Abstract
Activating transcription factor (ATF)/cAMP-response element (CRE)-binding (CREB) proteins induce the CRE-mediated gene transcription depending on the cAMP stimulation. cAMP-dependent signaling oscillates in a circadian manner, which in turn also sustains core oscillation machinery of the circadian clock. Here, we show that among the ATF/CREB family proteins, ATF4 is essential for the circadian expression of the Period2 (Per2) gene, a key component of the circadian clock. Transcription of the Atf4 gene was regulated by core components of the circadian clock, and its expression exhibited circadian oscillation in mouse tissues as well as embryonic fibroblasts. ATF4 bound to the CRE of the Per2 promoter in a circadian time-dependent manner and periodically activated the transcription of the Per2 gene. Consequently, the oscillation of the Per2 expression was attenuated in embryonic cells prepared from Atf4-null mice. Furthermore, the loss of ATF4 also disrupted the rhythms in the expression of other clock genes. These results suggest that ATF4 is a component responsible for sustaining circadian oscillation of CRE-mediated gene expression and also constitute a molecular link connecting cAMP-dependent signaling to the circadian clock.
Keywords: Cyclic AMP (cAMP), Gene Expression, Microarray, Signal Transduction, Transcription, Activating Transcription Factor-4, Circadian Rhythm, Clock Genes
Introduction
Genetic and molecular approaches have identified a basic mechanism of the circadian oscillator that is governed by interconnected transcriptional and translational feedback loops (1, 2). Gene products of Clock and Bmal1 (also known as Arntl) form a heterodimer that activates the transcription of Period (Per) and Cryptochrome (Cry) genes. Once PER and CRY proteins have reached a critical concentration, they attenuate CLOCK/BMAL1-mediated transactivation, thus generating circadian oscillation in their own transcription. Rev-erbα (known as Nrd1d1) is also activated by CLOCK/BMAL1 and transrepressed by PER and CRY, resulting in circadian oscillation in the expression of Rev-erbα. In turn, REV-ERBα periodically represses Bmal1 transcription, thereby interconnecting the positive and negative loops (3). Like the mechanism of Rev-erbα transcription, clock genes comprising the core oscillation loop transduce downstream events by regulating the expression of clock-controlled output genes (4). cAMP-dependent signaling is involved in the circadian output pathways, but the cAMP signaling subsequently sustains the core oscillation loops in the suprachiasmatic nucleus (SCN),3 the center of the mammalian circadian clock (5, 6). However, the regulation mechanism of cAMP to sustain the circadian oscillator remains to be elucidated.
The intracellular accumulation of cAMP induces CRE-mediated gene expression via ATF/CREB protein activation (7). ATF/CREB proteins belong to the bZIP transcription factor superfamily, and they are characterized by a conserved domain including highly charged basic amino acids that are required for DNA recognition at the TGACGT(C/A)(G/A) sequence (8). Although the phosphorylated states of CREB in the SCN vary in a circadian manner (5), the functional importance of the transcriptional factors in regulating the circadian clock system remains unknown.
ATF4, also known as CREB2 or tax-responsive enhancer element B67, is a member of ATF/CREB family, and is expressed ubiquitously in the whole body (8). ATF4 is involved in multiple intracellular signal pathways, including energy metabolism, amino acid transport, and osteogenesis (9–11). In this study, we characterized the circadian nature of ATF4 in mice and showed that transcription of the Atf4 gene was regulated by core components of the circadian clock. Because ATF4 positively or negatively regulates the transcription of its target genes via CRE sequences, we also investigated whether ATF4 was involved in the circadian regulation of CRE-mediated transcription.
EXPERIMENTAL PROCEDURES
Animals
Clock mutant (Clock/Clock) mice (C57BL/6J-Clockm1Jt/J) were purchased from The Jackson Laboratory (Bar Harbor, ME). Heterozygous Atf4-null (Atf+/−) mice and Per2::Luciferase (Per2::Luc) knock-in mice were provided by Dr. S. Akira (Osaka University, Osaka, Japan) and Dr. Y. Shigeyoshi (Kinki University, Osaka, Japan), respectively. The animals were housed in a temperature-controlled (24 ± 1 °C) room under a 12 h-light, 12-h dark cycle or constant darkness. Under the light/dark cycle, Zeitgeber time (ZT) 0 was designated as lights on and ZT12 as lights off. For mice housed under constant darkness, circadian time (CT) was used instead of Zeitgeber time, and CT0 was characterized as the beginning of the subjective light phase, and CT12 was defined as the beginning of the subjective dark phase. The animals were cared for in accordance with the guidelines established by the Animal Care and Use Committee of Kyushu University.
Cell Cultures
We maintained mouse embryonic fibroblasts (MEFs) prepared from wild-type and Clock/Clock mice in Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 10% FBS (AFC Biosciences, Lenexa, KS) at 37 °C under a humidified 5% CO2 atmosphere. The culture medium for Atf4−/− MEFs was supplemented with 20 μm β-mercaptoethanol and 1× nonessential amino acid mixture (10). The cells were then incubated with 10 μm forskolin (FSK; Sigma), 50% fetal bovine serum (FBS), or 10 nm dexamethasone (DEX) (Sigma) for 2 h. To assess the circadian oscillation of gene expression, semi-confluent cultured MEFs were stimulated with 10 nm DEX for 2 h; the medium was then replaced with DMEM supplemented with 2% FBS. Cells were harvested for RNA extraction at the indicated times after DEX stimulation. RNA was extracted using RNAiso (Takara Bio, Otsu, Japan), and mRNA levels were measured by RT-PCR.
Immunoblotting
Nuclear fractions containing 20 μg of protein were resolved by SDS-PAGE. Separated proteins were then transferred to a polyvinylidene difluoride membrane and reacted against ATF4 (C-20), CREB-1 (C-21), phosphorylated CREB-1 (pCREB; Ser-133), and actin (all from Santa Cruz Biotechnology, Santa Cruz, CA), as well as phosphorylated ATF4 (pATF4; phospho-Ser-245; Abcam Plc., Cambridge, UK) antibodies. Specific antigen-antibody complexes were visualized using horseradish peroxidase-conjugated secondary antibodies and Chemi-Lumi One (Nacalai Tesque Inc., Kyoto, Japan).
Quantitative RT-PCR
The cDNA was synthesized by reverse-transcribing 0.4 μg of RNA using oligo(dT)15 primer and reverse transcriptase (Invitrogen). The cDNA equivalent to 12 ng of RNA was amplified by PCR in a real-time PCR system (Invitrogen). Sequences for PCR primers are described in supplemental Table S1.
Gel Mobility Shift Assays
Gel mobility shift assays were performed using in vitro translated CLOCK and BMAL1 proteins and 32P-labeled native (E-box, 5′-CGCTGCGGTAGGATCACGTGACCACAGTGGCA-3′) or mutated E-box (ΔE-box, 5′-CGCTGCGGTAGGATGAGTCTACCACAGTGGCA-3′) probes derived from the sequence of the mouse Atf4 promoter. CLOCK and BMAL1 proteins were synthesized from their expression plasmids using the tnt T7 Quick-coupled transcription/translation system (Promega, Madison, WI). The protein-DNA complexes were detected by autoradiography. For supershift assays, the protein-DNA complexes were incubated for 30 min with 2 μl of antibodies against CLOCK (Alpha Diagnostic, San Antonio, TX) or BMAL1 (Novus Biologicals Llc., Littleton, CO) prior to electrophoresis.
Luciferase Reporter Assays
We seeded MEFs prepared from wild type at a density of 1 × 105/well in 24-well culture plates. The cells were transfected 18 h later with 100 ng/well of reporter vectors and 1–2 μg/well (total) of expression vectors. The pRL-TK vector (0.5 ng/well; Promega) was also co-transfected as an internal control reporter. The cells were then harvested, and the cell lysates were analyzed using a Dual-Luciferase reporter assay system (Promega). The ratio of firefly (expressed from reporter construct) to Renilla (expressed from pRL-TK) luciferase activities in each sample served as a measure of normalized luciferase activity.
Construction of Reporter and Expression Vectors
The mouse Atf4 promoter region spanning from bp −346 to +170 (the number is the distance in base pairs from the putative transcription start site, +1) was amplified by PCR, and the product was ligated into the pGL3-Basic luciferase reporter vector (Promega). Luciferase reporter constructs of the mouse Per2 promoter region spanning from bp −3410 to +130 (Per2::Luc) were a gift from Dr. Y. Shigeyoshi (Kinki University, Osaka, Japan). The CRE in the Per2 promoter was mutated from TGCCGTCA to TGAATTCA by use of the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Expression vectors for mouse CLOCK, CLOCK-Δ19, BMAL1, PER2, and CRY1 were constructed using a cDNA generated from mouse liver RNA by RT-PCR. All coding regions were ligated into the pcDNA3.1(+) vector (Invitrogen).
Microarray Analysis
RNA was extracted from wild-type and Atf4−/− MEFs using TRIzol reagent (Invitrogen). The quality was analyzed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). The cRNA was amplified and labeled using a low input quick amp labeling kit. Labeled cRNA was hybridized to a 44K Agilent 60-mer oligomicroarray (Whole Mouse Genome Microarray kit version .2.0) according to the manufacturer's instructions. All hybridized microarray slides were washed and scanned using an Agilent scanner. Relative hybridization intensities and background hybridization values were calculated using Agilent Feature Extraction software (version 9.5.1.1).
Raw signal intensities and flags for each probe were calculated from hybridization intensities and spot information, according to the procedures recommended by Agilent. The raw signal intensities of two samples were log2-transformed and normalized by a quantile algorithm in the “preprocessCore” library package of Bioconductor software. To identify up or down-regulated genes, we calculated Z-scores and ratios (non-log scaled fold-change) from the normalized signal intensities of each probe for comparison between wild-type and Atf4−/− samples. We then applied the following two criteria to the selection of differentially expressed genes between wild-type and Atf4−/− MEFs: 1) Z-score ≥2.0 and ratio ≥2-fold (up-regulated genes); 2) Z-score ≤ −2.0 and ratio ≤0.5 (down-regulated genes). The functional analysis of the differentially expressed genes using KEGG data base was performed using the DAVID system.
Chromatin Immunoprecipitation (ChIP) Analysis
Cross-linked chromatin from the liver was sonicated on ice, and nuclear fractions were obtained by centrifugation at 10,000 × g for 5 min. Supernatants were incubated with antibodies against ATF4, CREB, CLOCK, or rabbit IgG (Santa Cruz Biotechnology). DNA was isolated by use of the GeneElute mammalian genomic DNA kit (Sigma) and amplified by PCR for the surrounding CREs in the 5′-flanking region of Per1, Per2, Cry2, and Bmal1 genes and surrounding E-box in the Per2 promoter region. Primer sequences for amplification of the surrounding CREs are described in supplemental Table S2. The quantitative reliability of PCR was evaluated by kinetic analysis of the amplified products to ensure that signals were derived only from the exponential phase of amplification. ChIP proceeded in the absence of antibody or in the presence of rabbit IgG as negative controls. Ethidium bromide staining did not detect any PCR products in these samples.
Real Time Monitoring of Circadian Bioluminescence
MEFs prepared from wild-type and Atf4−/− were infected with adenoviral CRE::luciferase vectors (Ad CRE::Luc; Vector BioLabs, Philadelphia). We also tracked bioluminescence from wild-type MEFs transfected with native or CRE-mutated Per2::Luc reporter vectors. Thereafter, cells were stimulated with 10 nm DEX for 2 h to synchronize their circadian clocks. Bioluminescence from Ad CRE::Luc-infected cells or Per2::Luc-transfected cells was recorded using a real time monitoring system (Lumicycle, Actimetrics, Wilmette, IL), and its amplitude was calculated using Lumicycle analysis software (Actimetrics). To explore the role of ATF4 in the center of mammalian circadian oscillators, the SCN slice cultures were prepared from Per2::Luc knock-in mice. The slices were transfected with siRNA against Art4, and thereafter bioluminescence from the slices was recorded as described above.
Small Interfering RNA
siRNA of the mouse Atf4 gene was designed as described previously (12). The siRNA oligonucleotide sequences were as follows: Atf4 siRNA sense 5′-GAGCAUUCCUUUAGUUUAGUU-3′ and antisense 5′-CUAAACUAAAGGAAUGCUCUU-3′; control siRNA sense, 5′-UAGUGUGAGCACUGUGAUUCCUUGG-3′ and antisense 5′-CCAAGGAUCACAGUGCUCACACUA-3′. The oligonucleotides were transfected into SCN slices at a final concentration of 20 pmol/ml using HVJ envelope vectors (Ishihara Sangyo, Osaka, Japan) according to previous report (13).
Statistical Analysis
The significance of the 24-h variation in each parameter was tested by analysis of variance. The statistical significance of differences among groups was analyzed by analysis of variance and Tukey's multiple comparison tests. A 5% level of probability was considered significant.
RESULTS
Circadian Expression of ATF4 Is Dependent on CLOCK Protein
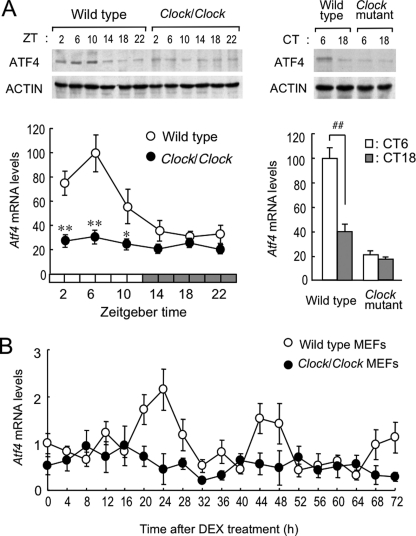
As observed previously (14–16), among the ATF/CREB family, Atf5 mRNA in the mouse liver significantly varied with a 24-h oscillation (supplemental Fig. S1). In this analysis, we found that Atf4 also exhibited a significant circadian oscillation in mouse liver. Under light/dark cycling conditions, levels of ATF4 protein and its mRNA in wild-type mice increased during the light phase and decreased in the dark phase (Fig. 1A, left panel). Oscillation in ATF4 protein levels was delayed by about 4 h relative to its mRNA rhythm. The expression of Atf4 mRNA was also rhythmic in the SCN as well as in the other tissues, including the cerebral cortex, kidney, and small intestine (supplemental Fig. S2). Rhythmic Atf4 expression in these tissues was similar to that in the liver.
FIGURE 1.
CLOCK is essential for circadian expression of ATF4. A, temporal expression profiles of ATF4 protein (upper panels) and mRNA (lower panels) in liver of wild-type and Clock/Clock mice housed under light/dark cycle (left) or constant darkness (right). Under the light/dark cycle, Zeitgeber time (ZT) 0 was designated as lights on and ZT12 as lights off. For mice housed under constant darkness, circadian time (CT) was used instead of Zeitgeber time. B, temporal expression profiles of Atf4 mRNA in MEFs generated from wild-type and Clock/Clock mice after incubation with 10 nm DEX for 2 h. All values are shown as means ± S.E. (n = 6–8). **, p < 0.01; *, p < 0.05 compared with wild-type group at corresponding Zeitgeber time. ##, p < 0.01 compared between groups.
Clock/Clock mice have a point mutation that produces a mutant protein lacking the residues encoded by exon 19 in the Clock gene, thus synthesizing a mutant CLOCK protein (CLOCKΔ19) with deficient transcriptional activity (1, 17). The levels of Atf4 mRNA and its protein in the liver of Clock/Clock mice constantly decreased throughout the day (Fig. 1A, left panel). The amplitude of hepatic Atf4 expression similarly decreased in the liver of Clock/Clock mice housed under constant darkness (Fig. 1A, right panel).
Circadian gene expression is caused not only by a cell-autonomous mechanism but also by time-dependent changes in extracellular stimuli (18, 19). To determine whether circadian expression of the Atf4 gene is cell autonomous, we examined Atf4 mRNA levels in MEFs prepared from wild-type and Clock/Clock mice. Cells were incubated with 10 nm DEX for 2 h to synchronize their circadian oscillators (20). Treatment of wild-type MEFs with DEX induced significant oscillation of the Atf4 mRNA levels with a period length about 24 h (p < 0.05, Fig. 1B). In contrast, variation in the Atf4 mRNA levels of DEX-treated Clock/Clock cells was not obviously time-dependent. These results suggest that the circadian expression of Atf4 is driven by a cell-autonomous mechanism and that CLOCK is required for the circadian control of Atf4 gene expression.
Transcriptional Regulation of Atf4 Gene by Clock Gene Products
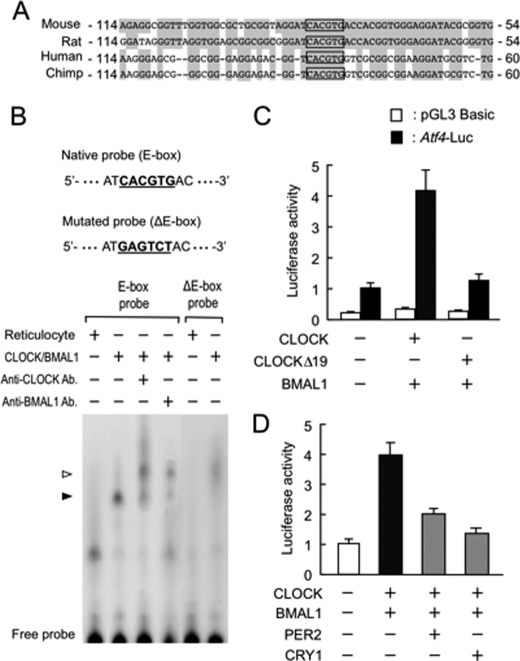
CLOCK forms heterodimers with BMAL1 and positively regulates the expression of its target genes through an E-box (CACGTG) element (2, 5). An E-box motif was found upstream of the Atf4 gene of mice and all mammals examined, including rats, chimpanzees, and humans (Fig. 2A). The results of electrophoretic mobility shift assays revealed that CLOCK/BMAL1 heterodimers bound to the E-box motif in the mouse Atf4 promoter (Fig. 2B). The supershifted bands were also detected when CLOCK/BMAL1 proteins were incubated with E-box probes in the presence of their antibodies. The specificity was further confirmed by the inability of CLOCK/BMAL1 proteins to bind to the E-box-mutated probe. As a consequence, luciferase reporter of the mouse Atf4 promoter (Atf4-Luc) that contains the E-box could respond to CLOCK/BMAL1 (Fig. 2C). However, the transcriptional activity of Atf4-Luc did not increase significantly when cells were transfected with both CLOCKΔ19 and BMAL1. Because CLOCKΔ19 is deficient in transactivation, despite retaining the ability to bind to DNA (1, 15), the mutated protein might have a subtle effect on the activity of the Atf4-Luc reporter.
FIGURE 2.
Transcriptional regulation of Atf4 by CLOCK. A, sequence comparison around the E-box in proximal Atf4 promoter regions from mouse, rat, human, and chimpanzee. Numbers at both sites indicate nucleotide residues from transcription initiation sites. Conserved nucleotide residues are shaded in gray, and E-boxes (CACGTG) are framed. B, gel mobility shift assays using in vitro translated CLOCK and BMAL1 proteins and 32P-labeled wild-type (E-box) or mutated E-box (ΔE-box) probes derived from sequence of mouse Atf4 promoter as indicated at top of panel. Closed arrowhead indicates CLOCK/BMAL1-binding bands. The open arrowhead shows supershift with antibodies. C and D, luciferase reporter assay of Atf4 promoter activity. Presence (+) or absence (−) of plasmids (0.1 μg each for pGL3-Basic (control), Atf4-Luc; 0.4 μg each for CLOCK, CLOCKΔ19, BMAL1, PER2, and CRY1) is denoted. Values are expressed as relative ratio to Atf4-Luc in the absence of expression of plasmids (set at 1.0). All values are shown as means ± S.E. (n = 4).
CLOCK/BMAL1-mediated transactivation of the Atf4-Luc was repressed by PER2 and CRY1 (Fig. 2D). The results of a ChIP analysis also revealed that the amount of endogenous PER2 and CRY1 binding to the promoter region of the Atf4 gene in the liver varied in a time-dependent manner (supplemental Fig. S3). The correlation between the binding activity of these negative regulators of the molecular loop and transcriptional regulation of the Atf4 gene suggests that PER and CRY proteins periodically repress CLOCK/BMAL1-mediated transactivation of the Atf4 gene.
Disrupted Rhythm in the Expression of Circadian Clock Genes in Atf4-null Cells
Mice lacking ATF4 have abnormal phenotypes such as microphthalmia, fetal anemia, and osteogenesis imperfects (11, 21–23). Furthermore, Atf4-null (Atf4−/−) mice died shortly after birth. We therefore prepared MEF cultures from Atf4−/− mice and attempted to identify genes that were under the circadian control of ATF4. Microarray analysis revealed that 1,047 genes were differentially expressed between wild-type and Atf4−/− MEFs. Because 339 of the 1,047 genes were uncharacterized, we analyzed the microarray data focusing on the known genes. Of the 708 known genes, 445 were highly expressed in Atf4−/− MEFs, whereas 263 were more highly expressed in wild-type MEFs (supplemental Table S3). The functional analysis of the differentially expressed genes using the KEGG data base showed that eight biological pathways were enriched in a statistically significant manner (p < 0.05; Table 1). During this analysis, we identified that “circadian rhythm” was the highly enriched pathway with the lowest p value. The expressions of Per2, Per3, Rev-erbα, Cry2, and Dec1 were found to be decreased in Atf4−/− MEFs, whereas Bmal1 was highly expressed in Atf4−/− cells (supplemental Table S4). These molecular findings prompted us to explore further the role of ATF4 in the regulation of circadian clock gene expression.
TABLE 1.
Statistically significant KEGG classifications of the differentially expressed genes in Atf4−/− MEFs
| Category | p value |
|---|---|
| Circadian rhythm | 6.1E-5 |
| Melanogenesis | 9.9E-3 |
| Retinol metabolism | 1.1E-2 |
| Arrhythmogenic right ventricular cardiomyopathy | 1.8E-2 |
| ABC transporter | 2.2E-2 |
| Xenobiotic metabolism | 3.2E-2 |
| Hypertrophic cardiomyopathy | 3.2E-2 |
| Hedgehog signaling pathway | 4.5E-2 |
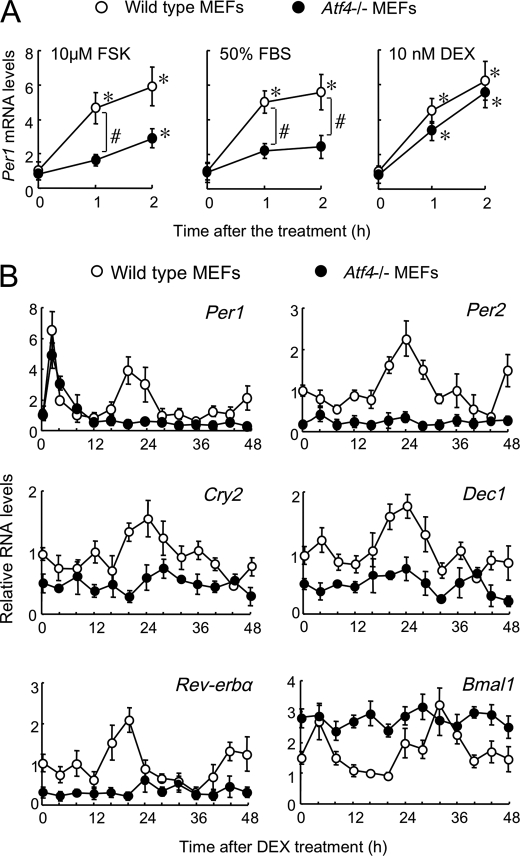
Exposing wild-type MEFs to 10 μm FSK, 50% FBS, or 10 nm DEX for 2 h resulted in significant induction of Per1 mRNA expression (all p < 0.05; Fig. 3A). Per1 mRNA in Atf4−/− cells also responded to these chemical compounds (all p < 0.05; Fig. 3A), but the induction of Per1 mRNA expression after incubation with FSK and FBS was significantly attenuated in Atf4−/− cells (p < 0.05, respectively). After incubation with DEX for 2 h, the mRNA levels of Per1, Per2, Cry2, Dec1, Rev-erbα, and Bmal1 in wild-type cells exhibited significant circadian oscillation (all p < 0.05; Fig. 3B) that was similar to that in NIH3T3 cells (24, 25). DEX also transiently induced Per1 mRNA in Atf4−/− cells, but the expression profiles of Per1 as well as of other clock genes in Atf4−/− cells were significantly different from those in wild-type MEFs (Fig. 3B). The mRNA levels of Per1, Per2, Cry2, Dec1, and Rev-erbα in Atf4−/− cells remained low, whereas Bmal1 mRNA levels increased constantly throughout the experimental period.
FIGURE 3.
Altered expression of circadian clock gene in Atf4−/− cells. A, induction of Per1 mRNA in wild-type and Atf4−/− MEFs incubated with 10 μm forskolin (FSK), 50% fetal bovine serum (FBS), and 10 nm dexamethasone (DEX) for 2 h. *, p < 0.05 compared with basal levels (0 h) of each genotype. #, p < 0.05 compared between groups. B, temporal expression profiles of clock genes in wild-type and Atf4−/− MEFs after incubation with 10 nm DEX for 2 h. All values are shown as means ± S.E. (n = 4).
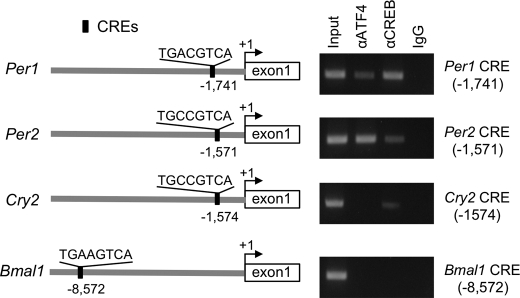
Computer-aided analysis of the 5′-flanking region of Per1, Per2, Rev-erbα, and Bmal1 genes up to −10,000 bp identified several CRE sequences (Fig. 4A). By contrast, no putative sequences resembling known CRE were found in the upstream region of Per3, Cry2, and Dec1 genes whose expression was decreased in Atf4−/− MEFs (supplemental Table S4). The results of a ChIP analysis also revealed that endogenous ATF4 substantially bound to the CRE in 5′-flanking region of the Per2 gene, but not to Cry2 and Bmal1 genes (Fig. 4A). Although the upstream region of the mouse Per1 gene also contains a consensus CRE sequence (26), the CRE within the Per1 gene promoter was preferentially bound by CREB rather than ATF4. These results suggest that ATF4 directly regulates the transcription of Per2 gene.
FIGURE 4.
ATF4 binding to the CRE within the Per2 promoter. Schematic representations of the location of CREs in the 5′-flanking region of the mouse Per1, Per2, Cry2, and Bmal1 genes and their corresponding binding affinities to ATF4 or CREB were analyzed by ChIP. The number of nucleotide residues indicates the distance from the putative transcription start site (+1).
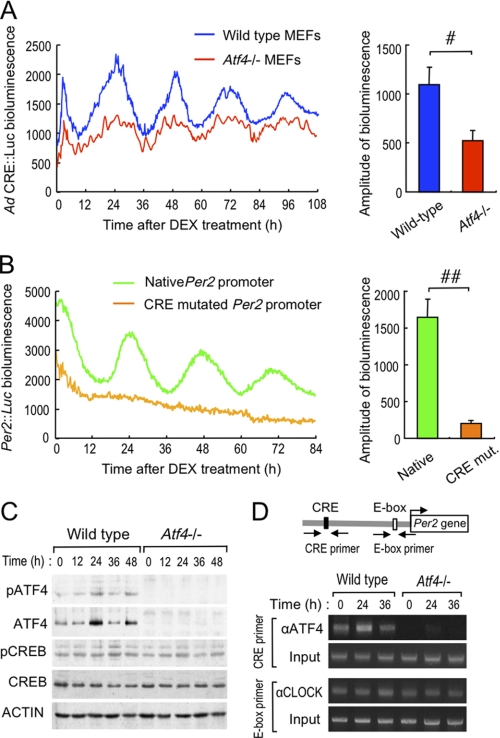
ATF4 Is Required for Circadian Regulation of CRE-mediated Transcription of Per2 Gene
To explore the role of ATF4 in the circadian regulation of CRE-mediated transcription, we tracked the temporal profile of luciferase activity driven by Ad CRE::Luc in wild-type and Atf4−/− MEFs. Bioluminescence oscillated with a period length of about 24 h in wild-type cells infected with Ad CRE::Luc after incubation with DEX (Fig. 5A, left panel). Bioluminescence also oscillated in Atf4−/− MEFs infected with Ad CRE::Luc, but with a significantly decreased amplitude (p < 0.05, Fig. 5A, right panel), suggesting that ATF4 acts as a positive regulator on CRE in adenoviral reporter vectors.
FIGURE 5.
Circadian control of CRE-mediated transcription of the Per2 gene by ATF4. A, left panel shows representative traces of bioluminescence oscillations driven by Ad CRE::Luc in wild-type and Atf4−/− MEFs after incubation with 10 nm DEX for 2 h. The right panel shows comparison of the amplitudes of bioluminescence oscillation between wild-type and Atf4−/− MEFs. Values are shown as means ± S.E. (n = 4). #, p < 0.05 compared between groups. B, representative traces of bioluminescent oscillations driven by luciferase reporter constructs of native Per2 promoter (Per2::Luc) and CRE-mutated Per2::Luc in wild-type MEFs after incubation with 10 nm DEX for 2 h. The right panel shows comparison of the amplitudes of bioluminescence oscillation between native and CRE-mutated Per2::Luc. Values are shown as means ± S.E. (n = 4). ##, p < 0.01 compared between groups. C, temporal profiles of pATF4, ATF4, pCREB, and CREB protein expression in wild-type and Atf4−/− MEFs after incubation with 10 nm DEX for 2 h. D, temporal binding profiles of ATF4 or CLOCK to Per2 promoter in wild-type and Atf4−/− MEFs after incubation with 10 nm DEX for 2 h. Solid line arrows represent PCR amplification areas of ATF4 binding to the CRE or of CLOCK binding to the E-box. C and D, results shown are representative of three independent experiments.
MEFs prepared from wild-type mice exhibited obvious circadian oscillation of reporter luciferase bioluminescence driven by the mouse Per2 promoter (Per2::Luc) containing CRE (Fig. 5B). Conversely, mutation of CRE resulted in a severely dampened bioluminescence oscillation of Per2::Luc even in wild-type cells, revealing that CRE-mediated regulation is indispensable for transcriptional oscillation of the Per2 gene.
The abundance of ATF4 protein, but not of CREB protein, in wild-type MEFs exhibited a circadian oscillation after DEX treatment (Fig. 5C). The amount of phosphorylated ATF4 (pATF4) as well as total (phosphorylated and nonphosphorylated) ATF4 protein increased when the oscillation of CRE-driven bioluminescence peaked. The results of ChIP analysis also showed that the endogenous ATF4 binding to the CRE within the Per2 promoter fluctuated in a circadian time-dependent manner (Fig. 5D). The amount of ATF4 binding in wild-type MEFs increased at the peak of Per2 mRNA expression (Fig. 3C) and bioluminescence oscillation (Fig. 5B). However, consistent with a previous report (27), CLOCK protein constitutively bound to a noncanonical E-box (CACGTT) located near the transcriptional start site of the Per2 gene (Fig. 5D). Taken together, these findings suggested that the binding of CLOCK to the E-box element was insufficient to generate cell-autonomous oscillation of the Per2 gene. ATF4 may play an important role as a positive regulator for the circadian expression of the Per2 gene.
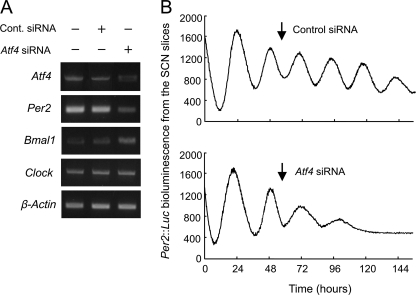
The mammalian circadian clock system is hierarchically organized; the master pacemaker in the SCN governs subsidiary oscillators in other brain regions and many peripheral tissues (5, 6). Transfection of Atf4 siRNA into the SCN slices resulted in the reduction of Atf4 mRNA levels (Fig. 6A). Transfection of Atf4 siRNA also decreased and increased the mRNA levels of Per2 and Bmal1, respectively, but had little effect on Clock mRNA expression. Bioluminescence oscillation driven by Per2::Luc in the SCN slice culture prepared from Per2::Luc knock-in mice was attenuated by transfection of Atf4 siRNA (Fig. 6B). The amplitude of bioluminescence oscillation of Per2::Luc after transfection with Atf4 siRNA was significantly lower than that after transfection with control siRNA (Atf4 siRNA, 114 ± 34; control, 487 ± 82, n = 3, means ± S.E., p < 0.05). These findings suggest that ATF4 acts as an additional component of mammalian circadian oscillator not only in peripheral cells but also in the center of mammalian circadian clock.
FIGURE 6.
ATF4 is required for circadian transcription of the Per2 gene in the SCN. A, influence of down-regulation of Atf4 by siRNA on the mRNA levels of Per2, Bmal1, and Clock in the SCN. The SCN slice cultures prepared from wild-type mice were transfected with Atf4 or control siRNA using HVJ envelope vectors. Two days after transfection, mRNA levels of Atf4 and clock genes were assessed. B, representative traces of bioluminescent oscillations driven by SCN slice culture prepared from Per2::Luc knock-in mice. The SCN slices were transfected with Atf4 or control siRNA using HVJ envelope vectors. Arrows indicate the application of HVJ envelope vectors encapsulated with control (upper) or Atf4 (lower) shRNAs. Data shown were confirmed in three independent siRNA-transfected slices. There was a significant difference in the amplitude of bioluminescent oscillation between Atf4 siRNA-transfected SCN and control slices (Atf4 siRNA, 114 ± 34; control siRNA, 487 ± 82, n = 3, means ± S.E., p < 0.05).
DISCUSSION
In this study, we have shown the role of ATF4 in the circadian clock system to regulate the oscillation of the CRE-mediated transcription of the Per2 gene. The CRE-mediated transactivation by ATF4 appeared to sustain core circadian oscillation machinery in cultured fibroblasts and SCN. It has been demonstrated that cAMP is essential for sustaining circadian oscillator in the SCN of mice (6). Because intracellular accumulation of cAMP ultimately induces the CRE-mediated gene expression via activation of ATF/CREB proteins, our findings indicate that ATF4 constitutes a molecular link connecting cAMP-dependent signaling to the core circadian oscillators.
Transcription of the Atf4 gene was controlled by the molecular components of the circadian clock. CLOCK/BMAL1 heterodimers transactivated the Atf4 gene through the proximal E-box located near the transcriptional start site. This CLOCK/BMAL1-mediated transactivation is repressed by PER and CRY proteins. Because ATF4 protein has a half-life of only 30–60 min (28), the time-dependent increase and decrease in Atf4 mRNA levels seem to cause the circadian fluctuation of its protein expression.
Bioluminescence in wild-type cells infected with Ad CRE::Luc exhibited a significant circadian oscillation, but the oscillation was significantly dampened in Atf4−/− MEFs. ATF4 positively or negatively regulate the transcription of its target genes through homo- or heterodimerization with Jun, AP-1, and CCAAT/enhancer-binding proteins (29, 30). The dimerization partners of ATF4 appear to determine the transcriptional direction of its target genes. In addition, the binding site sequence also affects the diverse actions of ATF4 (30). The adenoviral luciferase vectors contain tandem repeats of CREs located immediately upstream from the transcriptional start site of the luciferase gene. In cultured MEFs, ATF4 appeared to act as a positive regulator on the CREs of adenoviral reporter vectors. However, it is difficult to determine whether the circadian oscillation of CRE-mediated transcription is directly governed by ATF4, because the loss of ATF4 attenuated the circadian activation of CRE-mediated transcription accompanied by disruption of core clock function. In addition to circadian clock genes, no significant oscillations in the expression of typical clock-controlled genes, Dbp and Tef, were detected in Atf4−/− MEFs (supplemental Table S3 and supplemental Fig. S4). ATF4 may directly and/or indirectly regulate the circadian output pathways through CRE-mediated transcription.
Endogenous ATF4 was able to bind to the CRE in the 5′-flanking region of the Per2 gene, but not to the CREs in the upstream region of Cry2 and Bmal1 genes. Instead of ATF4, the CRE in the Per1 gene was mainly occupied by CREB. The sequence surrounding the CRE and its location may influence CRE binding affinity to ATF4. The difference in the binding affinity of Per1 and Per2 CREs to ATF4 could also account for the diverse induction kinetics of the two genes in response to cAMP stimuli (26). Although CLOCK protein constitutively bound to the E-box element of Per2 promoter in both wild-type and Atf4−/− MEFs, the mutation of the CRE resulted in the attenuation of bioluminescence oscillation driven by Per2::Luc even in wild-type cells. CLOCK protein is thought to be a dominant factor generating the transcriptional oscillation of the Per2 gene, because the amplitude of Per2 mRNA rhythm is reduced in Clock/Clock cells (27). However, the levels of Atf4 mRNA and its protein were decreased by the Clock gene mutation. The decreased levels of ATF4 in Clock/Clock MEFs may also contribute to the attenuated transcriptional oscillation of the Per2 gene. This notion is also supported by present findings that bioluminescence oscillation driven by Per2::Luc in the SCN slice culture was diminished by down-regulation of ATF4. Because the transcription of clock genes is regulated by other clock gene products, modulation in the expression of the Per2 gene induced by the loss of ATF4 would influence the oscillation of other clock genes. Alternatively, ATF4 may also directly regulate the circadian oscillation of other clock genes. Further studies are required to investigate this point.
It has been suggested that periodic phosphorylation of the CREB protein (pCREB) regulates the circadian oscillation of CRE-mediated transcription (5). However, the amount of pCREB protein did not exhibit circadian oscillation in either wild-type or Atf4−/− cells. Intracellular cAMP concentrations in MEFs were relatively higher than those in the organs of mice (supplemental Fig. S5). The high concentration of cellular cAMP attributable to culture conditions might have been sufficient to maintain constitutive CREB protein phosphorylation.
Mice lacking ATF4 display various abnormal phenotypes such as microphthalmia, fetal anemia, and osteogenesis imperfect (11, 21–23). However, Clock/Clock mice do not display such abnormal phenotypes despite low levels of ATF4. The expression of ATF4 is induced by various extracellular signals (31, 32), and its protein functions as a dominant regulator for development in response to extracellular cues (21, 33). Stimulation with fibroblast growth factor rapidly increased the mRNA levels of Atf4 in Clock/Clock MEFs (supplemental Fig. S6), suggesting that the Atf4 gene remains responsive to extracellular stimuli in Clock/Clock cells. The responsiveness of the Atf4 gene in Clock/Clock mice might prevent the abnormal phenotypes associated with Atf4−/− mice.
The present findings suggest that ATF4 acts as an output signal for regulating the circadian oscillation of CRE-mediated transcription, which, in turn, also sustains the rhythmic expression of the Per2 gene. The ATF4-mediated transactivation of the Per2 gene seems to sustain circadian oscillators in peripheral cells as well as in the center of mammalian circadian clock. Our results demonstrate a previously unknown role of ATF4 in the circadian timing system and revealed a molecular link between the cAMP-dependent signaling and core circadian clock machinery.
Supplementary Material
Acknowledgments
We are indebted to Dr. S. Akira (Osaka University, Japan) for providing Atf4-null mice. We are also grateful to Dr. Y. Shigeyoshi (Kinki University, Osaka, Japan) for providing luciferase reporter constructs of Per2 promoter and Per2::Luc knock-in mice. We thank Steven Sabotta for proofreading the manuscript.
This work was supported in part by Grant-in-aid for Scientific Research on Priority Areas (Cancer) 20014016 (to S. O.) from the Ministry of Education, Culture, Sport, Science, and Technology, Grant-in-aid for Scientific Research (B) 21390047 (to S. O.), and Grant-in-aid for Challenging Exploratory Research 21659041 (to S. O.) from the Japan Society for the Promotion of Science.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1–S4.
- SCN
- suprachiasmatic nucleus
- MEF
- mouse embryonic fibroblast
- ATF
- activating transcription factor
- CRE
- cAMP-response element
- CREB
- CRE-binding protein
- FSK
- forskolin
- DEX
- dexamethasone.
REFERENCES
- 1. Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., Takahashi J. S., Weitz C. J. (1998) Science 280, 1564–1569 [DOI] [PubMed] [Google Scholar]
- 2. Kume K., Zylka M. J., Sriram S., Shearman L. P., Weaver D. R., Jin X., Maywood E. S., Hastings M. H., Reppert S. M. (1999) Cell 98, 193–205 [DOI] [PubMed] [Google Scholar]
- 3. Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., Schibler U. (2002) Cell 110, 251–260 [DOI] [PubMed] [Google Scholar]
- 4. Cheng M. Y., Bullock C. M., Li C., Lee A. G., Bermak J. C., Belluzzi J., Weaver D. R., Leslie F. M., Zhou Q. Y. (2002) Nature 417, 405–410 [DOI] [PubMed] [Google Scholar]
- 5. Obrietan K., Impey S., Smith D., Athos J., Storm D. R. (1999) J. Biol. Chem. 274, 17748–17756 [DOI] [PubMed] [Google Scholar]
- 6. O'Neill J. S., Maywood E. S., Chesham J. E., Takahashi J. S., Hastings M. H. (2008) Science 320, 949–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin Y. S., Green M. R. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vallejo M., Ron D., Miller C. P., Habener J. F. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 4679–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoshizawa T., Hinoi E., Jung D. Y., Kajimura D., Ferron M., Seo J., Graff J. M., Kim J. K., Karsenty G. (2009) J. Clin. Invest. 119, 2807–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., Ron D. (2003) Mol. Cell 11, 619–633 [DOI] [PubMed] [Google Scholar]
- 11. Elefteriou F., Ahn J. D., Takeda S., Starbuck M., Yang X., Liu X., Kondo H., Richards W. G., Bannon T. W., Noda M., Clement K., Vaisse C., Karsenty G. (2005) Nature 434, 514–520 [DOI] [PubMed] [Google Scholar]
- 12. Zhang X., Yu S., Galson D. L., Luo M., Fan J., Zhang J., Guan Y., Xiao G. (2008) J. Cell Biochem. 105, 885–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shimamura M., Morishita R., Endoh M., Oshima K., Aoki M., Waguri S., Uchiyama Y., Kaneda Y. (2003) Biochem. Biophys. Res. Commun. 300, 464–471 [DOI] [PubMed] [Google Scholar]
- 14. Lemos D. R., Goodspeed L., Tonelli L., Antoch M. P., Ojeda S. R., Urbanski H. F. (2007) Endocrinology 148, 5811–5821 [DOI] [PubMed] [Google Scholar]
- 15. Panda S., Antoch M. P., Miller B. H., Su A. I., Schook A. B., Straume M., Schultz P. G., Kay S. A., Takahashi J. S., Hogenesch J. B. (2002) Cell 109, 307–320 [DOI] [PubMed] [Google Scholar]
- 16. Storch K. F., Lipan O., Leykin I., Viswanathan N., Davis F. C., Wong W. H., Weitz C. J. (2002) Nature 417, 78–83 [DOI] [PubMed] [Google Scholar]
- 17. Doi M., Hirayama J., Sassone-Corsi P. (2006) Cell 125, 497–508 [DOI] [PubMed] [Google Scholar]
- 18. Terazono H., Mutoh T., Yamaguchi S., Kobayashi M., Akiyama M., Udo R., Ohdo S., Okamura H., Shibata S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6795–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oishi K., Amagai N., Shirai H., Kadota K., Ohkura N., Ishida N. (2005) DNA Res. 12, 191–202 [DOI] [PubMed] [Google Scholar]
- 20. Nagoshi E., Saini C., Bauer C., Laroche T., Naef F., Schibler U. (2004) Cell 119, 693–705 [DOI] [PubMed] [Google Scholar]
- 21. Tanaka T., Tsujimura T., Takeda K., Sugihara A., Maekawa A., Terada N., Yoshida N., Akira S. (1998) Genes Cells 3, 801–810 [DOI] [PubMed] [Google Scholar]
- 22. Masuoka H. C., Townes T. M. (2002) Blood 99, 736–745 [DOI] [PubMed] [Google Scholar]
- 23. Ma Y., Hendershot L. M. (2003) J. Biol. Chem. 278, 34864–34873 [DOI] [PubMed] [Google Scholar]
- 24. Akashi M., Takumi T. (2005) Nat. Struct. Mol. Biol. 12, 441–448 [DOI] [PubMed] [Google Scholar]
- 25. Cavadini G., Petrzilka S., Kohler P., Jud C., Tobler I., Birchler T., Fontana A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12843–12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Travnickova-Bendova Z., Cermakian N., Reppert S. M., Sassone-Corsi P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7728–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yoo S. H., Ko C. H., Lowrey P. L., Buhr E. D., Song E. J., Chang S., Yoo O. J., Yamazaki S., Lee C., Takahashi J. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2608–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lassot I., Ségéral E., Berlioz-Torrent C., Durand H., Groussin L., Hai T., Benarous R., Margottin-Goguet F. (2001) Mol. Cell. Biol. 21, 2192–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kato Y., Koike Y., Tomizawa K., Ogawa S., Hosaka K., Tanaka S., Kato T. (1999) Mol. Cell. Endocrinol. 154, 151–159 [DOI] [PubMed] [Google Scholar]
- 30. Gachon F., Gaudray G., Thébault S., Basbous J., Koffi J. A., Devaux C., Mesnard J. (2001) FEBS Lett. 502, 57–62 [DOI] [PubMed] [Google Scholar]
- 31. Tan Y., Low K. G., Boccia C., Grossman J., Comb M. J. (1994) Mol. Cell. Biol. 14, 7546–7556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adams C. M. (2007) J. Biol. Chem. 282, 16744–16753 [DOI] [PubMed] [Google Scholar]
- 33. Sowa H., Karsenty G. (2007) J. Musculoskelet. Neuronal. Interact. 7, 326–367 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.