Abstract
In mammalian cells, the enzyme 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMGR), which catalyzes the rate-limiting step in the mevalonate pathway, is ubiquitylated and degraded by the 26 S proteasome when mevalonate-derived metabolites accumulate, representing a case of metabolically regulated endoplasmic reticulum-associated degradation (ERAD). Here, we studied which mevalonate-derived metabolites signal for HMGR degradation and the ERAD step(s) in which these metabolites are required. In HMGR-deficient UT-2 cells that stably express HMGal, a chimeric protein between β-galactosidase and the membrane region of HMGR, which is necessary and sufficient for the regulated ERAD, we tested inhibitors specific to different steps in the mevalonate pathway. We found that metabolites downstream of farnesyl pyrophosphate but upstream to lanosterol were highly effective in initiating ubiquitylation, dislocation, and degradation of HMGal. Similar results were observed for endogenous HMGR in cells that express this protein. Ubiquitylation, dislocation, and proteasomal degradation of HMGal were severely hampered when production of geranylgeranyl pyrophosphate was inhibited. Importantly, inhibition of protein geranylgeranylation markedly attenuated ubiquitylation and dislocation, implicating for the first time a geranylgeranylated protein(s) in the metabolically regulated ERAD of HMGR.
Keywords: Cholesterol Regulation, ER-associated Degradation, Protein Isoprenylation, Sterol, Ubiquitylation, HMG-CoA Reductase, Mevalonate Metabolism, Squalene
Introduction
Mammalian cells satisfy most of their cholesterol requirements through receptor-mediated endocytosis of cholesterol-rich plasma lipoproteins. However, when exogenous cholesterol supply is short, or when there is an increased demand for particular nonsterol isoprenoid(s), cells up-regulate the activity of the mevalonate (MVA)3 pathway (Fig. 1), in which sterols and essential MVA-derived nonsterol isoprenoids are produced (1, 2). Intracellular MVA levels are finely tuned and tightly regulated by a feedback mechanism that governs the rate of 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reduction into MVA, the major rate-limiting step for the entire pathway (3, 4). This reaction is catalyzed by HMG-CoA reductase (HMGR), an integral glycoprotein resident to the endoplasmic reticulum (ER). Each of the 97-kDa subunits of HMGR is embedded in the ER membrane via a ∼350 residue N-terminal noncatalytic region that spans the membrane 8 times (5–7), whereas the rest of the polypeptide faces the cytosol where it tetramerizes to form the active sites of the enzyme (5, 8, 9).
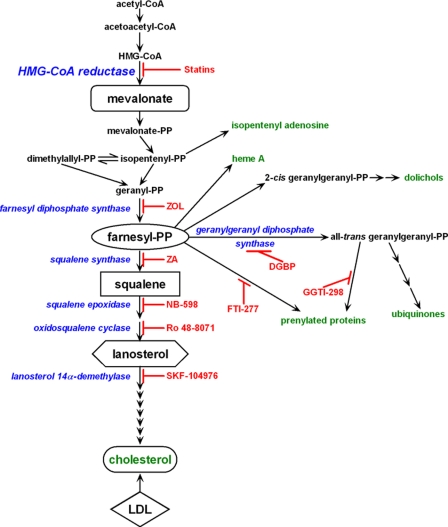
FIGURE 1.
The mevalonate pathway. Highlighted in blue are some enzymes of the MVA pathway and their inhibitors are in red. Key end products are highlighted in green.
To allow sufficient production of isoprenoids yet avoid overaccumulation of potentially toxic products, the activity of HMGR is stringently controlled and varies according to the cellular needs for sterol and the nonsterol products. HMGR activity increases when cells are starved for cholesterol or MVA. Conversely, abundant cholesterol and/or MVA acutely suppress the enzyme (4, 10). At the post-translational level, regulation of HMGR mainly involves modulation of its stability. Thus, in cells deprived of sterols and/or MVA, HMGR is a rather stable protein that turns over with a half-life of 10–15 h, whereas in sterols- and/or MVA-replete cells, the enzyme is rapidly degraded with half-life of 0.5–4 h. This conditional degradation is imparted onto the enzyme by its membrane region, which function as a metabolically regulated degron that is necessary to control the stability of HMGR and sufficient to confer regulated degradation onto many reporter proteins and immunogenic tags when fused to its C terminus (11–15).
The degradation of HMGR is mediated by the ubiquitin-proteasome system (16) and mandates, at least, the participation of either one of the polytopic ER Insig proteins (17). These proteins are thought to couple in a sterol-regulated manner between HMGR and a membrane-bound E3 protein ubiquitin ligase, reported to be gp78 (18). Thus, degradation of HMGR may be viewed as a special case of the more general cellular quality control mechanism, known as ER-associated degradation (ERAD). This mechanism protects against proteotoxicity by eliminating aberrant proteins from the ER (19, 20). ERAD also rids the cell of normal ER proteins when they are no longer needed under specific developmental or metabolic circumstances (21–23). En route to proteolysis by the cytoplasmic 26 S proteasome, HMGR is ubiquitylated on two lysine residues (Lys89 and Lys248) within the membrane region (15, 17) and extracted from the ER, along with the short-lived Insig-1, as an intact full-length polypeptide (24). The ATPase activity of AAA-ATPase p97 provides the driving force for the extraction of HMGR (24), a step considered to be the hallmark of ERAD known as retrotranslocation or dislocation (25).
Accelerated ubiquitin-dependent degradation of HMGR in mammalian cells can be elicited not only by sterols (lanosterol, cholesterol, and hydroxysterols) but also by tocotrienols (26, 27) and synthetic bisphosphonate esters, such as SR-12813 and Apomine (28, 29), which bear no structural resemblance to sterols. Importantly, none of these elicitors stimulate the turnover of HMGR in cells that are acutely deprived of MVA (e.g. by blocking HMGR activity with high concentrations of statins). Under such circumstances, the full potency of these elicitors comes to light only upon supplementing the cells with small amount of exogenous MVA, which, by itself, is not sufficient to stimulate degradation (10, 29–31). Moreover, the exogenous MVA must be metabolized in the pathway to synergize the action of sterols (31), indicating that at least two “metabolic signals” are required to stimulate the degradation of HMGR: a sterol (or a foreign exogenous compound such as tocotrienol or Apomine), and an as yet unknown MVA-derived nonsterol metabolite. Only through the synergistic action of both classes of molecules is the degradation of HMGR commenced (10, 29–31). Early studies, using free farnesol or its derivatives farnesyl acetate and ethyl farnesyl ether, suggested that this 15-carbon MVA-derived metabolite might be the nonsterol regulator for HMGR degradation (32–34). However, a more recent study has implicated the 20-carbon alcohol geranylgeraniol (GGOH), or a geranylgeraniol-derived metabolite, as the nonsterol that synergistically acts with sterols to promote HMGR degradation (17). Interestingly, it was previously demonstrated that nonsterol metabolites preceding squalene epoxide can efficiently accelerate HMGR degradation without the need for additional sterol-derived signal (31). In this study an attempt was made to further identify the MVA-derived metabolite(s) that are involved in the metabolically regulated degradation of HMGR and the ERAD step(s) in which these metabolite are required.
EXPERIMENTAL PROCEDURES
Reagents
Digeranyl bisphosphonate (DGBP) was generously provided by Raymond Hohl (University of Iowa) and Terpenoid Therapeutics. Lovastatin and zaragozic acid A (ZA) were provided by Merck. NB-598 was kindly provided by Banyu Pharmaceuticals, RO 48-8071 was a gift of Hoffmann-La Roche, and SKF 104976 was obtained from SmithKline Beecham Pharmaceuticals. Zoledronic acid (Zomera®, ZOL) was purchased from Novartis Pharma. Digitonin (high purity), ALLN, MG-132, GGTI-298, and FTI-277 were from Calbiochem. Mevalonolactone was from Fluka and cholesterol and 25-hydroxycholesterol from Steraloids. Polygram SIL G thin layer chromatography plates were obtained from Macherey-Nagel. Geneticin was from Invitrogen. [3H]Acetate and Expre35S35S protein labeling mix were from PerkinElmer Life Sciences. All other reagents were from Sigma. Fetal bovine lipoprotein-deficient serum (LPDS; d ≥ 1.25) was prepared by ultracentrifugation, as described (35).
Antibodies
Anti-β-galactosidase monoclonal antibody (clone Z378B) was purchased from Promega Corporation. Antibodies against Rap1A (c-17; SC-1482), Rap1 (c-121; SC-65), Rab6, (c-19; SC-310), and β-actin (AC-15; SC-69879) were from Santa Cruz Biotechnology. Anti-GAPDH (9484) was from Abcam. Rabbit anti-calnexin and anti-gp78 were generously provided by Ron Kopito (Stanford University) and Richard Wojcikiewicz (SUNY Upstate Medical University), respectively. Antiserum against the membrane region of HMGR was described previously (7). Horseradish peroxidase-conjugated secondary antibodies were from Jackson ImmunoResearch. Agarose-immobilized recombinant Protein A was purchased from RepliGen Corporation.
Cells and Media
All cells were maintained at 37 °C in a humidified 5% CO2 atmosphere and all media were based on minimal essential medium supplemented with 2 mm glutamine, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. UT-2 cells (36) were maintained in medium containing 5% (v/v) fetal calf serum (FCS) and 2 mm MVA (Medium A). The medium of UT-2/HMGal cells (31) also contained 250 μm geneticin. To starve UT-2 and UT-2/HMGal cells for MVA and sterols, the cells were washed once with phosphate-buffered saline (PBS) and re-fed with medium supplemented with 5% (v/v) dialyzed LPDS and 50 μm lovastatin MVA (Medium B) to block any residual HMGR activity in these cells (37). Met-18b-2 cells, which take up and metabolize MVA at 10–40 times greater rate than the progenitor CHO cells (38, 39), were grown in 5% (v/v) FCS. To starve Met-18b-2 cells for MVA and sterols, the cells were washed once with PBS and re-fed with medium supplemented with 5% (v/v) dialyzed LPDS and 2 μm lovastatin MVA (Medium C). Lovastatin-resistant LP-90 cells (40) were grown in the presence of 5% (v/v) LPDS and 90 μm lovastatin (Medium D).
Cell Fractionation, Immunoprecipitation, and Immunoblotting
Cell fractionation into 20,000 × g supernatant and pellet fractions was conducted as previously described (24), except the digitonin concentration was doubled to 0.05% (w/v). HMGal was immunoprecipitated with anti-HMGR membrane region antiserum (7). Immune complexes were resolved by 5–15% SDS-PAGE and blotted onto Optitran BAS-83 nitrocellulose membranes (Schleicher & Schuell). The membranes were probed with the appropriate primary antibodies followed by horseradish peroxidase-conjugated secondary antibodies and visualized by enhanced chemiluminescence reaction. Metabolic labeling with [35S]methionine/cysteine and immunoprecipitation of labeled proteins was previously described (15, 16, 31).
Incorporation of [3H]Acetate into Nonsaponifiable Lipids
LP-90 cells, grown in duplicate 60-mm dishes in Medium D, were washed twice with PBS and labeled in 1 ml of minimal essential medium containing 5% (v/v) LPDS and 50 μCi/dish of [3H]acetate (2–5 Ci/mmol). Lovastatin, ZA, or NB-598 were directly added to the labeling medium. The cells were labeled for 2 h at 37 °C after which they were washed three times and extracted for 15 min with 2.5 ml of hexane:isopropyl alcohol (3:2, v/v) while rotating at 4 °C. After addition of the lipid recovery mixture (10 μg each of cholesterol, lanosterol, squalene, and farnesol), the extracts were collected, the dishes were extracted again with 1.5 ml of hexane:isopropyl alcohol, and the extracts were combined. The extracted cells on the dish were dissolved in 0.5 ml of 0.5 m NaOH for estimation of protein content. The combined hexane:isopropyl alcohol extracts were evaporated under N2 and the dry residue was dissolved in 1 ml of 1 m KOH in ethanol and saponified at 80 °C for 60 min. After addition of 1 ml of water, the saponified material was extracted twice with 2.5 ml of petroleum ether. The combined extracts were dried under N2, re-dissolved in 200 μl of chloroform:methanol (2:1, v/v), and applied onto plastic-backed silica gel thin-layer chromatography plates. The plates were developed to a height of 15 cm in heptane:diethyl ether (90:60, v/v), with lipid standards running alongside, and stained in iodine vapors. The plates were cut into 1-cm segments and counted for 3H radioactivity. Radioactivity running at Rf 0.2–0.4 (migrating to the position of cholesterol and lanosterol standards) was taken cumulatively as “sterols,” whereas radioactivity running to Rf 0.9–1 (migrating to the position of squalene standard) is considered “squalene.”
RESULTS
Mevalonate Is Required for Sterol-stimulated HMGal Degradation in UT-2 Cells
Similar to other ERAD substrates, the pathway for HMGR degradation may be schematically divided into 4 distinct steps, which include: (i) substrate recognition; (ii) ubiquitylation; (iii) dislocation from the ER; and (iv) degradation in the cytosol by the 26 S proteasome. These steps involve the coordinated action of general constituents of the ubiquitin-proteasome machinery and the ERAD pathway, as well as additional cellular factors that are specific for the HMGR as a substrate. Unlike most ERAD substrates, which are aberrant proteins, HMGR is a conditional ERAD substrate whose rate of degradation is inversely correlated with the intracellular levels of products of the MVA pathway, exemplifying a case of “metabolically regulated ERAD.” To address which steps in ERAD of HMGR are regulated by which MVA-derived metabolites, we examined the degradation of HMGal that is stably expressed in UT-2 cells. UT-2 cells are deficient in HMGR protein (36) due to point mutations that abrogate the correct splicing of reductase mRNA (41), and depend on exogenous MVA for viability. Thus, intracellular MVA levels can be efficiently manipulated by changing MVA concentrations in the growth medium. HMGal is a fusion protein between β-galactosidase of Escherichia coli and the membrane region of HMGR (12). Similar to HMGR, HMGal expressed in CHO and UT-2 cells is ubiquitinated and rapidly degraded by the proteasome in a sterol-stimulated fashion, mirroring the fate of endogenous HMGR (16, 29).
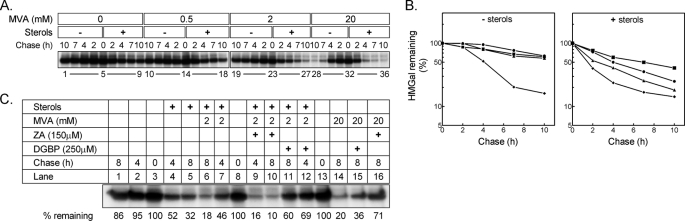
Fig. 2 shows an experiment in which UT-2/HMGal cells were acutely deprived of MVA, pulse-labeled with radioactive methionine/cysteine, and then chased in the absence or presence of sterols and increasing concentrations of MVA. As can be seen, HMGal is a fairly stable protein in MVA-depleted cells (Fig. 2, A, lanes 1–5, and B, left panel, squares) and, without added MVA, sterols only slightly accelerated its degradation (Fig. 2, A, lanes 5–9, and B, right panel, squares). Very low concentrations of exogenous MVA (Fig. 2A, 0.5 mm, lanes 10–14), which does not permit efficient sterol synthesis because of the poor uptake of MVA (38, 42), had no significant effect on HMGal stability (Fig. 2B, left panel, circles), yet it markedly potentiated the effect of sterols on HMGal turnover (Fig. 2, A, lanes 14–18, and B, right panel, circles). Likewise, a small effect on HMGal stability (Fig. 2, A, lanes 19–23, and B, left panel, triangles) and a stronger synergism with sterols was exerted by 2 mm MVA (Fig. 2, A, lanes 23–27 and B, right panel, triangles), whereas high (20 mm) concentrations of MVA led to robust degradation of HMGal (Fig. 2, A, lanes 28–32, and B, left panel, diamonds) that was only slightly accelerated further by exogenous sterols (Fig. 2, A, lanes 32–36, and B, right panel, diamonds). Thus, in agreement with previous reports (30, 31), these results indicate that minimal levels of MVA-derived nonsterol metabolite(s) are required to synergize the action of sterols in stimulating HMGal degradation.
FIGURE 2.
Sterols and MVA-derived nonsterol(s) act synergistically to promote HMGal degradation. A, UT-2/HMGal cells were incubated for 20 h in Medium B supplemented with 50 μm lovastatin. Cells were pulse-labeled and chased in Medium B in the absence or presence of sterols (2 μg/ml 25-hydroxycholesterol + 20 μg/ml of cholesterol) with the indicated concentrations of MVA. HMGal was immunoprecipitated with anti-HMGR membrane region antibodies. B, quantification of the results in A. ■, no MVA; ●, 0.5 mm MVA; ▴, 2 mm MVA; ⧫, 20 mm MVA. C, cells were set up for the experiment and labeled as in A. Cells were chased in the presence of the indicated additions, which were added to the chase medium. The remaining HMGal at the end of the chase (%) is indicated.
It can be formally argued that the sterol-accelerated degradation of HMGal observed at low MVA is not because of the synergistic effect of MVA-derived nonsterol isoprenoid(s) but rather due to the endogenous production of a specific sterol(s) that further promotes HMGal degradation. To distinguish between these possibilities, we inhibited the MVA pathway at a step prior to the sterologenic branch by treating the cells with the squalene synthase inhibitor ZA (see Fig. 1). In agreement with Fig. 2A, in MVA-depleted cells sterols alone only partially enhanced the degradation of HMGal (Fig. 2B, lanes 1–5) and addition of 2 mm MVA further stimulated HMGal elimination (Fig. 2B, compare lanes 3–5 to lanes 6–8). Interestingly, when ZA was also added to cells chased in the presence of 2 mm MVA, not only did HMGal degradation proceed unabated but in fact it was augmented (Fig. 2B, compare lanes 8–10 to lanes 6–8). That ZA effectively blocked sterol synthesis from MVA was demonstrated by the marked inhibition of HMGal degradation when the sole source of sterols was their endogenous synthesis from the exogenously added 20 mm MVA (Fig. 2B, compare lane 16 to lane 14 to lane 1).
A possible explanation for this augmentation is that farnesyl pyrophosphate (FPP) levels rise upon ZA-mediated inhibition (43), and therefore, FPP or FPP-derived metabolite(s) may synergize the action of sterols in HMGal degradation. Indeed, when the cells were chased in the presence of sterols, 2 mm MVA and the geranylgeranyl pyrophosphate (GGPP) synthase inhibitor DGBP (44), which blocks the conversion of FPP to GGPP (Fig. 1), HMGal degradation was nearly blocked (Fig. 2B, compare lanes 11-13 to 6–8). Similarly, DGBP halted the degradation stimulated by 20 mm MVA, which now resembled the degradation exerted by sterols alone (Fig. 2B, compare lane 15 to 14 to lane 1 to 5). Thus, GGPP or GGPP-derived nonsterol metabolite(s), rather than endogenously synthesized sterol(s), is the MVA-derived culprit that synergizes the sterol-accelerated degradation of HMGal.
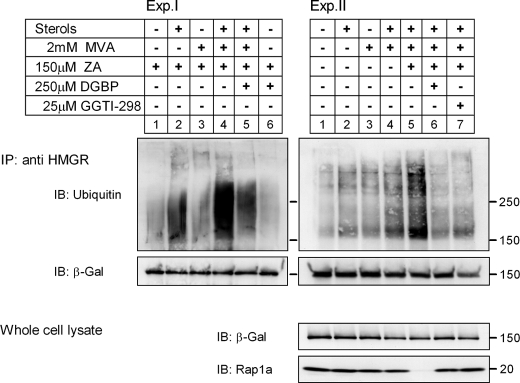
Geranylgeranylated Protein(s) Is Involved in Sterol-stimulated Ubiquitylation of HMGal
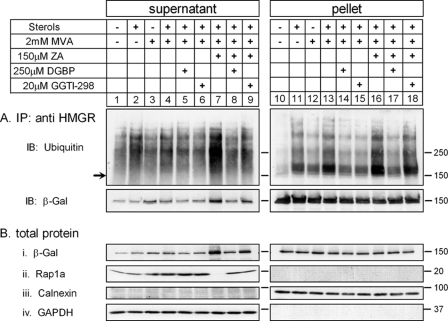
Sever et al. (17) have previously shown that sterols promote ubiquitylation of HMGR in MVA-depleted SV40-transformed human fibroblasts (SV-589 cells), and that exogenous MVA did not further enhance HMGR ubiquitylation. Also, evidence was presented that exogenously added geranylgeraniol (GGOH), but not farnesol, accelerated the sterol-induced degradation of endogenous reductase. This led DeBose-Boyd and colleagues (45, 46) to propose a model in which sterols act at the level of HMGR ubiquitylation, whereas GGOH is involved in post-ubiquitylation step(s). To test this model experimentally and determine whether newly synthesized MVA-derived metabolite(s) other than sterols are indeed not required for the sterol-enhanced ubiquitylation of HMGal, UT-2/HMGal cells were starved overnight for MVA after which sterols and low concentrations of MVA were added in the presence of a proteasome inhibitor, with or without ZA to block sterol synthesis. Specific ubiquitylation of HMGal was assessed by the ratio between the intensity of the ubiquitylated material trailing the 150-kDa HMGal band and the intensity of the immunoprecipitated HMGal. As expected, HMGal ubiquitylation was hardly detected in untreated cells (Fig. 3, Exp. II, lane 1), in cells treated only with ZA (Fig. 3, Exp. I, lane 1), or in cells treated with 2 mm MVA either with or without ZA (Fig. 3, Exp. I and II, lane 3). Unexpectedly, sterols stimulated only slightly the ubiquitylation of HMGal in the absence of exogenous MVA (Fig. 3, Exp. II, lane 2) but more so (1.8-fold) in the presence of ZA (Fig. 3, Exp. I, lane 2). Moreover, addition of sterols together with 2 mm MVA slightly enhanced HMGal ubiquitylation (Fig. 3, Exp. II, lane 4), and this effect was markedly augmented in ZA-treated cells (Fig. 3, Exp. I, lane 4, 2.5-fold increase relative to lane 1; Exp. II, lane 5, 4-fold increase relative to lane 1). As mentioned above, FPP levels rise upon ZA-mediated inhibition (43), thus FPP or FPP-derived GGPP, which synergize the action of sterols in HMGal degradation (Fig. 2) may synergize the action of sterols in HMGal ubiquitylation. Indeed, incubating the cells also with DGBP severely impaired ubiquitylation of HMGal in ZA-treated cells that were also challenged with sterols and low MVA (Fig. 3, Exp. I, lane 5; Exp. II, lane 6). Thus, in MVA-deprived cells sterols alone cannot stimulate HMGal ubiquitylation, and GGPP or a GGPP-derived metabolite is required for the efficient sterol-induced ubiquitylation as well as degradation of HMGal.
FIGURE 3.
Inhibition of protein geranylgeranylation blocks HMGal ubiquitylation. UT-2/HMGal cells were incubated for 20 h in Medium B supplemented with 50 μm lovastatin. The cells were then treated with ZA and/or DGBP and/or GGTI-298, as indicated. Following a 1-h incubation, all dishes received 5 μm MG-132 with or without sterols and/or MVA, as indicated, and were further incubated for 2 h. The cells were lysed in solution D, HMGal was immunoprecipitated (IP) with anti-HMGR membrane region antibodies and successively immunoblotted (IB) with mouse anti-ubiquitin and anti-β-galactosidase (β-Gal) monoclonal antibodies. Aliquots of whole cell lysates (lower panel) were immunoblotted with an antibody that specifically recognizes unprenylated Rap1a.
GGPP is covalently conjugated to the C terminus of many cellular proteins, many of which belong to the family of small GTP-binding proteins. This important modification is essential for the function of these proteins inasmuch as it dictates their correct intracellular localization and interaction(s) with other proteins. With the exception of members of the Rab family of proteins (47), attachment of geranylgeranyl group(s) to all other modified proteins is catalyzed by the enzyme protein gernaylgeranyl transferases I (GGTase I). To examine whether protein geranylgeranylation was required for the sterol-stimulated ubiquitylation of HMGal, the cells were treated with GGTI-298, a GGTase I peptidomimetic that selectively and efficiently inhibits GGTase I activity in cultured cells (48). As shown in Fig. 3, Exp. II, similar to the effect of DGBP, GGTI-298 attenuated >50% the sterol-stimulated ubiquitylation of HMGal in ZA-treated cells (compare lane 7 to 5). An antibody that recognizes only the unprenylated form of the Rap1a protein (SC-1482) was used to gauge the status of protein geranylgeranylation in the cells (Fig. 3, Exp. II, whole cell lysate). As can be seen, the low concentration of MVA, either alone or with sterols, was insufficient to efficiently prenylate Rap1a within 2 h so as to reduce appreciably the immunoreactivity of the unprenylated Rap1a (Fig. 3, Exp. II, lanes 3 and 4, respectively). However, when squalene synthase was inhibited with ZA, the increased levels of FPP were diverted to the synthesis of GGPP, resulting in its increased availability to GGTase-I and subsequently more efficient protein prenylation. This is indicated by the disappearance of the signal of unprenylated Rap1a (Fig. 3, Exp. II, lane 5). Accordingly, DGBP and GGTI-298 effectively inhibited protein geranylgeranylation and restored the immunoreactive signal of unprenylated Rap1a (Fig. 3, Exp. II, lanes 6 and 7). Taken together, these results indicate that a geranylgeranylated protein(s), rather than GGOH, is directly involved in the sterol-simulated ubiquitylation of HMGal.
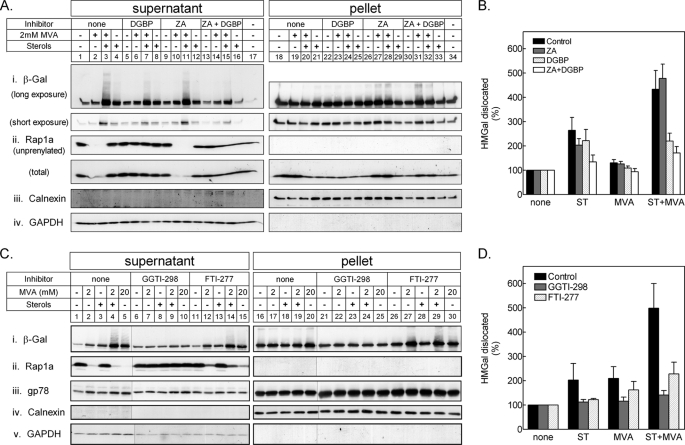
Dislocation of HMGal Requires Protein Prenylation
As with other ERAD substrates, HMGR must dislocate from the ER to the cytosol to be degraded by the proteasome. Based on the finding that GGPP synthesis was required for HMGal degradation (Fig. 2) and protein geranylgeranylation was required for HMGal ubiquitylation (Fig. 3), we next examined whether dislocation was also promoted by protein prenylation. Following incubation in the presence of proteasome inhibitor, the cells were permeabilized with digitonin, fractionated into supernatant (cytosol) and pellet (membrane) fractions, and the degree of HMGal dislocation was calculated as the amount of HMGal found in the supernatant out of the entire HMGal population both in the supernatant and pellet. In agreement with the pulse-chase and ubiquitylation results shown in Figs. 2 and 3, it was clearly seen that very little HMGal was dislocated to the supernatant in the cell that was starved for sterols and MVA (Fig. 4, A and C, i, lane 1; value set as unity). Likewise, low concentrations of MVA had only a minute effect on dislocation (Fig. 4, A and C, i, lane 2). In contrast, incubation with sterols increased HMGal dislocation 2–3-fold (Fig. 4, A and C, i, lanes 4 and 3, respectively), and treatment with sterols together with 2 mm MVA caused an impressive release of HMGal (∼5-fold increase, Fig. 4, A and C, i, lanes 3 and 4, respectively), which was trailed by a smear of higher molecular weight material indicative of ubiquitylated species (Fig. 4A, i, lane 3, see long exposure). Notably, at such low doses of MVA, protein geranylgeranylation was incomplete, as evidenced by the detection of unprenylated Rap1a in the supernatant (Fig. 4C, ii, lanes 2 and 4). Note that Rap1a was not detected in the pellet fraction, because only the prenylated form of this protein is associated with cellular membranes and, thus, not recognized by the antibody specific to the unprenylated Rap1a. Blotting the fractions with an antibody that is insensitive to the prenylation status of Rap1a (SC-65) demonstrated its abundant presence in the pellet as well (Fig. 4A, Rap1a total). Note that the decrease in the amount of unprenylated Rap1a in the supernatant was accompanied by a corresponding increase of total Rap1a in the pellet fraction, as only geranylgeranylated Rap1a partitioned to the membranes. Yet, the sum of Rap1a in the supernatant and the pellet fractions remained constant regardless of the treatment of the cells (data not shown). At 20 mm MVA, HMGal dislocation to the supernatant nearly equaled that obtained upon addition of exogenous sterols with low MVA (Fig. 4C, i, lane 5). Under these conditions, prenylation of Rap1a was complete (Fig. 4C, ii, lane 5). In ZA-treated cells, the release of HMGal to the supernatant following addition of sterols and low MVA was augmented (Fig. 4, A, i, lane 11, and B), in agreement with the effect of ZA on HMGal degradation, ubiquitylation, and protein prenylation (see Fig. 3, Exp. I, lane 4, and Exp. II, lane 5). Importantly, dislocation of HMGal in cells treated with sterols and MVA was severely impeded by DGBP, regardless of whether ZA was also present (Fig. 4, A, i compare lane 7 to 3, and lane 15 to 11, and B). Similarly, HMGal dislocation was nearly abolished in the presence of GGTI-298 (Fig. 4, C, i, compare lanes 9 and 10 to lanes 4 and 5, respectively, and D). The farnesyl transferase (FTase) inhibitor FTI-277 also impaired the release of HMGal to the supernatant but to a lesser extent than did GGTI-298 (Fig. 4C, i, lanes 11–15). Yet, in agreement with previous reports (49), this inhibitor was not very selective to FTase as it also inhibited the protein geranylgeranylation (Fig. 4C, ii, lanes 12, 14, and 15). Moreover, HMGal enhanced degradation in ZA-treated cells was counteracted by DGBP (Fig. 2B), indicating the importance of conversion of FPP to GGPP.
FIGURE 4.
Dislocation of HMGal requires protein geranylgeranylation. A, UT-2/HMGal cells were incubated for 20 h in Medium B supplemented with 50 μm lovastatin. The cells were then treated with ZA, DGBP, or both for 1 h before addition of 65 μm ALLN, sterols, and/or MVA, as indicated. Following an additional 7-h incubation, the cells were permeabilized in Solution C and fractionated by centrifugation, as described under “Experimental Procedures.” Aliquots of the supernatant and pellet fractions were immunoblotted with the indicated antibodies. Unprenylated and total Rap1a was detected with goat (SC-1482) and rabbit (SC-65) antibodies, respectively. B, densitometric analysis of the blots. For each inhibitor, the β-galactosidase immunoreactive signal in A, i (including trailing smear), was divided by the sum of β-galactosidase immunoreactive signals in pellet and supernatant, setting the value as “100%” for cells not treated with either sterols or MVA. Results are the mean ± S.E. of 8 independent experiments. C, UT-2/HMGal cells were set up as in A. Cells were treated for 1 h with either 50 μm GGTI-298 or 50 μm FTI-277 before an additional 3-h incubation with 65 μm ALLN and sterols, 2 mm MVA, or 20 mm MVA, as indicated. Cells were permeabilized and fractionated, as described. Aliquots of supernatant and pellet fractions were immunoblotted with the indicated antibodies. D, densitometric analysis of the blots as in B, except treatment with 20 mm MVA was not quantified. Results are the mean ± S.E. of 3 independent experiments.
Finally, to assess whether prenylated proteins were involved in dislocation of ERAD substrates in general, we examined the distribution between the supernatant and pellet fractions of gp78, which, in addition to being an E3 ubiquitin ligase, is also a short-lived polytopic ER protein (50, 51). As clearly shown in Fig. 4C, iii, the amount in the cytosol of this ERAD substrate was not affected by inhibition of protein geranylgeranylation (Fig. 4C, iii, lanes 6–10) or GGPP synthesis (data not shown). Taken together, these data indicate that a geranylgeranylated protein(s) is specifically involved in the dislocation of HMGal to the cytosol.
Geranylgeranyl-dependent Ubiquitylation Takes Place in the Membrane and Precedes HMGal Dislocation
The results presented in Figs. 3 and 4 indicated that protein geranylgeranylation is required both for ubiquitylation and dislocation of HMGal. In an attempt to dissect in which of these steps the putative geranylgeranylated protein(s) was directly involved, we examined which of these steps was most affected by inhibition of protein geranylgeranylation by evaluating the extent of HMGal dislocation and ubiquitylation in the supernatant, but more importantly in the pellet (membrane) fraction, the site of HMGal residence. As shown in Fig. 5, in MVA-deprived cells, HMGal in the pellet was poorly ubiquitylated (Fig. 5A, lane 10). Upon addition of sterols or a low concentration of MVA, the intensity of ubiquitin immunoreactive material associated with membrane-bound HMGal increased 2.1-fold (Fig. 5A, lane 11) and 1.7-fold (lane 12), respectively. Sterols together with MVA enhanced HMGal ubiquitylation in the pellet fraction nearly 3-fold (Fig. 5A, lane 13). This effect was further enhanced when the cells were also treated with ZA (Fig. 5A, lane 16), resulting in a 40% further increase in specific ubiquitylation relative to cells treated only with sterols and MVA. Moreover, a similar enhanced ubiquitylation of HMGal in cells treated with ZA, sterols, and MVA was also observed in the supernatant fraction. In agreement with Fig. 4, ZA-dependent accumulation of FPP was diverted toward increased synthesis of GGPP leading to the efficient prenylation of Rap1a (Fig. 5B, ii, lane 7). The ZA-stimulated ubiquitylation of HMGal was markedly hindered by DGBP, resulting in ubiquitylation levels similar to those observed in cells treated only with sterols (Fig. 5A, lane 17). GGTI-298 also inhibited HMGal ubiquitylation in the pellet fraction albeit less efficiently then DGBP (Fig. 5A, lane 18). Under all conditions, the amounts of HMGal that dislocated to the supernatant tightly correlated to the extent of HMGal ubiquitylation in the membrane. For example, the dislocation of HMGal in cells treated with sterols, MVA, and ZA was 2-fold more pronounced than in cells treated only with sterol and MVA (Fig. 5B, i, compare lane 7 to 4). Thus, it appeared that the ubiquitylation and dislocation of HMGal were tightly coupled and inseparable events that occur with similar kinetics. However, because the results indicated that HMGal ubiquitylation occurs in the membrane prior to dislocation and given this order of events, the putative geranylgeranylated protein is likely involved in HMGal ubiquitylation or a preceding step rather than in dislocation.
FIGURE 5.
Geranylgeranylation-dependent ubiquitylation of HMGal takes place in the membrane and precedes dislocation. UT-2/HMGal cells were incubated for 20 h in Medium B supplemented with 50 μm lovastatin. The cells were then treated with ZA, and/or DGBP, and/or GGTI-298 for 1 h before the addition of 5 μm MG-132, sterols, and/or MVA, as indicated. Cells were permeabilized and fractionated as described under “Experimental Procedures.” A, HMGal was immunoprecipitated (IP) from fractions with anti-HMGR membrane region antibodies and sequentially immunoblotted (IB) with mouse anti-ubiquitin and anti-β-galactosidase monoclonal antibodies. B, aliquots of the supernatant and pellet fractions were directly analyzed by immunoblotting with the indicated antibodies.
Dislocation and Degradation of HMGal Is Stimulated by a MVA-derived Nonsterol Metabolite Upstream of Squalene Epoxide but Downstream of Farnesyl Pyrophosphate
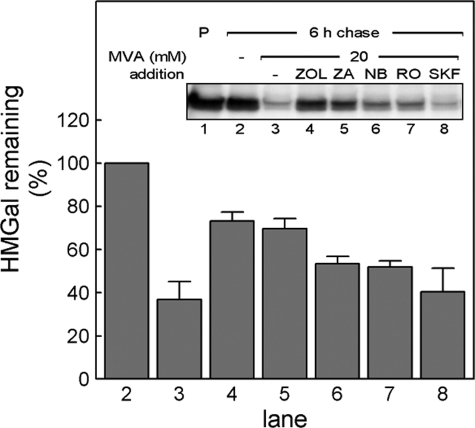
It has been shown that lanosterol, the first sterol intermediate in the MVA pathway (Fig. 1), induces ubiquitylation and degradation of HMGR (26). However, earlier evidence, using the squalene epoxidase inhibitor NB-598, indicated that MVA-derived intermediate(s) upstream of squalene epoxide is also involved in HMGR degradation (31, 52). In light of the findings regarding GGPP described above, we revisited these earlier results and examined which additional MVA-derived metabolites may be involved in HMGR turnover. To that end, UT-2/HMGal cells starved for sterols and MVA were pulse-labeled with [35S]methionine/cysteine and chased in the presence of high concentrations of MVA together with inhibitors specific to several MVA pathway enzymes. Again, 20 mm MVA stimulated the degradation of HMGal (Fig. 6, compare lane 3 to 2), whereas inhibition of farnesyl diphosphate synthase with ZOL (53) (see Fig. 1) significantly blocked the MVA-accelerated degradation of HMGal (Fig. 6, lane 4), indicating that metabolite(s) produced in the pathway downstream of FPP signal for HMGal degradation. Blocking squalene synthase activity with ZA (Fig. 6, lane 5) was nearly as effective in inhibiting HMGal degradation as was ZOL. Inhibiting squalene epoxidase activity with NB-598 (Fig. 1) also had some inhibitory effect on the MVA-accelerated degradation of HMGal (Fig. 6, NB, lane 6). Nevertheless, accumulation of squalene in the cells still allowed degradation of ∼50% of the newly synthesized HMGal population, consistent with previous reports (31, 52). Inhibiting enzymatic steps further downstream (Fig. 1), such as cyclization of squalene epoxide with RO 48-8071 (Fig. 6, RO, lane 7) (54) or demethylation of lanosterol with SKF 104976 (Fig. 6, SKF, lane 8) (55) was almost ineffective in inhibiting MVA-accelerated degradation of HMGal. The rapid MVA-accelerated elimination of HMGal in SKF 104976-treated cells is consistent with the intracellular buildup of 24,25-dihydrolanosterol, subsequent to inhibition of lanosterol 14α-demethylase, which leads to HMGR degradation (26, 45, 46, 56).
FIGURE 6.
MVA-derived nonsterol product(s) accelerate degradation of HMGal in UT-2 cells. UT-2/HMGal cells were depleted of sterols and MVA by 20 h incubation in Medium B containing 50 μm lovastatin. The cells were pulse-labeled and chased for 6 h in Medium B supplemented without (lane 2) or with 20 mm MVA (lane 3) plus the indicated inhibitors: 250 μm ZOL (lanes 4), 150 μm ZA (lane 5), 10 μm NB-598 (lane 6), 5 μm RO-48–8071 (lane 7), 10 μm SKF-104976 (lane 8). HMGal was immunoprecipitated with anti-HMGR membrane region polyclonal antibodies. Densitometric analysis (bars) represents HMGal remaining at the end of the chase relative to cells chased without MVA (lane 2). Results are the mean ± S.E. of 4 independent experiments.
That MVA-derived metabolites downstream of FPP but upstream of lanosterol were capable of promoting HMGR degradation was further examined in two additional cell lines, met-18b-2 and LP-90. The first, met-18b-2, is a CHO cell-derived line that was originally isolated by Faust and Krieger (38). These cells express a gain-of-function mutation in the MCT-1 monocarboxylate transporter that converts it into a high capacity transporter for MVA (39). Therefore, met-18b-2 cells allow studying MVA-dependent processes at much lower concentrations of exogenous MVA than parental CHO cells (38). Indeed, compared with HMGal in UT-2 cells or endogenous HMGR in wild-type CHO cells (data not shown and Ref. 40), accelerated degradation of endogenous HMGR in met-18b-2 cells was readily commenced by as little as 5 mm MVA (Fig. 7A, lanes 5–8). Whereas ZA inhibited the MVA-stimulated turnover of HMGR altogether (Fig. 7A, lanes 9–12), NB-598 only partially inhibited this degradation (Fig. 7A, lanes 14–17).
FIGURE 7.
Blocking the MVA pathway with squalene epoxidase inhibitor NB-598 permits ubiquitylation and degradation of HMGR. A, sterol-depleted met-18b-2 cells were pulse-labeled and chased in Medium B supplemented with 5 mm MVA in the absence or presence of 150 μm ZA or 10 μm NB-598, as indicated. HMGR was immunoprecipitated (IP) with anti-HMGR membrane region antibodies. B, LP-90 cells were pulse labeled in the presence of 90 μm lovastatin. The cells were washed and chased in the absence (lanes 6–20) or presence (lanes 1–5) of 90 μm lovastatin. Cells in lanes 11–15 were chased in the presence of 150 μm ZA, and cells in lanes 16–20 were chased in the presence of 10 μm NB-598. C, LP-90 cells, maintained in 90 μm lovastatin, were washed with PBS and further incubated for the indicated time in fresh medium without lovastatin, in the absence or presence of 10 μm NB-598 or 150 μm ZA. Cells were lysed in Solution D and HMGR was immunoprecipitated with anti-HMGR membrane region antibodies. Immune complexes were analyzed by immunoblotting (IB) sequentially with mouse anti-ubiquitin and anti-HMGR monoclonal antibodies.
The second cell line we have examined is LP-90, a line of lovastatin-resistant CHO-derived cells that continuously grow in the presence of lovastatin. These cells express extremely high levels of endogenous HMGR protein, most of which is catalytically inactive because of the lovastatin in their medium (40). When inhibition is abruptly alleviated by washing the drug away, the intracellular surge in MVA and subsequent high flux of intermediates through the pathway causes rapid ubiquitylation and degradation of the endogenous HMGR (16). Clearly, relative to cells that were continuously incubated with lovastatin (Fig. 7B, lanes 1-5), when lovastatin was washed off endogenous HMGR was rapidly degraded (Fig. 7B, lanes 6–10), and treating these “lovastatin off” cells with ZA severely inhibited this degradation (Fig. 7B, lanes 11–15). Again, addition of NB-598 had only a minor effect on HMGR turnover (Fig. 7B, lanes 16–20). These disparate effects of NB-598 and ZA were also evident in the ubiquitylation and elimination of the entire pool of HMGR in the lovastatin off LP-90 cells (Fig. 7C, lanes 5–8 and 9–12, respectively). To ascertain that NB-598 effectively prevented sterol synthesis by inhibiting squalene epoxidase, we measured the incorporation of [3H]acetate into nonsaponifiable lipids in lovastatin off LP-90 cells. These experiments demonstrated that NB-598 inhibited sterols synthesis more than 98% with a corresponding quantitative accumulation of squalene (see Table 1). Collectively, these results, which are in full agreement with similar data obtained in wild-type CHO cells (31), indicate that although endogenously synthesized sterols contribute to the degradation of HMGR, nonsterol MVA-derived product(s) downstream of FPP but upstream of squalene epoxide, which is not produced in ZA-treated cells but accumulates in NB-598-treated cells, can signal effectively for HMGR degradation. It appears that this nonsterol MVA-derived product is squalene, yet attempts to test whether direct addition of squalene to intact cells can lead to HMGR degradation have failed (data not shown), likely because squalene is totally insoluble in aqueous solutions.
TABLE 1.
Effect of ZA or NB-598 on synthesis of sterols and squalene in LP-90 cells
LP-90 cells were labeled with [3H]acetate and nonsaponifiable lipids were analyzed, as described under “Experimental Procedures.” Results, given in nmole [3H]acetate incorporated/mg of cell protein/h, are the mean of duplicate incubations.
| Experiment | Lovastatin on | Lovastatin off |
||
|---|---|---|---|---|
| ZA | NB-598 | |||
| 1 | ||||
| Sterols | 2.30 | 15.28 | 0.45 | 0.24 |
| Squalene | 0.00 | 0.02 | 0.03 | 18.47 |
| 2 | ||||
| Sterols | 0.92 | 21.76 | 1.17 | 0.36 |
| Squalene | 0.04 | 0.05 | 0.04 | 18.28 |
| 3 | ||||
| Sterols | 0.91 | 28.16 | 1.01 | |
| Squalene | 0.01 | 0.04 | 23.48 | |
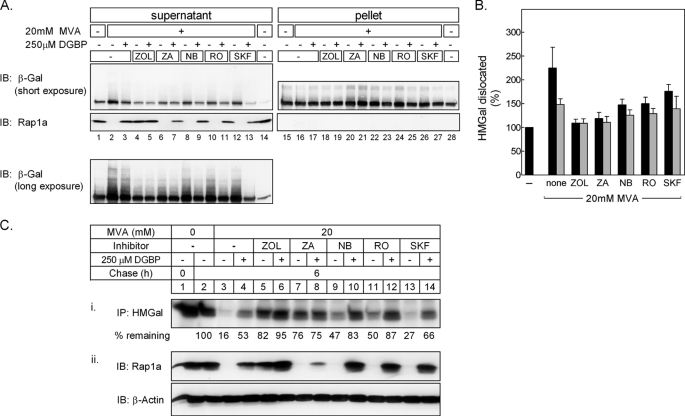
Finally, we examined whether the apparent squalene-stimulated degradation of HMGR correlated with its enhanced ubiquitylation, dislocation, and degradation. Again, in these experiments, we used the UT-2/HMGal cells. Fig. 8 shows that 20 mm MVA caused nearly 2.5-fold increase in HMGal dislocation to the supernatant, and as shown above (Figs. 4 and 5), this dislocation was ∼50% inhibited by DGBP (Fig. 8, A, supernatant, lanes 1–3, and B). These changes correlated with the prenylation status of Rap1a, further supporting the notion that GGPP production is required for HMGal dislocation. Notably, the dislocated HMGal was polyubiquitylated, as demonstrated by the smear of high molecular mass HMGal species that trail the main 150-kDa band (Fig. 8A, see long exposure, lower panel). Moreover, inhibition of FPP synthesis with ZOL blocked ∼90% the MVA-induced ubiquitylation and dislocation of HMGal (Fig. 8, lanes 4 and 5), resembling HMGal dislocation in MVA-depleted cells (Fig. 8A, lane 1). Indeed, ZOL significantly inhibited the MVA-stimulated degradation of HMGal (Fig. 8C, i, compare lane 5 to 3), and this effect was augmented by DGBP (Fig. 8C, i, lane 6), in correlation with the inhibition of Rap1a geranylgeranylation (Fig. 8C, ii, compare lane 5 to 6). In cells treated with ZA, only 10% more HMGal was extracted from the ER compared with MVA-starved cells (Fig. 8, A, compare lane 6 to 1, and B). Relative to the stronger effect of ZOL, the slightly enhanced dislocation of HMGal in ZA-treated cells likely stems from deflecting the accumulating FPP, which is not formed in ZOL-treated cells, toward GGPP production. However, simultaneous addition of ZA and DGBP only slightly decreased HMGal dislocation, in comparison to cells treated only with ZA (Fig. 8, A, compare lane 7 to 6, and B). This likely reflects the inability of DGBP to inhibit GGPP production under such massive amounts of accumulating FPP. This finding was supported by the lack of DGBP effect on the degree of inhibition of HMGal degradation (Fig. 8C, i, lanes 7 and 8), and its minor effect on the inhibition of Rap1a geranylgeranylation (Fig. 8C, ii, lane 8) in cells treated with ZA. In agreement with the effects of NB-598, RO 48-8071, and SKF 104976 on HMGal degradation (Fig. 6), these MVA pathway inhibitors allowed significant MVA-stimulated HMGal ubiquitylation, dislocation, and degradation, which were all inhibited by DGBP (Fig. 8). Hence, in addition to lanosterol that can induce rapid ubiquitylation and degradation of HMGR (26), our results show that a nonsterol(s) synthesized upstream of lanosterol, more precisely upstream of squalene epoxide and downstream of FPP, can also signal for HMGal ubiquitylation and dislocation. The diminishing effect of simultaneous inhibition of GGPP synthesis on HMGal dislocation shown here (Fig. 8) and throughout this study indicates that GGPP production and geranylgeranylation of protein(s) are required for HMGal ubiquitylation, dislocation, and degradation, regardless of whether the ERAD steps synergized by GGPP are triggered by nonsterols, such as squalene, or by sterols.
FIGURE 8.
Geranylgeranyl-PP synthesis is required for nonsterol-stimulated dislocation and degradation of HMGal. UT-2/HMGal cells were depleted of sterols and MVA by 20 h incubation in Medium B containing 50 μm lovastatin. The cells were treated with 250 μm ZOL, 150 μm ZA, 10 μm NB-598, 5 μm RO-48-8071, or 10 μm SKF-104976 with or without 250 μm DGBP for 1 h before adding 65 μm ALLN and 20 mm MVA, as indicated. Following an additional 7-h incubation, the cells were permeabilized and fractionated. A, aliquots of supernatant and solubilized pellet fraction were immunoblotted (IB) with anti-β-galactosidase and anti-unprenylated Rap1a antibodies. B, densitometric analysis of the blots. The β-galactosidase immunoreactive signal in supernatants in upper panel A (including trailing smear) was divided by the sum of β-galactosidase immunoreactive signal in supernatant and pellet, setting the value for untreated cells as “100% dislocation.” Black bars, cells not treated with DGBP; gray bars, DGBP-treated cells. Results are the mean ± S.E. of 3 independent experiments. C, UT-2/HMGal cells were set up as in A. The cells were pulse-labeled and chased for 6 h in Medium B supplemented without (lane 2) or with 20 mm MVA (lanes 3–14) in the absence or presence of DGBP and the indicated inhibitors: 250 μm ZOL, 150 μm ZA, 10 μm NB-598, 5 μm RO-48–8071, or 10 μm SKF-104976. (i) HMGal was immunoprecipitated (IP) with anti-HMGR membrane region polyclonal antibodies. The remaining HMGal at the end of the chase (%) is indicated. (ii) Aliquots of the labeled cell lysates were immunoblotted with the indicated antibodies.
DISCUSSION
In this study, we concentrated on the MVA-derived metabolite(s) that signal for HMGR degradation and their effect on ubiquitylation and dislocation. Following the fate of HMGal stably transfected in the MVA auxotrophic UT-2 cells, we show that under conditions of acute MVA deprivation this protein turned over slowly even when excess exogenous sterols were supplied. This supports the notion that a nonsterol isoprenoid(s) is required for, and cooperates with, sterols in HMGR degradation (30). Our results indicate that this putative nonsterol promotes degradation by facilitating both ubiquitylation and dislocation. Only when exogenous MVA was metabolized intracellularly, even at concentrations too low to allow efficient sterol synthesis, was HMGal ubiquitylated and dislocated out of the ER membrane and degraded. To ensure that the exogenous MVA was not converted to sterols, we inhibited the activity of squalene synthase with ZA (43). Under these conditions, HMGal ubiquitylation, dislocation, and degradation, as well as protein geranylgeranylation in general, was more efficient than in cells treated only with sterols and limiting MVA. These effects were markedly diminished upon inhibiting the activity of GGPP synthase with DGBP (44). Together, these results indicate that GGPP is the nonsterol that acts synergistically with sterols to promote HMGR degradation. Moreover, because inhibition of protein geranylgeranylation with the GGTase I peptidomimetic GGTI-298 substantially blocked HMGal ubiquitylation and dislocation, the results strongly suggest that a geranylgeranylated protein(s) is the sterols' accomplice in accelerating HMGR degradation, providing the first experimental evidence for the 20-year-old hypothesis on the involvement of prenylated proteins in HMGR turnover in mammalian cells (2). Although its identity is yet to be revealed, this putative geranylgeranylated protein appears to be specifically involved in HMGR turnover and not in the degradation of another ERAD substrate, gp78. Blocking protein farnesylation with FTI-277 also inhibited the sterol-stimulated dislocation of HMGal, but these results could be attributed to nonselective inhibition of GGTase I as prenylation of Rap1a, a strict substrate of GGTase I, was hampered by FTI-277 (Fig. 4; see also Ref. 49). Protein geranylgeranylation by GGTase-II (Rab GGTase) was not involved in HMGR degradation because inhibition of this enzyme by specific inhibitors (57, 58) had no effect on HMGal degradation (data not shown).
Because GGPP is involved in both ubiquitylation and dislocation of HMGal, our results differ from the model presented by DeBose-Boyd and colleagues (17, 45, 46), which suggests that sterols stimulate HMGR ubiquitylation, whereas MVA-derived GGOH is involved in the dislocation of the ubiquitylated HMGR out of the ER membrane. These differences may be due to our use of HMGR-deficient cells, which are strictly dependent on exogenous MVA, whereas DeBose-Boyd and colleagues (17) studied cells that express endogenous HMGR, making it more difficult to block synthesis and deplete endogenous MVA stores to observe processes that require only trace amounts of GGPP. We were unable to dissociate between ubiquitylation and dislocation and, in our hands, every manipulation that affected HMGal ubiquitylation also affected its dislocation and vice versa, suggesting that these steps are tightly coupled. Yet, our findings that ubiquitylated HMGal species accumulated in the membrane fraction of ZA-treated cells, and that this was largely prevented by DGBP (Fig. 5) strongly suggest that ubiquitylation takes place while HMGal is still in the ER membrane. This is also supported by the results in Fig. 7C where HMGR is heavily ubiquitylated but remains stable in ZA-treated lovastatin off LP-90 cells. Thus, GGPP is evidently required for ubiquitylation or a preceding step. Interestingly, in a recent study Garza et al. (59) have demonstrated that degradation of Hmg2p in Saccharomyces cerevisiae is stimulated by GGPP rather than GGOH, and that GGPP is involved in Hmg2p ubiquitylation but not dislocation. However, in the yeast, prevention of protein geranylgeranylation did not hamper the effect of GGPP, indicating that prenylated protein(s) is(are) not involved in Hmg2p turnover (59). Although these results appear to contrast the data presented here, it should be emphasized that Hmg2p stability in yeast is controlled mainly by FPP-derived nonsterol (GGPP) rather than by sterols (59, 60). Thus, unlike the mammalian reductase, degradation of Hmg2p in yeast does not involve co-signaling by different MVA-derived metabolites, and GGPP may fulfill all the requirements as a metabolic degradation signal. Sever et al. (17) also reported that the degradation of HMGR was accelerated by the free alcohol geranylgeraniol (GGOH). Through the activity of a salvage pathway, in which isoprenols are converted to their respective pyrophosphates (61), cells can utilize GGOH as a source for protein geranylgeranylation, including prenylation of Rap1a (48, 62). Thus, by conversion to GGPP and attachment to protein(s), GGOH may well support HMGR degradation.
It has been reported that lanosterol, the first sterol intermediate of the MVA pathway, can stimulate the degradation of HMGR (26). Therefore, a significant finding in our study is that acceleration of HMGR and HMGal degradation by excess exogenous MVA proceeded even when synthesis of lanosterol, but not of squalene, was blocked by inhibitors of either squalene epoxidase (NB-598) or squalene epoxide cyclase (RO 48-8071). These results, which reproduce earlier observations (31), indicate that metabolite(s) in the MVA pathway downstream of FPP but upstream of lanosterol (i.e. presqualene diphosphate and/or squalene) can signal for HMGR degradation. Inhibition of squalene epoxidation with NB-598 may cause a buildup of presqualene diphosphate, a potent intracellular signaling molecule (63, 64). Consistent with our results, Giron et al. (52) have also reported that inhibition of squalene conversion to downstream sterols did not prevent MVA-mediated suppression of HMGR activity in perforated CHO cells. Moreover, Song and DeBose-Boyd (27) have shown that HMGR ubiquitylation and degradation is stimulated by γ- and δ-tocotrienols, and that the crucial determinant for the action of these molecules is their GGPP-derived isoprenoid side chain, which resembles the structure of squalene and therefore may mimic its action. Tocopherols, which possess a saturated side chain, have no effect on HMGR ubiquitylation or degradation. Remarkably, acceleration of HMGR degradation by tocotrienols also required MVA for their action (27). Likewise, our results indicate that GGPP synergizes the activity of endogenously synthesized squalene (Fig. 8). Based on these observations and the findings that stimulation of HMGR ubiquitylation and degradation by unnatural molecules, such as Apomine, also depends on low concentrations of MVA, we predict that the action of any elicitor of HMGR degradation depends on GGPP, most likely on the function of a geranylgeranylated protein.
Squalene is the first precursor for the sterologenic branch of the MVA pathway (Fig. 1) and metabolic conversion of one molecule of squalene to cholesterol requires expensive investment of 15 reducing equivalents, in the form of NAD(P)H, and 11 molecules of O2 (65). For certain eukaryotes, such as insects but not mammals, the sterologenic branch is entirely dispensable, and sterols are obtained from the diet (66–68). Recently it has been demonstrated that the ER enzyme squalene epoxidase, which catalyzes the first oxygenation step in cholesterol synthesis, is subject to cholesterol-stimulated ERAD, leading to accumulation of squalene (69). Therefore, it is plausible that intracellular squalene levels would provide a reliable readout for the flux through the sterologenic branch and negative feedback on the major rate-limiting enzyme HMGR so as to block further energetic investment in the synthesis of sterologenic precursors when they are not required. This, of course, does not preclude regulation of HMGR degradation by other sterol intermediates produced further down the sterologenic branch (26). It is therefore very appealing that a central biosynthetic pathway such as the MVA pathway is feedback regulated through degradation of ER proteins at two critical points: HMGR, the point of entry to the pathway where essential metabolites must be produced intracellularly, and squalene epoxidase, the point of entry to an energy costly branch that can be dispensed with by obtaining its products from the diet. Because production of MVA must take into account all its derived metabolites, ERAD of HMGR is regulated by integrating a nonsterol signal, in the form of the geranylgeranyl moiety of a protein, and a sterologenic signal in the form of squalene.
Acknowledgments
We thank Merck Laboratories, Banyu Pharmaceuticals, Terpenoid Therapeutics, Roche Applied Science, and SmithKline Beecham Pharmaceuticals for their generous gifts of inhibitors. We also thank Raymond Hohl for DGBP, Miguel Seabra for GGT-II inhibitors, Ron Kopito and Richard Wojcikiewicz for antibodies, Patrick Casey (Duke University) for good advice, and Shoshana Bar-Nun (Tel Aviv University) for fruitful discussions and critical reading of the manuscript.
This work was supported in part by a grant from the Chief Scientist of the Israel Ministry of Health.
- MVA
- mevalonate
- ALLN
- N-acetyl-leucinyl-leucinyl-norleucinal
- DGBP
- digeranyl bisphosphonate
- ER
- endoplasmic reticulum
- ERAD
- ER-associated protein degradation
- FPP
- farnesyl pyrophosphate
- FTase
- farnesyl transferase
- GGPP
- geranylgeranyl pyrophosphate
- GGTase
- geranylgeranyl transferase
- HMG-CoA
- 3-hydroxy-3-methylglutaryl coenzyme A
- HMGR
- 3-hydroxy-3-methylglutaryl-coenzyme A reductase
- HMGal
- fusion protein between HMGR membrane region and β-galactosidase
- LPDS
- lipoprotein-deficient serum
- ZA
- zaragozic acid
- ZOL
- zoledronic acid.
REFERENCES
- 1. Schroepfer G. J., Jr. (1981) Annu. Rev. Biochem. 50, 585–621 [DOI] [PubMed] [Google Scholar]
- 2. Goldstein J. L., Brown M. S. (1990) Nature 343, 425–430 [DOI] [PubMed] [Google Scholar]
- 3. Siperstein M. D., Guest M. J. (1960) J. Clin. Invest. 39, 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Siperstein M. D., Fagan V. M. (1966) J. Biol. Chem. 241, 602–609 [PubMed] [Google Scholar]
- 5. Liscum L., Cummings R. D., Anderson R. G., DeMartino G. N., Goldstein J. L., Brown M. S. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 7165–7169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brown D. A., Simoni R. D. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 1674–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roitelman J., Olender E. H., Bar-Nun S., Dunn W. A., Jr., Simoni R. D. (1992) J. Cell Biol. 117, 959–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liscum L., Finer-Moore J., Stroud R. M., Luskey K. L., Brown M. S., Goldstein J. L. (1985) J. Biol. Chem. 260, 522–530 [PubMed] [Google Scholar]
- 9. Istvan E. S., Palnitkar M., Buchanan S. K., Deisenhofer J. (2000) EMBO J. 19, 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brown M. S., Goldstein J. L. (1980) J. Lipid Res. 21, 505–517 [PubMed] [Google Scholar]
- 11. Gil G., Faust J. R., Chin D. J., Goldstein J. L., Brown M. S. (1985) Cell 41, 249–258 [DOI] [PubMed] [Google Scholar]
- 12. Skalnik D. G., Narita H., Kent C., Simoni R. D. (1988) J. Biol. Chem. 263, 6836–6841 [PubMed] [Google Scholar]
- 13. Hampton R. Y., Koning A., Wright R., Rine J. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng H. H., Xu L., Kumagai H., Simoni R. D. (1999) J. Biol. Chem. 274, 17171–17178 [DOI] [PubMed] [Google Scholar]
- 15. Doolman R., Leichner G. S., Avner R., Roitelman J. (2004) J. Biol. Chem. 279, 38184–38193 [DOI] [PubMed] [Google Scholar]
- 16. Ravid T., Doolman R., Avner R., Harats D., Roitelman J. (2000) J. Biol. Chem. 275, 35840–35847 [DOI] [PubMed] [Google Scholar]
- 17. Sever N., Song B. L., Yabe D., Goldstein J. L., Brown M. S., DeBose-Boyd R. A. (2003) J. Biol. Chem. 278, 52479–52490 [DOI] [PubMed] [Google Scholar]
- 18. Song B. L., Sever N., DeBose-Boyd R. A. (2005) Mol. Cell 19, 829–840 [DOI] [PubMed] [Google Scholar]
- 19. Bonifacino J. S., Weissman A. M. (1998) Annu. Rev. Cell Dev. Biol. 14, 19–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Elkabetz Y., Kerem A., Tencer L., Winitz D., Kopito R. R., Bar-Nun S. (2003) J. Biol. Chem. 278, 18922–18929 [DOI] [PubMed] [Google Scholar]
- 22. Hampton R. Y., Garza R. M. (2009) Chem. Rev. 109, 1561–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wojcikiewicz R. J., Pearce M. M., Sliter D. A., Wang Y. (2009) Cell Calcium 46, 147–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leichner G. S., Avner R., Harats D., Roitelman J. (2009) Mol. Biol. Cell 20, 3330–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bar-Nun S. (2005) Curr. Top. Microbiol. Immunol. 300, 95–125 [DOI] [PubMed] [Google Scholar]
- 26. Song B. L., Javitt N. B., DeBose-Boyd R. A. (2005) Cell Metab. 1, 179–189 [DOI] [PubMed] [Google Scholar]
- 27. Song B. L., DeBose-Boyd R. A. (2006) J. Biol. Chem. 281, 25054–25061 [DOI] [PubMed] [Google Scholar]
- 28. Berkhout T. A., Simon H. M., Patel D. D., Bentzen C., Niesor E., Jackson B., Suckling K. E. (1996) J. Biol. Chem. 271, 14376–14382 [DOI] [PubMed] [Google Scholar]
- 29. Roitelman J., Masson D., Avner R., Ammon-Zufferey C., Perez A., Guyon-Gellin Y., Bentzen C. L., Niesor E. J. (2004) J. Biol. Chem. 279, 6465–6473 [DOI] [PubMed] [Google Scholar]
- 30. Nakanishi M., Goldstein J. L., Brown M. S. (1988) J. Biol. Chem. 263, 8929–8937 [PubMed] [Google Scholar]
- 31. Roitelman J., Simoni R. D. (1992) J. Biol. Chem. 267, 25264–25273 [PubMed] [Google Scholar]
- 32. Correll C. C., Ng L., Edwards P. A. (1994) J. Biol. Chem. 269, 17390–17393 [PubMed] [Google Scholar]
- 33. Bradfute D. L., Simoni R. D. (1994) J. Biol. Chem. 269, 6645–6650 [PubMed] [Google Scholar]
- 34. Meigs T. E., Simoni R. D. (1997) Arch. Biochem. Biophys. 345, 1–9 [DOI] [PubMed] [Google Scholar]
- 35. Goldstein J. L., Basu S. K., Brown M. S. (1983) Methods Enzymol. 98, 241–260 [DOI] [PubMed] [Google Scholar]
- 36. Mosley S. T., Brown M. S., Anderson R. G., Goldstein J. L. (1983) J. Biol. Chem. 258, 13875–13881 [PubMed] [Google Scholar]
- 37. Engfelt W. H., Shackelford J. E., Aboushadi N., Jessani N., Masuda K., Paton V. G., Keller G. A., Krisans S. K. (1997) J. Biol. Chem. 272, 24579–24587 [DOI] [PubMed] [Google Scholar]
- 38. Faust J., Krieger M. (1987) J. Biol. Chem. 262, 1996–2004 [PubMed] [Google Scholar]
- 39. Garcia C. K., Goldstein J. L., Pathak R. K., Anderson R. G., Brown M. S. (1994) Cell 76, 865–873 [DOI] [PubMed] [Google Scholar]
- 40. Ravid T., Avner R., Polak-Charcon S., Faust J. R., Roitelman J. (1999) J. Biol. Chem. 274, 29341–29351 [DOI] [PubMed] [Google Scholar]
- 41. Engfelt W. H., Masuda K. R., Paton V. G., Krisans S. K. (1998) J. Lipid Res. 39, 2182–2191 [PubMed] [Google Scholar]
- 42. Kim C. M., Goldstein J. L., Brown M. S. (1992) J. Biol. Chem. 267, 23113–23121 [PubMed] [Google Scholar]
- 43. Bergstrom J., Kurtz M., Rew D., Amend A., Karkas J. D., Bostedor R. G., Bansal V., Dufresne C., VanMiddlesworth F., Hensens O., Liesch J., Zink D., Wilson K., Onishi J., Milligan J., Bills G., Kaplan L., Omstead M., Jenkins R., Huang L., Meinz M., Quinn L., Burgh R., Kong Y., Mochales S., Mojena M., Martin I., Pelaez F., Diez M., Alberts A. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wiemer A. J., Tong H., Swanson K. M., Hohl R. J. (2007) Biochem. Biophys. Res. Commun. 353, 921–925 [DOI] [PubMed] [Google Scholar]
- 45. DeBose-Boyd R. A. (2008) Cell Res. 18, 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jo Y., Debose-Boyd R. A. (2010) Crit. Rev. Biochem. Mol. Biol. 45, 185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Leung K. F., Baron R., Seabra M. C. (2006) J. Lipid Res. 47, 467–475 [DOI] [PubMed] [Google Scholar]
- 48. McGuire T. F., Qian Y., Vogt A., Hamilton A. D., Sebti S. M. (1996) J. Biol. Chem. 271, 27402–27407 [DOI] [PubMed] [Google Scholar]
- 49. Sebti S. M., Hamilton A. D. (2000) Methods Enzymol. 325, 381–388 [DOI] [PubMed] [Google Scholar]
- 50. Shen Y., Ballar P., Apostolou A., Doong H., Fang S. (2007) Biochem. Biophys. Res. Commun. 352, 919–924 [DOI] [PubMed] [Google Scholar]
- 51. Shmueli A., Tsai Y. C., Yang M., Braun M. A., Weissman A. M. (2009) Biochem. Biophys. Res. Commun. 390, 758–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Giron M. D., Havel C. M., Watson J. A. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 6398–6402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dunford J. E., Thompson K., Coxon F. P., Luckman S. P., Hahn F. M., Poulter C. D., Ebetino F. H., Rogers M. J. (2001) J. Pharmacol. Exp. Ther. 296, 235–242 [PubMed] [Google Scholar]
- 54. Morand O. H., Aebi J. D., Dehmlow H., Ji Y. H., Gains N., Lengsfeld H., Himber J. (1997) J. Lipid Res. 38, 373–390 [PubMed] [Google Scholar]
- 55. Mayer R. J., Adams J. L., Bossard M. J., Berkhout T. A. (1991) J. Biol. Chem. 266, 20070–20078 [PubMed] [Google Scholar]
- 56. Lange Y., Ory D. S., Ye J., Lanier M. H., Hsu F. F., Steck T. L. (2008) J. Biol. Chem. 283, 1445–1455 [DOI] [PubMed] [Google Scholar]
- 57. Coxon F. P., Ebetino F. H., Mules E. H., Seabra M. C., McKenna C. E., Rogers M. J. (2005) Bone 37, 349–358 [DOI] [PubMed] [Google Scholar]
- 58. Baron R. A., Tavaré R., Figueiredo A. C., Błazewska K. M., Kashemirov B. A., McKenna C. E., Ebetino F. H., Taylor A., Rogers M. J., Coxon F. P., Seabra M. C. (2009) J. Biol. Chem. 284, 6861–6868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Garza R. M., Tran P. N., Hampton R. Y. (2009) J. Biol. Chem. 284, 35368–35380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gardner R. G., Shan H., Matsuda S. P., Hampton R. Y. (2001) J. Biol. Chem. 276, 8681–8694 [DOI] [PubMed] [Google Scholar]
- 61. Crick D. C., Andres D. A., Waechter C. J. (1997) Biochem. Biophys. Res. Commun. 237, 483–487 [DOI] [PubMed] [Google Scholar]
- 62. Ownby S. E., Hohl R. J. (2003) Lipids 38, 751–759 [DOI] [PubMed] [Google Scholar]
- 63. Levy B. D., Petasis N. A., Serhan C. N. (1997) Nature 389, 985–990 [DOI] [PubMed] [Google Scholar]
- 64. Levy B. D., Serhan C. N. (2002) Cell Mol. Life Sci. 59, 729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Faust J. R., Trzaskos J. M., Gaylor J. L. (1988) in Biology of Cholesterol (Yeagle P. L. eds) pp. 19–38, CRC Press, Boca Raton, FL [Google Scholar]
- 66. Clark A. J., Bloch K. (1959) J. Biol. Chem. 234, 2578–2582 [PubMed] [Google Scholar]
- 67. Behmer S. T., Nes D. W. (2003) Adv. Insect Physiol. 31, 1–72 [Google Scholar]
- 68. Bellés X., Martín D., Piulachs M. D. (2005) Annu. Rev. Entomol. 50, 181–199 [DOI] [PubMed] [Google Scholar]
- 69. Gill S., Stevenson J., Kristiana I., Brown A. J. (2011) Cell Metab. 13, 260–273 [DOI] [PubMed] [Google Scholar]