Abstract
Cell cycle progression for the spherical microbe Staphylococcus aureus requires the coordinated synthesis and remodeling of peptidoglycan. The majority of these rearrangements takes place at the mid-cell, in a compartment designated the cross-wall. Secreted polypeptides endowed with a YSIRK-G/S signal peptide are directly delivered to the cross-wall compartment. One such YSIRK-containing protein is the murein hydrolase LytN. lytN mutations precipitate structural damage to the cross-wall and interfere with staphylococcal growth. Overexpression of lytN also affects growth and triggers rupture of the cross-wall. The lytN phenotype can be reversed by the controlled expression of lytN but not by adding purified LytN to staphylococcal cultures. LytN harbors LysM and CHAP domains, the latter of which functions as both an N-acetylmuramoyl-l-alanine amidase and d-alanyl-glycine endopeptidase. Thus, LytN secretion into the cross-wall promotes peptidoglycan separation and completion of the staphylococcal cell cycle.
Keywords: Bacteria, Cell Division, Cell Wall, Mass Spectrometry (MS), Peptidases, LytN, Staphylococcus aureus, YSIRK/GS Signal Peptide, Cross-wall, Murein Hydrolase
Introduction
Staphylococcus aureus is a frequent cause of skin and soft tissue infections, sepsis, endocarditis, or pneumonia (1). Most antibiotics in current clinical use fail to treat infections with methicillin-resistant S. aureus (2); vancomycin and daptomycin are considered antibiotics of last resort for methicillin-resistant S. aureus infection; however, these compounds are also frequently associated with therapeutic failure (3, 4). The identification of new targets for antibiotic therapy and the development of new drugs for methicillin-resistant S. aureus infections are therefore urgently needed. S. aureus is a small spherical bacterium with a diameter of 800–1,000 nm and a surrounding thick layer of peptidoglycan cell wall (5). Staphylococcal cell wall synthesis begins in the cytoplasm where a sequence of reactions generates lipid II (C55-PP-MurNAc(-l-Ala-d-iGlu-(NH2-Gly5)l-Lys-d-Ala-d-Ala)-GlcNAc). This product is translocated across the plasma membrane and polymerized into peptidoglycan strands with repeating disaccharide structure (MurNAc-GlcNAc)n (6–8). Neighboring peptidoglycan strands are cross-linked by penicillin-binding proteins (9), which form the amide bond between d-Ala at position four and the amino group of the pentaglycine (Gly5) cross-bridge within the wall peptides of peptidoglycan strands (Fig. 1A) (10). The end product of the cell wall synthesis pathway, the murein sacculus, can be thought of as a large macromolecule that protects staphylococci from osmotic lysis (11). Peptidoglycan also serves as a scaffold for the anchoring and display of proteins, teichoic acids, and carbohydrates on the staphylococcal surface (12).
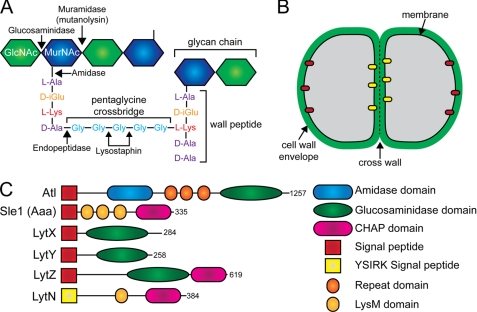
FIGURE 1.
Staphylococcal peptidoglycan and murein hydrolases. A, illustration of staphylococcal peptidoglycan with selected enzymatic cleavage sites. GlcNAc, N-acetylglucosamine; MurNAc, N-acetylmuramic acid. B, schematic of a dividing staphylococci illustrating the location of the cell wall envelope, membrane, and cross-wall. Yellow and red ovals represent translocons for the secretion of YSIRK/GS (yellow) and conventional signal peptide precursors (red), respectively. C, diagram of chromosomally encoded murein hydrolases with N-terminal signal peptides of S. aureus strain Newman. Functional domains predicted to exert specific hydrolytic activities are listed.
Shortly following cell division and separation, staphylococci assume a cell size that does not significantly increase prior to the next cell division (13, 14). Staphylococci synthesize a new layer of peptidoglycan at mid-cell in a compartment enclosed by membrane invaginations, which are formed following the constriction of FtsZ rings (5). This peptidoglycan synthesis compartment is designated the cross-wall (Fig. 1B). Once peptidoglycan synthesis in the cross-wall is completed, it separates two daughters that are half the size of the mother cell. Staphylococci cut the cross-wall peptidoglycan layer and separate their daughters (15). Because of high intracellular pressure, the former cross-wall section of each new daughter cell assumes the spherical shape of staphylococci. Daughter cells often remain connected through small segments of peptidoglycan. This pattern of incomplete cell wall separation produces the characteristic clusters of S. aureus cells (15). Incomplete separation of staphylococcal cells is exacerbated by mutations in either atl or sle1. Variants carrying deletions of atl, sle1, or both genes form abnormally large conglomerates of incompletely separated staphylococci, in agreement with the hypothesis that Atl and Sle1 contribute to the splitting of S. aureus cross-walls (16–18).
Atl is a prepro-protein. Following signal peptide removal, the pro-protein is cleaved, separating Atl amidase and glucosaminidase domains (19). Both domains possess repeat sequences that direct Atl enzymes to the vicinity of the cross-wall (19) (Fig. 1C). Sle1 is known to display N-acetylmuramoyl-l-Ala amidase activity (18). Unlike the Atl amidase, Sle1 harbors a cysteine, histidine-dependent amidohydrolase/peptidase (CHAP) domain (20, 21). Three LysM domains may be involved in directing Sle1 to the staphylococcal cell wall; however, the specific location of Sle1 within the staphylococcal cell wall envelope has not been revealed (18). Analyses of the genome sequence of S. aureus Newman identified four additional murein hydrolases, LytX (NWMN1667), LytY (NWMN2207), LytZ (NWMN2543), and LytN. LytX, LytY, and LytZ all possess an N-terminal signal peptide and glycosaminidase domain, with LytZ also containing a CHAP domain (Fig. 1C) (22). Other murein hydrolases lack signal peptides and likely require bacteriophage holins for export from the cytoplasm (23). All S. aureus isolates sequenced to date encode the lytN gene whose translational product is endowed with a YSIRK/GS-type signal peptide, CHAP and LysM domains. Precursor proteins with YSIRK/GS-type signal peptides are secreted into the cross-wall of S. aureus (24). Most of these proteins carry C-terminal sorting signals and, following cleavage by sortase A, are covalently linked to pentaglycine cross-bridges in the cross-wall (25, 26). The lytN gene was first described as being located immediately adjacent to fmhC, a homolog of femA, whose translational product transfers glycyl residues from glycyl-tRNA to the ϵ-amino group of l-Lys within lipid II (7, 27, 28). The contribution of lytN to staphylococcal growth and cell wall synthesis or degradation was previously not studied. In this study, we report that disruption of lytN results in a retardation of growth, gross abnormalities to the cell wall, in particular the cross-wall of dividing staphylococci, and biochemically characterizes the muralytic properties of LytN.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Reagents
S. aureus Newman (22) and its variant carrying a bursa aurealis insertion in lytN are part of the Phoenix library (29). The lytN mutational lesion was transduced with bacteriophage φ85 into the wild-type S. aureus Newman isolate (30). Plasmid DNA was first electroporated into S. aureus RN4220 (31) and then into the S. aureus Newman or the lytN mutant strain. S. aureus was grown in tryptic soy broth or on tryptic soy agar plates supplemented with appropriate antibiotics. Erythromycin and chloramphenicol were used at a concentration of 10 μg/ml. Escherichia coli was grown in LB broth or on LB agar plates supplemented with either ampicillin (100 μg/ml) or kanamycin (50 μg/ml). All chemicals used were purchased from either Sigma or Fisher, unless otherwise stated. Lysostaphin was purchased from AMBI Products.
lytN Expression Vectors and Protein Purification
For inducible expression of lytN in staphylococci, primers 5′-ggCCTAGGAGGAGGacagctatgtttttatattattgtaaggagtgtttcatc-3′ and 5′-gatcCCGCGGttatgcttttttaaatggtctaataaaaatc-3′ were used, which incorporate AvrII and SacII restriction sites, respectively, and permit amplification of lytN using S. aureus Newman genomic DNA as template. The PCR products, along with pMF312 (harboring the anhydrotetracycline-inducible promoter, iTet, on a replicating plasmid) (32), were digested with AvrII/SacII and ligated. Plasmid DNA was sequenced to confirm an error-free construct and introduced into S. aureus RN4220, followed by electroporation into wild-type S. aureus Newman or the isogenic lytN mutant.
A LytN-mCherry construct for expression in S. aureus was generated by SOE4 PCR. lytN was amplified using 5′-ggCCTAGGaggaggacagctatgtttttatattattgtaaggagtgtttcatc-3′ and 5′-ctcgcccttgcttgcttttttaaatggtctaataaaaatcat-3′, whereas mCherry was amplified using 5′-tttaaaaaagcaagcaagggcgaggaggataacatgg-3′ and 5′-gatcCCGCGGttacttgtacagctcgtccatgcc-3′. Underlined sequence denotes complementary sequence for the SOE reaction. Uppercase sequence indicates AvrII and SacII restriction sites, respectively. The two PCR products were isolated using the Qiaquick PCR cleanup kit (Qiagen) and combined for the SOE reaction using the 5′ primer of lytN and the 3′ primer of mCherry. The PCR product and pCL55itet (33, 34) were cut with AvrII and SacII and ligated as described above.
For expression and purification of recombinant LytN (rLytN), the signal peptide-less gene was amplified using primers 5′-ccGGATCCGgatgaaattgataaatctaaagattttacaagag-3′ and 5′-ggCTCGAGtgcttttttaaatggtctaataaaaatcat-3′, incorporating BamHI and XhoI sites, respectively. The PCR product, along with the pET24b vector, were cut with the corresponding enzymes and ligated, generating a C-terminal six histidyl tag for affinity chromatography. Plasmid prLytN was analyzed by DNA sequencing and transformed into E. coli BL21 (DE3).
To generate a C-terminal mCherry hybrid to LytN, SOE PCR was used and cloned as described above using primers 5′-ccGGATCCGgatgaaattgataaatctaaagattttacaagag-3′ and 5′-ctcgcccttgcttgcttttttaaatggtctaataaaaatcat (for lytN), and 5′-tttaaaaaagcaagcaagggcgaggaggataacatgg-3′ and 5′-ggCTCGAGcttgtacagctcgtccatgcc-3′ (mCherry). The SOE reaction product was ligated into pET24b using BamHI and XhoI restriction sites. Truncation of the CHAP domain was performed using the primers 5′-ccGGATCCGgatgaaattgataaatctaaagattttacaagag-3′ and 5′-ggCTCGAGactattacttttattatttgaagacactgtttttg-3′ and cloned as described above. LytN amino acid substitutions were performed by QuikChange mutagenesis (Stratagene) using primers 5′-ggtagttatggatggcaaGCTttcgatttagttaatgta-3′ and 5′-tacattaactaaatcgaaAGCttgccatccataactacc-3′ for the C266A mutation or 5′-tttggtggaggatatggtGCTacagctattgtcttaaat-3′ and 5′-atttaagacaatagctgtAGCaccatatcctccaccaaa-3′ for the H329A mutation. prLytN was used as template for mutagenesis.
To purify rLytN, E. coli BL21 (DE3, prLytN) was grown to an A600 0.6 and rLytN, expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at 37 °C. Cells were sedimented by centrifugation, suspended in column buffer (50 mm NaH2PO4, 300 mm NaCl, 10 mm imidazole, pH 8.0), and lysed by two passages in a French pressure cell (14,000 pounds/inch2). Lysates were cleared by centrifugation at 30,000 × g and subjected to affinity chromatography on nickel-nitrilotriacetic acid-Sepharose. The column was washed with column buffer containing 10 or 20 mm imidazole. rLytN was eluted in column buffer with 250 mm imidazole and dialyzed against column buffer without imidazole.
Purification and Digestion of Peptidoglycan
Peptidoglycan purification was performed as described previously (35). Briefly, 2 liters of staphylococci were grown to an A600 0.6, quickly cooled to 4 °C in an ice bath, and centrifuged. Cells were washed in water, suspended in 4% SDS, and boiled for 30 min. Detergent was removed by washing cells extensively in water. Staphylococci were subjected to bead beating, and glass beads were removed, and cell debris was sedimented by centrifugation. The extract was incubated with amylase for 2 h, followed by the addition of DNase and RNase for 2 h, and finally trypsin was added and incubated for 16 h at 37 °C. Peptidoglycan extracts were centrifuged, washed with water, and suspended in 1% SDS and boiled for 15 min to heat-inactivate all enzymes. Peptidoglycan was extensively washed with water to remove all traces of SDS, followed by washing with 8 m LiCl, 100 mm EDTA, and acetone. Cell walls were then washed with water and lyophilized. Hydrofluoric acid was then added and incubated for 48 h at 4 °C to remove teichoic acid. Peptidoglycan was neutralized, and the pellet was treated with alkaline phosphatase for 16 h at 37 °C. Purified peptidoglycan was boiled for 5 min, washed with water, and stored at 4 °C.
Peptidoglycan, adjusted for concentration by A600, was incubated for 16 h at 37 °C with 100 μl of enzyme:lysostaphin (2 mg/ml), mutanolysin (7,000 units/ml), or rLytN (300 μg/ml). Peptidoglycan samples were incubated in 12.5 mm phosphate buffer, pH 5.5 (mutanolysin), 50 mm phosphate buffer, pH 5.5 (rLytN), or 50 mm Tris-HCl, pH 7.5, 150 mm NaCl (lysostaphin). After 16 h of incubation, peptidoglycan samples were boiled for 10 min and centrifuged at 15,000 × g for 15 min. Soluble material was either incubated with rLytN or reduced via the addition of 0.5 m sodium borate and 3–5 mg of sodium borohydride. Samples were incubated for 30 min and inactivated by the addition of 20% phosphoric acid to reach pH <4.0.
Purification of Muropeptides and Mass Spectrometry
Reduced muropeptides were separated by rpHPLC on 250 × 4.6-mm ODS hypersil C18 column (Thermo Scientific). Individual peaks were desalted as described previously (36). Desalted muropeptides were dried under vacuum, suspended in 5–30 μl of 30% acetonitrile, and 0.5 μl were co-spotted with 0.5 μl of matrix, α-cyano-4-hydroxycinnamic acid, at 10 mg/ml in 50% acetonitrile, 0.1% trifluoroacetic acid (TFA). Samples were subjected to MALDI-TOF mass spectrometry using a Autoflex Speed Bruker MALDI instrument. Ions were detected in reflectron positive mode. For Edman degradation, purified muropeptides were desalted by rpHPLC, dried under vacuum, and analyzed by a Procise 494 HT sequencer (PerkinElmer Life Sciences and University of Illinois at Urbana-Champaign Protein Sciences Facility).
Immunoblotting and Microscopy
Staphylococci were grown to mid log phase, sedimented by centrifugation, and incubated with mutanolysin, lysostaphin, or rLytN. Proteins were precipitated with 7.5% trichloroacetic acid and washed with ice-cold acetone. Protein samples were dried, solubilized in sample buffer, and separated on 10% SDS-PAGE. Following electro-transfer to PVDF membrane (Millipore), residual binding sites on the filter were blocked with 5% milk in PBS, 0.1% Tween 20. For SpA detection, monoclonal antibodies (SPA-27; Sigma) were used. Secondary antibodies were HRP-conjugated (Cell Signaling Technology) and visualized following incubation with ECL solution.
Fluorescent microscopy was performed by first fixing cells in 2.5% paraformaldehyde, 0.006% glutaraldehyde, 30 mm NaPO4, pH 7.4, for 20 min. Cells were blocked in PBS containing 3% BSA and stained with BODIPY-vancomycin as described previously (37). Cells were applied to polylysine-treated glass coverslips and mounted onto glass slides containing a drop of SlowFade anti-fading reagent (Invitrogen). Images were captured on a Leica TCS SP2 AOBS laser scanning confocal microscope with 100× oil objective.
Electron Microscopy
Transmission electron microscopy was performed essentially as described previously (34). For scanning electron microscopy, staphylococci were suspended in H2O, washed twice, fixed for 30 min in 2% glutaraldehyde in phosphate-buffered saline (PBS) at room temperature, and post-fixed for 30 min in glutaraldehyde onto freshly prepared poly-l-lysine-coated glass coverslips. Samples were washed twice with PBS and subsequently serially dehydrated by consecutive incubations in 25 and 50% ethanol/PBS, 75 and 90% ethanol/H2O, 2× 100% ethanol, followed by 50% ethanol/hexamethyldisilazane and finally with 100% hexamethyldisilazane. After overnight evaporation of hexamethyldisilazane at room temperature, samples were mounted onto specimen mounts (Ted Pella, Inc., Redding, CA) and coated with 80% platinum, 20% palladium to 8 nm using a Cressington 208HR Sputter Coater at 20 mA prior to examination with a Fei Nova NanoSEM 200 scanning electron microscope (FEI Co., Hillsboro, OR). The S.E. was operated with an acceleration voltage of 5 kV, and samples were viewed at a distance of 5 mm (38).
Circular Dichroism
Recombinant proteins were purified as described above and dialyzed against 10 mm sodium phosphate buffer, pH 6.0, 50 mm sodium sulfate. Proteins were then analyzed with a AVIV 202 CD Spectrometer in the far-UV range. Readings were taken in triplicate, buffer subtracted, and plotted.
RESULTS
lytN Is Required for Proper Staphylococcal Growth
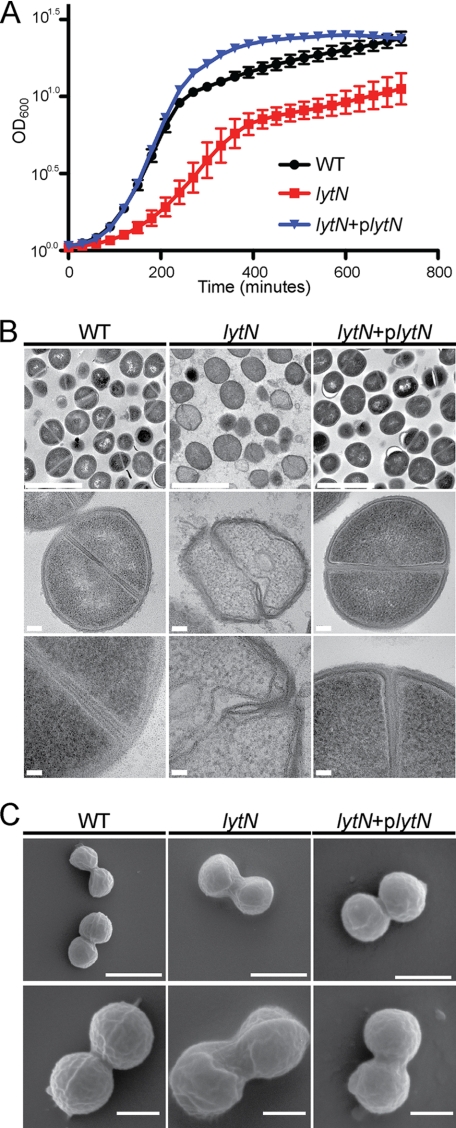
The lytN open reading frame is composed of 384 codons (22). A bursa aurealis insertion at codon 258 of lytN from the Phoenix library (29) was transduced with bacteriophage φ85 into the wild-type strain S. aureus Newman. The mutational lesion was verified by DNA sequencing. Wild-type and lytN strains were cultured in tryptic soy broth at 37 °C, and growth was monitored as an increase in absorbance (A600). Compared with S. aureus Newman, the lytN mutant multiplied more slowly and, during stationary phase, failed to reach the same density (Fig. 2). We also measured staphylococcal growth by plating culture aliquots on agar media followed by enumeration of colony-forming units; the lytN mutant displayed a similar growth defect in this assay (data not shown). The lytN growth defect was restored to wild-type levels when a copy of the intact lytN gene was supplied on a plasmid under the control of the anhydrotetracycline-inducible promoter (Fig. 2). Expression of lytN in the resulting strain, lytN (Ptet::lytN), can be induced with anhydrotetracycline, although the regulation of expression is incomplete (34). The phenotype of lytN mutants was restored to wild-type levels by growth of lytN (Ptet::lytN) staphylococci in the absence of the anhydrotetracycline inducer (Fig. 2).
FIGURE 2.
lytN mutation results in S. aureus growth defects and altered cellular morphology. A, growth rate of wild-type S. aureus Newman, its lytN variant, and the complemented mutant was interrogated by monitoring the absorbance at A600 over 12 h at 37 °C. Experiments were performed in triplicate, and error bars represent the means ± S.E. B, transmission electron micrograph of wild-type (WT), lytN mutant, and the lytN complemented cells grown to mid-logarithmic phase. Scale bar for top panels is 2 μm; middle panels is 100 nm; bottom panels is 50 nm. C, scanning electron micrographs of WT, lytN, and the lytN complemented mutant cells as described above. Scale bar for top panels is 1 μm, and bottom panels is 500 nm.
Staphylococci were grown to mid-logarithmic phase, fixed, embedded in paraffin, thin-sectioned, stained with uranyl acetate, and examined for their cell wall envelope structure by transmission electron microscopy. As expected, S. aureus Newman elaborated murein sacculi and cross-walls with characteristic thickness and uniform integrity (5). In contrast, the cell wall envelope of lytN mutants appeared irregular in structure with discrete layers of peptidoglycan that were disintegrated and distorted in their spherical shape (Fig. 2). Scanning electron microscopy experiments corroborated these findings. Images of wild-type staphylococci revealed the characteristic round cell shape with equatorial surface rings, the binding sites for the cell separation enzyme Atl. In contrast, the surface of lytN mutants appeared irregular, and the spherical shape of mutant cells appeared deformed. Cell division sites and equatorial surface rings could not be clearly discerned in images of lytN mutant cells. Moreover, daughter cell separation appeared impaired, implying a role for LytN in cell separation. Introduction of the wild-type lytN gene in trans restored the structural features of peptidoglycan in the cell wall envelope and cross-wall of lytN mutants to those observed in wild-type staphylococci (Fig. 2).
Overexpression of lytN Triggers Lysis of the Staphylococcal Cross-wall
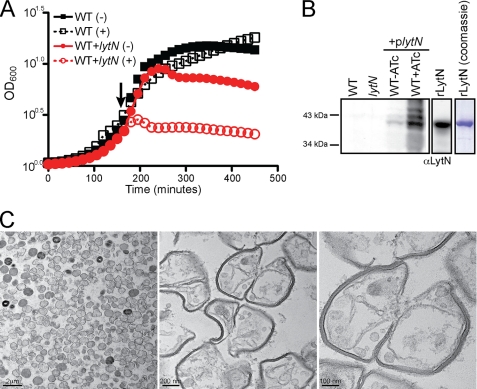
Anhydrotetracycline induction of Ptet::lytN in the strain lytN (Ptet::lytN) inhibited bacterial growth (data not shown). We generated a variant of wild-type S. aureus Newman carrying a multicopy plasmid harboring lytN, again under the control of anhydrotetracycline (wild-type (Ptet::lytN)). In the absence of the anhydrotetracycline inducer, wild-type (Ptet::lytN) staphylococci grew during exponential phase but failed to reach the same density as the wild-type parent (Fig. 3). This is due to the incomplete suppression of expression in this multicopy vector (Fig 3B). In the presence of the inducer, wild-type (Ptet::lytN) cells ceased growth and declined in absorbance (A600) (Fig. 3). To assess whether growth inhibition was caused by the increased expression of lytN, extracts derived from wild-type staphylococci, lytN and wild-type strains harboring (Ptet::lytN), were subjected to immunoblotting experiments using LytN-specific rabbit antisera (Fig. 3). LytN was detected in wild-type strains harboring (Ptet::lytN), but not in the wild-type parent or lytN mutant staphylococci lacking inducible expression of lytN. The overall abundance of LytN was increased in wild-type (Ptet::lytN) cells that had been grown in the presence of the inducer (Fig. 3). Wild-type (Ptet::lytN) staphylococci incubated in the presence of anhydrotetracycline were fixed and thin-sectioned, and uranyl acetate stained samples were viewed by transmission electron microscopy. These images revealed that the cell wall envelope of Ptet::lytN staphylococci was largely intact but the cross-wall peptidoglycan had in part disintegrated. Almost all staphylococci in these samples were lysed, and the sites of their rupture were located in the cross-wall compartment. In sum, we presume physiological levels of lytN expression are low, as the enzyme could not be detected with LytN-specific antibodies. Growth defect and structural damage to the cross-wall of lytN mutants could be complemented with Ptet::lytN. Overexpression of lytN triggered rupture of the cross-wall and staphylococcal lysis.
FIGURE 3.
Overexpression of lytN causes cellular lysis at the cross-wall. A, wild-type staphylococci with or without a multicopy vector expressing lytN under the control of the anhydrotetracycline-inducible promoter were grown for 2.5 h at 37 °C. At this point, inducer was added to cells, and absorbance at A600 was monitored for growth over time. Experiments were performed in triplicate, and error bars represent the means ± S.E. B, immunoblot analysis of lytN production using specific rabbit antisera. Wild-type, lytN mutant, and wild-type containing plytN were grown as described above, and total proteins were harvested. rLytN was loaded alongside staphylococcal lysates as a control, and samples were either examined by immunoblot or stained with Coomassie Brilliant Blue. C, transmission electron micrographs of wild-type staphylococci induced for overexpression of lytN for 30 min. Cells were fixed, thin-sectioned, and negatively stained prior to electron microscopic examination.
Localization of LytN in the Staphylococcal Envelope
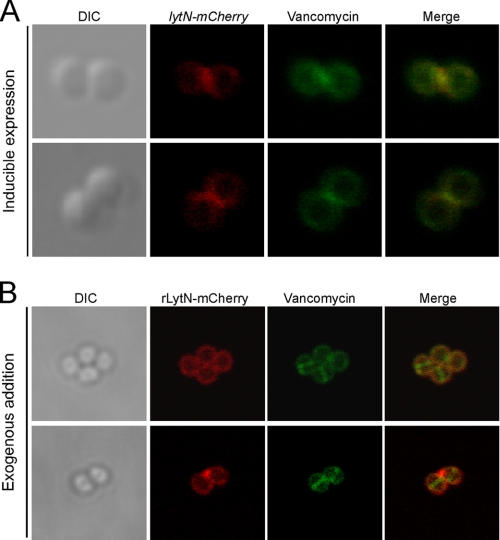
Anhydrotetracycline-induced expression of the translational hybrid lytN-mCherry resulted in lysis of wild-type cells (Ptet::lytN-mCherry) (data not shown), indicating that LytN-mCherry is functionally active. To circumvent this issue and to more carefully control the expression of lytN-mCherry, the translational hybrid was expressed from the integrative vector, pCL55iTET, again under the control of anhydrotetracycline but allowing a more tight regulation (Refs. 32, 33 and data not shown). BODIPY-vancomycin, which binds to the terminal d-Ala within the wall peptide of peptidoglycan, was used to visualize the staphylococcal cell wall envelope and its cross-wall in fluorescence microscopy experiments (37). Upon brief anhydrotetracycline induction of Ptet::lytN-mCherry cells, red fluorescent staining initially appeared near the cross-wall and the BODIPY-vancomycin fluorescent compartment between adjacent daughter cells (Fig. 4A). Over time, LytN-mCherry appeared throughout the cell wall as peptidoglycan synthesis and cell division continued, resulting in cell lysis (data not shown). rLytN-mCherry was purified from recombinant E. coli and added to staphylococcal cells. Cells were washed, stained with BODIPY-vancomycin, and viewed by fluorescence microscopy. Red fluorescence staining was observed throughout the cell wall envelope with some accumulation in cell wall envelope sections that surround the perpendicular cross-wall (Fig. 4B). Of note, red fluorescence signals were not detected in the staphylococcal cross-wall, despite BODIPY-vancomycin staining at this site (Fig. 4B). These data suggest that vancomycin, but not exogenously added polypeptides (rLytN-mCherry), penetrate the cross-wall of staphylococci. Conversely, the LytN-mCherry precursor, which is endowed with a YSIRK/GS signal peptide, appears to be secreted into the cross-wall compartment.
FIGURE 4.
LytN is directly trafficked to the cross-wall. A, wild-type S. aureus encoding lytN-mCherry under the control of an anhydrotetracycline-inducible promoter from an integrative vector was grown to mid-logarithmic phase, and inducer was briefly added. Shortly thereafter, cells were fixed and stained with BODIPY-vancomycin. Micrographs are arranged in the following order from left to right: differential interference contrast (DIC), mCherry fluorescence, BODIPY-vancomycin fluorescence, and the merged fluorescent images. Two representative images are presented. B, recombinant LytN-mCherry was added to staphylococci and allowed to bind for 15 min, and samples were washed with PBS, fixed with paraformaldehyde, and stained with BODIPY-vancomycin. Images were acquired as described in A.
Purified rLytN Cleaves Staphylococcal Peptidoglycan
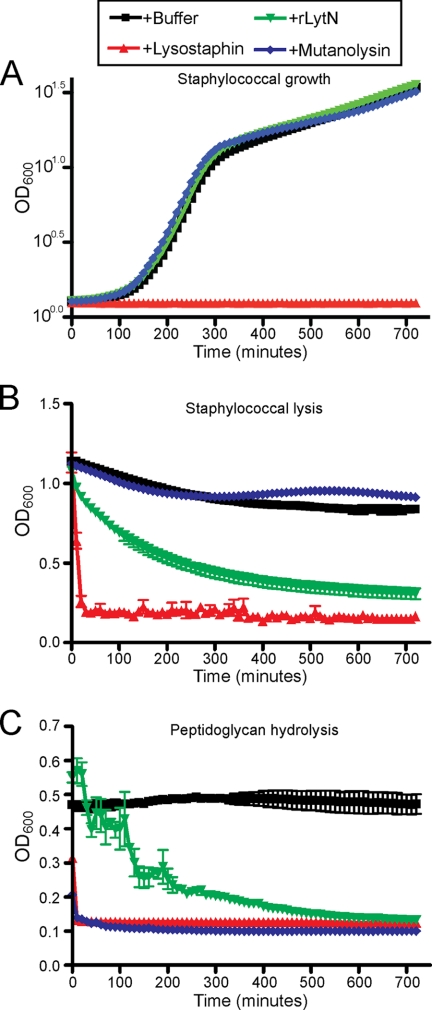
Purified rLytN was added to staphylococcal cultures, and growth was monitored as an increase in absorbance at 600 nm light (A600). As controls, lysostaphin, the staphylolytic glycyl-glycine endopeptidase (39), prevented bacterial growth, whereas mutanolysin, a streptomycetal muramidase (40), did not (Fig. 5A). Purified rLytN also did not interfere with staphylococcal growth (Fig. 5A). Incubation of stationary phase S. aureus cells with chaotropic agents or murein hydrolases may cause staphylococcal lysis, detected as a decline in A600. Addition of lysostaphin to stationary phase staphylococci triggered rapid lysis, whereas mutanolysin had no effect (Fig. 5B). The addition of rLytN to stationary phase cultures caused partial staphylococcal lysis (Fig. 5B). Peptidoglycan of S. aureus was isolated and degraded with either lysostaphin or mutanolysin, which was measured as a decline in A600 (Fig. 5C). rLytN also displayed peptidoglycan hydrolase activity (Fig. 5C). Compared with similar amounts of lysostaphin or mutanolysin, rLytN degraded less peptidoglycan. Taken together, these data indicate that rLytN, which does not display autolysin activity, functions as a murein hydrolase that cleaves S. aureus peptidoglycan.
FIGURE 5.
Comparison of rLytN with other murein hydrolases. A, an overnight culture of S. aureus Newman was diluted into fresh media supplemented with buffer, lysostaphin, mutanolysin, or recombinant LytN (rLytN), and growth was monitored over 12 h at 37 °C by A600 readings. Conditions were performed in triplicate, and error bars represent the means ± S.E. B, staphylococci grown to stationary phase were washed in buffer and subsequently incubated in the presence of buffer alone, lysostaphin, mutanolysin, or rLytN. Cell lysis was monitored by A600 over time as in A. C, purified staphylococcal peptidoglycan was incubated with buffer alone, lysostaphin, mutanolysin, or rLytN, and hydrolysis was measured by monitoring A600 over 12 h.
Muropeptide Profile of Wild-type and lytN Mutant Staphylococci
We wondered whether lytN mutants harbor changes in peptidoglycan structure when compared with wild-type staphylococci. Murein sacculi of S. aureus Newman and the isogenic lytN mutant were isolated, and peptidoglycan was purified. The peptidoglycan was treated with mutanolysin, which cleaves the glycosidic bond between MurNAc-(β1–4)-GlcNAc, releasing disaccharide muropeptides that were reduced with sodium borohydride and separated by rpHPLC. As expected, muramidase treatment of wild-type peptidoglycan released disaccharide tetrapeptide, disaccharide pentapeptide, and tetrasaccharide peptidoglycan fragments (supplemental Fig. S1). Similar results were observed when the cell wall of the lytN mutant was cleaved with muramidase, indicating that the overall structure of peptidoglycan is not altered by the insertional disruption of the lytN gene (supplemental Fig. S1).
Characterization of Peptidoglycan Fragments Released with rLytN
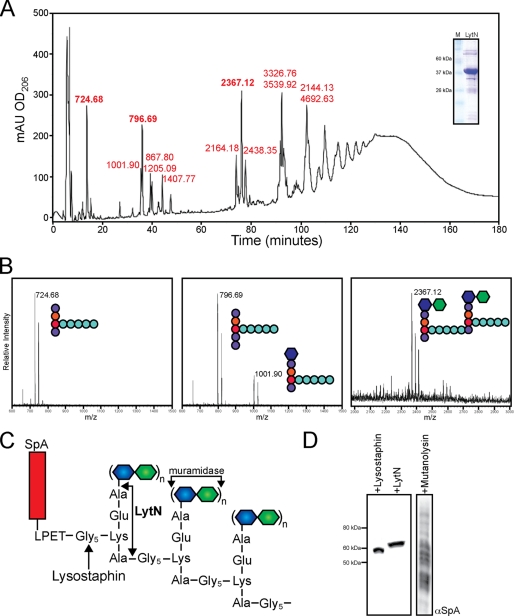
Purified peptidoglycan of wild-type staphylococci was treated with rLytN, and soluble fragments were subjected to rpHPLC (Fig. 6A). Isolated compounds were dried, again subjected to rpHPLC for salt removal, and analyzed by matrix-assisted laser desorption ionization time of flight (MALDI-TOF) mass spectrometry. Ion signals at m/z 724.68 (13 min) and 796.69 (35 min) were explained as the sodium ions of NH2-l-Ala-iGln-l-Lys-(NH2-Gly5)-d-Ala-COOH (calculated mass of 724.72) and NH2-l-Ala-iGln-l-Lys-(NH2-Gly5)-d-Ala-d-Ala-COOH (calculated mass 795.80). The ion at m/z of 2367.12 (75 min) corresponded to the cross-linked disaccharide peptidoglycan fragment MurNAc-(l-Ala-iGln-l-Lys-(NH2-Gly5)-d-Ala-COOH)-(β1–4)-GlcNAc (calculated mass 2367.38) (Fig. 6B). A complete list of peptidoglycan fragments with their observed mass and predicted structure is provided in Table 1. To confirm the identity of sodiated ions, CsCl (cesium m/z 132.90) was added to samples. The addition of cesium can displace sodium ions (m/z 22.99) and should consequently increase the mass to charge ratio by m/z 110. Indeed, we observed increases in m/z 110 for the sodium ions of rLytN-released peptidoglycan fragments (supplemental Fig. S2). Together, these data suggest that rLytN cleaves the staphylococcal peptidoglycan at two positions, the amide bonds between MurNAc and l-Ala as well as d-Ala-Gly. If so, Edman degradation of peptidoglycan fragments should release phenylthiohydantoin-Ala (PT-Ala) and phenylthiohydantoin-Gly (PT-Gly), respectively. This was tested, and compounds with m/z 724.68 and 796.69 released equimolar amounts of PT-Ala and PT-Gly after the first cleavage cycle, followed by four cycles of PT-Gly release (the amide bond between d-iGln-l-Lys is not subject to Edman degradation). In contrast, Edman degradation of m/z 2367.38 released five cycles of PT-Gly without releasing PT-Ala (Table 2). Thus, treatment of peptidoglycan with rLytN-generated fragments with either one (NH2-Gly) or two (NH2-Gly and NH2-Ala) amino groups, in agreement with the proposed functions of LytN as MurNAc-l-Ala amidase and d-Ala-Gly endopeptidase.
FIGURE 6.
rLytN functions as an amidase and endopeptidase. A, S. aureus wild-type peptidoglycan was treated with recombinant LytN for 16 h at 37 °C, reduced, and resolved by rpHPLC as described under “Experimental Procedures.” Individual peaks were desalted and subjected to MALDI-TOF MS, with observed m/z listed in red above each peak tested. Inset, Coomassie-Brilliant Blue-stained SDS-PAGE of rLytN. B, example mass spectra obtained from the three most prominent peaks shown in A (m/z 724.68, 796.69, and 2367.12), with the proposed structure for each compound (see Fig. 1 for color coding). C, diagram of cell wall anchored protein A (SpA). The cleavage sites of lysostaphin, mutanolysin, and LytN are noted with arrows. D, immunoblot with SpA-specific monoclonal antibody from protein lysates derived from staphylococci treated with lysostaphin, rLytN, or mutanolysin. mAU, milliabsorbance units.
TABLE 1.
MALDI-TOF MS of rLytN-released peptidoglycan fragments
See Fig. 7A for absorption peaks of rpHPLC of rLytN-treated S. aureus peptidoglycan. Observed and calculated m/z values are for sodiated ions.
| Compound |
m/z |
Δ (observed and calculated) | Predicted structure | |
|---|---|---|---|---|
| Observed | Calculated | |||
| 1 | 724.68 | 724.72 | −0.04 | AQKG5A |
| 2 | 796.69 | 795.80 | 0.89 | AQKG5AA |
| 3 | 867.80 | 867.41 | −0.039 | AQKG5A3 |
| 4 | 1001.90 | 1001.01 | 0.89 | MurNAc-AQKG5A |
| 5 | 1205.09 | 1205.20 | 0.11 | GlcNAc-MurNAc-AQKG5A |
| 6 | 1407.77 | 1407.27 | −0.41 | GlcNAc2-MurNAc-AQKG5A |
| 7 | 2164.18 | 2163.92 | −0.26 | GlcNAc-MurNAc2-(AQKG5A)2 |
| 8 | 2367.12 | 2367.38 | −0.026 | (GlcNAc-MurNAc)2-(AQKG5A)2 |
| 9 | 2438.35 | 2438.03 | −0.32 | (GlcNAc-MurNAc)2-(AQKG5A)3 |
TABLE 2.
Edman degradation of rLytN released muropeptides
| Cycle | Amino acid |
||
|---|---|---|---|
| m/z 724.68 | m/z 796.69 | m/z 2367.12 | |
| pmol | |||
| 1 | Ala (7.87); Gly (6.85) | Ala (5.39); Gly (6.69) | Gly (98.78) |
| 2 | Gly (10.88) | Gly (8.51) | Gly (91.47) |
| 3 | Gly (7.87) | Gly (6.56) | Gly (80.81) |
| 4 | Gly (6.10) | Gly (5.46) | Gly (72.95) |
| 5 | Gly (6.75) | Gly (3.72) | Gly (63.28) |
Release of Cell Wall Anchored Protein A with Murein Hydrolases
Surface proteins of S. aureus and other Gram-positive bacteria are linked to peptidoglycan by a transpeptidation reaction (41). Sortase A cleaves the C-terminal sorting signal of surface proteins between the threonine and the glycine residues of the LPXTG motif (42) and forms an amide bond between the C-terminal threonine and the amino group of the pentaglycine cross-bridge (43). The cell wall anchor of surface proteins is embedded within peptidoglycan, i.e. anchor peptides are cross-linked to wall peptides of neighboring peptidoglycan strands, and the MurNAc-GlcNAc disaccharide of each anchor unit is located within long glycan chains (44, 45). Treatment of peptidoglycan with muramidase (mutanolysin) solubilizes cell wall anchored protein A as a spectrum of fragments linked to C-terminal peptidoglycan with variable numbers of cross-linked subunits (Fig. 6, C and D) (45). Conversely, treatment of peptidoglycan with lysostaphin, a glycyl-glycine endopeptidase, solubilizes protein A with two to three C-terminal glycine residues (average mass increase of 57–114 Da) (43). Lysostaphin-solubilized protein A migrates as a single species on SDS-PAGE, because the mass difference of one glycine residue (57 Da) cannot be resolved with this technology. Similar to lysostaphin, rLytN treatment solubilized protein A also as a single species on SDS-PAGE (Fig. 6C). rLytN-solubilized protein A migrated more slowly on SDS-PAGE than the lysostaphin-released counterpart (Fig. 6C). This can be explained by the MurNAc-l-Ala amidase and d-Ala-Gly endopeptidase activities of LytN, which released surface protein attached to a C-terminal anchor unit (NH2-l-Ala-iGln-l-Lys-(NH2-Gly5)-d-Ala-COOH, 725 Da) (23).
Treatment of Cross-linked Murein Disaccharides with LytN
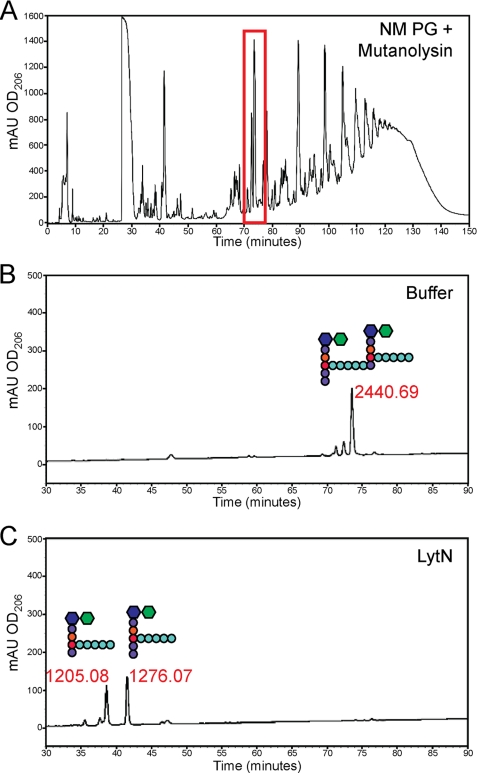
Purified peptidoglycan was treated with mutanolysin and subjected to rpHPLC. Cross-linked murein disaccharide MurNAc-(l-Ala-iGln-l-Lys-(NH2-Gly5)-d-Ala-Gly5)-d-Ala-d-Ala-COOH)-(β1–4)-GlcNAc and MurNAc-(l-Ala-iGln-l-Lys-(NH2-Gly5)-d-Ala-COOH)-(β1–4)-GlcNAc was isolated, and mass was measured (m/z 2440.69). The sample was treated with rLytN, and cleavage products were resolved by rpHPLC and analyzed by MALDI-TOF mass spectrometry (Fig. 7). Two products were isolated, which generated ion signals with m/z 1205.08 and 1276.07. As a control, incubation with buffer alone did not affect the substrate at m/z 2440.69 (Fig. 7). These data indicated that rLytN cut m/z 2440.69 at the amide bond between d-Ala-Gly to generate MurNAc-(l-Ala-iGln-l-Lys-(Gly5)-d-Ala-d-Ala-COOH)-(β1–4)-GlcNAc (m/z 1276.07) and MurNAc-(l-Ala-iGln-l-Lys-(NH2-Gly5)-d-Ala-COOH)-(β1–4)-GlcNAc (m/z 1205.08). Of note, MurNAc-l-Ala amidase activity was not observed when cross-linked murein disaccharides were incubated with rLytN. Similar results were observed when rLytN was incubated with other cross-linked disaccharides (supplemental Fig. S3). Even upon incubation of the monomeric peptidoglycan fragment, MurNAc-(l-Ala-iGln-l-Lys-(Gly5)-d-Ala-d-Ala-COOH)-(β1-4)-GlcNAc, no rLytN cleavage products were observed (supplemental Fig. S3).
FIGURE 7.
rLytN-specific activity using muramidase-derived muropeptide substrates. A, representative chromatograph of peptidoglycan from wild-type staphylococci treated with mutanolysin. The peak at 74 min (highlighted with a red box) represents a cross-linked peptidoglycan subunit. B, sample of peak 74 was split into two, and one half was treated with buffer alone and resolved again by rpHPLC. The peak was desalted and subjected to MALDI-TOF MS, with the mass listed above as well the structure of the muropeptide. C, sample of peak 74 was treated with rLytN for 4 h and examined as described in B. mAU, milliabsorbance units.
Treatment of Muramidase-digested Peptidoglycan with rLytN
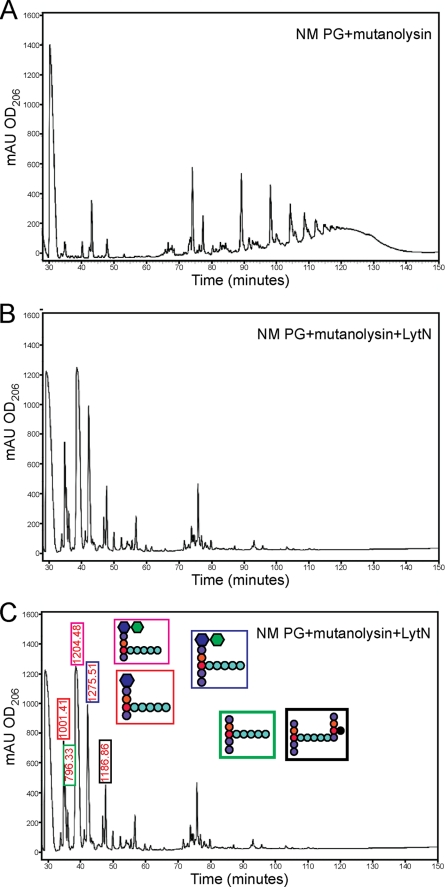
We asked whether rLytN MurNAc-l-Ala amidase activity occurred not only with murein sacculi but also when muramidase-treated peptidoglycan was used as substrate. Purified peptidoglycan was first treated with mutanolysin. As a control, cleavage products were analyzed by rpHPLC. The sample was then split and either incubated with rLytN or left untreated. Reaction products were separated by rpHPLC (Fig. 8). rLytN treatment converted cross-linked peptidoglycan of mutanolysin digested murein to monomeric peptidoglycan. Most of the rLytN-generated peptidoglycan fragments represented disaccharide (MurNAc-GlcNAc) or monosaccharide (MurNAc) peptides that had been liberated by d-Ala-Gly endopeptidase cleavage (Fig. 8). For example, the most abundant ion, m/z 1204.48, represented MurNAc-(l-Ala-iGln-l-Lys-(NH2-Gly5)-d-Ala-COOH)-(β1–4)-GlcNAc. Nevertheless, we also observed m/z 796.44, a cleavage product with the structure NH2-l-Ala-iGln-l-Lys-(NH2-Gly5)-d-Ala-d-Ala-COOH (calculated m/z 795.80). A complete listing of the observed ions and their proposed structure is presented in Table 3. As small amounts of wall peptide without disaccharide have been reported in muramidase-treated peptidoglycan samples, we presume that LytN cannot cleave the MurNAc-l-Ala amide bond of murein disaccharides. Nevertheless, rLytN is able to cleave this bond within fully assembled peptidoglycan.
FIGURE 8.
Sequential digestion of peptidoglycan using muramidase and rLytN. A, S. aureus wild-type peptidoglycan was treated with mutanolysin. B, wild-type peptidoglycan was first treated with mutanolysin, and the soluble muropeptides were then digested with rLytN and resolved by rpHPLC. C, muropeptides derived in B were desalted and subjected to MALDI-TOF MS. Shown above particular peaks are the m/z values of ions detected. Color-coded boxes demonstrate proposed structures for selected compounds. mAU, milliabsorbance units.
TABLE 3.
MALDI-TOF mass spectrometry of mutanolysin and rLytN treated peptidoglycan fragments
| Compound |
m/z |
Δ (observed and calculated) | Predicted structure | |
|---|---|---|---|---|
| Observed | Calculated | |||
| 1 | 796.33 | 795.80 | 0.53 | AQKG5AA |
| 2 | 1001.41 | 1001.01 | 0.40 | MurNAc-AQKG5A |
| 3 | 1204.48 | 1205.20 | −0.72 | GlcNAc-MurNAc-AQKG5A |
| 4 | 1275.51 | 1275.24 | 0.27 | GlcNAc-MurNAc-AQKG5AA |
Characterization of the CHAP Domain of LytN
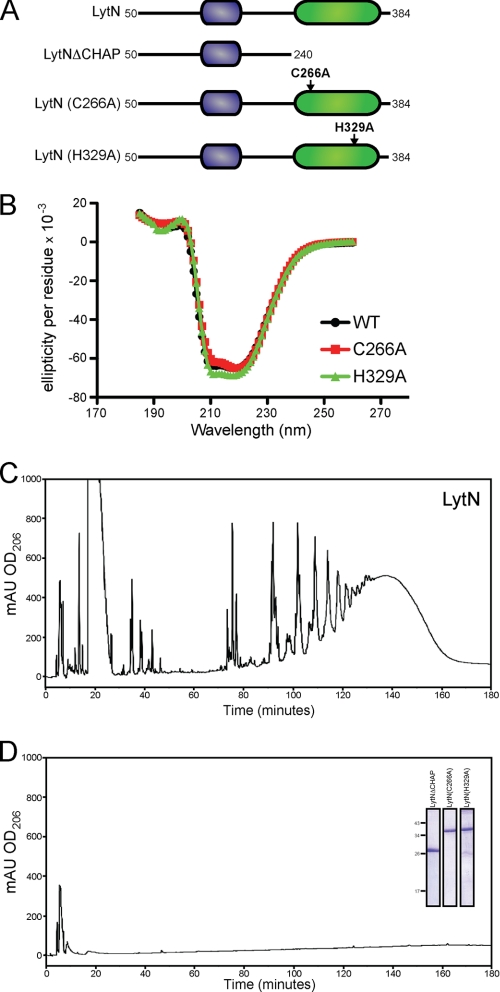
The CHAP domains of other murein hydrolases have been shown to display single activities, either amidase or endopeptidase. Our results suggest that LytN may be able to perform both. We queried whether these functions may be performed by the CHAP domain or whether LytN harbored a second functional module. CHAP domains harbor conserved cysteine and histidine residues essential for activity (20, 21, 46). To determine the role of the CHAP domain of LytN, we generated a rLytN variant without the C-terminal CHAP domain (rLytNΔCHAP). Treatment of purified peptidoglycan with rLytNΔCHAP did not solubilize peptidoglycan fragments or degrade the murein sacculi. Similarly, variants with alanine substitutions at Cys266 (rLytNC266A) or His329 (rLytNH329A) did not display murein hydrolase activity (Fig. 9). To examine whether loss of function was caused by improper folding, rLytN and its variants were subjected to circular dichroism, which produced superimposable spectra in agreement with the hypothesis that the mutations do not affect folding and that Cys266 and His329 are required for catalysis (Fig. 9). Together, these data suggest that the conserved residues of the CHAP domain are necessary for rLytN activity and that the CHAP domain contributes to both the amidase and endopeptidase functions of LytN.
FIGURE 9.
CHAP domain of LytN is required for hydrolytic activity. A, diagram of the primary structure of rLytN constructs in which the CHAP domain is removed or specific amino acid substitutions have been introduced. Mutated amino acids are indicated with arrows. B, circular dichroism spectra of rLytN or its C266A and H329A substitution variants were analyzed in the far-UV range. C, rpHPLC of soluble rLytN-treated muropeptides. D, rpHPLC of muropeptides generated upon incubation of peptidoglycan with rLytNΔCHAP. The C266A and H329A variants also failed to cleave staphylococcal peptidoglycan. Inset, Coomassie Brilliant Blue-stained SDS-PAGE samples of purified rLytNΔCHAP, rLytNC266A, and rLytNH329A. mAU, milliabsorbance units.
DISCUSSION
Homologs of LytN, a murein hydrolase whose YSIRK/GS signal peptide directs precursor secretion to the cross-wall of S. aureus, are found only in staphylococcal species. These bacteria replicate by a distinctive program, whereby cell division planes are built perpendicular to previous planes (13) and bulk de novo peptidoglycan synthesis occurs at the cross-wall, the peptidoglycan layer that separates a mother cell into two daughters (47). S. aureus mutants lacking lytN display delayed growth as well as structural defects to the cell wall envelope in that the otherwise homogeneous appearance of peptidoglycan is perturbed, giving rise to disordered layers of cell wall and irregular cell shapes. Synthesis and secretion of LytN must be carefully controlled, as increased expression of lytN triggered the rupture of cross-walls as well as lysis of staphylococci. Although overexpressed LytN could be detected by immunoblot, this technology did not reveal the overall abundance or the stability of the murein hydrolase under physiological conditions. Of note, intracellular LytN precursor with its YSIRK/GS signal peptide, but not extracellular purified rLytN, can fully complement the growth phenotype of lytN mutants. Future research must resolve whether synthesis, secretion, and degradation of LytN occur constitutively or in a regulated manner, accompanying discrete steps in staphylococcal cytokinesis.
LytN harbors a CHAP domain. Similar domains have been found in murein hydrolases from several different Gram-positive bacteria (21). Although conserved throughout bacteria and even eukaryotes, the function of this domain is not definitively known. Examples exist within parasitic species whereby the CHAP domain is not at all related to peptidoglycan metabolism (19). Where tested, bacterial enzymes with CHAP domains have been reported to cleave peptidoglycan either as a MurNAc-l-Ala amidase (18) or a d-Ala-Gly endopeptidase (23). To the best of our knowledge, we report here for the first time that LytN, an enzyme with a CHAP domain, can exert both activities. The evidence for this is as follows. Treatment of purified staphylococcal peptidoglycan with rLytN released uncross-linked wall peptides (l-Ala-d-iGln-l-Lys(NH2-Gly5)-d-Ala) both with and without linked disaccharide (MurNAc-GlcNAc). Additionally, rLytN treatment of staphylococcal cells liberated protein A attached to a single peptidoglycan fragment, not a spectrum of cross-linked fragments. This result can only be explained as the release of surface protein anchor structures from both the glycan strands (MurNAc-l-Ala amidase activity) and the cross-linked peptidoglycan (d-Ala-Gly endopeptidase activity) of the staphylococcal cell wall (48). Curiously, pretreatment of peptidoglycan with muramidase (mutanolysin) did not permit detection of LytN amidase activity without impacting the endopeptidase activity of the enzyme. One plausable explanation for these data is that LytN amidase requires either intact glycan strands or MurNAc reducing ends, a product of glucosaminidase cleavage. GlcNAc reducing ends, generated via muramidase treatment of peptidoglycan, apparently do not support LytN amidase activity. The genome of S. aureus encodes for several murein hydrolases with glucosaminidase or CHAP domains but not for muramidases (22). We wonder whether the observed glycan substrate requirements for dual enzyme activity represent a general feature of murein hydrolases with CHAP domains. For S. aureus, this attribute could apply to Sle1 (18), LytA, φ11 hydrolase (23), and LytN.
Two staphylococcal murein hydrolases harbor LysM domains, Sle1 (23) and LytN. A third protein, Atl, harbors three repeat domains that are similar in size but lack sequence similarity with LysM domains (49). All three proteins are directly involved in cleaving the cross-wall and enabling daughter cell separation following the completion of staphylococcal cytokinesis (5). Two secreted proteins, Atl and Sle1, accomplish this task by associating with the bacterial peptidoglycan. For Atl, this involves binding to the envelope in the vicinity of the cross-wall or cell division site (14, 49). We presume, but do not know, that Sle1 may also target to this site (18). In contrast to Atl and Sle1, LytN harbors a single LysM domain (22). When added exogenously to staphylococci, rLytN binds to the cell wall envelope. Unlike Atl (15), however, rLytN appears to be distributed all over the envelope, i.e. it is not restricted to the cell division site. Nevertheless, murein hydrolases with LysM domains appear specifically involved in the separation of cell wall during cytokinesis (50). For secreted proteins, this involves multiple LysM domains (50–53), whereas LytN harbors only one of these domains. Thus, identification of the cell wall ligand for the LysM domain may provide important insights into the physiology of bacterial cell wall separation and cytokinesis. We hypothesize that the LysM domain, rather than conferring substrate specificity as has been postulated for Sle1, provides LytN, which has been delivered to the cross-wall via its YSIRK signal peptide, peptidoglycan binding, and envelope association of this enzyme. In conclusion, we propose a revised model for staphylococcal separation following cytokinesis, whereby Atl and Sle1 are initially secreted into the extracellular milieu and bind the cell wall envelope in the vicinity of the cross-wall. Atl and Sle1 then split the cross-wall from the outside in. In contrast, LytN is secreted directly into the cross-wall and splits the cross-wall from the inside out, thereby separating staphylococcal daughter cells.
Supplementary Material
Acknowledgments
We thank Justin Kern and Stefan Richter for expert assistance with the HPLC and mass spectrometry analysis. We also thank Valerie Anderson for helpful discussions and experimental assistance and all members of the laboratory for critical comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant AI038897 from Infectious Disease Branch, NIAID (to O. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- SOE
- splicing by overlap extension
- rpHPLC
- reversed-phase HPLC
- PT
- phenylthiohydantoin
- rLytN
- recombinant LytN.
REFERENCES
- 1. Lowy F. D. (1998) N. Engl. J. Med. 339, 520–532 [DOI] [PubMed] [Google Scholar]
- 2. Klevens R. M., Edwards J. R., Gaynes R. P., and National Nosocomial Infections Surveillance System (2008) Clin. Infect. Dis. 47, 927–930 [DOI] [PubMed] [Google Scholar]
- 3. Hiramatsu K., Hanaki H., Ino T., Yabuta K., Oguri T., Tenover F. C. (1997) J. Antimicrob. Chemother. 40, 135–136 [DOI] [PubMed] [Google Scholar]
- 4. Chang S., Sievert D. M., Hageman J. C., Boulton M. L., Tenover F. C., Downes F. P., Shah S., Rudrik J. T., Pupp G. R., Brown W. J., Cardo D., Fridkin S. K. (2003) N. Engl. J. Med. 348, 1342–1347 [DOI] [PubMed] [Google Scholar]
- 5. Giesbrecht P., Kersten T., Maidhof H., Wecke J. (1998) Microbiol. Mol. Biol. Rev. 62, 1371–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chatterjee A. N., Park J. T. (1964) Proc. Natl. Acad. Sci. U.S.A. 51, 9–1614106674 [Google Scholar]
- 7. Berger-Bächi B. (1994) Trends Microbiol. 2, 389–393 [DOI] [PubMed] [Google Scholar]
- 8. Strominger J. L., Izaki K., Matsuhashi M., Tipper D. J. (1967) Fed. Proc. 26, 9–22 [PubMed] [Google Scholar]
- 9. Tipper D. J., Strominger J. L. (1965) Proc. Natl. Acad. Sci. U.S.A. 54, 1133–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tipper D. J., Strominger J. L. (1968) J. Biol. Chem. 243, 3169–3179 [PubMed] [Google Scholar]
- 11. Ghuysen J. M. (1968) Bacteriol. Rev. 32, 425–464 [PMC free article] [PubMed] [Google Scholar]
- 12. Navarre W. W., Schneewind O. (1999) Microbiol. Mol. Biol. Rev. 63, 174–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tzagoloff H., Novick R. (1977) J. Bacteriol. 129, 343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamada S., Sugai M., Komatsuzawa H., Nakashima S., Oshida T., Matsumoto A., Suginaka H. (1996) J. Bacteriol. 178, 1565–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Komatsuzawa H., Sugai M., Nakashima S., Yamada S., Matsumoto A., Oshida T., Suginaka H. (1997) Microbiol. Immunol. 41, 469–479 [DOI] [PubMed] [Google Scholar]
- 16. Oshida T., Sugai M., Komatsuzawa H., Hong Y. M., Suginaka H., Tomasz A. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sugai M., Yamada S., Nakashima S., Komatsuzawa H., Matsumoto A., Oshida T., Suginaka H. (1997) J. Bacteriol. 179, 2958–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kajimura J., Fujiwara T., Yamada S., Suzawa Y., Nishida T., Oyamada Y., Hayashi I., Yamagishi J., Komatsuzawa H., Sugai M. (2005) Mol. Microbiol. 58, 1087–1101 [DOI] [PubMed] [Google Scholar]
- 19. Baba T., Schneewind O. (1998) Trends Microbiol. 6, 66–71 [DOI] [PubMed] [Google Scholar]
- 20. Rigden D. J., Jedrzejas M. J., Galperin M. Y. (2003) Trends Biochem. Sci. 28, 230–234 [DOI] [PubMed] [Google Scholar]
- 21. Bateman A., Rawlings N. D. (2003) Trends Biochem. Sci. 28, 234–237 [DOI] [PubMed] [Google Scholar]
- 22. Baba T., Bae T., Schneewind O., Takeuchi F., Hiramatsu K. (2008) J. Bacteriol. 190, 300–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Navarre W. W., Ton-That H., Faull K. F., Schneewind O. (1999) J. Biol. Chem. 274, 15847–15856 [DOI] [PubMed] [Google Scholar]
- 24. DeDent A., Bae T., Missiakas D. M., Schneewind O. (2008) EMBO J. 27, 2656–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bae T., Schneewind O. (2003) J. Bacteriol. 185, 2910–2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mazmanian S. K., Ton-That H., Su K., Schneewind O. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2293–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sugai M., Fujiwara T., Komatsuzawa H., Suginaka H. (1998) Gene 224, 67–75 [DOI] [PubMed] [Google Scholar]
- 28. Berger-Bächi B., Barberis-Maino L., Strässle A., Kayser F. H. (1989) Mol. Gen. Genet. 219, 263–269 [DOI] [PubMed] [Google Scholar]
- 29. Bae T., Banger A. K., Wallace A., Glass E. M., Aslund F., Schneewind O., Missiakas D. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12312–12317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bae T., Baba T., Hiramatsu K., Schneewind O. (2006) Mol. Microbiol. 62, 1035–1047 [DOI] [PubMed] [Google Scholar]
- 31. Kreiswirth B. N., Löfdahl S., Betley M. J., O'Reilly M., Schlievert P. M., Bergdoll M. S., Novick R. P. (1983) Nature 305, 709–712 [DOI] [PubMed] [Google Scholar]
- 32. Frankel M. B., Wojcik B. M., DeDent A. C., Missiakas D. M., Schneewind O. (2010) Mol. Microbiol. 78, 238–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee C. Y., Buranen S. L., Ye Z. H. (1991) Gene 103, 101–105 [DOI] [PubMed] [Google Scholar]
- 34. Gründling A., Schneewind O. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 8478–8483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. de Jonge B. L., Chang Y. S., Gage D., Tomasz A. (1992) J. Biol. Chem. 267, 11248–11254 [PubMed] [Google Scholar]
- 36. Glauner B., Höltje J. V., Schwarz U. (1988) J. Biol. Chem. 263, 10088–10095 [PubMed] [Google Scholar]
- 37. DeDent A. C., McAdow M., Schneewind O. (2007) J. Bacteriol. 189, 4473–4484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cheng A. G., Kim H. K., Burts M. L., Krausz T., Schneewind O., Missiakas D. M. (2009) FASEB J. 23, 3393–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schindler C. A., Schuhardt V. T. (1964) Proc. Natl. Acad. Sci. U.S.A. 51, 414–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yokogawa K., Kawata S., Nishimura S., Ikeda Y., Yoshimura Y. (1974) Antimicrob. Agents Chemother. 6, 156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mazmanian S. K., Liu G., Ton-That H., Schneewind O. (1999) Science 285, 760–763 [DOI] [PubMed] [Google Scholar]
- 42. Navarre W. W., Schneewind O. (1994) Mol. Microbiol. 14, 115–121 [DOI] [PubMed] [Google Scholar]
- 43. Schneewind O., Fowler A., Faull K. F. (1995) Science 268, 103–106 [DOI] [PubMed] [Google Scholar]
- 44. Ton-That H., Faull K. F., Schneewind O. (1997) J. Biol. Chem. 272, 22285–22292 [DOI] [PubMed] [Google Scholar]
- 45. Navarre W. W., Ton-That H., Faull K. F., Schneewind O. (1998) J. Biol. Chem. 273, 29135–29142 [DOI] [PubMed] [Google Scholar]
- 46. Donovan D. M., Foster-Frey J., Dong S., Rousseau G. M., Moineau S., Pritchard D. G. (2006) Appl. Environ. Microbiol. 72, 5108–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Giesbrecht P., Wecke J., Reinicke B. (1976) Int. Rev. Cytol. 44, 225–318 [DOI] [PubMed] [Google Scholar]
- 48. Marraffini L. A., Dedent A. C., Schneewind O. (2006) Microbiol. Mol. Biol. Rev. 70, 192–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Baba T., Schneewind O. (1998) EMBO J. 17, 4639–4646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mesnage S., Chau F., Dubost L., Arthur M. (2008) J. Biol. Chem. 283, 19845–19853 [DOI] [PubMed] [Google Scholar]
- 51. Buist G., Kok J., Leenhouts K. J., Dabrowska M., Venema G., Haandrikman A. J. (1995) J. Bacteriol. 177, 1554–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Carroll S. A., Hain T., Technow U., Darji A., Pashalidis P., Joseph S. W., Chakraborty T. (2003) J. Bacteriol. 185, 6801–6808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Steen A., Buist G., Horsburgh G. J., Venema G., Kuipers O. P., Foster S. J., Kok J. (2005) FEBS J. 272, 2854–2868 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.