Abstract
DNA polymerase (pol) ϵ is thought to be the leading strand replicase in eukaryotes, whereas pols λ and β are believed to be mainly involved in re-synthesis steps of DNA repair. DNA elongation by the human pol ϵ is halted by an abasic site (apurinic/apyrimidinic (AP) site). In this study, we present in vitro evidence that human pols λ, β, and η can perform translesion synthesis (TLS) of an AP site in the presence of pol ϵ, likely by initiating the 3′OHs created at the lesion by the arrested pol ϵ. However, in the case of pols λ and β, this TLS requires the presence of a DNA gap downstream from the product synthesized by the pol ϵ, and the optimal gap for efficient TLS is different for the two polymerases. The presence of gaps did not affect the TLS capacity of human pol η. Characterization of the reaction products showed that pol β inserted dAMP opposite the AP site, whereas gap filling synthesis by pol λ resulted in single or double deletions opposite the lesion. The synthesis up to the AP site by pol ϵ and the subsequent TLS by pols λ and β are not influenced by human processivity factor proliferating cell nuclear antigen and human single-stranded DNA-binding protein replication protein A. The bypass capacity of pol λ at the AP site is greatly reduced when a truncated form of the enzyme, which has lost the BRCA1 C-terminal and proline-rich domains, is used. Collectively, our in vitro results support the existence of a mechanism of gap-directed TLS at an AP site involving a switch between the replicative pol ϵ and the repair pols λ and β.
Keywords: DNA Damage, DNA Polymerase, DNA Repair, DNA Replication, DNA-Protein Interaction
Introduction
Chromosomal DNA replication in eukaryotic cells requires three DNA polymerases (pols)3: pol α, pol δ, and pol ϵ. pol α is the only polymerase that has an associated activity for synthesis of RNA primers and is able to extend from such primers by synthesizing short stretches of DNA (1, 2). Subsequently, processive DNA synthesis is resumed by pol δ and/or pol ϵ. Recent work in yeast supports a model wherein, during normal DNA replication, pol ϵ is primarily responsible for copying the leading strand, and pol δ is primarily responsible for copying the lagging strand (3).
Abasic sites (AP sites) arise frequently by spontaneous hydrolysis of purines in DNA, represent a common intermediate of numerous DNA repair systems, and are among the most common endogenous DNA lesions generated during normal cell growth (4, 5).
An AP site poses a serious problem to the advancement of a pol because the modified base has lost its coding capacity. Accordingly, its replication requires the intervention of one or more Y family polymerases in a process called translesion synthesis or TLS (for reviews see Refs. 6, 7). Recent publications have shown that among these polymerases human pol η was able to insert nucleotides opposite the AP site and extend primers further past the lesion in vitro (8). Moreover, pol η showed higher abasic lesion bypass capacity in vivo than pols ι, κ, and Rev1 (9). Furthermore, it has also been reported that an AP site could be bypassed in vitro by polymerases of other families such as pol α (10), pol δ in the presence of the processivity factor PCNA (11), and pols λ and β (12). Concerning pol ϵ, a limited capacity of TLS of an AP site has been reported for the yeast enzyme (13) but not for its human counterpart, which appeared to be blocked mainly at the base preceding the lesion with minor incorporation opposite to it (14).
A widely accepted model of DNA lesion bypass is the polymerase-switching model that is believed to act at the replication fork to enable replication to continue by bypassing DNA lesions that halt the progression of the replicative polymerases. In this model, protein-protein interactions mediate a pol handoff at the template-primer terminus from the replicative pol to one or more specialized polymerases. In eukaryotes, this switching appears to be mediated by a monoubiquitinated form of the processivity clamp factor PCNA. A further switch restores the replicative pol to the primer terminus, and accurate synthesis resumes (reviewed in Ref. 7).
Conversely, a second model, named gap-filling model, can be envisaged to account for TLS-assisted bypass of DNA lesions outside the context of the replication fork, and its purpose would be to seal gaps containing lesions resulting from re-priming events or processing of closely spaced lesions on opposite DNA strands (7). In contrast to the polymerase-switching model, the molecular mechanism(s) underlying the gap-filling model are still largely unknown.
In this work, we report in vitro DNA gap-dependent TLS at an AP site by human DNA repair polymerases, pol λ and pol β, in the presence of the replicative human pol ϵ. Human pol η can also perform TLS that does not appear to depend on DNA gaps. We also present evidence that TLS by pols λ and β is not influenced by the human processivity factor PCNA and the human single-stranded DNA-binding protein RPA. We also show that the capacity of pol λ to bypass an AP site is greatly reduced when a truncated form of the enzyme, which has lost the BRCT and proline-rich domains, is used. Taken together, our in vitro results may suggest the existence of a novel pathway of DNA repair, gap-directed TLS involving human pols ϵ, λ, and β.
EXPERIMENTAL PROCEDURES
Proteins
Recombinant human pol λ, RPA, and PCNA were expressed and purified as described previously (15–17). Recombinant pol λ(244–575) mutant was expressed as described previously (18) and purified as described previously (19). Recombinant human pol β was from Trevigen Inc. (Gaithersburg, MD). Recombinant human pol η was from Enzymax (Lexington, KY). Human pol ϵ was purified from HeLa cells through six purification steps as described previously (14, 20). The glycerol gradient fraction used in this study had a specific activity of 24,000 units/mg. Its purity was estimated to be >50%, and the fraction was devoid of other replicative polymerases (14).
DNA Substrates and Chemicals
The 100-mer oligonucleotide templates, either undamaged or containing a synthetic AP site (tetradroxyfuran moiety), and the oligonucleotide primers were from Eurogentec. The oligonucleotides complementary to the 5′ end of the templates were from Sigma, and all molecules carried a 5′-phosphate. All oligonucleotides were purified by PAGE. The DNA substrates used in this study are indicated in Table 1. Primers were 5′-labeled with T4 polynucleotide kinase (New England Biolabs) in the presence of [γ-32P]ATP according to the manufacturer's protocol. Each primer was mixed with the templates at an equimolar ratio. When necessary, the oligonucleotides complementary to the 5′ end of templates were added to the reaction at an oligonucleotide/template ratio of 2:1 to ensure complete hybridization to all templates. [γ-32P]ATP was from PerkinElmer Life Sciences; dNTPs were from Fermentas Life Science, and ddGTP was from GE Healthcare. 20× glycerol tolerant gel (GTG) buffer was from United States Biochemical Corp.
TABLE 1.
DNA templates
X indicates the synthetic abasic site on the damaged template or a guanine on the undamaged template. All the oligonucleotides complementary to the 5′ end of the 100-mer templates bear a 5-phosphate.
Primer Extension Assays
Reaction solutions of 10 μl were incubated at 37 °C and contained 0.15 pmol of DNA templates, 50 mm Hepes, pH 7.5, 5 mm MgCl2, 1 mm DTT, 200 μg/ml BSA, and 100 μm each of dATP, dCTP, dGTP, and dTTP. The incubation times and the amount of proteins used are indicated in the legends of the figures. The reactions were stopped by adding 5 μl of stop solution containing 0.1% xylene cyanol and 0.1% bromphenol blue in 90% formamide. Before loading onto the gel, samples were denatured by heating at 100 °C for 3 min. The reaction products were resolved on denaturing PAGE (7 m urea, 10% acrylamide) run in GTG buffer (90 mm Tris, pH 9, 30 mm taurine, and 5 mm EDTA) and visualized and quantified using phosphorimager (GE Healthcare) and ImageQuant software. The percentage of TLS was calculated as the ratio of the intensity of bands present at the position opposite the lesion or beyond to the intensity of these bands plus the intensity of the band present one nucleotide before the lesion.
DNA Sequencing of Reaction Products
Reaction solutions of 10 μl were incubated at 37 °C and contained 50 mm Hepes, pH 7.5, 5 mm MgCl2, 1 mm DTT, 200 μg/ml BSA; 100 μm each of dATP, dCTP, dGTP, and dTTP; and 0.15 pmol of DNA templates containing a single AP site within either a 2- or a 4-nucleotide gap. The 2- and 4-nucleotide gap templates were replicated by pol ϵ for 15 min followed by addition of pol β or pol λ for 5 min, respectively. Gap-filled products were converted to 100-mer by addition of 10 units of T4 DNA ligase and incubation at 37 °C for 10 min. Ligated products were amplified by 30 cycles of PCR in the presence of 1.25 units of Pfu DNA polymerase (Fermentas) and 25 pmol of both 5′-ACTACATTTACTTTCAATTACATAATTTCAAATCCTAATAATCT-3′ and 5′-TAAGGTAGTAGTATTATAAATTATG-3′ primers. PCR-amplified products from three independent reactions were pooled and purified. 29 and 28 individual clones were sequenced after TOPO cloning of the purified products for pols β and λ, respectively (Millegen, Toulouse, France).
Gel Mobility Shift Assay
Reaction of 10 μl contained 0.3 pmol of DNA substrates and 1.5 pmol of pol β in 50 mm Hepes, pH 7.5, 5 mm MgCl2, 1 mm DTT, and 200 μg/ml BSA. After an incubation time of 15 min at 37 °C, 1 μl of loading buffer (50% glycerol, 0.20% bromphenol blue, 0.20% xylene cyanol, and 0.2 m EDTA) was added, and the mixture was loaded on an 8% native gel in 0.5× TBE running buffer (89 mm Tris borate, pH 8.3, and 2 mm EDTA) and electrophoresed at 4 °C at 10 V/cm for 3 h.
RESULTS
DNA Polymerase λ Requires a DNA Gap with a Specific Length to Perform Translesion Synthesis in the Presence of DNA Polymerase ϵ
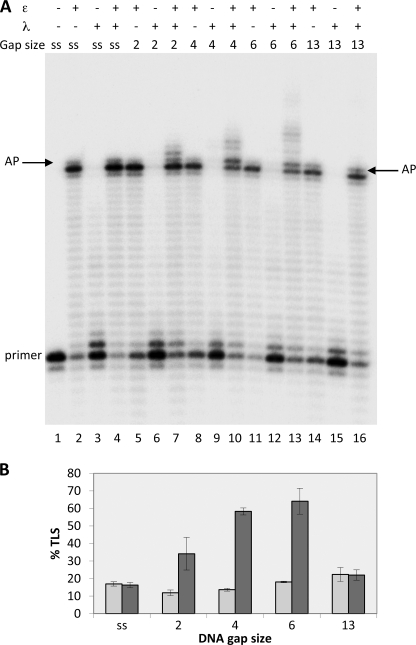
DNA elongation by human pol ϵ is severely blocked by an abasic site (14), although human pol λ can bypass such a DNA lesion (12). We therefore investigated whether pol λ could resume DNA synthesis when pol ϵ was stalled at an AP site. For initial experiments, the 100-mer template shown in Table 1a was used. The template contains a unique synthetic AP site at a defined position, and it was annealed to a primer of 44-mer, because a minimal primer length of ≈40 bp is required to maximize the binding and processivity of pol ϵ (21). As reported previously (14), when incubated with the 100- to 44-mer template-primer (which is defined in this work as single-stranded template-primer), pol ϵ was unable to replicate past the abasic site and stopped primarily at the base preceding the lesion, with some incorporation opposite the lesion (lane 2 of Fig. 1A and quantified in Fig. 1B). Addition of pol λ (0.25 pmol) at this stage and further incubation for 5 min did not resume DNA synthesis (Fig. 1A, lane 4).
FIGURE 1.
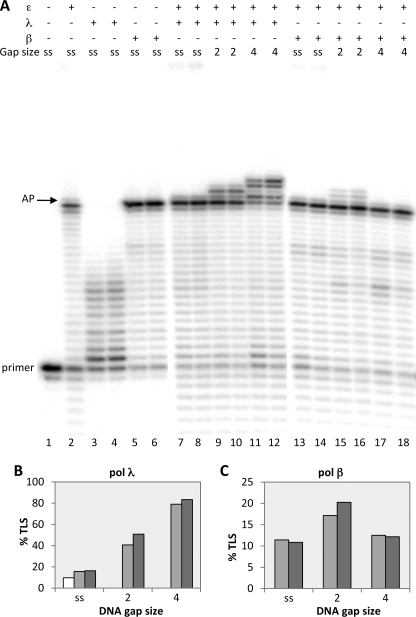
Ability of DNA pol λ to perform translesion synthesis of an AP site in the presence of DNA pol ϵ depends on the gap size. Experiments were performed with templates shown in Table 1, part a. The enzymes and the DNA substrates used are indicated at the top. ss (single-stranded) stands for template-primer with no oligonucleotides hybridized downstream from the AP site; 2, 4, 6, and 13 indicate the length of gap regions between the AP site and the oligonucleotide hybridized downstream. Assays were carried out as described under “Experimental Procedures.” A, lane 1, no polymerase present. Lanes 2, 5, 8, 11, and 14, reactions incubated for 35 min with 0.025 pmol of pol ϵ. Lanes 3, 6, 9, 12, and 15, reactions incubated for 5 min with 0.25 pmol of pol λ. Lanes 4, 7, 10, 13, and 16, reactions were incubated with 0.025 pmol of pol ϵ for 30 min; then 0.25 pmol of pol λ were added, and incubation was continued for 5 min. The positions of the primer and of the AP site are indicated. B, quantification of the percentage of TLS calculated as described under “Experimental Procedures.” Mean ± S.D. values for three independent experiments are indicated. Light gray bars, pol ϵ alone. Dark gray bars, pols ϵ and λ together.
pol λ is a family X pol that is involved in DNA repair and has higher incorporation efficiency on gapped than single-stranded DNA (see Ref. 22 and references therein). We therefore constructed template-primers containing gaps of different lengths around the abasic site and tested the capacity of pol λ to perform TLS in presence of pol ϵ. To this purpose, we hybridized the 100:44-mer single-stranded template-primer with oligonucleotides of different lengths placed downstream from the abasic site. With these template-primers, arrest of the elongation catalyzed by pol ϵ at the AP site results in gaps of a size from 1 to 13 nucleotides starting from the base that follow the abasic site (Fig. 1A). Because pol λ possesses a 5′-deoxyribose-phosphate lyase activity and it has been shown that a 5′-phosphate present in a gap strengthens the binding of the enzyme (23), all oligonucleotides placed downstream from the AP site were synthesized with a 5′-phosphate.
As can be seen in Fig. 1A, DNA gaps of 2, 4, and 6 nucleotides starting from the lesion radically changed the behavior of pol λ that now acquired the capacity to perform TLS of the AP site in the presence of pol ϵ (see lanes 7, 10, and 13). The gaps enabled pol λ to catalyze both incorporation opposite the lesion and beyond and to fill the gaps with no or little strand displacement synthesis consistent with the known limited strand displacement capacity of the enzyme (Fig. 2) (19). The TLS capacity of pol λ appears to increase from gaps of 2 nucleotides to gaps of 4–6, but it is dramatically reduced with a gap of 13 nucleotides (see quantifications in Fig. 1B), where the enzyme almost behaved as with the single-stranded template-primer in lane 4. Thus it appears that the TLS capacity of pol λ is strongly modulated by the size of the DNA gap.
FIGURE 2.
DNA pol λ can extend both from a nucleotide preceding the AP site and from an A incorporated opposite to it when placed within a DNA gap, although DNA pol ϵ cannot. Experiments were performed with the templates shown in Table 1c. The enzymes and the DNA substrates used are indicated at the top. ss stands for a template-primer with no oligonucleotides hybridized downstream from the AP site, and 2, 4, 6, and 13 indicate the length of gap regions between the AP site and the oligonucleotide hybridized downstream. Assays were carried out as described under “Experimental Procedures.” A, experiments conducted with the template primed with the 45-mer terminating one nucleotide before the AP site. Lane 1, no polymerase present. Lanes 2–6, reactions incubated for 35 min with 0.025 pmol of pol ϵ. Lane 7, no polymerase present. Lanes 8–12, reactions incubated for 5 min with 0.25 pmol of pol λ. The positions of the primer and of the AP site are indicated. B, experiments conducted with the template primed with the 45-mer terminating with an A opposite to the AP site. Lane 1, no polymerase present. Lanes 2–6, reactions incubated for 35 min with 0.025 pmol of pol ϵ. Lane 7, no polymerase present. Lanes 8–12, reactions incubated for 5 min with 0.25 pmol of pol λ. The positions of the primer and of the AP site are indicated.
As seen in Fig. 1, when 0.25 pmol of pol λ were incubated for 5 min with the substrates in the absence of pol ϵ, the products synthesized were all too short to reach the lesion (lanes 3, 6, 9, 12, and 15). This finding suggested that pol λ catalyzed TLS by using the 3′OHs created by pol ϵ, arrested either at the base preceding the lesion or opposite it, rather than using shorter primers generated during its own synthesis.
To further clarify this point, an experiment was devised in which the template-primers shown in Table 1b were used. If one compares template 1a to 1b in Table 1, it can be seen that in the latter the sequence between the 3′OH of the primer and the AP site has been changed, so that the only cytosines present in the template are now within this sequence. This allows specific inhibition of any elongation from 3′OHs in this region when the chain elongation inhibitor ddGTP is used. Oligonucleotides were hybridized to this template-primer to create gaps of 4 and 13 nucleotides. The rationale of this approach is that simultaneous addition of both pol λ and ddGTP in the presence of the stalled pol ϵ will abolish any priming contribution not starting from a pre-existing 3′OH. The result of this experiment is shown in supplemental Fig. 1. Note that to maximize the inhibitory effect of ddGTP, a concentration of 1 pmol of pol λ instead of the usual 0.25 pmol was used. As expected, incubation of 1 pmol of pol λ for 5 and 10 min in the absence of pol ϵ resulted in increased synthesis compared with that seen previously with 0.25 pmol (compare lanes 3 and 4 of supplemental Fig. 1 with lane 3 of Fig. 1). Samples were incubated for 5 and 10 min, and at 10 min one can see some TLS because of the intrinsic AP bypass activity of pol λ. Addition of ddGTP restrained incorporation to the first G following the 3′OH of the primer (supplemental Fig. 1, lanes 5 and 6). On single-stranded template-primer, addition of 1 pmol of pol λ in the presence of pol ϵ induced some TLS, particularly at 10 min, and this is seen also for the template-primer containing the 13-nucleotide gap (supplemental Fig. 1, lanes 7 and 8 and 15 and 16). However, addition of ddGTP before addition of pol λ completely abolished this TLS, indicating that it was due to elongation by pol λ of pre-existing primers and not of those created by pol ϵ (supplemental Fig. 1, lanes 9, 10 and 17, 18). Interestingly, the situation appeared different with the template-primer containing the 4-nucleotide gap, where a substantial part of the TLS by a high amount of pol λ took place also in the presence of ddGTP (supplemental Fig. 1, compare lanes 11 and 12 with 13 and 14), indicating that in this case pol λ could effectively use 3′OHs created at the lesion by pol ϵ.
To directly confirm that pol λ could extend past the lesion in presence of a gap starting from 3′OH generated by pol ϵ, as suggested by the experiments shown in Fig. 1 and supplemental Fig. 1, we created two template-primers in which the primer is either a 45-mer bearing a 3′OH at the base preceding the AP site or a 45-mer bearing the 3′OH at an A placed opposite the lesion (Table 1, template-primers 1c). The choice of the latter template-primer was motivated by our previous results that showed that either A or C is incorporated by pol ϵ opposite the AP site (14). To these template-primers, appropriate oligonucleotides were hybridized to generate gaps of 2, 4, 6, and 13 nucleotides, and they were used in reactions with 0.025 pmol of pol ϵ and 0.25 pmol of pol λ.
Fig. 2A shows the results of reactions using the primer ending at the nucleotide preceding the AP site. As expected, pol ϵ can incorporate in front of the lesion but is unable to elongate past it with all the substrates tested (lanes 2–6). On the contrary, pol λ alone can replicate past the AP site when gaps are present and, importantly, with a gap preference similar to that observed when the 3′OHs were generated by the arrest of pol ϵ at the lesion (compare lanes 9–12 of Fig. 2A with lanes, 7, 10, 13, and 16 of Fig. 1A).
Fig. 2B shows the results of reactions with the primer bearing an A opposite the AP site. As can be seen, pol ϵ is unable to elongate from this nucleotide, likely because its 3′-5-′-exonuclease continuously excised the A, as indicated by the increased intensity of the band preceding the lesion (compare lanes 2–6 of Fig. 2A with lanes 2–6 of B). Unlike pol ϵ, pol λ can also replicate from the A opposite the AP site, again with a gap preference similar to the one observed in Fig. 1. It should be noted that, as shown in Fig. 1, the amount of λ used in our study essentially fills the gaps during TLS with no significant strand displacement synthesis.
Taken together these data strongly suggest that the presence of gaps of defined size is a major determinant allowing pol λ to substitute for pol ϵ to bypass the AP site (see also under “Discussion”). Related to our finding, it is interesting to note that a recent work has shown that the polymerization activity of human pol λ increases with DNA gaps from 1 to 4, remains constant for gaps from 4 to 7, and then drops for gaps from 7 to 10 nucleotides (22).
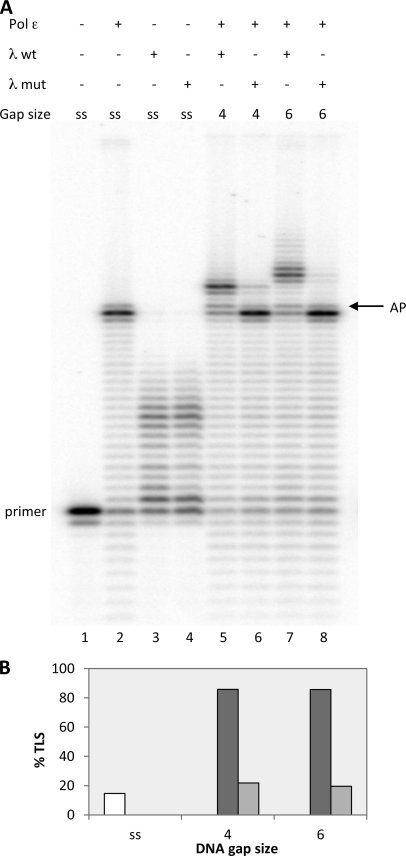
Full-length DNA Polymerase λ Is Required for the AP Site Translesion Synthesis in the Presence of DNA Polymerase ϵ
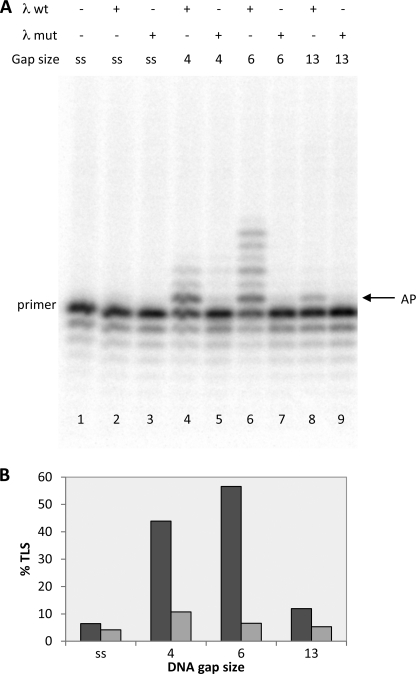
pol λ has two nonenzymatic domains at its N terminus, a BRCA1 C-terminal (BRCT) domain and a proline-rich domain (for review see Ref. 23). Little is known about the functions of these domains, but BRCT domains are known to mediate protein-protein and protein-DNA interactions (24), and both domains have been suggested to up-regulate or down-regulate fidelity of pol λ during gap filling activity, depending on the length of the gaps (22, 25). We therefore compared the AP site TLS capacities of pol λ WT and of the mutant form missing the BRCT and proline-rich domains (pol λ(244–575)) in the presence or absence of pol ϵ. The results of this comparison are shown in Figs. 3 and 4. Fig. 3A, quantified in B, shows that the λ(244–575), with both the 4- and 6-nucleotide gaps depicted in Table 1, had a severely reduced capacity to perform AP site translesion synthesis in the presence of pol ϵ compared with the WT (compare lanes 5 with 6 and lanes 7 with 8). Note that the two enzymes displayed the same activity on single-stranded template-primer (lanes 3 and 4 of Fig. 3A). Next, we examined the capacity of the two forms of pol λ to replicate past the AP site when initiating elongation from a 3′OH preceding the lesion, using the 100/45 template-primer shown in Table 1c. As can be seen in Fig. 4, A and B, pol λ(244–575) had also a clearly diminished intrinsic capacity to bypass the AP lesion compared with the WT (compare lanes 4 with 5 and lanes 6 with 7). Note that neither WT nor mutant pol λ could replicate past the AP site on single-stranded template-primer or on a template bearing a gap as long as 13 nucleotides (see lanes 2, 3, 8, and 9 of Fig. 4A and quantified in B), in agreement with the results previously shown.
FIGURE 3.
Full-length DNA pol λ is required for the AP site translesion synthesis in the presence of DNA pol ϵ. Experiments were performed with templates shown in Table 1a. The enzymes and the DNA substrates used are indicated at the top. ss stands for template-primer with no oligonucleotides hybridized downstream from the AP site, whereas 4 and 6 indicate the length of gap regions between the AP site and the oligonucleotide hybridized downstream. Assays were carried out as described under “Experimental Procedures.” A, lane 1, no polymerase present. Lane 2, reaction incubated for 35 min with 0.025 pmol of pol ϵ. Lane 3, reaction incubated with 0.25 pmol of pol λ for 5 min. Lane 4, reaction incubated with 0.25 pmol of pol λ(244–575) for 5 min. Lane 5, reaction incubated with 0.025 pmol of pol ϵ for 30 min and then 0.25 pmol of pol λ was added and incubation continued for 5 min. Lane 6, reaction incubated with 0.025 pmol of pol ϵ for 30 min, and 0.25 pmol of pol λ(244–575) was then added and incubation continued for 5 min. Lanes 7 and 8, were as for lanes 5 and 6, respectively. The positions of the primer and of the AP site are indicated. B, quantification of the data from A, expressed as percentage of TLS calculated as described under “Experimental Procedures.” White bar, pol ϵ alone. Dark gray bar, pol ϵ plus pol λ. Light gray bar, pol ϵ plus pol λ(244–575).
FIGURE 4.
Capacity of the λ(244–575)-truncated form of DNA pol λ to extend from the nucleotide preceding the AP site is impaired. Experiments were performed with templates shown in Table 1c. The enzymes and the DNA substrates used are indicated at the top. ss stands for template-primer with no oligonucleotides hybridized downstream from the AP site, whereas 4, 6, and 13 indicate the length of gap regions between the AP site and the oligonucleotide hybridized downstream. Assays were carried out as described under “Experimental Procedures.” A, lane 1, no polymerase present. Lane 2, reaction incubated for 5 min with 0.25 pmol of pol λ. Lane 3, reaction incubated for 5 min with 0.25 pmol of pol λ(244–575). Lane 4, reaction incubated for 5 min with 0.25 pmol of pol λ. Lane 5, reaction incubated for 5 min with 0.25 pmol of pol λ(244–575). Lane 6, reaction incubated for 5 min with 0.25 pmol of pol λ. Lane 7, reaction incubated for 5 min with 0.25 pmol of pol λ(244–575). Lane 8, reaction incubated for 5 min with 0.25 pmol of pol λ. Lane 9, reaction incubated for 5 min with 0.25 pmol of pol λ(244–575). The positions of the primer and of the AP site are indicated. B, quantification of the data from A, expressed as percentage of TLS calculated as described under “Experimental Procedures.” Dark gray bar, pol λ. Light gray bar, pol λ(244–575).
These experiments show the following: 1) a pol λ mutant lacking its BRCT and proline-rich domains has an impaired capacity to perform TLS of an AP site in the presence of pol ϵ, and 2) this defect can be attributed to an intrinsic diminished capacity to bypass the lesion, therefore suggesting a role of these domains in facilitating TLS of an AP site by pol λ.
DNA Polymerase β and DNA Polymerase λ Show Different DNA Gap Size Preference for Translesion Synthesis Past an AP Site in the Presence of DNA Polymerase ϵ
Next we tested whether DNA gap sizes could influence, in the presence of pol ϵ, TLS of an AP site by pol β, another X family repair polymerase that displays high affinity for very short DNA gaps. A direct comparison between the TLS capacity of β and λ as a function of DNA gap size is presented in Fig. 5.
FIGURE 5.
DNA pol β favors smaller gap size than DNA pol λ in TLS of an AP site in the presence of DNA pol ϵ. Experiments were performed with templates shown in Table 1a. The polymerases and the DNA substrates used are indicated at the top. ss stands for template-primer with no oligonucleotides hybridized downstream from the AP site, whereas 2 and 4 indicate the length of gap regions between the AP site and the oligonucleotide hybridized downstream. Assays were carried out as described under “Experimental Procedures.” A, lane 1, no polymerase present. Lane 2, reaction incubated for 35 min with 0.025 pmol of pol ϵ. Lanes 3 and 4, reactions incubated with 0.25 pmol of pol λ for 5 and 10 min, respectively. Lanes 5 and 6, reactions incubated with 0.25 pmol of pol β for 5 and 10 min, respectively. Lanes 7 and 8, reactions were incubated with 0.025 pmol of pol ϵ for 30 min, then 0.25 pmol of pol λ were added and the incubation continued for 5 and 10 min, respectively. Lanes 9–12: as for lanes 7 and 8. Lanes 13 and 14, reactions were incubated with 0.025 pmol of pol ϵ for 30 min, then 0.25 pmol of pol β were added and the incubation continued for 5 and 10 min, respectively. Lanes 15–18: as for lanes 13 and 14. The positions of the primer and of the AP site are indicated. B, quantification of the data from A with pol λ, expressed as percentage of TLS calculated as described under “Experimental Procedures.” White bar, pol ϵ alone. Light gray bars, incubation with pol ϵ and then pol λ added for 5 min. Dark gray bars, incubation with pol ϵ and then pol λ added for 10 min. C, quantification of the data from A with pol β, expressed as percentage of TLS and calculated as described under “Experimental Procedures.” Light gray bars, incubation with pol ϵ and pol β added for 5 min. Dark gray bars, incubation with pol ϵ and pol β added for 10 min.
First, it should be noted that on a single-stranded template-primer and at the same protein concentration (0.25 pmol) pol β could synthesize up to the AP site but pol λ could not (compare lanes 5 and 6 with lanes 3 and 4 of Fig. 5A). However, the elongation was blocked at the base preceding the lesion. Therefore, it appears that pol β cannot bypass the AP site when acting alone on a single-stranded template-primer. Furthermore, Fig. 5A also shows that neither pol λ nor β can perform TLS by utilizing primers created by pol ϵ in a single-stranded context (lanes 7, 8, 13, and 14 and quantified in Fig. 5B). In the presence of pol ϵ and in agreement with the data shown in Fig. 1, pol λ could easily replicate past the AP site when the gaps were 2 or 4 nucleotides long (lanes 9–12).
Interestingly, pol β also showed the capacity to bypass an AP site in the presence of pol ϵ, but only when the gap was 2 nucleotides long; enlarging the gap to 4 nucleotides abolished the TLS capacity of pol β (Fig. 5A, lanes 15–18). pol β appeared to be less efficient than pol λ in TLS of a 2-nucleotides gap (see quantifications in Fig. 5, B and C), which could be due to the superior intrinsic capacity of pol λ to replicate an AP site (12).
The different gap size dependence of TLS between pols λ and β (see also Figs. 7 and 8) corresponds to the preferences of the respective polymerases for undamaged substrates. pol β fills preferentially the gaps of one nucleotide, and its incorporation efficiency decreases with the increase in gap size from 1 to 4 nucleotides (22). To directly investigate the affinity of pol β to the templates-primers used, we measured its binding capacity to substrates containing AP sites in gaps of 1–4, 6, and 8 nucleotides. These substrates were created by first annealing a 66-mer to the 100-mer primer containing the AP site, so that the primer ended at the base preceding the AP site, as depicted in Table 1d. Then the appropriate oligonucleotides were annealed downstream from the lesion to create the substrates mentioned above. The experiment in supplemental Fig. 2 shows that the amount of pol β bound to the substrate was highest with a 1-nucleotide gap and diminished with increasing gap size. The reduction in binding efficiency parallels the decrease of TLS by pol β in the presence of pol ϵ, indicating that the TLS capacity of the enzyme is directly correlated with its binding capacity at gaps.
FIGURE 7.
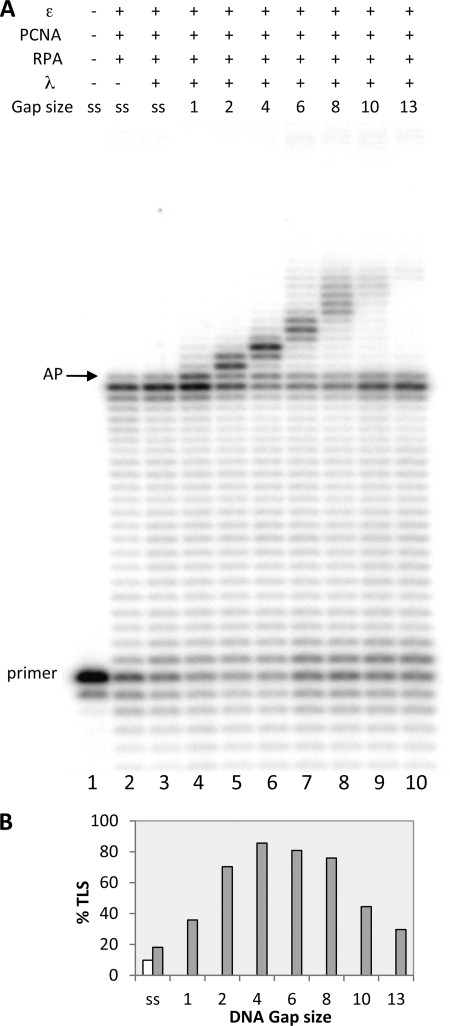
PCNA and RPA have no effect on gap size specificity of TLS of an AP site by DNA pol λ in presence of DNA pol ϵ. Experiments were performed with templates shown in Table 1a. The proteins and the DNA substrates used are indicated at the top. ss stands for template-primer with no oligonucleotides hybridized downstream from the AP site, whereas 1, 2, and 4, 6, 8, 10, and 13 indicate the length of gap regions between the AP site and the oligonucleotide hybridized downstream. Assays were carried out as described under “Experimental Procedures.” A, lane 1, no polymerase present. Lane 2, reaction incubated for 35 min with 0.025 pmol of pol ϵ, 1.2 pmol of PCNA, and 0.25 pmol of RPA. Lanes 3–10, the reactions were incubated for 30 min with 0.025 pmol of pol ϵ, 1.2 pmol of PCNA, and 0.25 pmol of RPA, and then 0.25 pmol of pol λ were added, and the incubation was continued for 5 min. The positions of the primer and of the AP site are indicated. B, quantification of data from Fig. 6A, expressed as percentage of TLS calculated as described under “Experimental Procedures.” White bar, reaction without pol λ. Gray bar, complete reaction.
FIGURE 8.
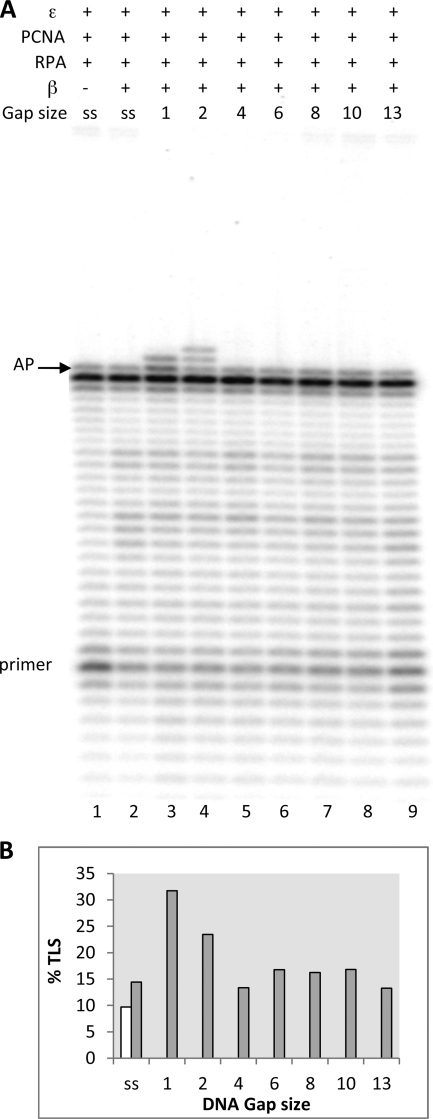
PCNA and RPA have no effect on gap size specificity of TLS of an AP site by DNA pol β in the presence of DNA pol ϵ. Experiments were performed with templates shown in Table 1a. The proteins and the DNA substrates used are indicated at the top: ss stands for template-primer with no oligonucleotides hybridized downstream from the AP site, whereas 1, 2, and 4, 6, 8, 10, and 13 indicate the length of gap regions between the AP site and the oligonucleotide hybridized downstream. Assays were carried out as described under “Experimental Procedures.” A, lane 1, reaction incubated for 35 min with 0.025 pmol of pol ϵ, 1.2 pmol of PCNA, and 0.25 pmol of RPA. Lanes 2–9: the reactions were incubated for 30 min with 0.025 pmol of pol ϵ, 1.2 pmol of PCNA, and 0.25 pmol of RPA, and then 0.25 pmol of pol β were added, and the incubation was continued for 5 min. The positions of the primer and of the AP site are indicated. B, quantification of data from Fig. 7A, expressed as percentage of TLS calculated as described under “Experimental Procedures.” White bar, reaction without pol β. Gray bar, complete reaction.
Nucleotide Insertion Opposite the AP Site by Polymerases β and λ during Gap-directed Translesion Synthesis
To determine the identity of the nucleotide inserted opposite the synthetic abasic site by pol β and λ, we have ligated the elongated primers to the downstream oligonucleotide and amplified the full-length products by PCR (see “Experimental Procedures”). Because Pfu DNA polymerase is blocked by the synthetic AP site (data not shown), only the newly synthesized DNA strand is amplified during the PCR. As shown in supplemental Table 1, sequencing of individual clones revealed that pol β TLS of the AP site in a 2-nucleotide DNA gap context resulted in incorporation of dAMP opposite the AP site in all the products sequenced, according to the proposed model of bypass of an abasic lesion by pol β (26). However, a 4-nucleotide DNA gap filling reactions by pol λ resulted in single or double deletions opposite the lesion, according to the known misalignment capacity of the enzyme (27). It should be noted that, in our template sequence, the AP site is followed by a run of 4 thymine residues; therefore, incorporation of dAMP by pol λ opposite the lesion followed by template-primer slippage and annealing to a downstream thymine can lead to the observed pattern.
Translesion Synthesis of an AP Site by DNA Polymerase η, Acting Alone or in the Presence of DNA Polymerase ϵ, Is Not Influenced by the Presence of DNA Gaps
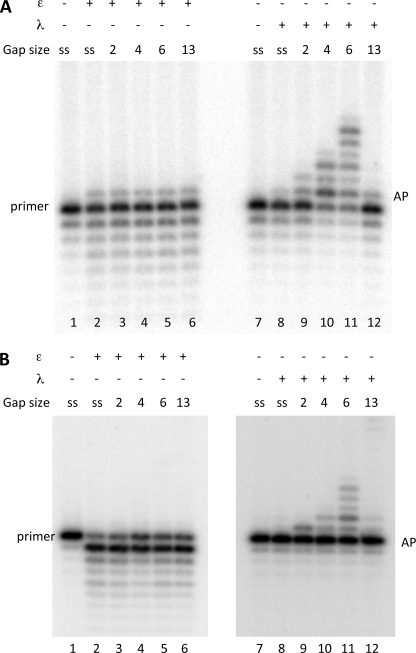
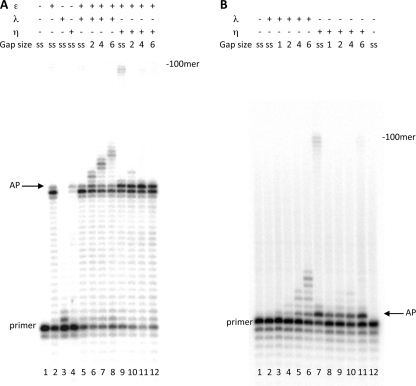
Next, we investigated the capacity of the human pol η to perform TLS of an AP site either in the presence of pol ϵ or when acting alone (Fig. 6). 0.25 pmol of pol η, incubated with single-stranded template-primer in a running start reaction, synthesized up to the AP site and, differently from pol ϵ, efficiently incorporated opposite the lesion (compare lane 4 with lane 2 of Fig. 6A and data not shown). When the reaction was performed in the presence of pol ϵ, pol η performed TLS with the single-stranded template primer, as indicated by the increase in incorporation in front of the lesion and the appearance of some full-length products (see lane 9 of Fig. 6A). However, this TLS was diminished in the presence of gaps of 2, 4, and 6 nucleotides, and extension past the lesion was not affected by the increase in their size (see lanes 10 to 12 of Fig. 6A). This result was different from what was observed with pol λ, where no TLS was observed in the presence of pol ϵ with a single-stranded template-primer (see lane 5 of Fig. 6A) whereas efficient gap filling was performed with DNA gap sizes of 2, 4, and 6 nucleotides (see lanes 6 to 8 of Fig. 6A).
FIGURE 6.
Influence of DNA gap sizes on translesion synthesis of an AP site by DNA pol λ and η. Experiments were performed with templates shown in Table 1. The enzymes and the DNA substrates used are indicated at the top. ss (single-stranded) stands for template-primer with no oligonucleotides hybridized downstream from the AP site, whereas 1, 2, 4, and 6 indicate the length of gap regions between the AP site and the oligonucleotide hybridized downstream. Assays were carried out as described under “Experimental Procedures.” A, experiments with templates in Table 1a. Lane 1, no polymerase present. Lane 2, reaction incubated for 35 min with 0.025 pmol of pol ϵ. Lane 3, reaction incubated for 5 min with 0.25 pmol of pol λ. Lane 4, reaction incubated for 5 min with 0.25 pmol of pol η. Lanes 5–8, reaction was incubated with 0.025 pmol of pol ϵ, and then 0.25 pmol of pol λ were added, and the reaction was continued for 5 min. Lanes 9–12, reaction was incubated with 0.025 pmol of pol ϵ, and 0.25 pmol of pol η were then added, and the reaction was continued for 5 min. B, experiments with templates in Table 1c primed with the 45-mer terminating one nucleotide before the Ap site. Lanes 1 and 12, no polymerase present. Lanes 2–6, reactions incubated 5 min with pol λ. Lanes 7–11, reactions incubated 5 min with pol η. The positions of the primers, of the AP site, and of the 100-mer full-length product are indicated.
We have also studied the TLS capacity of pol η when acting alone on gaps created in a template-primer where the primer is a 45-mer with a 3′OH at the base preceding the lesion. As shown in Fig. 6B, pol η can perform TLS on the single-stranded template (lane 7) but to a lesser extent with gaps of 1, 2, 4, and 6 nucleotides and with no increase in efficiency with the increasing gap size (see lanes 8–11). Conversely, TLS by pol λ was stimulated with gap size increasing from 2 to 6 nucleotides (Fig. 6B, lanes 4–6). Note that pol η showed little TLS also with gap size of 1 and 2 nucleotides that are optimal for TLS by pol β (see Fig. 8). Taken together, these results show that, differently from pol λ, TLS of an AP site by pol η is not stimulated by the presence of DNA gaps.
PCNA and RPA Do Not Influence the Gap Size Preference of DNA Polymerases λ and β to Perform Translesion Synthesis of an AP Site in the Presence of DNA Polymerase ϵ
The processivity factor PCNA and the single-stranded DNA-binding protein RPA play a fundamental role in DNA replication, repair, and recombination (2, 5). Therefore, we studied whether the TLS catalyzed by pols λ and β at DNA gaps in the presence of pol ϵ could be influenced by human PCNA and RPA. To this aim we performed experiments by adding 1.2 pmol of PCNA and 0.25 pmol of RPA. With respect to the 0.15 pmol of template-primer used in the study, the RPA concentration corresponds to roughly one molecule of RPA for 30 nucleotides of single-stranded DNA.
Fig. 7 shows the results with pol λ. As can be seen with a wide range of DNA gaps (Fig. 7A), pol λ performed TLS with an efficiency dictated by the size of the DNA gaps. This efficiency increased with gaps from 1 to 4 nucleotides, remained constant with 6 and 8 nucleotides, and then declined to an almost undetectable level with the gap of 13 nucleotides, as quantified in Fig. 7B. As shown previously, also in this reaction containing both PCNA and RPA, the TLS products length essentially matched the size of the gaps, although low levels of strand displacement could also be detected.
Fig. 8 shows the results with pol β. Ast can be seen, the TLS by pol β is restricted to gaps of 1 and 2 nucleotides, becoming almost undetectable with a gap of 4 nucleotides or longer (Fig. 8A). In agreement with the experiments described previously, the efficiency of TLS by pol β was lower when compared with that observed with pol λ (quantified in Fig. 8B), and no strand displacement was observed.
DISCUSSION
pols δ and ϵ are the two eukaryotic polymerases that replicate DNA. Evidence in yeast supports the conclusion that pol δ is primarily responsible for copying the lagging strand, whereas pol ϵ primarily copies the leading strand (3). In addition, these polymerases also participate in DNA repair processes such as nucleotide excision repair (NER), long patch base excision repair (LP-BER), and mismatch repair (2).
Apurinic or apyrimidinic sites are the most frequent spontaneous lesions in DNA. A number of polymerases, belonging mainly but not exclusively to the Y family polymerases, can perform in vitro TLS of an AP site (9, 28). pols λ and β, two polymerase of the X family believed to be implicated in re-synthesis steps of DNA repair, can also bypass an abasic site (12).
Although much information is available in the literature concerning the capacity of pol δ to deal with an AP site (11, 29), such information is scarce for pol ϵ, particularly for the human enzyme. In a recent publication, we have shown that in vitro elongation by human pol ϵ stopped predominantly at the base preceding the lesion with roughly 10% of residual incorporation opposite to it (14).
As indicated in the Introduction, it is generally accepted that during TLS specialized polymerases replace arrested replicative polymerases at lesions to allow bypass. We have investigated the ability of human pols λ, β, and η to perform TLS of an AP site in the presence of short DNA gaps created by the arrest of human pol ϵ at the lesion. To this aim we have set up a system in which pol ϵ synthesizes on a DNA oligonucleotide template-primer leaving gaps with lengths spanning from 1 to 13 nucleotides because of stalling at the AP site (Table 1).
pol λ cannot bypass an AP site in the presence of pol ϵ if a single-stranded stretch of 35 nucleotides is present downstream from the lesion, but it can do it with gaps from 1 to 10 nucleotides, with a maximum efficiency around 4 to 6 and with almost no TLS with a gap of 13 nucleotides (Figs. 1, 3, 5, and 7). Furthermore, our data indicate the following: (a) pol λ can bypass an AP site by using the 3′OHs generated by the arrest of pol ϵ at the lesion (supplemental Fig. 1); (b) it can act alone to bypass the AP site only when the lesion is present in a gap context and, importantly, with a gap preference similar to the one observed in the presence of pol ϵ (Fig. 2); (c) the BRCT and proline-rich domains of the pol λ are required for efficient TLS of the lesion (Figs. 3 and 4). These results show that pol λ can replace pol ϵ and bypass an AP site utilizing 3′OHs created by the arrest of pol ϵ at the base preceding the lesion or opposite to it. Most interestingly, this switch can take place only on short DNA gaps, whose size strongly influences the efficacy of the process, and bypass of an AP site requires the presence of the BRCT and proline-rich domain of the pol λ. We attempted to further characterize this scenario by investigating directly the binding capacity of pol λ with the gapped DNA substrates used in this study. In contrast to experiments with pol β (see below), we were unable to detect stable interaction between pol λ and DNA substrates under a variety of conditions. However, our results are in full agreement with the gap size preference of pol λ on undamaged DNA recently published (22).
pol β can also catalyze TLS of an AP site in the presence of pol ϵ, but only if the gaps have a size of 1–3 nucleotides, with an efficacy of 1 > 2 > 3 nucleotides and with no bypass of an AP site at gaps longer than 3 nucleotides (Figs. 5 and 8 and data not shown). This preference parallels the binding affinity of pol β for the DNA substrates, as can be seen in supplemental Fig. 2, and it is in agreement with the fact that its incorporation efficiency for undamaged substrates is the highest with 1 nucleotide gap and then decreases with the increase of gap size (22). The efficiency of TLS by pol β appears to be reduced compared with the one displayed by pol λ, likely because of the superior intrinsic capacity of pol λ to replicate an abasic site (12).
Sequencing of the replication products revealed that pol β exclusively inserts dAMP in front of the abasic site in the context of a 2-gap substrate, whereas pol λ bypass of an AP site in a 4-gap substrate induces single or double deletions (supplemental Table 1). Insertion of dAMP by pol β is in accordance with a previous model of pol β translesion synthesis of an abasic site that revealed its predisposition to inserting a nucleotide complementary to the first downstream templating base, which is a thymine in our template sequence (26). The observed behavior of pol λ fits with the scrunching gap-filling model derived from crystal structures of the ternary DNA-pol λ-dNTP complex (30).
pol η was also proficient in TLS of an AP site when acting alone or in the presence of pol ϵ. However its TLS capacity was not stimulated by the presence of DNA gaps (Fig. 6).
PCNA increases the processivity of the replicative pol δ (2), but its capacity to stimulate or not the processivity of pol ϵ remains controversial, possibly depending on the type of DNA substrates and experimental conditions used (31–34). Furthermore, PCNA has been shown to stimulate TLS of an AP site by pol λ when placed on a 73-mer template, 31 nucleotides away from the 17-mer primer (35).
Because PCNA and RPA play a fundamental role in DNA transactions such as DNA replication, repair, and recombination, we set up to study whether the TLS catalyzed by pols λ and β at DNA gaps in the presence of pol ϵ would be influenced by these proteins. When compared with previous figures, Figs. 7 and 8 show that PCNA and RPA had no effect. In addition, Figs. 7 and 8 summarize the major findings of this work by showing that human pols λ and β can perform TLS of an AP site in the presence of human pol ϵ, PCNA, and RPA only in DNA gaps no longer than 10 nucleotides. However, the two polymerases show distinctly different DNA gap size preference and efficiency in performing such TLS.
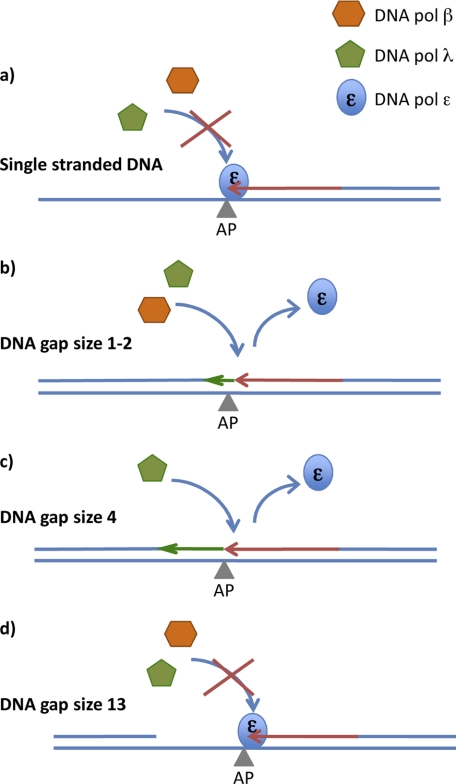
Accordingly, our results suggest the simple model shown in Fig. 9, in which the different role of pol β and λ in extending primers generated by pol ϵ arrested at an AP site can be visualized. When long single-stranded DNA stretch is present downstream from the lesion, neither pol has sufficient affinity for the DNA substrate to displace pol ϵ and continue DNA synthesis past the AP site (Fig. 9a). When the gap downstream from the lesion is only 1 or 2 nucleotides long, both pols β and λ could use their affinity for such gaps to bind and perform TLS (Fig. 9b). If the gap size is between 4 and 10 nucleotides, only pol λ would have the capacity to bind and extend past the lesion (Fig. 9c). If the gap size is larger than 13 nucleotides, neither λ nor β could bind productively, and we are back to the initial situation with single-stranded DNA downstream from the AP site (Fig. 9d).
FIGURE 9.
Tentative model of TLS over an AP site by DNA pol λ and β in the presence of DNA pol ϵ. For details see text.
What might be the physiological significance of our in vitro observations? At least two possibilities exist. The first concerns DNA repair, namely NER and LP-BER pathways. In mammalian cells, it has been shown that pol ϵ can fill the gap of about 27 nucleotides that is produced during NER in a reaction that includes PCNA and RPA (36). The redundant roles of pols δ and ϵ have been confirmed during NER of a defined lesion reconstituted with recombinant or highly purified factors in vitro (37). The requirement of pol ϵ and RPA in the re-synthesis step of NER in human cells has been further demonstrated recently, together with another pathway involving pols δ and κ (38). Because AP sites are among the most frequent endogenous lesions, it is possible that the removal of the damaged 27-mer during NER uncovers an AP site in the DNA template sequence of the gap to be filled. This gap filling will be performed first by pol ϵ up to the lesion and subsequently by pol λ or β depending on the distance of the AP site from the 5′ end of the gap.
A similar situation can arise during the long patch BER pathway. It has been suggested that either pol ϵ or pol δ catalyzed elongation during long patch BER synthesis (39), and it has been shown that both polymerases can participate in the re-synthesis step of long patch BER (40). Furthermore, it is now known that two clustered DNA lesions enhance the mutagenicity of individual lesions (for review see Ref. 41). This observation suggests that the delayed repair at one lesion because of the initiation of repair on the opposite strand can lead to mutagenic TLS of the unrepaired damage. In mammalian cells, pols ϵ, β, or λ could sequentially perform the replication of an AP site during LP-BER of clustered DNA lesions in the way suggested by our model.
The second possibility concerns gap-directed TLS in connection with DNA replication where small single-stranded DNA gaps accumulate along replicated duplexes. Such gaps may arise either by re-priming of the leading strand (42) or the initiation of a new Okazaki fragment (42, 43). Gap-directed lesion bypass has the advantage that the TLS may be separated from the fork progression, and in fact, recent results suggest that a considerable fraction of TLS occurs in the G2/M phase of the cell cycle, when DNA replication has essentially completed (44). As in repair-dependent, gap-directed TLS, the nature of the lesion and the size of the gap may determine which polymerase could be best suited for the bypass.
In summary, although based on in vitro experiments, our study suggests the existence of DNA gap-directed TLS of an abasic site involving human pols ϵ, λ, and β, and it might serve as a working model for further investigations.
Supplementary Material
Acknowledgment
We thank Dr. Neil Johnson for critical reading of the manuscript.
This work was supported in part by a grant from Electricité de France (to G. V.) and by Grants 106986 (to H. P.) and 123082 (to J. E. S.) from the Academy of Finland.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Table 1.
- pol
- polymerase
- TLS
- translesion synthesis
- PCNA
- proliferating cell nuclear antigen
- BRCT
- BRCA1 C-terminal domain
- NER
- nucleotide excision repair
- LP-BER
- long patch base excision repair
- AP
- apurinic/apyrimidinic site
- RPA
- replication protein A.
REFERENCES
- 1. Kornberg A., Baker T. A. (1992) DNA Replication, 2nd Ed., W. H. Freeman & Co., New York [Google Scholar]
- 2. Hübscher U., Spadari S., Villani G., Maga G. (2010) DNA Polymerases: Discovery, Characterization, and Functions in Cellular DNA Transactions, 1st Ed., World Scientific Publishing Co., Singapore [Google Scholar]
- 3. Kunkel T. A., Burgers P. M. (2008) Trends Cell Biol. 18, 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lindahl T. (1993) Nature 362, 709–715 [DOI] [PubMed] [Google Scholar]
- 5. Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., Ellenberger T. (2005) DNA Repair and Mutagenesis, 2nd Ed., ASM Press, Washington, D. C [Google Scholar]
- 6. Prakash S., Johnson R. E., Prakash L. (2005) Annu. Rev. Biochem. 74, 317–353 [DOI] [PubMed] [Google Scholar]
- 7. Waters L. S., Minesinger B. K., Wiltrout M. E., D'Souza S., Woodruff R. V., Walker G. C. (2009) Microbiol. Mol. Biol. Rev. 73, 134–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi J. Y., Lim S., Kim E. J., Jo A., Guengerich F. P. (2010) J. Mol. Biol. 404, 34–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sherrer S. M., Fiala K. A., Fowler J. D., Newmister S. A., Pryor J. M., Suo Z. (2011) Nucleic Acids Res. 39, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shibutani S., Takeshita M., Grollman A. P. (1997) J. Biol. Chem. 272, 13916–13922 [DOI] [PubMed] [Google Scholar]
- 11. Mozzherin D. J., Shibutani S., Tan C. K., Downey K. M., Fisher P. A. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 6126–6131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blanca G., Villani G., Shevelev I., Ramadan K., Spadari S., Hübscher U., Maga G. (2004) Biochemistry 43, 11605–11615 [DOI] [PubMed] [Google Scholar]
- 13. Sabouri N., Johansson E. (2009) J. Biol. Chem. 284, 31555–31563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Locatelli G. A., Pospiech H., Tanguy Le Gac N., van Loon B., Hubscher U., Parkkinen S., Syväoja J. E., Villani G. (2010) Biochem. J. 429, 573–582 [DOI] [PubMed] [Google Scholar]
- 15. Shevelev I., Blanca G., Villani G., Ramadan K., Spadari S., Hübscher U., Maga G. (2003) Nucleic Acids Res. 31, 6916–6925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Henricksen L. A., Umbricht C. B., Wold M. S. (1994) J. Biol. Chem. 269, 11121–11132 [PubMed] [Google Scholar]
- 17. Jónsson Z. O., Hindges R., Hübscher U. (1998) EMBO J. 17, 2412–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blanca G., Shevelev I., Ramadan K., Villani G., Spadari S., Hübscher U., Maga G. (2003) Biochemistry 42, 7467–7476 [DOI] [PubMed] [Google Scholar]
- 19. García-Díaz M., Bebenek K., Sabariegos R., Domínguez O., Rodríguez J., Kirchhoff T., García-Palomero E., Picher A. J., Juárez R., Ruiz J. F., Kunkel T. A., Blanco L. (2002) J. Biol. Chem. 277, 13184–13191 [DOI] [PubMed] [Google Scholar]
- 20. Syvaoja J., Linn S. (1989) J. Biol. Chem. 264, 2489–2497 [PubMed] [Google Scholar]
- 21. Asturias F. J., Cheung I. K., Sabouri N., Chilkova O., Wepplo D., Johansson E. (2006) Nat. Struct. Mol. Biol. 13, 35–43 [DOI] [PubMed] [Google Scholar]
- 22. Brown J. A., Pack L. R., Sanman L. E., Suo Z. (2011) DNA Repair 10, 24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garcia-Diaz M., Bebenek K., Gao G., Pedersen L. C., London R. E., Kunkel T. A. (2005) DNA Repair 4, 1358–1367 [DOI] [PubMed] [Google Scholar]
- 24. Bork P., Hofmann K., Bucher P., Neuwald A. F., Altschul S. F., Koonin E. V. (1997) FASEB J. 11, 68–76 [PubMed] [Google Scholar]
- 25. Fiala K. A., Duym W. W., Zhang J., Suo Z. (2006) J. Biol. Chem. 281, 19038–19044 [DOI] [PubMed] [Google Scholar]
- 26. Efrati E., Tocco G., Eritja R., Wilson S. H., Goodman M. F. (1997) J. Biol. Chem. 272, 2559–2569 [DOI] [PubMed] [Google Scholar]
- 27. Bebenek K., Garcia-Diaz M., Foley M. C., Pedersen L. C., Schlick T., Kunkel T. A. (2008) EMBO Rep. 9, 459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seki M., Masutani C., Yang L. W., Schuffert A., Iwai S., Bahar I., Wood R. D. (2004) EMBO J. 23, 4484–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Daube S. S., Tomer G., Livneh Z. (2000) Biochemistry 39, 348–355 [DOI] [PubMed] [Google Scholar]
- 30. Garcia-Diaz M., Bebenek K., Larrea A. A., Havener J. M., Perera L., Krahn J. M., Pedersen L. C., Ramsden D. A., Kunkel T. A. (2009) Nat. Struct. Mol. Biol. 16, 967–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee S. H., Pan Z. Q., Kwong A. D., Burgers P. M., Hurwitz J. (1991) J. Biol. Chem. 266, 22707–22717 [PubMed] [Google Scholar]
- 32. Chilkova O., Stenlund P., Isoz I., Stith C. M., Grabowski P., Lundström E. B., Burgers P. M., Johansson E. (2007) Nucleic Acids Res. 35, 6588–6597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Syväoja J., Suomensaari S., Nishida C., Goldsmith J. S., Chui G. S., Jain S., Linn S. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 6664–6668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kanuri M., Minko I. G., Nechev L. V., Harris T. M., Harris C. M., Lloyd R. S. (2002) J. Biol. Chem. 277, 18257–18265 [DOI] [PubMed] [Google Scholar]
- 35. Maga G., Villani G., Ramadan K., Shevelev I., Tanguy Le Gac N., Blanco L., Blanca G., Spadari S., Hübscher U. (2002) J. Biol. Chem. 277, 48434–48440 [DOI] [PubMed] [Google Scholar]
- 36. Shivji M. K., Podust V. N., Hübscher U., Wood R. D. (1995) Biochemistry 34, 5011–5017 [DOI] [PubMed] [Google Scholar]
- 37. Araújo S. J., Tirode F., Coin F., Pospiech H., Syväoja J. E., Stucki M., Hübscher U., Egly J. M., Wood R. D. (2000) Genes Dev. 14, 349–359 [PMC free article] [PubMed] [Google Scholar]
- 38. Ogi T., Limsirichaikul S., Overmeer R. M., Volker M., Takenaka K., Cloney R., Nakazawa Y., Niimi A., Miki Y., Jaspers N. G., Mullenders L. H., Yamashita S., Fousteri M. I., Lehmann A. R. (2010) Mol. Cell 37, 714–727 [DOI] [PubMed] [Google Scholar]
- 39. Dianov G. L., Sleeth K. M., Dianova I. I., Allinson S. L. (2003) Mutat. Res. 531, 157–163 [DOI] [PubMed] [Google Scholar]
- 40. Stucki M., Pascucci B., Parlanti E., Fortini P., Wilson S. H., Hübscher U., Dogliotti E. (1998) Oncogene 17, 835–843 [DOI] [PubMed] [Google Scholar]
- 41. Sage E., Harrison L. (2011) Mutat. Res. 711, 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lopes M., Foiani M., Sogo J. M. (2006) Mol. Cell 21, 15–27 [DOI] [PubMed] [Google Scholar]
- 43. Heller R. C., Marians K. J. (2006) Nature 439, 557–562 [DOI] [PubMed] [Google Scholar]
- 44. Karras G. I., Jentsch S. (2010) Cell 141, 255–267 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.