Abstract
Typically as a result of phosphorylation, OmpR/PhoB response regulators form homodimers through a receiver domain as an integral step in transcriptional activation. Phosphorylation stabilizes the ionic and hydrophobic interactions between monomers. Recent studies have shown that some response regulators retain functional activity in the absence of phosphorylation and are termed atypical response regulators. The two currently available receiver domain structures of atypical response regulators are very similar to their phospho-accepting homologs, and their propensity to form homodimers is generally retained. An atypical response regulator, ChxR, from Chlamydia trachomatis, was previously reported to form homodimers; however, the residues critical to this interaction have not been elucidated. We hypothesize that the intra- and intermolecular interactions involved in forming a transcriptionally competent ChxR are distinct from the canonical phosphorylation (activation) paradigm in the OmpR/PhoB response regulator subfamily. To test this hypothesis, structural and functional studies were performed on the receiver domain of ChxR. Two crystal structures of the receiver domain were solved with the recently developed method using triiodo compound I3C. These structures revealed many characteristics unique to OmpR/PhoB subfamily members: typical or atypical. Included was the absence of two α-helices present in all other OmpR/PhoB response regulators. Functional studies on various dimer interface residues demonstrated that ChxR forms relatively stable homodimers through hydrophobic interactions, and disruption of these can be accomplished with the introduction of a charged residue within the dimer interface. A gel shift study with monomeric ChxR supports that dimerization through the receiver domain is critical for interaction with DNA.
Keywords: Bacterial Genetics, Bacterial Signal Transduction, DNA Transcription, Protein-DNA Interaction, Protein Structure, Protein-Protein interactions, X-ray Crystallography
Introduction
Two-component signal transduction systems are important mechanisms that mediate many physiological functions within an organism. The output response of these systems is generally an alteration of gene expression (1, 2). The prototypical two-component system consists of a membrane-bound sensor histidine kinase that transfers a phosphoryl group to a cognate response regulator (3). Phosphorylation of the response regulator stabilizes the active form of the protein, which promotes oligomerization through a receiver domain. Oligomerization facilitates an effector domain to interact with DNA and the transcriptional machinery.
Although the genes regulated by these transcription factors vary, a highly conserved protein architecture and residue composition (i.e. structure and sequence) appears to be critical for a canonical mechanism of activation. The topology of the receiver domain (β1-α1-β2-α2-β3-α3-β4-α4-β5-α5) is highly conserved among response regulators (3, 4). In addition to the conserved domain architecture, the current understanding of the mechanism of activation within these proteins is derived from comparisons of multiple structures of these proteins in both the inactive (unphosphorylated) and active (phosphorylated) state (5–9). The cognate sensor kinase transfers a phosphoryl group to a conserved phospho-accepting Asp in the receiver domain of the response regulator. An essential Mg2+ ion and a Lys residue assist in the transfer and retention of the phosphoryl group within the binding site (8, 9). The activation signal is then transduced to the dimer interface (α4-β5-α5) through the reorientation of two conformational switch residues (Thr/Ser and Tyr/Phe) toward the phosphoryl group. The reorientation of these two residues and subtle conformational changes throughout the protein dramatically enhances homodimer formation by properly aligning residues within the dimer interface that are involved in ionic and hydrophobic interactions between monomers. Homodimer formation through the receiver domain enhances the ability of the effector domain to bind to DNA and regulate transcription (10).
An increasing number of response regulators have been identified that appear to not rely on a sensor kinase or a phosphorylation event for activation. These atypical response regulators do not retain many of the residues critical to the canonical phosphorylation (activation) process (4). For example, HP1043, from Helicobacter pylori, lacks the canonical phospho-accepting Asp but is still capable of forming homodimers and interacting with DNA in the absence of phosphorylation (11–13). Two experimentally determined structures of atypical receiver domains have shown that the conserved structural topology of the typical response regulators is largely retained (11, 14). Functional studies of these proteins, albeit limited, have determined that the propensity to form homodimers is also generally retained (11, 14–16). The structural elements that maintain their phosphorylation-independent activity, however, are poorly understood due to the paucity of functional and structural studies of these proteins.
The medically important bacteria Chlamydia encodes a transcriptional regulator termed ChxR. Transcriptional regulation has been determined to be a key factor in the development and pathogenesis of Chlamydia (17, 18). Largely due to a lack of a system for directed gene disruption in Chlamydia, the ability to determine a specific biological role for ChxR has been impeded; however, prior studies support that the function of ChxR is exerted during the middle and late stages of the developmental cycle, which includes generation of the infectious form of Chlamydia (i.e. elementary body) (16, 19). Given the relative paucity of alternative transcription factors encoded by Chlamydia, the number of putative ChxR binding sites throughout the genome (16), and the restricted growth conditions (obligate intracellular), it is expected that ChxR plays an important role in biology of Chlamydia (20).
ChxR was identified from primary sequence homology to be a member of the OmpR/PhoB subfamily of response regulators (21). Computational analysis indicated that ChxR lacks the phospho-accepting Asp, and a cognate sensor kinase was not identified within the chlamydial genome, suggesting that the function of ChxR is not directly controlled by phosphorylation (19). Functional studies have reported that ChxR exists as a stable homodimer and could activate transcription in the absence of phosphorylation (16, 19). Although these results support the conclusion that ChxR is an atypical OmpR/PhoB transcriptional regulator, the critical structural features and molecular interactions that permit the protein to be maintained in an active state and thereby mimic phosphorylated response regulators have yet to be identified. We hypothesize that the intra- and intermolecular interactions involved in activation and dimerization of ChxR are distinct from the canonical phosphorylation (activation) paradigm in the OmpR/PhoB response regulator subfamily. To test this hypothesis, structural and functional studies were performed with the receiver domain of ChxR to identify the residues and structural features necessary for dimerization.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of ChxRRec2
DNA encoding the receiver domain of ChxR (ChxRRec, residues 2–113) was PCR-amplified using Chlamydia trachomatis LGV (L2/434/Bu) genomic DNA and primers for ChxRRec (5′-GGAATTCCATATGCAGGGCCTAAACATGTG-3′ and 5′-CCGCTCGAGATGTAGCGAATGCTGAGAAAG-3′) (Integrated DNA Technologies, Coralville, IA). The PCR product was digested with NdeI/XhoI and inserted into the N-terminal polyhistidine tag encoding pET28b vector (Novagen, San Diego, CA). ChxRRec was expressed and purified as described for full-length ChxR (ChxRFL) (16). Briefly, the protein was initially purified using Co2+ affinity chromatography (Clontech, Mountain View, CA) equilibrated with 50 mm Tris-HCl, pH 8.0, 400 mm NaCl. ChxRRec was further purified by size exclusion chromatography using a Sephacryl S-200 16/60 column (GE Healthcare) equilibrated with 50 mm Tris-HCl, pH 8.0, 400 mm NaCl.
Crystallization of ChxRRec
Purified ChxRRec, concentrated to 10 mg/ml in 20 mm NaH2PO4/K2HPO4, pH 7.0, 400 mm NaCl, was screened for crystallization in Compact Jr. (Emerald BioSystems, Bainbridge Island, WA) sitting drop vapor diffusion plates by mixing 1 μl of protein and 1 μl of crystallization solution equilibrated against 100 μl of the latter. Prismatic ChxRRec crystals were obtained from two crystallization conditions. ChxRRec crystals, belonging to a C-centered monoclinic lattice (space group C2), grew in ∼2 days at 4 °C from the Wizard 3 screen (Emerald Biosystems) condition #10 (20% (w/v) PEG 3350, 0.2 m sodium thiocyanate). A tetragonal crystal form (space group I41) grew in ∼2 days at 4 °C from the Precipitant Synergy screen (Emerald Biosystems) condition #7 and pHat screen (Emerald Biosystems) condition #42 (4 m NaCl, 5% isopropyl alcohol, and 100 mm NaH2PO4/K2HPO4, pH 7.0). Single crystals were transferred to a cryoprotectant solution containing 80% crystallization solution and 20% ethylene glycol before flash-cooling in liquid nitrogen for data collection. For SIRAS phasing, a crystal belonging to the C2 form was soaked for 5 min in 50 mm 5-amino-2,4,6-triiodoisophthalic acid (I3C, Hampton Research, Aliso Viejo, CA) (22) dissolved in crystallization solution before the transfer to the cryoprotectant solution.
Data Collection and Processing
Diffraction data for structure solution using the SIRAS phasing method were collected at 93 K at the University of Kansas Protein Structure Laboratory using a Rigaku RU-H3R rotating anode generator (Cu-Kα) equipped with an R-axis IV2+ image plate detector and osmic blue focusing mirrors. The exposure time for each 1° oscillation image was 8 min at a detector distance of 150 mm. Intensities were integrated and scaled using the HKL2000 package (23). Structure solution was carried out using the SIRAS phasing method with the SHELX C/D/E software package (24) via the CCP4 interface (25). Iodine positions corresponding to three I3C sites were identified using SHELXC and SHELXD that yielded correlation coefficient all/weak of 38.80/27.17. Calculation of initial phase angles and density modification were conducted with SHELXE and yielded a pseudo-free correlation coefficient of 69.28% and an estimated mean figure of merit of 0.653 for the inverted substructure. BUCCANNER (26) was used to generate a Cα trace of the model for future molecular replacement against the high resolution native data.
High resolution native ChxRRec (C2 space group) data were collected at 100 K at the IMCA-CAT beamline 17BM at the Advanced Photon Source using an ADSC Quantum 210r CCD detector at a wavelength of 1.0 Å. The exposure time for each 1° oscillation image was 5 s. Intensities were integrated and scaled using D*TREK (27). High resolution native ChxRRec (I41 space group) data were collected at 100 K at Stanford Synchrotron Radiation Laboratory beamline 9-2 using an MAR325 detector at a distance of 170 mm, a wavelength of 1.54 Å, an exposure time of 5 s and a 1° oscillation per image. Intensities were integrated and scaled using MOSFLM and SCALA (28), respectively. A Cα model obtained from SIRAS phasing with I3C was used as the search model for molecular replacement with PHASER (29) against the high resolution synchrotron data. Initial automated model building was carried out using ARP/wARP (30). Anisotropic atomic displacement parameters were modeled by TLS refinement 7 groups as generated by the TLSMD server (31). Final model building and structure refinement were performed with COOT (32) and PHENIX (33), respectively. Data collection and processing statistics are listed in Table 1. Figures were created using CCP4 Molecular Graphics Program (34).
TABLE 1.
Data collection and refinement statistics
| Apo (C2) | SIRAS (I3C) | Apo (I41) | |
|---|---|---|---|
| Data collection | |||
| Unit cell parameters (Å, o) | a = 149.9 b = 41.3 | a = 149.8 b = 41.1 | a = 53.7 b = 53.7 |
| c = 45.2, α = γ = 90 β = 105.5 | c = 45.1, α = γ = 90 β = 106.1 | c = 190.1, α = β = γ = 90 | |
| Space group | C2 | C2 | I41 |
| Resolution (Å)a | 23.66-1.6 (1.66-1.6) | 30.0-2.1 (2.18-2.1) | 30.0-2.15 (2.27-2.15) |
| Wavelength (Å) | 1.0 | 1.54 | 1.54 |
| Observed reflections | 128,193 | 52,247 | 54,653 |
| Unique reflections | 35,320 | 15,613 | 14,580 |
| 〈I/σI〉a | 13.5 (3.1) | 13.1 (2.9) | 9.5 (2.7) |
| Completeness (%)a | 99.8 (100) | 98.8 (96.2) | 99.9 (100) |
| Redundancya | 3.63 (3.63) | 3.3 (3.2) | 3.7 (3.7) |
| Rmerge (%)a,b | 3.9 (29.6) | 12.2 (46.7) | 8.1 (45.6) |
| Refinement | |||
| Resolution (Å) | 23.66-1.60 | 28.14-2.09 | 29.67-2.15 |
| Rfactor/Rfree (%)c | 18.83/21.02 | 19.60/25.51 | 20.50/24.69 |
| No. of atoms (protein/water) | 1,802/108 | 1,700/77 | 1,631/41 |
| Model quality | |||
| r.m.s deviations | |||
| Bond lengths (Å) | 0.015 | 0.018 | 0.008 |
| Bond angles (o) | 1.513 | 1.698 | 1.079 |
| Average B factor (Å2) | |||
| Protein | 29.5 | 37.6 | 40.1 |
| Water | 30.9 | 35.6 | 39.4 |
| I3C | − | 43.4 | − |
| Coordinate error based on maximum likelihood (Å) | 0.21 | 0.30 | 0.25 |
| Ramachandran plot | |||
| Favored (%) | 99.5 | 99.5 | 99.5 |
| Allowed (%) | 0.5 | 0.0 | 0.5 |
| Disallowed (%) | 0.0 | 0.5 | 0.0 |
| PDB ID | 3Q7R | 3Q7S | 3Q7T |
a Values in parenthesis are for the highest resolution shell.
b Rmerge = ΣhklΣi|Ii(hkl) − 〈I(hkl)〉|/ΣhklΣiIi(hkl), where Ii(hkl) is the intensity measured for the ith reflection, and 〈I(hkl)〉 is the average intensity of all reflections with indices hkl.
c Rfactor = Σhkl‖Fobs(hkl)| − |Fcalc(hkl)‖/Σhkl|Fobs(hkl)|; Rfree is calculated in an identical manner using 5% of randomly selected reflections that were not included in the refinement.
For the 1.6 Å resolution model, monomer A comprises residues 2–111, and monomer B comprises residues 4–37, 43–60, and 68–110. The model contained two ethylene glycol molecules and 106 water molecules. For the 2.15 Å resolution model, monomer A comprises residues 4–54 and 66–110, and monomer B comprises residues 4–60 and 67–109. The model contained two sodium ions and 39 water molecules. For the 2.1 Å resolution structure, monomer A comprises residues 3–111, and monomer B comprises residues 4–37, 43–60, and 68–109. The model contained 75 water molecules and three I3C molecules. One I3C molecule is located at the interface between monomer A in the asymmetric unit and monomer B of a symmetric-related molecule. I2 is 2.8 Å away from the amide O of Leu-23, and O8 of one carboxyl group of the I3C molecule forms a hydrogen bond with the symmetry-related Lys-101 NZ (2.7Å) and Arg98 NH1 (3.0Å). The second I3C molecule is located at the solvent-exposed surface of monomer B. Hydrogen bonds are observed between four water molecules and N1, O8, O9, and O12 of the I3C molecule. Additionally, O9 forms a hydrogen bond with His-13 ND (2.8Å). The third I3C molecule is located at the solvent-exposed surface of monomer B. O8 of one carbonyl group forms a hydrogen bond with the amide N of Gln-35 (2.9Å) and a water molecule (3.0 Å). Additionally, O9 forms a hydrogen bond with Gln-35 NE2 (3.1 Å).
Far-UV CD Spectroscopy
CD analysis was performed with a Chirascan-plus circular dichroism spectrometer equipped with a Peltier temperature controller and a four-position cuvette holder (Applied Photophysics Ltd, Leatherhead, UK). Far UV spectra of YycFRec and ChxRRec and ChxRRecW89E at 0.1 mg/ml in CD buffer (20 mm NaH2PO4, pH 7.5, 20 mm NaCl) were collected in the range of 190–260 nm using a 0.1-cm path length cuvette sealed with a Teflon stopper. A sampling time-per-point of 2 s and a bandwidth of 1 nm were used. The secondary structure components were estimated by the CDNN CD spectra deconvolution software (35). CDNN is a neural networks method-based program that can be used to analyze data to determine the content of α-helix, parallel and anti-parallel β-structure, turns, and random coil. The results from the CDNN analysis were sorted automatically in five regions (190–260, 195–260, 200–260, 205–260, and 210–260 nm) for each secondary structure component. For each secondary component, results of five regions were averaged, and S.D. were calculated.
Site-directed Mutagenesis
Mutations were introduced into the ChxRFL and ChxRRec plasmids using the QuikChange II XL site-directed mutagenesis kit and following the manufacturer's protocol (Agilent Technologies, La Jolla, CA). All clones were verified by DNA sequencing analysis (ACGT, Inc., Wheeling, Il). The proteins were overexpressed in pET28b and purified as described previously for ChxRFL (16) or above for ChxRRec.
Analytical Size Exclusion Chromatography
After purification, ChxRRec was concentrated to 100 μm using an Amicon Ultra centrifugal filter (Millipore, Billerica, MA). ChxRRec was then diluted to 10 μm and 1 μm in 50 mm Tris-HCl, pH 8.0, 400 mm NaCl. The proteins were applied to a Superdex 75 10/300 GL analytical size exclusion column (GE Healthcare) equilibrated with 50 mm Tris-HCl, pH 8.0, 400 mm NaCl. For analytical size exclusion chromatography of ChxRRec, ChxRRecW89E, ChxRFL, and ChxRFLW89E, the proteins were concentrated to 10 μm and applied to a Superdex 75 10/300 GL analytical size exclusion column (GE Healthcare) equilibrated with 50 mm Tris-HCl, pH 7.5, 100 mm NaCl, and 250 mm KCl. A protein standard solution containing bovine serum albumin (66 kDa), chicken ovalbumin (44 kDa), horse myoglobin (17 kDa), and vitamin B12 (1.35 kDa) (Bio-Rad) was used to generate a standard curve.
Analytical Ultracentrifugation
ChxRFL and ChxRFLW89E in 20 mm NaH2PO4, pH 7.5, 20 mm NaCl were subjected to analytical ultracentrifugation at a concentration of 23 and 27 μm, respectively. Each protein was loaded into two-channel, 12-mm optical path length cells and analyzed using a Beckman Coulter XL-I analytical ultracentrifuge (Palo Alto, CA). A four-hole An60 Ti rotor was used for housing the cells; each sample cell was run at 10 °C for 450 continuous scans. The speed of rotor was 120,000 × g for the sedimentation velocity analysis and was conducted using SEDFIT (Version 12.1b) (36). Continuous c(s) distribution analysis was used by employing nonlinear regression during the data fit. In addition, viscosities and densities of the buffers were measured using the SVM 3000 viscometer (Anton Parr USA Inc., Ashland, VA) for accurate parameters used in the analytical ultracentrifugation fitting process. Each experiment was performed in duplicate.
Electrophoretic Mobility Shift Assay
An EMSA to test DNA binding by ChxR was performed as previously described (16) with IR800-labeled DNA corresponding to the high affinity (DR2) binding site within the chxR promoter. The assays were performed at a protein concentration of 44 nm, which is the approximate dissociation constant for this binding site (i.e. 50% of this DNA sequence is shifted at this protein concentration) and would thus permit maximal variation in the amount of DNA bound with the substitutions (16). The binding reactions contained 1 nm DNA and either 44 nm wild-type or variant ChxRFL. The DNA was visualized and quantified using an Odyssey Infrared Imaging System (LI-COR Biosciences, Lincoln, NE). The effector domain of ChxR used in this assay was purified as described previously (37).
Phosphorylation Assays
Purified ChxRFL in 50 mm Tris-HCl, pH 7.5, 50 mm KCl, and 50 mm MgCl was incubated with 1, 10, or 100 mm acetyl phosphate (Sigma). After incubation for 1 h at 37 °C, the reaction was subjected to native PAGE, and protein bands were visualized through Coomassie staining. For DNA binding analysis, ChxRFL was incubated with 100 mm acetyl phosphate for 1 h before the analysis. An EMSA was performed in triplicate with 1 nm DR2 DNA and 44 mm concentrations of either untreated or acetyl phosphate-treated ChxRFL. The amount of DNA shifted was quantified as described above.
RESULTS
Primary Sequence Comparison of ChxR and Other OmpR/PhoB Subfamily Members
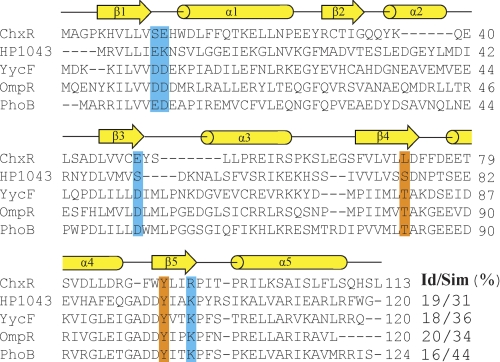
A prior study reported that ChxR could activate transcription despite the absence of a phospho-accepting Asp (16, 19); however, a comprehensive analysis of other residues that may be important to its function has not been performed. To determine whether six residues important for activation in phosphorylation-dependent homologs are retained in ChxR, the primary sequence of ChxRRec from C. trachomatis L2/434/Bu was aligned with an atypical (HP1043) and three well characterized phospho-accepting (PhoB, OmpR, and YycF) subfamily members using the multiple sequence alignment program ClustalW (Fig. 1) (38). Only one (Tyr-90; using ChxR numeration) of the six highly conserved residues is retained in ChxR. This suggests that these residues may not contribute to the function of ChxR in Chlamydia.
FIGURE 1.
Primary sequence alignment of ChxR and other OmpR/PhoB subfamily members. The primary sequence of ChxRRec was aligned with an atypical OmpR/PhoB subfamily member (HP1043) and three well characterized phosphorylation-dependent OmpR/PhoB subfamily members (YycF, OmpR, and PhoB). The secondary structure elements correspond to PhoB (PDB ID 1ZES). The catalytic and conformational switch residues important for activation are highlighted in blue and orange, respectively. The percent sequence identity and similarity for ChxRRec with each homolog is listed. The proteins used for the primary sequence alignment are ChxR from C. trachomatis (UniProt: B0B8K5), HP1043 from H. pylori (NCBI: NP_207833.1), YycF from B. subtilis (UniProt: P37478), OmpR from Escherichia coli (GenBankTM: CAQ33726.1), and PhoB from E. coli (GenBankTM: ACJ50526.1).
Another observation from this comparison was that 13 residues in total are absent in ChxR in two regions corresponding to α2 and part of α3 in other OmpR/PhoB subfamily members (Fig. 1). Additionally, a primary sequence comparison using ChxRRec from other serovars of C. trachomatis (A and D) and another species of Chlamydia (C. pneumoniae) with the OmpR/PhoB subfamily members listed above gave similar results (data not shown). The absence of these 13 residues in ChxR may have a large impact on the number and length of secondary structure elements in ChxRRec.
Secondary Structure Analysis
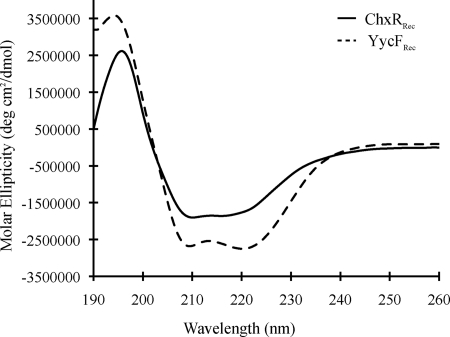
The absence of many residues in ChxR relative to other OmpR/PhoB subfamily members could affect the number and length of secondary structure elements in ChxRRec. Therefore, CD was employed to determine the relative secondary structure content of ChxRRec (Fig. 2). The receiver domain of YycF (YycFRec), an OmpR/PhoB homolog from Bacillus subtilis, was used as a reference protein for the analysis because its structure has been determined and is very similar to the five-α/β-fold topology of the OmpR/PhoB subfamily (39). YycFRec is also one of the closest homologs of ChxR based upon primary sequence homology (Fig. 1). The CD analysis indicated that a majority (37.6 ± 0.013%) of ChxRRec is random coil and that 26.4 ± 0.005, 23.0 ± 0.029, and 18.2 ± 0.002% of the protein is α-helical, β-sheet, and β-turn, respectively. In contrast, CD analysis of YycFRec indicated that the protein is 32.1 ± 0.005% random coil, 32.5 ± 0.014% α-helical, 16.9 ± 0.008% β-sheet, and 16.6 ± 0.003% β-turn. Comparing the relative estimated percentages of the secondary structures in the two proteins indicates that the α-helical content of ChxRRec is reduced relative to YycFRec.
FIGURE 2.
Comparative CD spectra of ChxRRec and a closely related response regulator (YycF). An estimation of the secondary structure elements was determined from a CD analysis of ChxRRec (solid line). The receiver domain of YycF (YycF1aRec), one of the closest homologs of ChxRRec based upon primary sequence comparison, was used as a reference protein (dotted line).
ChxRRec Is a Stable Homodimer
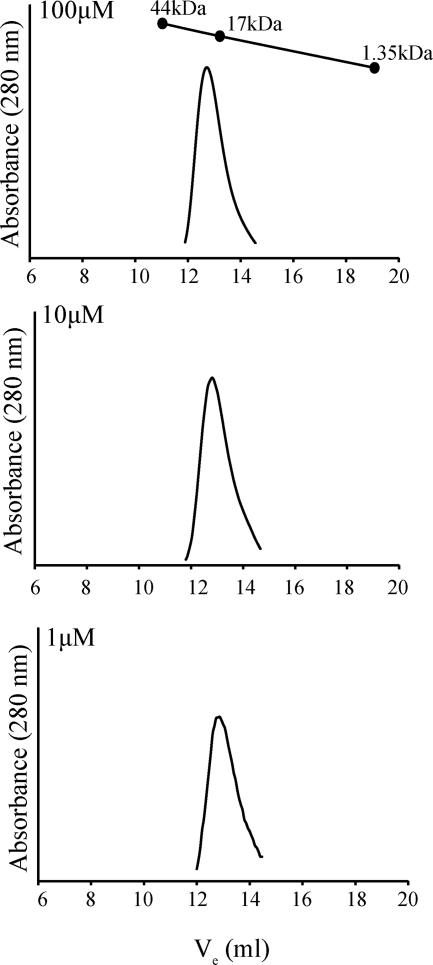
The physiological concentrations of some members of the OmpR/PhoB subfamily have been reported to be ∼1–15 μm (40, 41). Although the physiological concentration of ChxR in Chlamydia is unknown, our prior studies with full-length ChxR indicated that it is a stable homodimer at 1 μm (16). Because dimerization occurs through the receiver domain, we determined the oligomeric state of the ChxR receiver domain at a relatively high concentration (100 μm) and at two concentrations (10 and 1 μm) within the reported physiological concentrations of OmpR/PhoB subfamily members (Fig. 3). The calculated molecular mass of a monomer of ChxRRec is 13.8 kDa. ChxRRec eluted from the analytical size exclusion column as a single population with an approximate molecular mass of 21 kDa, independent of concentration, corresponding to a compact homodimer. These results indicate that ChxRRec is a stable homodimer at the physiological concentrations of other members of the OmpR/PhoB subfamily.
FIGURE 3.
Influence of protein concentration on the stability of ChxRRec. To determine the oligomeric state of the receiver domain of ChxR, recombinant ChxRRec was subjected to analytical size exclusion chromatography at 100, 10, and 1 μm. The calculated molecular mass of monomeric and dimeric ChxR is 13.8 and 27.6 kDa, respectively. Chicken ovalbumin (44 kDa), horse myoglobin (17 kDa), and vitamin B12 (1.35 kDa) were used to generate the standard curve.
ChxRRec Structure
Structural studies were performed with ChxRRec to elucidate the residues and structural elements that contribute to the constitutive activity of the protein. Crystallization screening with recombinant ChxRRec and commercially available sparse matrix screens resulted in multiple crystal forms from which two high resolution data sets were obtained (Table 1). The space groups of these two crystals were monoclinic C2 and tetragonal I41. Molecular replacement with the current collection of known receiver domain structures of OmpR/PhoB subfamily members was unsuccessful. Therefore, the structure of ChxRRec was solved using SIRAS phasing with the recently developed compound I3C. I3C has proven to be a remarkable compound for phasing because it gives a strong anomalous signal using in-house x-ray instrumentation (Cu-Kα) from its three iodine atoms, and its carboxylic acid and amino groups facilitate hydrogen bonding with a protein (22, 42). Protein crystals were soaked in the crystallant supplemented with I3C. Using the I3C for initial phasing, the final ChxRRec models of the two crystal forms were refined using data to 2.15 and 1.6 Å resolution for the I41 and C2 crystal forms, respectively.
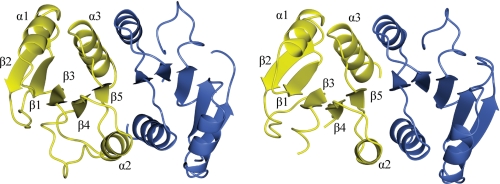
The molecular topology of the two ChxRRec models is β1-α1-β2-β3-β4-α2-β5-α3 with two large random coils between β2-β3 and β3-β4 (Fig. 4). The r.m.s. deviations between the Cα for 188 of a total of 194 residues was 0.95 Å, indicating a high degree of structural similarity between the two models. The biggest regions of dissimilarity between the two models are in the random coils between β2-β3 and β3-β4; however, the Cα r.m.s. deviations for the residues in these regions is <2 Å. The asymmetric unit of both crystal lattices consisted of two ChxRRec monomers, which formed a homodimer with a similar interface (α2-β5-α3). The dimer interface surface area of each monomer was ∼1095 Å2.
FIGURE 4.
Ribbon diagram of ChxRRec. ChxRRec crystallized in two distinct crystal forms. High resolution data sets from each crystal form were refined to 2.15 Å (I41 space group) and 1.6 Å (C2 space group). The asymmetric unit of both crystals contained two protein molecules. The molecular topology of each monomer is β1-α1-β2-β3-β4-α2-β5-α3. The two molecules from the C2 data set (left) and the I41 data set (right) are shown in yellow and blue, respectively.
Structural Comparison of ChxRRec with Other OmpR/PhoB Subfamily Members
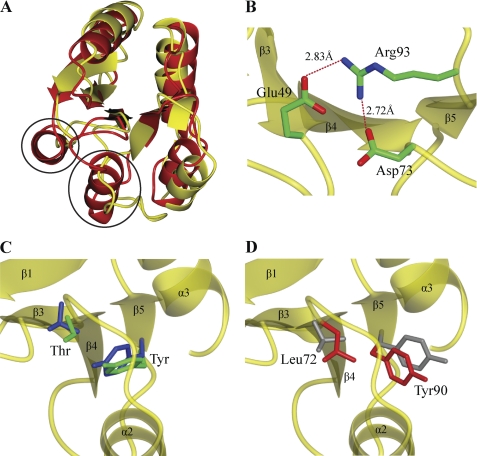
The structure of ChxRRec is distinct from other subfamily members. A superimposition of ChxRRec and YycFRec indicated that the r.m.s. deviations between the Cα of 94 residues in each monomer was 2.05 Å (Fig. 5A). Despite the overall structural conservation, the α2 and α3 of YycFRec correspond to random coils in ChxRRec (Fig. 5B). The absence of these helices in the ChxRRec structure may suggest that these two regions are random coils in endogenous ChxR.
FIGURE 5.
Comparison of ChxRRec structural features to other OmpR/PhoB subfamily members. A, the structure of ChxRRec (yellow) was superimposed on the receiver domain of the OmpR/PhoB subfamily member YycF (red) (PDB ID 3F6P). Two α-helices that are present in YycFRec are absent in ChxRRec (black circles). B, the conserved phospho-accepting Asp and coordinating Lys in other OmpR/PhoB subfamily members is a Glu (Glu-49) and Arg (Arg-93), respectively, in ChxR. Arg-93 forms a salt bridge with Glu-49 and Asp-73. C, two key conformational switch residues, typically a Ser/Thr and Phe/Tyr, reorient toward the active site upon phosphorylation (blue; PhoP (PDB ID 2PL1)). These two residues in an atypical OmpR/PhoB homolog (green; HP1043 (PDB ID 2PLN)) have a similar orientation as an activated subfamily member. D, the conformational switch residues in ChxRRec (red; Leu-72 and Tyr-90) are oriented similar to an inactive homolog (gray; PhoP (PDB ID 2PKX)).
In addition to the distinct molecular topology of ChxRRec, the structure also revealed that the architecture and residue composition of canonical site of phosphorylation is unique. As mentioned previously (Fig. 1), none of the residues that coordinate the divalent cation and phosphoryl group in phospho-accepting homologs was retained in ChxR. In ChxRRec, Glu-49 and Arg-93 replace the phospho-accepting Asp and coordinating Lys, respectively (Fig. 5B). Interestingly, Arg-93 forms a salt bridge with Glu-49; therefore, these residues mimic the positions of the Asp and Lys in phosphorylated OmpR/PhoB homologs. This interaction could be important in maintaining ChxR in a constitutively active state.
The rotomeric state of the two conformational switch residues reflects the activation state of OmpR/PhoB response regulators (3). Because recombinant ChxRRec was shown to exist as a stable homodimer (Fig. 3), we hypothesized that the orientation of the residues (Leu-72 and Tyr-90) in the same position as the canonical conformational switch residues would be in a similar conformation to that of an activated homolog (i.e. toward the site of phosphorylation) (Fig. 5C). In fact, the ChxR receiver domain structure revealed the exact opposite. These two residues had similar orientations to those in inactive subfamily members (Fig. 5D). This suggests that these two residues possibly contribute to the oligomeric state of ChxR in a different fashion than they do in other subfamily members.
ChxRRec Dimer Interface
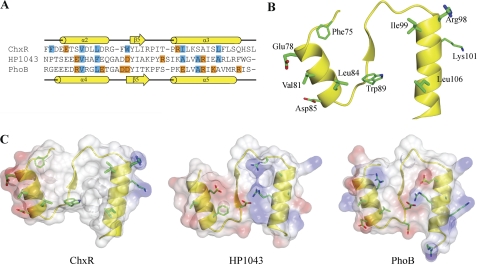
Based on a sequence alignment of residues comprising the dimer interface of ChxR, HP1043, and PhoB (Fig. 6A), two of the three hydrophobic residues in PhoB (Val-91, Leu-94, and Ala-112) are retained in ChxR (Val-81 and Leu-84). Additionally, four of the residues involved in hydrophobic interaction in an HP1043 dimer are retained in ChxR (Val-81, Leu-84, Iso-99, and Leu-106). An analysis of the accessible surface area supports that these four residues comprise a relatively large percentage (∼27%) of the interface surface area. In addition, the accessible surface area analysis indicated that Phe-75 and Trp-89 (Fig. 6B), which comprise 7 and 12% of the accessible surface area, respectively, contribute to the hydrophobic core of the interface.
FIGURE 6.
Comparison of the hydrophobic and charged residues at the dimer interface of OmpR/PhoB subfamily members. A, shown is sequence alignment of the residues comprising the dimer interface of ChxR, HP1043 (atypical), and PhoB (typical). The secondary structures of ChxRRec and PhoB are indicated above and below the alignment, respectively. Blue and orange highlights represent residues involved in hydrophobic and ionic interactions, respectively. B, hydrophobic interaction between ChxRRec monomers occurs through Phe-75, Val-81, Leu-84, Trp-89, Ile-99, and Leu-106 in each monomer, whereas ionic interactions occur through Glu-78 and Arg-98 and potentially through Asp-85 and Lys-101 between each monomer. C, shown is the electrostatic potential surface of the dimer interface of ChxR, HP1043, and PhoB.
In contrast to the relative conservation of hydrophobic residues between ChxR and other subfamily members, the locations of the residues involved in ionic interactions between the two ChxR receiver domains are distinct from the subfamily. The residues involved in these ionic interactions in other subfamily members are positioned throughout the dimer interface (Fig. 6C). However, the structure of ChxRRec revealed only two salt bridges that occur through Glu-78 and Arg-98 of each monomer. These residues are located within the α2 and α3, respectively (Fig. 6B). The ChxRRec structure also suggested a potential second intermolecular salt bridge between Lys-101 and Asp-85 (Fig. 6B). These two residues are ∼5 Å apart in the crystal structure. Although the distance between the two residues is slightly outside the limit of a salt bridge (4 Å) (43), the position of these two residues could be closer in solution. However, a salt bridge between these two residues is likely not essential for dimerization given the length between the residues and that solvent ions would compete to interact with these residues.
A comparison of the surface area and the residue composition of the dimer interface from many members of the OmpR/PhoB subfamily support that the dimer interface of ChxR is unique within the subfamily. As Table 2 indicates, the interface surface area of activated or inactivated phosphorylation-dependent OmpR/PhoB response regulators generally ranges from 1090 to 807 Å2. Furthermore, the residues that comprise their intermolecular interface are 27–39% nonpolar, 8–32% polar, and 36–56% charged. Despite the activity of HP1043 in the absence of phosphorylation, the dimer interface surface area and the residue composition are similar to typical OmpR/PhoB response regulators. The intermolecular surface area of ChxR is 1095Å2 and the percentage of nonpolar, polar, and charged residues is 52, 26, and 22%, respectively. The relatively large percentage of hydrophobic residues within the interface of ChxR and its larger surface area than most OmpR/PhoB response regulators provide further support that intermolecular interactions between ChxR monomers are distinct from both typical and atypical OmpR/PhoB response regulators.
TABLE 2.
Surface area and residue composition of the OmpR/PhoB dimer interface
| Protein | PDB ID | Interface surface areaa | Interface residuesa |
||
|---|---|---|---|---|---|
| Non-polar | Polar | Charged | |||
| Å2 | % | ||||
| ChxR (atypical) | 3Q7R | 1095 | 52 | 26 | 22 |
| YycF (inactive) | 3F6P | 1087 | 36 | 11 | 53 |
| PhoP (BeF3-activated) | 2PL1 | 1027 | 32 | 32 | 36 |
| TorR (inactive) | 1ZGZ | 981 | 28 | 24 | 48 |
| DrrD (Inactive) | 3NNN | 978 | 39 | 13 | 48 |
| PhoB (BeF3-activated) | 1ZES | 977 | 36 | 8 | 56 |
| PhoP (inactive) | 2PKX | 936 | 35 | 25 | 39 |
| ArcA (BeF3-activated) | 1XHF | 887 | 27 | 23 | 50 |
| ArcA (inactive) | 1XHE | 876 | 30 | 22 | 48 |
| HP1043 (atypical) | 2PLN | 837 | 38 | 17 | 45 |
| DrrB (inactive) | 3NNS | 802 | 38 | 19 | 43 |
a The dimer interface surface area and residue composition were calculated using PROTORP (51).
Rational for Residue Substitutions and Functional Analysis
As evident from the structure of ChxRRec, the architecture and residue composition of the canonical site of phosphorylation in OmpR/PhoB response regulators is not conserved in ChxR. Additionally, the noncovalent interactions between monomers are distinct in ChxR compared with typical and even other atypical OmpR/PhoB response regulators. Substitutions were generated within the residues comprising these two regions to begin elucidating their contribution to the constitutive active state of ChxR. The ability of each full-length protein to form homodimers was tested at a relatively low concentration (10 μm) by analytical size exclusion chromatography.
Glu-49 is the site in ChxR where the phospho-accepting Asp is typically located. Glu-49 in ChxRFL was previously substituted to an Asp (E49D) to test if the native Glu was solely responsible for the proteins constitutive activity (16). ChxRFLE49D retained homodimer formation and the ability to interact with DNA in vitro, suggesting that additional factors contribute to the protein constitutive activity. Upon solving the structure of ChxRRec, however, it was evident that Glu-49 forms an intramolecular salt bridge with Arg-93 (Fig. 5B) and that converting this residue to an Asp could potentially still retain the ionic interaction. Therefore, Glu-49 was substituted with an Ala (E49A) to disrupt the interaction with Arg-93. After purification, ChxRFLE49A was subjected to analytical size exclusion chromatography. The protein eluted at an approximate molecular mass of 45 kDa, corresponding to a homodimer (data not shown). This indicates that the Glu-49–Arg-93 interaction is not essential for dimerization.
Structural studies with inactive and activated OmpR/PhoB subfamily members have identified the conformation changes that occur in response to phosphorylation. The two conformational switch residues reorient toward the site of phosphorylation, which moves the β4-α4 loop and the N terminus of α4 toward the site of phosphorylation (9). This structural repositioning, along with other subtle structural changes (i.e. Cα r.m.s. deviations of ∼1 Å (6)) is thought to enhance the interactions between receiver domains thus promoting dimerization. Within the corresponding loop (β4-α2) in ChxR, Asp-73 forms a salt bridge with Arg-93 (Fig. 5B). The interaction between Asp-73 and Arg-93 may stabilize this loop in ChxR, which could be important in positioning α2 for dimerization. But when Asp-73 was substituted to an Ala (D73A), the full-length protein eluted from the column as a homodimer (data not shown), suggesting that the Asp-73–Arg-93 interaction is not critical for dimerization. As mentioned above, Glu-49 is also in the position to interact with Arg-93; therefore, a double substitution was also generated (E49A/D73A). This double substitution was expected to completely disrupt the interactions in ChxR in the region of the canonical site of phosphorylation in typical OmpR/PhoB subfamily members. Similar to the E49A or D73A single substitutions, the double substitution had no detectable effect on dimer stability as the protein was again determined to be a homodimer (data not shown).
Recently, Hong et al. (11) proposed that ionic interactions between receiver domains of the atypical OmpR/PhoB homolog HP1043 could be the initial intermolecular interaction and that hydrophobic interactions then stabilize dimerization. We hypothesized that disrupting these interactions in ChxR through Ala substitutions at Glu-78 (E78A), Lys-101 (K101A), or in combination (E78A/K101A) would have little effect on the oligomeric state of the protein as the intermolecular interface is primarily comprised of hydrophobic residues. Our functional analysis supports this hypothesis, as each full-length protein was determined to exist as a homodimer in solution (data not shown).
In addition to disrupting the ionic interactions between ChxR monomers, we introduced a substitution within the hydrophobic center of the interface to determine the importance of this interaction in dimer stability. A candidate for this substitution was Trp-89, a residue that is oriented toward the opposing monomer within the dimer interface, which contributes the most surface area to the interface, and Trp-89 in monomer B forms a hydrogen bond (2.72 Å) with Gln-110 in monomer A. Additionally, Trp-89 is located within the hydrophobic core of the dimer interface, packed closely with hydrophobic residues Leu-69 (3.8 Å; distance away from Trp-89), Leu8–4 (3.8 Å), and Phe-107 (4.7 Å) in the same monomer and Leu-69 (4.0 Å), Trp-89 (3.0 Å), Leu-91 (3.5 Å), Phe-107 (3.9 Å), Ala-103 (4.6 Å), and Leu-106 (4.6 Å) in the opposing monomer. We hypothesized that substituting Trp-89 to a Glu (W89E) would destabilize the dimer by disrupting the hydrophobic interaction between the two monomers. Trp-89 was also substituted to an Ala (W89A) to determine whether the orientation and/or relatively bulky size of Trp-89 is an important factor in the hydrophobic interactions at the dimer interface.
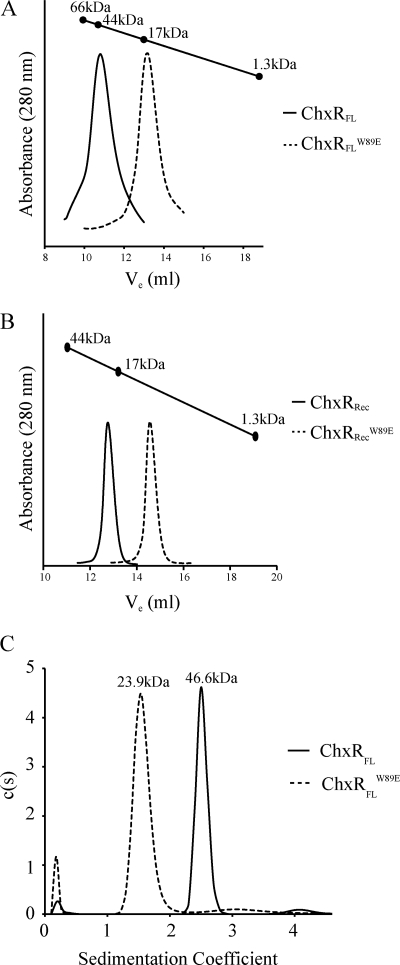
ChxRFLW89E eluted from the analytical size exclusion column as a monomer with an approximate molecular mass of 16 kDa (Fig. 7A). Additionally, no higher-ordered species were present in the chromatogram. Further supporting that this substitution disrupts dimerization was that ChxRRecW89E was subjected to analytical size exclusion chromatography, which eluted as a monomer with an approximate molecular mass of 9.8 kDa (Fig. 7B). ChxRFLW89E was also analyzed through analytical ultracentrifugation, which further supports that the W89E substitution abolishes dimer formation (Fig. 7C). In contrast, the W89A substitution had no detectable effect on dimer stability as it eluted from the column as a homodimer (data not shown). These results provide strong evidence that dimer destabilization can be accomplished with the introduction of a charged residue within the hydrophobic core between ChxR monomers.
FIGURE 7.
Dimerization analysis of the W89E substitution. Analytical size exclusion chromatography and ultracentrifugation was utilized to determine the oligomeric state of the W89E substitution. The calculated molecular mass of a ChxRFL monomer or dimer is 25.8 or 51.6 kDa, respectively. A, wild-type ChxRFL (solid line) eluted from an analytical size exclusion column as a dimer with an approximate molecular mass of 45 kDa, whereas ChxRFLW89E (dashed line) eluted from the column as a compact monomer with an approximate molecular mass of 16 kDa. B, wild-type ChxRRec (solid line) and ChxRRecW89E (dashed line) eluted from an analytical size exclusion column as a dimer (21 kDa) and monomer (9.8 kDa), respectively. C, wild-type ChxRFL (solid line) was determined to exist as a dimer through analytical ultracentrifugation with an approximate molecular mass of 46.6 kDa, whereas ChxRFLW89E (dashed line) was determined to exist as a monomer with an approximate molecular mass of 23.9 kDa.
Monomeric ChxR Binds Weakly to DNA in Vitro
A study with PhoP from B. subtilis reported that a residue substitution at the dimer interface prevented the protein from forming dimers and interacting with DNA in vitro (44). In contrast, the atypical response regulator NblR was reported to bind to DNA as a monomer (45). To determine whether monomeric ChxR can bind to DNA, EMSAs were performed with ChxRFLW89E and the previously reported high affinity binding site (DR2) in the chxR promoter (Fig. 8) (16). Although the other substitutions (W89A, E49A, D73A, E49A/D73A, E78A, K101A, and E78A/K101A) did not affect dimer stability, as determined through analytical size exclusion chromatography, their ability to interact with DNA was also quantified to determine whether they influence overall protein conformation and/or an interaction between the receiver and effector domain. The effector domain of ChxR was included in the assay as a control for DNA interaction in the absence of the receiver domain.
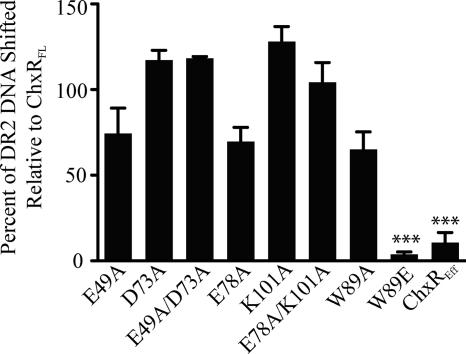
FIGURE 8.
DNA binding analysis of activation and dimer interface substitutions in ChxR. To determine the effect of the substitutions on the protein ability to bind DNA, EMSAs were performed with 1 nm concentrations of the high affinity DR2 site from the chxR promoter and 44 nm concentrations of each protein. The percent of DNA shifted was calculated for each substitution relative to wild-type ChxRFL. Substitutions that had a significant (p < 0.001) effect on DNA interaction are denoted by an asterisk (***). Error bars represent the S.D, from triplicate experiments.
As evident in Fig. 8, disrupting the hydrophobic interaction within the dimer interface had a significant impact on ChxR-DNA interaction. The amount of DNA shifted by the dimer mutants W89A, E49A, D73A, E49A/D73A, E78A, K101A, and E78A/K101A was found to not be statistically significant from wild-type ChxR. In contrast, the W89E substitution had the lowest affinity (3%) for DNA relative to wild-type ChxRFL. The dramatic reduction in DNA affinity with the W89E substitution suggests that dimerization through the receiver domain is critical for stable ChxR-DNA interaction, albeit within the given in vitro experimental conditions.
DISCUSSION
The structure of ChxRRec supports the hypothesis that the intra- and intermolecular interactions in ChxR are distinct from its phosphorylation-dependent homologs. The ChxRRec structure revealed that the canonical site of phosphorylation is composed of three resides (Glu-49, Asp-73, and Arg-93; Fig. 5B). When Glu-49 was substituted to an Ala, the protein was still able to form a homodimer, and the amount of DNA electrophoretically shifted with this substitution was not statistically significant from that of wild-type ChxR (Fig. 8). Similar to the Glu-49 substitution, the Asp-73 and Glu-49/Asp-73 substitutions resulted in proteins that formed homodimers and interacted with DNA in a manner similar to wild-type ChxR. These results were expected as the two conformational switch residues in ChxR are oriented away from the canonical site of phosphorylation (Fig. 5D), and therefore, modifications to this region would likely not be transduced to the dimer interface. These results also indicate that the residues in ChxR in the same positions as the residues critical to the coordination of the phosphoryl group in other OmpR/PhoB subfamily members do not significantly influence overall protein stability, homodimerization, or interaction with DNA.
ChxR Dimer Stability
Receiver domains from phosphorylation-dependent OmpR/PhoB subfamily members primarily exist in a monomeric state in the absence of phosphorylation (6, 7). Analytical size exclusion chromatography indicated that ChxRRec is a dimer, even at a concentration (1 μm; Fig. 3) similar to the physiological concentration of other OmpR/PhoB subfamily members (41). This observation implies that ChxR receiver domains have a higher propensity to form dimers than typical OmpR/PhoB subfamily members. The structure of the receiver domain supports this observation as six residues contribute to the hydrophobic interaction between monomers. This is twice the number of hydrophobic residues found in the dimer interface of homologs (5). Furthermore, a comparison of the ChxR dimer interface and other OmpR/PhoB response regulators indicated that the primarily hydrophobic dimer interface of ChxR is very distinct from other subfamily members (Table 2). Additionally, the only substitution that rendered ChxR monomeric was to a residue (W89E) within the hydrophobic region of the dimer interface (Fig. 7). In combination, these results strongly support the conclusion that ChxR is a stable homodimer in solution and that this interaction is accomplished largely through hydrophobic interactions.
Significance of ChxR Dimerization
Dimerization through the receiver domain of ChxR is essential for stable interaction with DNA. ChxR was previously reported to interact with tandem repeat sequences, but mutations to either recognition site greatly reduced ChxR-DNA interaction (16). These results suggested that optimal ChxR-DNA interaction requires that ChxR bind to DNA as a homodimer. EMSAs with a dimer-deficient ChxR support this observation as the amount of DNA shifted with the W89E substitution was reduced >95% compared with wild-type ChxR (Fig. 8). In support of this observation, the effector domain of ChxR alone binds to DNA with ∼10-fold less affinity than full-length ChxR.3 Currently, it is unknown whether the ChxR receiver domain is responsible for positioning the effector domain for optimal interaction with DNA through a direct interaction, but these data strongly support that dimerization increases DNA affinity by binding cooperatively to adjacent binding sites.
DNA Binding Characteristics of Atypical Response Regulators
Interestingly, the significant reduction in DNA binding by monomeric ChxR is in stark contrast to other atypical OmpR/PhoB response regulators, which may be a result of the DNA sequences recognized by these proteins. NblR was reported to exist as a monomer in vivo and likely binds to DNA as a monomer (45). A dimer-deficient HP1043 protein was reported to bind to DNA with an apparent similar affinity as dimeric HP1043 (11). This suggests that dimerization of these atypical response regulators is not essential for DNA interaction. These differing DNA binding characteristics between ChxR, HP1043, and NblR are likely a result of their affinity and specificity for DNA. Although the DNA sequence recognized by NblR is currently unknown, HP1043 was determined to bind to relatively conserved DNA sequence (46). In contrast, the frequency of specific nucleotides in the DNA sequence recognized by ChxR is relatively low (16). This suggests that HP1043 forms dimers primarily to increase DNA specificity, whereas ChxR forms dimers to increase both DNA affinity and specificity.
Regulatory Mechanisms of Atypical Response Regulators
Recent studies have begun to identify potential mechanisms that regulate atypical response regulators, which include ligand and protein-protein interaction-based mechanisms. NarB, a nitrogen reductase from Synechococcus elongatus PCC 7942, has been shown to interact with NblR and possibly inhibit its transcriptional regulatory activity (47). Additionally, a recent study reported that the DNA binding activity of an atypical response regulator, JadR1 from Streptomyces venezuelae, is severely reduced in the presence of a compound (Jadomycin B), which led the authors to speculate that small ligands might regulate the activity of other atypical response regulators (48).
Our structural and functional observations presented here as well as previous reports (16, 19) strongly support that ChxR exists in a constitutively active state that is not influenced directly by phosphorylation. This conclusion is further supported by the absence of enhanced DNA binding when ChxR is treated with acetyl phosphate in conditions previously reported to phosphorylate OmpR/PhoB subfamily members (data not shown) (49). Similarly, the mobility of ChxR within native PAGE was not affected after incubation with increasing concentrations of acetyl phosphate (data not shown). In the absence of direct phosphorylation, it is highly expected that ChxR is regulated by an alternative mechanism similar to those observed in atypical response regulators. Whether a ligand or protein partner is employed is unknown; however, the two large random coils in the receiver domain of ChxR may serve as a site of regulation. Many studies have reported that random coils in proteins can undergo conformational transitions into more rigid structures in the presence of binding partners (50). The most commonly observed random coil-to-structured transition occurs in short (8–12 residues), hydrophilic regions that form a helix upon binding. Similarly, the two random coils in the ChxR receiver domain are short (8 and 11 residues, respectively) and contain few hydrophobic residues (1 and 2, respectively). If ChxR is post-transcriptionally regulated, these similarities suggest that the two random coils in the receiver domain may participate in the regulation.
Acknowledgments
Use of the IMCA-CAT beamline 17-ID at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-06CH11357. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the United States Department of Energy Office of Science by Stanford University. The Stanford Synchrotron Radiation Laboratory Structural Molecular Biology Program is supported by the Department of Energy Office of Biological and Environmental Research and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program (P41RR001209) and the NIGMS. Use of the KU COBRE Protein Structure Laboratory was supported by National Institutes of Health Grant P20 RR-17708 from the National Center for Research Resources. We thank Drs. Liang Tang and Haiyan Zhao for the recombinant YycF. We are extremely appreciative of critical comments and suggestions provided by Dr. Audrey Lamb.
This work was supported, in whole or in part, by National Institutes of Health Grants P20RR17708 and AI079083 (to J. M. H. and P. S. H.).
The atomic coordinates and structure factors (codes 3Q7R, 3Q7S, and 3Q7T) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
J. M. Hickey, A. Anbanandam, and P. S. Hefty, manuscript in preparation.
- ChxRRec
- ChxR receiver domain
- ChxRFL
- full-length ChxR
- YycFRec
- YycF receiver domain
- I3C
- 5-amino-2,4,6-triiodoisophthalic acid
- r.m.s.
- root mean square.
REFERENCES
- 1. Stock A. M., Robinson V. L., Goudreau P. N. (2000) Annu. Rev. Biochem. 69, 183–215 [DOI] [PubMed] [Google Scholar]
- 2. Wang S., Engohang-Ndong J., Smith I. (2007) Biochemistry 46, 14751–14761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gao R., Stock A. M. (2009) Annu. Rev. Microbiol. 63, 133–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bourret R. B. (2010) Curr. Opin. Microbiol. 13, 142–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Toro-Roman A., Mack T. R., Stock A. M. (2005) J. Mol. Biol. 349, 11–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bachhawat P., Stock A. M. (2007) J. Bacteriol. 189, 5987–5995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Toro-Roman A., Wu T., Stock A. M. (2005) Protein Sci. 14, 3077–3088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Solá M., Gomis-Rüth F. X., Serrano L., González A., Coll M. (1999) J. Mol. Biol. 285, 675–687 [DOI] [PubMed] [Google Scholar]
- 9. Bachhawat P., Swapna G. V., Montelione G. T., Stock A. M. (2005) Structure 13, 1353–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mack T. R., Gao R., Stock A. M. (2009) J. Mol. Biol. 389, 349–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hong E., Lee H. M., Ko H., Kim D. U., Jeon B. Y., Jung J., Shin J., Lee S. A., Kim Y., Jeon Y. H., Cheong C., Cho H. S., Lee W. (2007) J. Biol. Chem. 282, 20667–20675 [DOI] [PubMed] [Google Scholar]
- 12. Schär J., Sickmann A., Beier D. (2005) J. Bacteriol. 187, 3100–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delany I., Spohn G., Rappuoli R., Scarlato V. (2002) J. Bacteriol. 184, 4800–4810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fraser J. S., Merlie J. P., Jr., Echols N., Weisfield S. R., Mignot T., Wemmer D. E., Zusman D. R., Alber T. (2007) Mol. Microbiol. 65, 319–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O'Connor T. J., Nodwell J. R. (2005) J. Mol. Biol. 351, 1030–1047 [DOI] [PubMed] [Google Scholar]
- 16. Hickey J. M., Weldon L., Hefty P. S. (2011) J. Bacteriol. 193, 389–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Belland R. J., Zhong G., Crane D. D., Hogan D., Sturdevant D., Sharma J., Beatty W. L., Caldwell H. D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 8478–8483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nicholson T. L., Olinger L., Chong K., Schoolnik G., Stephens R. S. (2003) J. Bacteriol. 185, 3179–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koo I. C., Walthers D., Hefty P. S., Kenney L. J., Stephens R. S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hickey J. M., Spedding L., Jaafar Z. A., Hefty P. S. (2010) Chlamydial Infections: Proceedings of the 12th International Symposium of Human Chlamydial Infections, Hof bei Salzburg, Austria, June 20–25, 2010, (Schachter J., Byrne G., Caldwell H., Chernesky M. A., Clarke I. N., Mabey D. C., Paavonen J., Saikku P., Starnbach M., Stary A., Stephens R. S., Timms P., Wyrick P. B. eds) pp. 313–316, University Press, San Francisco, CA [Google Scholar]
- 21. Stephens R. S., Kalman S., Lammel C., Fan J., Marathe R., Aravind L., Mitchell W., Olinger L., Tatusov R. L., Zhao Q., Koonin E. V., Davis R. W. (1998) Science 282, 754–759 [DOI] [PubMed] [Google Scholar]
- 22. Beck T., Krasauskas A., Gruene T., Sheldrick G. M. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 1179–1182 [DOI] [PubMed] [Google Scholar]
- 23. Otwinowski Z., Minor W. (1997) Method Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 24. Sheldrick G. M. (2008) Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 25. Collaborative Computational Project, N (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 26. Cowtan K. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 27. Pflugrath J. W. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 1718–1725 [DOI] [PubMed] [Google Scholar]
- 28. Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 29. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Painter J., Merritt E. A. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 32. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 35. Böhm G., Muhr R., Jaenicke R. (1992) Protein Eng. 5, 191–195 [DOI] [PubMed] [Google Scholar]
- 36. Schuck P. (2000) Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hickey J. M., Hefty P. S., Lamb A. L. (2009) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 791–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 39. Zhao H., Heroux A., Sequeira R. D., Tang L. (2009) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 65, 719–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cai S. J., Inouye M. (2002) J. Biol. Chem. 277, 24155–24161 [DOI] [PubMed] [Google Scholar]
- 41. Lejona S., Castelli M. E., Cabeza M. L., Kenney L. J., García Véscovi E., Soncini F. C. (2004) J. Bacteriol. 186, 2476–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sippel K. H., Robbins A. H., Reutzel R., Domsic J., Boehlein S. K., Govindasamy L., Agbandje-McKenna M., Rosser C. J., McKenna R. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 1172–1178 [DOI] [PubMed] [Google Scholar]
- 43. Jelesarov I., Karshikoff A. (2009) Methods Mol. Biol. 490, 227–260 [DOI] [PubMed] [Google Scholar]
- 44. Chen Y., Birck C., Samama J. P., Hulett F. M. (2003) J. Bacteriol. 185, 262–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ruiz D., Salinas P., Lopez-Redondo M. L., Cayuela M. L., Marina A., Contreras A. (2008) Microbiology 154, 3002–3015 [DOI] [PubMed] [Google Scholar]
- 46. Delany I., Spohn G., Pacheco A. B., Ieva R., Alaimo C., Rappuoli R., Scarlato V. (2002) Mol. Microbiol. 46, 1107–1122 [DOI] [PubMed] [Google Scholar]
- 47. Kato H., Chibazakura T., Yoshikawa H. (2008) Biosci. Biotechnol. Biochem. 72, 1072–1079 [DOI] [PubMed] [Google Scholar]
- 48. Wang L., Tian X., Wang J., Yang H., Fan K., Xu G., Yang K., Tan H. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8617–8622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McCleary W. R., Stock J. B. (1994) J. Biol. Chem. 269, 31567–31572 [PubMed] [Google Scholar]
- 50. Sandhu K. S., Dash D. (2007) Proteins 68, 109–122 [DOI] [PubMed] [Google Scholar]
- 51. Reynolds C., Damerell D., Jones S. (2009) Bioinformatics 25, 413–414 [DOI] [PubMed] [Google Scholar]