the IC50 of the Bcr-Abl inhibitor imatinib mesylate. Degradation of Bcr-Abl via PTP1B inhibition is also observed in human CML cell lines K562 and LAMA-84. These results suggest that inhibition of PTP1B may be a useful strategy to explore in the development of novel therapeutic agents for the treatment of CML, particularly because host drugs currently used in CML such as imatinib focus on inhibiting the kinase activity of Bcr-Abl.
Keywords: Leukemia, Protein Phosphorylation, Tyrosine Protein Kinase (Tyrosine Kinase), Tyrosine Protein Phosphatase (Tyrosine Phosphatase), Ubiquitination, Bcr-Abl, CML, PTP1B
Introduction
Chronic myeloid leukemia (CML)3 is a myeloproliferative disorder characterized by deregulated growth and apoptosis of hematopoietic stem cells in the bone marrow (1). It is associated with a characteristic chromosomal translocation between chromosomes 9 and 22, known as the Philadelphia chromosome (2). The resulting hybrid gene, bcr-abl, codes for a fusion protein with constitutive tyrosine kinase activity that activates several downstream signal transduction pathways. Bcr-Abl has several distinct tyrosine phosphorylation sites, but the role played by these tyrosine phosphorylations is to an extent largely unknown at present (3).
Protein tyrosine phosphorylation plays a significant role in a wide range of cellular processes such as cell cycle, cell adhesion, and cell survival and at a molecular level regulating the activity and stability of proteins (4). The state of tyrosine phosphorylation depends on the balance between the protein-tyrosine kinases and the protein-tyrosine phosphatases (PTPs). An imbalance leads to altered tyrosine phosphorylation, which has been shown to be a feature of several human diseases, including cancer (5). So far, PTPs have been best known as down-regulators of protein-tyrosine kinase signaling, but their functions and regulations are only recently beginning to be understood.
Protein-tyrosine phosphatase 1B (PTP1B) is a prototype of the family of PTPs. It has been shown to act as a negative regulator of intracellular signaling driven by several receptor tyrosine kinases such as the receptors for insulin (6), platelet-derived growth factor (7), and hepatocyte growth factor (8). It has also been demonstrated that its overexpression suppresses cell transformation by oncogenes that increase tyrosine phosphorylation such as ErbB2 (9), Src (10), Crk, and Ras (11). Therefore, these findings suggest that PTP1B may block proliferative and metabolic signaling. On the other hand, some proliferative pathways that are associated with the activation of the small GTPases Ras (12) and Rac (13) and the Src protein kinase (14) require PTP1B to function. PTP1B expression is altered in human breast (15), ovarian (16), and epithelial carcinomas (17), but it is reduced in esophageal cancer (18). Taken together, these results imply that PTP1B may play a critical role in multiple signaling networks involved in oncogenesis.
Studies carried out in Bcr-Abl model cell systems and in CML cell lines show that PTP1B is up-regulated (19, 20) and that Bcr-Abl may be a substrate of this phosphatase (19). Furthermore, overexpression of PTP1B prevents Bcr-Abl-induced transformation of fibroblast cells (21). To clarify its role in CML, we employed a Bcr-Abl expression model in a murine pro-B cell line (22). We show that PTP1B is required for stabilization of Bcr-Abl. When PTP1B activity is inhibited, Bcr-Abl is degraded by the ubiquitin lysosomal pathway.
EXPERIMENTAL PROCEDURES
Cell Lines, Culture Conditions, Treatment, and Reagents
TonB.210 cells were kindly provided by Dr. George Daley (Massachusetts Institute of Technology (MIT), Cambridge, MA). TonB.210 cells are derived from the interleukin-3 (IL-3)-dependent murine pro-B cell line BaF3 and contain a doxycycline-responsive promoter whereby Bcr-Abl p210 can be conditionally induced (20, 22). TonB.210 cells were maintained in RPMI 1640 containing 10% fetal calf serum, 2 mm l-glutamine, 1% penicillin/streptomycin, and 10% Wehi-3B conditioned medium as a source of murine IL-3, and 1 mg/ml G418 sulfate for TonB.210 cells, in a humidified incubator at 37 °C with 5% CO2. K562, LAMA-84, and MV4–11 were maintained in RPMI 1640 containing 10% fetal calf serum, 2 mm l-glutamine and 1% penicillin/streptomycin. TonB.210 cells were routinely incubated with 1 μg/ml doxycycline hyclate (DOX) in full IL-3 supplemented medium for the indicated times to induce Bcr-Abl expression. For PTP1B inhibition, cells were treated with 1 μg/ml DOX for 48 h and 35 μm 3-(3,5-dibromo-4-hydroxy-benzoyl)-2-ethyl-benzofuran-6-sulfonic acid-(4-(thiazol-2-ylsulfanyl)-phenyl)-amide (PTP1B inhibitor) for 2 h (Calbiochem, Nottingham, UK) prior to cell harvest. Cells were also treated with proteasome inhibitor lactacystin (5 μm for 3 h) (Calbiochem) and lysosome inhibitor chloroquine diphosphatase salt (100 μm for 4 h). Imatinib mesylate was from Novartis (Basel, Switzerland). Unless otherwise stated, all reagents were purchased from Sigma-Aldridge (Dublin, Ireland).
Measurement of Intracellular ROS Levels
Following treatments, ROS levels were determined using the cell-permeable fluorescent probe 2,7-dichlorodihydrofluorescein diacetate (Invitrogen Dublin, Ireland). Cells were treated as described above with DOX and/or different inhibitors. Following treatment, 50 μm 2,7-dichlorodihydrofluorescein diacetate was added for 15 min in the dark. Cells were then analyzed for the mean fluorescent intensity of 10,000 events counted in the FL-1 channel on a FACSCalibur (BD Biosciences Europe) using the CellQuest Pro software (BD Biosciences).
Immunoblotting and Immunoprecipitation
Immunoblotting was carried out under conditions previously described by us (20). We used Alexa Fluor 680 or 800 coupled with anti-rabbit or anti-mouse secondary antibodies (LI-COR Biosciences, Cambridge, UK) for detection with the Odyssey® infrared imaging system (LI-COR Biosciences).
Immunofluorescence and Confocal Microscopy
Following treatment, cells (1 × 105) were adhered to glass slides using a cytocentrifuge. Cells were fixed with 3% paraformaldehyde and permeabilized in PBS containing 0.2% BSA and 0.05% saponin (23). Slides were then incubated with the appropriate antibodies diluted in 5% FBS/PBS for 2 h at room temperature, washed in PBS three times, and incubated with an appropriate secondary antibody conjugated to Alexa Fluor 488 or Alexa Fluor 594 for 1 h at room temperature. Following washes with PBS, slides were mounted with coverslips using Mowiol and dried overnight before imaging. Images of the cells were visualized and acquired using an Olympus FluoView 1000 inverted confocal laser scanning microscope. Cells were examined and imaged with an UPlanSAPO 60× oil immersion objective (1.35 numerical aperture). Images were obtained at a 1024 × 1024-pixel resolution in a frame-scan mode.
Antibodies
Primary antibodies used for immunoblotting, immunoprecipitation, or immunofluorescence were anti-c-Abl Ab-3 (Merck Biosciences), c-Abl (BD Biosciences) for immunofluorescence, c-Cbl and Bcr (Cell Signaling Technology, Danvers, MA), anti-β-Actin, anti-GAPDH (Advanced ImmunoChemical, Long Beach, CA), lysobisphosphatidic acid (LBPA) (Echelon Biosciences, Salt Lake City, UT), PTP1B (Millipore, Dublin, Ireland), phosphotyrosine PY20 (BD Biosciences), ubiquitin (Dako, Glostrup, Denmark), and ubiquitin clone Ubi1 (Millipore, Bedford, MA). Secondary antibodies for immunoblotting were Alexa Fluor 680 or 800 conjugated to anti-mouse or anti-rabbit (LI-COR Biosciences, Bad Homburg, Germany). Secondary antibodies for immunofluorescence were goat anti-mouse or goat anti-rabbit conjugated to Alexa Fluor 488 or Alexa Fluor 594 (Invitrogen and Bio Sciences Ltd.). Densitometric analysis was carried out using the ImageJ software (rsbweb.nih.gov/ij).
PTP1B siRNA and Transfection
RNA interference mediated by duplexes of 21-nucleotide RNAs was performed in TonB.210 and K562 cells. Negative control 1 siRNA and two Silencer® predesigned siRNAs (Ambion, Warrington, UK) were used for silencing PTP1B. The siRNAs chosen for PTP1B were negative control 1 siRNA ID: s72430 and negative control 2 siRNA ID: s72431. The sequences are available from the manufacturer's website. The transfection of siRNAs used the Amaxa Nucleofector technology with the Amaxa cell optimization kit V (Amaxa, Cologne, Germany) and followed the Amaxa guidelines. Expression of PTP1B and Bcr-Abl was examined by immunoblot after siRNA knockdown.
Reverse Transcription-Polymerase Chain Reaction Analysis
Total RNA was extracted from 2 × 106 TonB.210 cells untreated or treated with PTP1B inhibitor using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. cDNA was synthesized using Moloney murine leukemia virus reverse transcriptase (Promega, Southampton, UK) according to the manufacturer's instructions using oligo(dT)15 primer for GAPDH (Promega), and for Bcr-Abl, the primers used were (24) sense, 5′-GAAGAAGTGTTTCAGAAGCTTCTCCC-3′; and antisense, 5′-TGTGATTATAGCCTAAGACCCGGAG-3′. PCR for Bcr-Abl and GAPDH was performed using Taq (GoTaq, Promega) according to the manufacturer's instructions.
Two-dimensional Gel Electrophoresis
Samples containing 100 μg of total protein in radioimmune precipitation buffer were resuspended in 10% TCA and acetone to remove residual salts. The suspensions were incubated overnight at −20 °C and then centrifuged at 15,000 × g for 30 min (4 °C). The resulting pellet was resuspended in acetone containing 0.2% DTT and incubated for 1 h at −20 °C. The suspension was centrifuged at 15,000 × g for 30 min (4 °C), and proteins present in the pellets were resuspended in 90 μl of sample buffer (7 m urea, 2 m thiourea, 30 mm Tris base, 1.2% (w/v) CHAPS (Anachem), 0.4% (w/v) amidosulfobetaine-14 (ASB14) (Merck Biosciences), 10% (v/v) carrier ampholytes (pH 3–10), and 43 mm DTT and a trace of bromphenol blue), applying a modified Taguchi method (25). The first dimension was performed on 7-cm pH 3–10 non-linear immobilized pH gradient strips (GE Healthcare Biosciences) that were rehydrated without protein extract in 135 μl of rehydration buffer (7 m urea, 2 m thiourea, 1% CHAPS, 0.5% carrier ampholytes (pH 3–10), 1.5% of 2-hydroxyethyl disulfide (or DeStreak reagent), and a trace amount of bromphenol blue) at least for 16 h at room temperature. The protein solution was applied by the anodic cup-loading method and focused on a Protean isoelectric focusing cell (Bio-Rad) at 20 °C with rapid voltage increases: 250 V for 30 min and then up to a maximum voltage of 6000 V for 20 h. Strips were equilibrated as previously described (26) loaded on 8% SDS-PAGE, and transferred to nitrocellulose membranes. The blots were probed with the anti-c-Abl antibody (Cell Signaling Technology) or anti-phosphotyrosine PY20 antibody. For spot detection, the Odyssey® infrared imaging system was used.
Cell Viability
The CellTiter-Blue® cell viability assay (Promega) was used to monitor call viability. Briefly, cells were plated in black-walled clear-bottomed 96-well plates at 1–5 × 105 cells/well in 100 μl volumes with quadruplicate wells for each treatment. Following treatments, 20 μl of CellTiter-Blue reagent was added to each well, and plates were analyzed after 1 h on a FlexStation II benchtop scanning fluorometer (Molecular Devices) at excitation/emission wavelengths of 560/590 nm.
Statistical Analysis
Data are given as mean ± S.D. for at least three experiments. Statistical significance was evaluated by Student's t test for comparison between groups with p < 0.05 considered significant. All data were generated from at least three independent experiments.
RESULTS
Bcr-Abl Levels Decrease When PTP1B Activity or Expression Is Inhibited
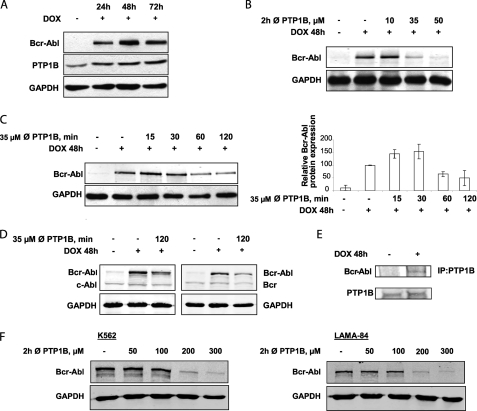
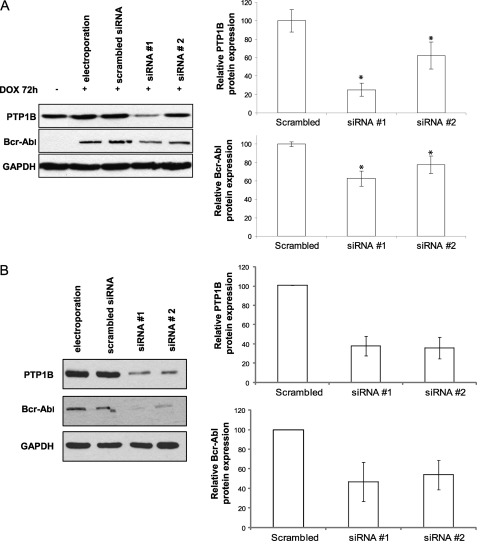
To elucidate the interactions between PTP1B and Bcr-Abl, we initially analyzed the expression of PTP1B and Bcr-Abl proteins. In agreement with previous studies (19, 20), when Bcr-Abl is expressed in TonB.210 cells, there is an increase in PTP1B protein levels (Fig. 1A). Further experiments were performed to evaluate the appropriate concentration of the PTP1B inhibitor to inhibit the activity of this phosphatase and minimize its cytotoxic effects in TonB.210 cells. Surprisingly, it was found that treatment with increasing concentrations of the inhibitor for 2 h led to Bcr-Abl degradation. A dose of 35 μm of the PTP1B inhibitor for 2 h was optimal for reducing Bcr-Abl protein levels (Fig. 1B), and these conditions were adopted to study the effects of the inhibition of PTP1B on Bcr-Abl. PTP1B expression levels remained unchanged following treatment with PTP1B inhibitor (data not shown). To characterize the down-regulation of Bcr-Abl by the inhibition of PTP1B, the expression of Bcr-Abl was examined from 30 to 180 min by Western blotting. The levels of Bcr-Abl protein appeared to increase within 30 min but were barely detectable at 120 min (Fig. 1C). To explore this further, we showed that inhibition of PTP1B had no effect on Bcr and c-Abl protein levels (Fig. 1D), suggesting that the inhibition of PTP1B induced specific down-regulation of Bcr-Abl, perhaps by a mechanism that recognizes a post-translational modification of Bcr-Abl chimeric protein. Additionally, immunoprecipitation of PTP1B from TonB.210 cells followed by Western blot for Bcr-Abl demonstrated that Bcr-Abl and PTP1B bind to each other (Fig. 1E). To confirm that the degradation of Bcr-Abl by PTP1B inhibition occurs in human CML cell lines with endogenous Bcr-Abl expression, the K562 and LAMA-84 cell lines were also examined. Decreased Bcr-Abl protein was observed in both these cell lines following a 2-h treatment with PTP1B inhibitor, although a higher dose of up to 200 μm was required (Fig. 1F). Although this is a considerably higher dose, this may be due to cell-specific differences that may mean a higher dose of inhibitor is required to inhibit PTP1B. To determine whether this down-regulation of Bcr-Abl is due only to the specific inhibition of PTP1B and that the inhibitor is not affecting other PTPs, we used siRNAs to specifically suppress PTP1B. We transfected TonB.210 cells with scrambled siRNA, PTP1B siRNA 1, or PTP1B siRNA 2 for 72 h, and the expression levels of PTP1B and Bcr-Abl were determined by Western blot analysis. When PTP1B levels were knocked down by ∼75% (siRNA 1) and 40% (siRNA 2), the expression of Bcr-Abl was decreased by ∼40% (siRNA 1) and 20% (siRNA 2), respectively (Fig. 2A). Inhibition of PTP1B using siRNAs was also performed in the human K562 cell line, and a similar decrease in Bcr-Abl was observed (Fig. 2B), further demonstrating that the effect is specific to PTP1B inhibition. These results suggest that stabilization of the Bcr-Abl protein in TonB.210 requires the presence of functional PTP1B.
FIGURE 1.
Inhibition of PTP1B activity decreases Bcr-Abl levels. A, TonB.210 cells, a clone of BaF3 cells that induce overexpression of Bcr-Abl in the presence of DOX, were cultured for the indicated times with DOX. Cells were lysed and subjected to Western blot analysis of PTP1B and Bcr-Abl. GAPDH was blotted as a protein-loading control. B and C, effect of different doses (B) and effect of duration of PTP1B inhibition (C) on the expression of Bcr-Abl by Western blotting in TonB.210 cells. GAPDH was used as a protein-loading control. In C, the bar chart in the right panel represents densitometric analysis of Bcr-Abl Western blot. ∅, PTP1B, PTP1B inhibitor. D, TonB.210 cells were treated with 35 μm of the PTP1B inhibitor for 2 h, and the expression of c-Abl and Bcr proteins in treated and untreated cells was determined by Western blotting. GAPDH was blotted as a protein-loading control. E, PTP1B was immunoprecipitated (IP) from TonB.210 cells treated with or without DOX for 48 h followed by Western blot for Bcr-Abl and PTP1B expression. F, K562 and LAMA-84 cells were treated with indicated doses of PTP1B inhibitor for 2 h, and protein lysates were analyzed for Bcr-Abl expression. GAPDH is shown as loading control.
FIGURE 2.
siRNA-mediated inhibition of PTP1B decreases Bcr-Abl levels. A, TonB.210 cells were transfected with scrambled siRNA and two different PTP1B siRNAs (named siRNA 1 and siRNA 2) and treated with DOX for 72 h. Proteins from lysates were immunoblotted for PTP1B and Bcr-Abl. GAPDH was used as a protein-loading control. Bar charts indicate the results of semiquantitative band measures. The asterisk indicates significant differences between siRNA versus scrambled (p < 0.05 by Student's t test). B, K562 cells were transfected with scrambled siRNA and PTP1B siRNAs (1 and 2) for 72 h followed by immunoblotting for PTP1B and Bcr-Abl with GAPDH as loading control.
The Ubiquitin Proteolytic System Is Involved in the Degradation of Bcr-Abl
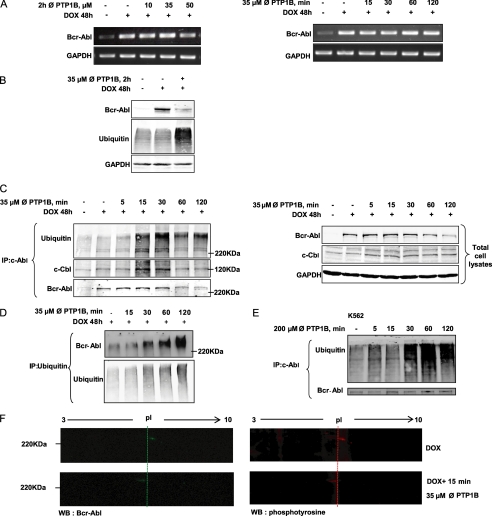
To investigate the underlying mechanism by which PTP1B inhibition affects Bcr-Abl protein expression, we evaluated whether the effects were mediated at the RNA or protein level. Following the same treatment conditions used in the previous experiment (Fig. 1B), Bcr-Abl mRNA levels were similar in TonB.210 cells treated with or without inhibition of PTP1B as analyzed by RT-PCR (Fig. 3A). This suggests that Bcr-Abl down-regulation is due to a post-translational modification rather than a transcriptional inhibition. Previous studies (27) have shown that non-receptor tyrosine kinases undergo an activation-dependent degradation through the ubiquitin proteasome system. To determine whether the reduced expression of Bcr-Abl resulted from the activation of the ubiquitin proteasome system, the levels of ubiquitination in total cell lysates following PTP1B inhibition were examined. Fig. 3B shows an increase in ubiquitinated proteins when cells were treated with 35 μm of the PTP1B inhibitor for 2 h. Immunoprecipitation experiments were carried out to investigate whether Bcr-Abl was ubiquitinated prior to its degradation. TonB.210 cells in the presence of DOX were treated with 35 μm of the PTP1B inhibitor for 5, 15, 30, 60, or 120 min. Increased amounts of ubiquitin were detected following immunoprecipitation of Bcr-Abl between 15 and 30 min of treatment, and a band was detected at the same molecular weight as Bcr-Abl in the ubiquitin Western blot (Fig. 3C). Furthermore, this ubiquitination of Bcr-Abl was associated with an increase in the binding between c-Cbl, an ubiquitin E3 ligase that recognizes activated protein kinases (27), and Bcr-Abl (Fig. 3C). To confirm the ubiquitination of Bcr-Abl, ubiquitin was immunoprecipitated from DOX-treated TonB.210 cells following treatment with the PTP1B inhibitor and examined for the presence of Bcr-Abl (Fig. 3D). In addition to increased Bcr-Abl recovered in the ubiquitin immunoprecipitate following PTP1B inhibition, a shift in the molecular weight of Bcr-Abl was observed, which further indicates that a post-translational modification such as ubiquitination has occurred. To further demonstrate that ubiquitination of Bcr-Abl is important in its degradation following PTP1B inhibition, K562 cells were treated with PTP1B inhibitor, and Bcr-Abl was examined. Following immunoprecipitation of Bcr-Abl, a marked increase in ubiquitin was also observed in K562 cells (Fig. 3E). Together these results indicate that the ubiquitination is an important step in the PTP1B-mediated degradation of Bcr-Abl. Recently, cross-talk between different types of post-translational modification has been shown as important in eukaryotic biology, particularly the multiple connections between phosphorylation and ubiquitination (28). We hypothesized that the mechanism by which PTP1B inhibitor affects Bcr-Abl protein levels is due to a tyrosine phosphorylation of Bcr-Abl residues that triggers its ubiquitination and subsequent degradation. This increase in Bcr-Abl phosphorylation may be a consequence of loss of PTP1B activity caused by PTP1B inhibition. Examination of overall tyrosine phosphorylation of Bcr-Abl by one-dimensional electrophoresis did not reveal any significant change following PTP1B inhibition (data not shown), which was not unexpected as Bcr-Abl is phosphorylated at several tyrosine residues and changes at specific residues may not be detected this way. In an attempt to identify any difference in post-translational modification of Bcr-Abl protein between cells in the presence of DOX with the PTP1B inhibitor or untreated cells, a two-dimensional gel electrophoresis was carried out. The combination of two-dimensional gel electrophoresis and Western blotting demonstrated that PTP1B inhibition caused an acidic isoelectric point shift of Bcr-Abl (at 15 min), which is suggestive of increased Bcr-Abl phosphorylation (Fig. 3F, left panel). Furthermore when this two-dimensional gel electrophoresis was followed by Western blot using a phosphotyrosine antibody, a spot was observed at the same position as Bcr-Abl, and this also underwent an acidic isoelectric point shift following a 15-min treatment with the PTP1B inhibitor (Fig. 3F, right panel). This may indicate the accumulation of a hyperphosphorylated form of Bcr-Abl following PTP1B inhibition priming the protein for ubiquitination. These results show that PTP1B could stabilize Bcr-Abl by post-translational modification, probably preventing its phosphorylation and subsequent ubiquitination and degradation.
FIGURE 3.
Bcr-Abl is degraded at the protein level through the ubiquitin proteolytic system. A, RT-PCR analysis of Bcr-Abl mRNA extracted from the TonB.210 cells was carried out to examine the effect of different doses and times of PTP1B inhibition on transcription of the bcr-abl gene. GAPDH is shown as loading control. ∅, PTP1B, PTP1B inhibitor. B, TonB.210 cells were treated with 35 μm of the PTP1B inhibitor for 2 h, and the expression of Bcr-Abl and ubiquitin proteins in treated and untreated cells was determined by Western blotting. GAPDH was blotted as a protein-loading control. C, TonB.210 cells were treated with 35 μm of the PTP1B inhibitor for the indicated times. After this, c-Abl was immunoprecipitated (IP) from the cell lysates and immunoblotted with anti-c-Abl antibody, anti-c-Cbl antibody, or anti-ubiquitin antibody. Total cell lysates were resolved by SDS-PAGE, transferred to membranes, and subjected to immunoblotting with indicated antibodies. GAPDH was used as a protein-loading control. Bcr-Abl was detected at predicted size. D, TonB.210 cells were treated with 35 μm of the PTP1B inhibitor for the indicated times. Immunoprecipitation of ubiquitin was followed by immunoblotting for Bcr-Abl and ubiquitin. E, K562 cells were treated with 200 μm PTP1B inhibitor for the indicated times. Following this, c-Abl was immunoprecipitated from the cell lysates and immunoblotted with anti-c-Abl antibody and anti-ubiquitin antibody. Bcr-Abl was detected at predicted size. F, TonB.210 cells, in presence of DOX, were left untreated or were treated for 15 min with 35 μm PTP1B inhibitor. Total cell lysates were resolved by two-dimensional gel electrophoresis, transferred to membranes, and immunoblotted (WB) with anti-c-Abl antibody (green) or anti-phosphotyrosine antibody (red). The membranes were cropped to highlight the Bcr-Abl point shift. pl, isoelectric point.
PTP1B Inhibition Leads to Bcr-Abl Degradation by Lysosomal Pathway
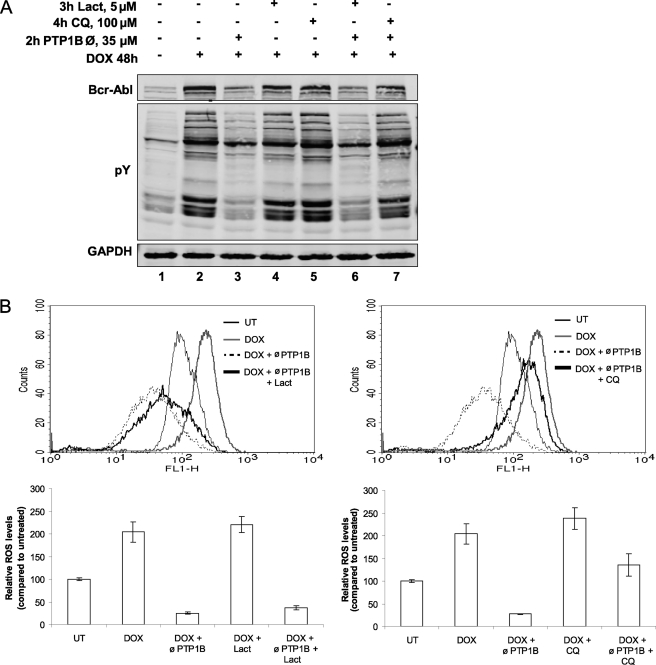
We have shown here (Fig. 3C) an important role for the ubiquitination of Bcr-Abl in its degradation. To check that PTP1B inhibition mediates the degradation of Bcr-Abl through the proteasome pathway, TonB.210 cells were treated with lactacystin, a proteasome inhibitor. As shown in Fig. 4A, pretreatment with lactacystin at 5 μm for 3 h did not block the degradation of Bcr-Abl (compare lane 3 with lane 6). Moreover, we did not observe an increase in the tyrosine phosphorylation of the proteins or in the ROS levels, both features of Bcr-Abl signaling. Inhibition of Bcr-Abl-induced ROS signaling is shown in Fig. 4B, left panel, and remains unaffected by lactacystin pretreatment. These results indicate that Bcr-Abl down-regulation by PTP1B inhibition is proteasome-independent. It has been described that ubiquitination targets receptor tyrosine kinases and other receptors to the lysosome for its degradation (29). We evaluated the contribution of lysosomal proteases to Bcr-Abl degradation by PTP1B inhibition through the use of chloroquine, a lysosomotropic agent. Cells were treated with PTP1B inhibitor in the presence or absence of chloroquine. As shown in Fig. 4A, pretreatment with 100 μm chloroquine for 4 h prevented the degradation of Bcr-Abl induced by PTP1B inhibitor (compare lane 3 with lane 7). Furthermore, treatment with chloroquine caused maintenance of the hallmarks of Bcr-Abl expression, such as an increased tyrosine phosphorylation of proteins (Fig. 4A) and increased ROS levels (Fig. 4B, right panel). These results suggest that the degradation of Bcr-Abl induced by PTP1B inhibition functions through the lysosomal pathway.
FIGURE 4.
PTP1B inhibitor promotes lysosomal degradation of Bcr-Abl. A, TonB.210 cells were untreated (lane 1) or treated with DOX for 48 h, resulting in the expression of Bcr-Abl (lanes 2–7). Cells were treated with 35 μm of the PTP1B inhibitor for 2 h (lane 3) or lactacystin (Lact, lane 4), chloroquine (CQ, lane 5), or PTP1B inhibitor (∅, PTP1B) for 2 h in addition to pretreatment with lactacystin (lane 6) or chloroquine (lane 7). Protein lysates were analyzed for Bcr-Abl, Tyr(P) (pY), and GAPDH by Western blot. B, cells were analyzed for intracellular ROS levels as measured by 2,7-dichlorodihydrofluorescein diacetate fluorescence on the FL-1 channel of a flow cytometer. The histogram shows relative fluorescence of treated cells in comparison with control cells. The data represent the mean ± S.D. of four independent experiments. UT, untreated.
Altered Localization of Bcr-Abl in TonB.210 Cells Is Related to the Inhibition of PTP1B
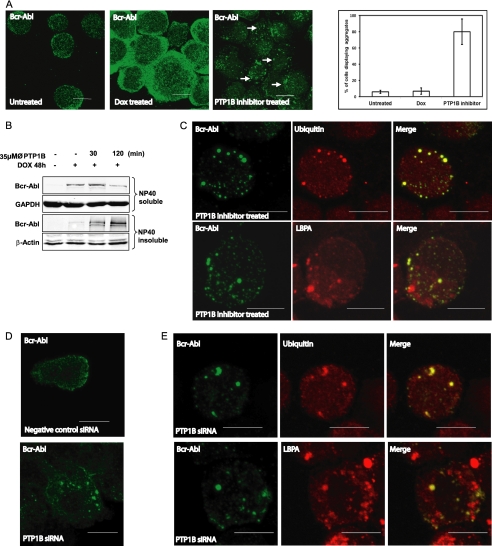
We performed immunofluorescence microscopy to examine the subcellular distribution of Bcr-Abl in TonB.210 cells in the absence or presence of PTP1B inhibition. In contrast to DOX-treated TonB.210 cells, where Bcr-Abl was localized to the cytoplasm, Bcr-Abl was localized predominantly in aggregates that are dispersed in the cytoplasm in cells treated with the PTP1B inhibitor (Fig. 5A). Quantification of the percentage of cells displaying these aggregates is represented in the bar chart in Fig. 5A. It has been reported that in neurodegenerative diseases, polyubiquitylation can induce accumulation of proteins as aggregates in the Nonidet P-40 detergent-insoluble cellular fraction prior its degradation (30). Treatment with the PTP1B inhibitor led to a decrease in Bcr-Abl in the Nonidet P-40 detergent-soluble fraction concomitant with an increase of the Bcr-Abl protein in the Nonidet P-40 detergent-insoluble fraction before degradation by the lysosome (Fig. 5B). Colocalization studies between Bcr-Abl and ubiquitin (Fig. 5C, top panel) demonstrated that the aggregates are ubiquitinated bodies, and colocalization between Bcr-Abl and the late endosome marker LBPA further indicated that these aggregates are degraded via a lysosomal pathway (Fig. 5C, bottom panel). To confirm the effects observed following pharmacological inhibition of PTP1B, immunofluorescence studies were then performed on TonB.210 cells following siRNA inhibition of PTP1B. Cells treated with negative control siRNA and analyzed for Bcr-Abl by immunofluorescence displayed cytoplasmic staining in 91.6 ± 5.3% of cells (Fig. 5D, top panel), whereas treatment with PTP1B siRNA resulted in 63.4 ± 5.2% of cells displaying aggregates similar to those observed following treatment with the PTP1B inhibitor (Fig. 5D, bottom panel). The remaining cells showed cytoplasmic staining similar to that observed in negative control cells. These Bcr-Abl protein aggregates formed following PTP1B siRNA also colocalized with ubiquitin and the late endosome marker LBPA (Fig. 5E). We concluded from these results that Bcr-Abl forms multiubiquitinated aggregates that are incorporated into the lysosome in TonB.210 cells in the presence of PTP1B inhibition.
FIGURE 5.
Bcr-Abl forms ubiquitinated aggregates following PTP1B inhibition. A, TonB.210 cells were untreated (left panel) or treated with DOX for 48 h to induce Bcr-Abl expression (middle panel). DOX-treated cells were also treated with the PTP1B inhibitor for 2 h (right panel). Cells were processed for immunofluorescence microscopy and immunostained with the anti-c-Abl antibody (green). The graph on the right displays quantitation of the number of cells displaying Bcr-Abl aggregates following PTP1B inhibition. A minimum of 100 cells was analyzed per experiment, and results are expressed a mean percentage ±S.D. of at least four independent experiments. B, TonB.210 cells were treated with PTP1B inhibitor (∅, PTP1B) for 30 and 120 min. After treatment with the drug, cells were lysed with radioimmune precipitation buffer to obtain the Nonidet P-40 (NP40)-soluble fraction. The Nonidet P-40-insoluble pellet was further treated with SDS buffer to obtain the Nonidet P-40-insoluble fraction. Detergent-soluble or -insoluble fractions were subjected to SDS-PAGE and Western blot analyses using c-Abl. GAPDH and β-actin served as the loading controls. C, TonB.210 cells treated with DOX for 48 h and PTP1B for 2 h were immunostained for Bcr-Abl (left panels) and co-stained with ubiquitin (top middle panel) and LBPA (bottom middle panel) with the merged images displayed on the right-hand panels. Bcr-Abl is stained green, and ubiquitin and LBPA are stained red. D, TonB.210 cells were treated with DOX and either negative control siRNA (top panel) or PTP1B siRNA (bottom panel) for 72 h followed by immunostaining with anti-c-Abl antibody. E, TonB.210 cells treated with DOX and PTP1B siRNA for 72 h were immunostained for Bcr-Abl (right panels) and ubiquitin (top middle) or LBPA (bottom middle) with merged images displayed on the right. Scale bar is 10 μm.
PTP1B Inhibition Causes Decreased Viability in Bcr-Abl-expressing Cells
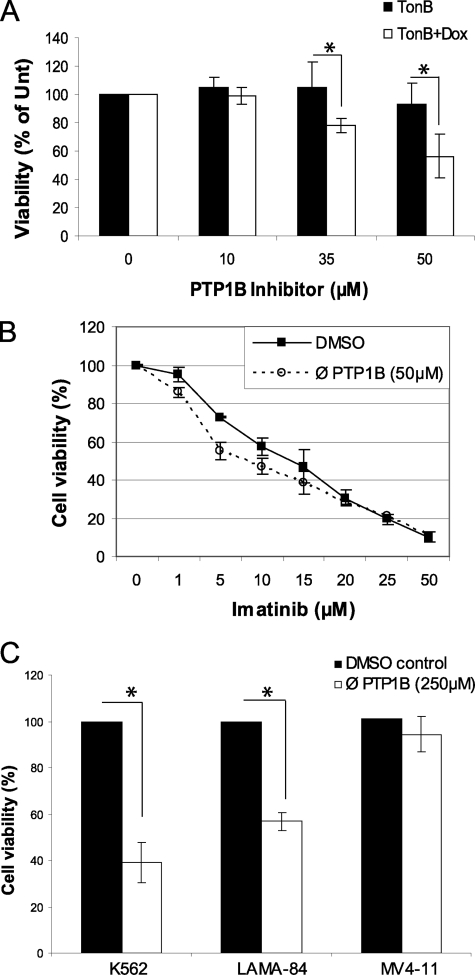
To demonstrate that degradation of Bcr-Abl following PTP1B inhibition had a negative effect on survival, viability was measured following a 24-h treatment with the PTP1B inhibitor. Treatment with 35 and 50 μm doses of the inhibitor caused decreased viability of Bcr-Abl-induced TonB.210 cells (reduction of 23 ± 5 and 48 ± 15% below that of untreated cells, respectively), whereas these doses had no significant effect on uninduced cells (Fig. 6A). Furthermore, it was observed that the IC50 of the tyrosine kinase inhibitor imatinib mesylate, currently used for treatment of CML, was reduced when co-incubated with the PTP1B inhibitor. Treatment with 50 μm PTP1B inhibitor reduced the IC50 of imatinib mesylate treatment over 24 h from 9.9 ± 0.6 to 4.8 ± 0.5 μm (Fig. 6B). Furthermore, when cell viability was examined in human CML cell lines following a 24-h treatment with the PTP1B inhibitor, a significant decrease was observed in both K562 and LAMA-84 (Fig. 6C). However, the AML cell line MV4–11, which lacks Bcr-Abl expression, was unaffected. This demonstrates that it is through the degradation of Bcr-Abl that the PTP1B inhibitor exerts a negative effect on cell viability.
FIGURE 6.
PTP1B inhibition affects viability of Bcr-Abl-expressing cells and increases efficiency of imatinib mesylate. A, TonB.210 cells were treated with or without DOX for 48 h. PTP1B inhibitor was included for the final 24 h at stated doses. Cell viability was measured using the CellTiter-Blue viability assay (* represents p < 0.05 as analyzed by Student's t test). % of Unt, result expressed as percentage of untreated cells. B, TonB.210 were treated as in A with the addition of indicated doses of imatinib mesylate for the final 24 h followed by CellTiter-Blue viability assay. IC50 values were calculated using a four-parameter sigmoidal model. ∅, PTP1B, PTP1B inhibitor. C, K562, LAMA-84, and MV4–11 cells were treated with PTP1B inhibitor (250 μm) or dimethyl sulfoxide (DMSO) vehicle for 24 h and then assayed for cell viability using the CellTiter-Blue viability assay (* represents p < 0.005 as analyzed by Student's t test).
DISCUSSION
In the present study, we used both pharmacological inhibition and siRNA knockdown methods to examine the relationship between PTP1B and Bcr-Abl-expressing cells. TonB.210 cells expressing Bcr-Abl showed an elevated level of PTP1B protein. This may reflect a homeostatic adaptation to enhanced kinase activity. However, the results of other groups (31) have shown that Bcr-Abl induces up-regulation of PTP1B transcript levels by regulating transcription factors, leading to an increase of PTP1B protein levels. Intriguingly, when we characterized the optimal experimental conditions for the inhibition of PTP1B, we detected a down-regulation of Bcr-Abl. This result suggests that PTP1B acts as a tumor promoter in CML, and it is consistent with previously published data from several laboratories that have reported that PTP1B functions as a positive regulator of signaling events associated with breast tumorigenesis (32, 33). Until recently, the only PTP that had been demonstrated to function as an oncogene in certain tumors was SHP-2 (34). In fact, SHP-2 is required for hematopoietic cell transformation by Bcr-Abl, and this function of SHP-2 is attributed to its role in stabilizing Bcr-Abl as well as in downstream signal transduction of Bcr-Abl kinase (35). Moreover, it has been reported that mutations that activate this PTP are present in several leukemias (36). To confirm that the decreases in Bcr-Abl protein levels are likely due to a specific loss of PTP1B function and avoid the possible off-target effects of pharmaceutical approach, we knocked down PTP1B by two different PTP1B siRNAs, resulting in a reduction of Bcr-Abl protein levels in TonB.210 and K562 cells. The difference between the two PTP1B siRNAs used in down-regulating Bcr-Abl, where an increased knockdown of PTP1B correlates with higher down-regulation of Bcr-Abl, suggests that the down-regulation of Bcr-Abl is due to a reduction in PTP1B activity. In summary, these results indicate that PTP1B regulates stabilization of Bcr-Abl protein and that its activity is required for its stabilization.
Bcr-Abl protein levels were decreased in cells treated with PTP1B inhibitor, but Bcr-Abl mRNA levels were similar in TonB.210 cells treated or untreated with PTP1B inhibitor, suggesting that rapid Bcr-Abl down-regulation is mediated by post-transcriptional mechanisms that lead to its degradation. Initially, the treatment with PTP1B inhibitor stabilized and increased Bcr-Abl protein levels, but at later time points, they induced its degradation. Previous studies have reported that activation of many kinases triggers their subsequent down-regulation through the ubiquitin proteasome system pathway (27). Activation-dependent degradation and signaling termination usually involve members of the CBL family of E3 ligases. In the present study, we have demonstrated that treatment with PTP1B inhibitor causes a shift of Bcr-Abl species to a more acidic isoelectric point, which is consistent with an increase in phosphorylation. We suggest that this phosphorylation creates a recognition signal for binding of a ubiquitin ligase such as c-Cbl to Bcr-Abl that causes ubiquitination of Bcr-Abl prior to its degradation. These data are consistent with findings that other oncogenes, such as c-Myc, avoid degradation and increase stabilization by mutations that prevent phosphorylation and subsequent ubiquitination and proteasomal degradation (37). In fact, the inhibition of PTP1B leads to the translocation of Bcr-Abl from the Nonidet P-40 detergent-soluble fraction to the Nonidet P-40 detergent-insoluble fraction, which is known to contain polyubiquitinated proteins (30). This result was confirmed by immunofluorescence where PTP1B inhibitor induced a redistribution of Bcr-Abl into punctate, cytosolic aggregates prior to its degradation.
In mammalian cells, there are two main pathways for protein degradation: the ubiquitin-proteasome pathway and the autophagy-lysosome pathway. Depending on the cell type, its metabolic activity, and the specific protein to be degraded, the cell will use one or the other. Both catabolic pathways share the use of ubiquitin. The structural complexity of different polyubiquitin chains may be sufficient to maintain selectivity and specificity of the ubiquitin proteasome system and autophagy toward their substrates (38). We have evaluated which degradative pathway was activated when TonB.210 cells were treated with PTP1B inhibitor (Fig. 7). Pretreatment with lactacystin, a proteasome inhibitor, did not attenuate the down-regulation. This result indicates that Bcr-Abl down-regulation by PTP1B inhibitor is proteasome-independent. Moreover, PTP1B inhibition induces a destabilization of Bcr-Abl independent of the Hsp90/Hsp70 pathway, molecular chaperones well known as facilitators of protein folding and stability (39). The down-regulation of Bcr-Abl was abolished by pretreatment with chloroquine, a lysosome inhibitor. We show that treating TonB.210 cells with the inhibitor of PTP1B induced rapid down-regulation of Bcr-Abl through the autophagy-lysosome pathway. It has been shown that aggregates of polyubiquitinated proteins formed in the cytoplasm are selectively removed from cells by autophagy (40) or the aggresome pathway (41).
FIGURE 7.
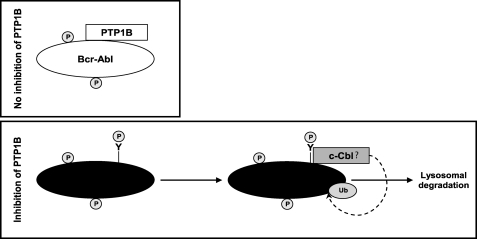
Model illustrating the proposed mechanism of Bcr-Abl destabilization in response to inhibition of PTP1B. Expression of Bcr-Abl protein can be altered by inhibition of PTP1B. In the presence of PTP1B (top panel), Bcr-Abl associates with PTP1B, which provides stability to Bcr-Abl. The treatment with PTP1B inhibitor (bottom panel) elicits tyrosine phosphorylation (P) of critical residues of Bcr-Abl, and this acts as a trigger for Bcr-Abl to be ubiquitinated and degraded in the lysosome. Ub, ubiquitin.
The importance of PTP1B stabilization of Bcr-Abl for oncogenic signaling is confirmed by the decreased viability of Bcr-Abl-expressing cell lines following PTP1B inhibition, whereas the effect on viability is significantly less in non-Bcr-Abl cell lines. In addition to this, the IC50 of the imatinib mesylate was decreased by PTP1B inhibition. This raises the possibility that PTP1B inhibitors, which are currently the subject of intensive research, may be useful in the treatment of CML, either in conjunction with existing treatments or possibly as an alternative therapy.
Although several investigations have elucidated mechanisms governing Bcr-Abl regulation (42), identification of the cellular protein components that can regulate its activity or stability may reveal potential pharmacological targets that could inhibit Bcr-Abl signaling. Our studies show that PTP1B is required for the stabilization of Bcr-Abl and functions as a tumor promoter in TonB.210 cells. Inhibition of PTP1B activity leads to down-regulation of Bcr-Abl through ubiquitination and lysosomal degradation. Manipulating the stability of PTP1B and modulating the ubiquitin system might offer interesting therapeutic approaches for the treatment of CML.
This work was supported by grants from the Health Research Board of Ireland, the Children's Leukemia Research Project, and the Irish Cancer Society.
- CML
- chronic myeloid leukemia
- PTP
- protein-tyrosine phosphatase
- PTP1B
- protein-tyrosine phosphatase 1B
- ROS
- reactive oxygen species
- LBPA
- lysophosphatidic acid
- DOX
- doxycycline hyclate.
REFERENCES
- 1. Deininger M. W., Goldman J. M., Melo J. V. (2000) Blood 96, 3343–3356 [PubMed] [Google Scholar]
- 2. Melo J. V. (1996) Leukemia 10, 751–756 [PubMed] [Google Scholar]
- 3. Meyn M. A., 3rd, Wilson M. B., Abdi F. A., Fahey N., Schiavone A. P., Wu J., Hochrein J. M., Engen J. R., Smithgall T. E. (2006) J. Biol. Chem. 281, 30907–30916 [DOI] [PubMed] [Google Scholar]
- 4. Monteiro H. P., Arai R. J., Travassos L. R. (2008) Antioxid. Redox Signal. 10, 843–889 [DOI] [PubMed] [Google Scholar]
- 5. Ostman A., Hellberg C., Böhmer F. D. (2006) Nat. Rev. Cancer 6, 307–320 [DOI] [PubMed] [Google Scholar]
- 6. Bourdeau A., Dubé N., Tremblay M. L. (2005) Curr. Opin. Cell Biol. 17, 203–209 [DOI] [PubMed] [Google Scholar]
- 7. Haj F. G., Markova B., Klaman L. D., Bohmer F. D., Neel B. G. (2003) J. Biol. Chem. 278, 739–744 [DOI] [PubMed] [Google Scholar]
- 8. Kakazu A., Sharma G., Bazan H. E. (2008) Invest. Ophthalmol. Vis. Sci. 49, 2927–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown-Shimer S., Johnson K. A., Hill D. E., Bruskin A. M. (1992) Cancer Res. 52, 478–482 [PubMed] [Google Scholar]
- 10. Woodford-Thomas T. A., Rhodes J. D., Dixon J. E. (1992) J. Cell Biol. 117, 401–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu F., Sells M. A., Chernoff J. (1998) Mol. Cell. Biol. 18, 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dubé N., Cheng A., Tremblay M. L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 1834–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dadke S., Chernoff J. (2003) J. Biol. Chem. 278, 40607–40611 [DOI] [PubMed] [Google Scholar]
- 14. Bjorge J. D., Pang A., Fujita D. J. (2000) J. Biol. Chem. 275, 41439–41446 [DOI] [PubMed] [Google Scholar]
- 15. Wiener J. R., Kerns B. J., Harvey E. L., Conaway M. R., Iglehart J. D., Berchuck A., Bast R. C., Jr. (1994) J. Natl. Cancer Inst. 86, 372–378 [DOI] [PubMed] [Google Scholar]
- 16. Wiener J. R., Hurteau J. A., Kerns B. J., Whitaker R. S., Conaway M. R., Berchuck A., Bast R. C., Jr. (1994) Am. J. Obstet. Gynecol. 170, 1177–1183 [DOI] [PubMed] [Google Scholar]
- 17. Nanney L. B., Davidson M. K., Gates R. E., Kano M., King L. E., Jr. (1997) J. Cutan. Pathol. 24, 521–532 [DOI] [PubMed] [Google Scholar]
- 18. Warabi M., Nemoto T., Ohashi K., Kitagawa M., Hirokawa K. (2000) Exp. Mol. Pathol. 68, 187–195 [DOI] [PubMed] [Google Scholar]
- 19. LaMontagne K. R., Jr., Flint A. J., Franza B. R., Jr., Pandergast A. M., Tonks N. K. (1998) Mol. Cell. Biol. 18, 2965–2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Naughton R., Quiney C., Turner S. D., Cotter T. G. (2009) Leukemia 23, 1432–1440 [DOI] [PubMed] [Google Scholar]
- 21. LaMontagne K. R., Jr., Hannon G., Tonks N. K. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 14094–14099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klucher K. M., Lopez D. V., Daley G. Q. (1998) Blood 91, 3927–3934 [PubMed] [Google Scholar]
- 23. Bhatt L., Horgan C. P., Walsh M., McCaffrey M. W. (2007) Biochem. Biophys. Res. Commun. 359, 277–284 [DOI] [PubMed] [Google Scholar]
- 24. Perego R. A., Costantini M., Cornacchini G., Gargantini L., Bianchi C., Pungolino E., Rovida E., Morra E. (2000) Eur. J. Cancer 36, 1395–1401 [DOI] [PubMed] [Google Scholar]
- 25. Khoudoli G. A., Porter I. M., Blow J. J., Swedlow J. R. (2004) Proteome Sci. 2, 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tedesco S., Doyle H., Blasco J., Redmond G., Sheehan D. (2010) Comp Biochem. Physiol. C Toxicol. Pharmacol. 151, 167–174 [DOI] [PubMed] [Google Scholar]
- 27. Lu Z., Hunter T. (2009) Annu. Rev. Biochem. 78, 435–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hunter T. (2007) Mol. Cell 28, 730–738 [DOI] [PubMed] [Google Scholar]
- 29. Marmor M. D., Yarden Y. (2004) Oncogene 23, 2057–2070 [DOI] [PubMed] [Google Scholar]
- 30. de Pril R., Fischer D. F., Maat-Schieman M. L., Hobo B., de Vos R. A., Brunt E. R., Hol E. M., Roos R. A., van Leeuwen F. W. (2004) Hum. Mol. Genet. 13, 1803–1813 [DOI] [PubMed] [Google Scholar]
- 31. Fukada T., Tonks N. K. (2001) J. Biol. Chem. 276, 25512–25519 [DOI] [PubMed] [Google Scholar]
- 32. Julien S. G., Dubé N., Read M., Penney J., Paquet M., Han Y., Kennedy B. P., Muller W. J., Tremblay M. L. (2007) Nat. Genet. 39, 338–346 [DOI] [PubMed] [Google Scholar]
- 33. Bentires-Alj M., Neel B. G. (2007) Cancer Res. 67, 2420–2424 [DOI] [PubMed] [Google Scholar]
- 34. Chan G., Kalaitzidis D., Neel B. G. (2008) Cancer Metastasis Rev. 27, 179–192 [DOI] [PubMed] [Google Scholar]
- 35. Chen J., Yu W. M., Daino H., Broxmeyer H. E., Druker B. J., Qu C. K. (2007) Blood 109, 778–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tartaglia M., Niemeyer C. M., Shannon K. M., Loh M. L. (2004) Curr. Opin. Hematol. 11, 44–50 [DOI] [PubMed] [Google Scholar]
- 37. Junttila M. R., Westermarck J. (2008) Cell Cycle 7, 592–596 [DOI] [PubMed] [Google Scholar]
- 38. Welchman R. L., Gordon C., Mayer R. J. (2005) Nature Rev. 6, 599–609 [DOI] [PubMed] [Google Scholar]
- 39. Nimmanapalli R., O'Bryan E., Bhalla K. (2001) Cancer Res. 61, 1799–1804 [PubMed] [Google Scholar]
- 40. Kaniuk N. A., Kiraly M., Bates H., Vranic M., Volchuk A., Brumell J. H. (2007) Diabetes 56, 930–939 [DOI] [PubMed] [Google Scholar]
- 41. Kawaguchi Y., Kovacs J. J., McLaurin A., Vance J. M., Ito A., Yao T. P. (2003) Cell 115, 727–738 [DOI] [PubMed] [Google Scholar]
- 42. Hantschel O., Superti-Furga G. (2004) Nat. Rev. Mol. Cell Biol. 5, 33–44 [DOI] [PubMed] [Google Scholar]