Abstract
Altered expression of RNA-binding proteins modulates gene expression in association with mRNAs encoding many proto-oncogenes, cytokines, chemokines, and proinflammatory factors. Hu antigen R (HuR), a ubiquitously expressed protein, controls a range of cellular functions such as tumor progression, apoptosis, invasion, and metastasis by stabilizing the AU-rich element located at the 3′-untranslated region (UTR) of target mRNAs. Although significant progress has been made in understanding HuR regulation in gene expression, little is known about how HuR undergoes post-translational modifications and recruits target mRNAs during hypoxic stress. Here, we report that during CoCl2-induced hypoxic stress, HuR is significantly overexpressed and undergoes caspase-dependent cleavage in head and neck squamous cell carcinoma cells. Unexpectedly, the HuR-cleavage product 1 (HuR-CP1) was found to strongly associate with the 3′-UTR of c-myc mRNA and block mRNA translation. The binding efficiency of HuR to the 3′-UTR of c-myc mRNA was confirmed using ribonucleoprotein immunoprecipitation and site-directed mutagenesis at the AU-rich element sequences of the c-myc mRNA. Overexpression of a non-cleavable isoform, HuR-D226A, revealed a potent dominant-negative effect, repressing cleavage of endogenous HuR and promoting cell viability. Surprisingly, under hypoxia, siRNA knockdown of HuR elevated c-Myc protein expression. These findings suggest an important role for HuR in hypoxia, and we may have revealed a novel post-transcriptional mechanism that controls c-Myc expression in oral cancer progression.
Keywords: Apoptosis, Caspase, Post-translational Modification, RNA-binding Protein, RNA Turnover, HuR Cleavage, c-Myc Expression
Introduction
Gene expression is controlled by multiple biological networks, and post-transcriptional gene regulation determines mRNA fate in association with RNA-binding proteins and microRNAs (1, 2). Several RNA-binding proteins associate with the 3′-untranslated region (UTR) of target mRNAs and regulate their expression via alteration of the half-life and/or rate of translation of target mRNAs. An RNA-binding protein that increases mRNA stability through this mechanism is the Hu antigen R (HuR,3 also named ELAVL1) (3). HuR is expressed ubiquitously in most cancers, including head and neck squamous cell carcinoma (HNSCC) (4, 5). HuR associates with AU- and U-rich elements (AREs) in the 3′-UTR of target mRNAs to control transcript stability and protein translation (6). In addition, HuR has been shown to be involved in various cancer processes such as cellular proliferation, differentiation, invasion, metastasis, apoptosis, and angiogenesis in association with target mRNAs (7, 8). HuR also increases the translation of certain mRNAs and represses the translation of other transcripts (9, 10). HuR is predominantly localized to the nucleus and, in response to cellular stress, is exported to the cytoplasm where it elicits its post-transcriptional influence on target mRNAs. During stress, HuR has been shown to undergo several post-translational modifications including phosphorylation (11, 12), methylation (13), and ubiquitination (14) and cleaved by caspase-3 (15). However, the molecular consequences of these modifications and cleavage products of HuR are not fully understood. Furthermore, although the cell proliferative effects of HuR-associated target genes have been studied extensively, few apoptosis-related functions have been analyzed in detail. HuR acts as a proliferative protein in response to moderate stress and in the early stages of severe stress (11, 16, 17). However, under extended stress conditions, it augments apoptosis and is cleaved in a caspase-dependent manner, yielding cleavage products 1 and 2 (HuR-CP1 and HuR-CP2) (15).
In mammalian cells, proliferation, growth, and differentiation processes are well orchestrated in response to the local microenvironment such as oxygen deficiency and nutrient deprivation. In response to oxygen insufficiency, HuR potently elevates the expression of two major hypoxia-inducible proteins, hypoxia-inducible factor 1α (HIF-1α) and VEGF (18, 19). In addition, HuR has been proposed to regulate the levels and/or translation of other hypoxia-inducible target mRNAs such as GLUT1, TGF-β, and p53 (6). The proto-oncogene c-myc has been shown to sensitize cells to a variety of cellular death stimuli, including serum (20) and oxygen deprivation (21). Dysregulated expression of c-Myc occurs frequently in human cancers, altering cell proliferation and occasionally inducing cell death (22–24), and the mRNA of the proto-oncogene c-myc was previously identified as a target of HuR (25). Recently it has been shown that HuR inhibits c-myc translation by recruiting the Let-7-associated RNA microRNA-induced silencing complex (RISC) to the c-myc 3′-UTR under non-stress conditions (26). Although HuR was shown to interact strongly with the 3′-UTR of c-myc, the influence of stress-induced HuR on c-myc expression has not been studied in depth. Therefore, we sought to examine the effects of HuR cleavage on c-Myc expression during hypoxia in HNSCC cells.
Here we report how HuR regulates c-myc mRNA stability and translation, both of which contribute to HNSCC progression. Consistent with the down-regulation of c-myc expression during hypoxia, we observed three events that contribute to the fate of c-myc mRNA during hypoxic conditions. First, we observed cleavage of HuR in HNSCC tissues. Interestingly, CoCl2-induced hypoxia also induces HuR expression and cleavage in HNSCC cells. Second, the HuR cleavage product stabilizes a subset of mRNAs including c-myc and represses c-myc translation. Third, the cleavage mutant isoform HuR-D226A acts as a dominant-negative mutant and represses the cleavage of endogenous HuR. These observations suggest that HuR cleavage at least partially regulates c-Myc expression during tumorigenesis.
EXPERIMENTAL PROCEDURES
Patient Tissue Samples
Frozen tongue tumor samples with adjacent normal tissue samples were obtained from patients surgically treated in the Department of Otolaryngology-Head and Neck Surgery at the Medical University of South Carolina using the appropriate informed consent procedure approved by the Medical University of South Carolina institutional review board. Frozen tumor tissue were microdissected to assure that >80% of tumor tissues contained oral squamous cell carcinomas.
Cell Lines, Constructs, and Transfection Experiments
The oral cancer cell line UM74B was maintained in DMEM with high glucose, 10% fetal bovine serum, penicillin-streptomycin-l-glutamine in a humidified 5% CO2 environment at 37 °C. The GFP plasmid vectors containing full-length HuR, HuR-D226A, HuR-CP1, and HuR-CP2 (a kind gift from I. Gallouzi, McGill University) have been previously described (27) (supplemental Fig. S2). The GFP reporter constructs containing the 3′-UTR of c-myc (A–D, AB, and AB mutant; see supplemental Fig. S3) were a kind gift from Myriam Gorospe, National Institute of Aging, Baltimore, MD, and have been described earlier (26). The plasmid transfections were carried out using Lipofectamine (Invitrogen). For HuR knockdown analysis, cells were transfected with siRNAs (20 nm) in medium containing 2% fetal bovine serum. Then either the control siRNA (20 nm; GTTCAATTGTCTACAGCTA) or siRNAs targeting HuR (20 nm; mixture of siRNA oligos AATCTTAAGTTTCGTAAGTTA, TTCGTAAGTTATTTCCTTTAA, and AGTGCAAAGGGTTTGGCTTT) (28) were used for the experiments. The siRNA transfections were carried out using HiPerFect (Qiagen).
Cell Viability and Apoptosis Assays
Cell viability and apoptosis assays were performed as described previously (29, 30). Cell viability was determined using an MTT colorimetric assay. Briefly, 2 × 104 cells were inoculated into each well of a 96-well plate (well area = 0.32 cm2). After 24 h of culture, medium was replaced with experimental medium (100 μl). MTT solution (Sigma) was prepared fresh (5 mg/ml in H2O), filtered through a 0.22-μm filter, and kept for 5 min at 37 °C. The MTT solution (10 μl) was added to each well, and plates were incubated in the dark for 4 h at 37 °C. Then, absorbance was measured at A595 nm using a plate reader (Bio-Rad). The cell apoptosis assay was performed according to the manufacturer's protocol (Annexin V-EGFP apoptosis detection kit; Biovision). UM74B cells with or without CoCl2 treatment (48 h) were analyzed to determine whether hypoxia regulates cell apoptosis. Cells were trypsinized and centrifuged at 300 × g for 5 min and resuspended in 500 μl of 1× binding buffer. Annexin V-EGFP (5 μl) and propidium iodide (5 μl; 0.05 mg/ml) were added, and the suspension was incubated for 5 min in the dark and filtered through a nylon mesh to remove cell clusters. The fluorescence was measured using FACSCalibur Flow Cytometer (BD Biosciences).
Immunohistochemistry, Immunofluorescence, and Western Blot Analysis
Frozen tissue sections from tongue tumors were cut using cryostat and fixed in acetone for 5 min, washed in PBS for 5 min, and then processed for immunostaining. The primary antibodies directed against HuR (1:500, Santa Cruz), VEGF (1:50, Santa Cruz), and HIF-1α (1:200, Abcam) were diluted with DAKO antibody diluents, added to the slides, and incubated for 60 min at room temperature. A biotinylated link antibody plus streptavidin biotin peroxidase kit (DAKO LSAB+ System-HRP) was then used along with a 3,3′-diaminobenzidine chromogen and peroxide substrate to detect the bound antibody complexes. The slides were briefly counterstained with hematoxylin and dehydrated through graded alcohols to xylene. Finally, slides were cover-slipped with permanent mounting media. For immunofluorescence, UM74B cells were grown on coverslips overnight. They were treated as reported (31) by using the following antibody dilutions: anti-HuR (1:300) and anti-β-actin (1:100; Santa Cruz).
For Western blotting, protein extracts resolved by SDS/PAGE were analyzed as described below. Cells were lysed by vortexing 4–5 times in radioimmune precipitation assay buffer (2 mm Tris-Cl (pH 7.6), 30 mm NaCl, 1 mm EDTA, 1 mm DTT, 1 mm PMSF, complete protease inhibitor mixture (Roche Applied Science) and 1% Nonidet P-40) at 10-min intervals each at 4 °C followed by centrifugation at 12,000 × g for 30 min at 4 °C. Supernatants were mixed with an equal volume of 2× Laemmli buffer and heated for 5 min at 95 °C. Total protein was estimated using a Bradford assay (32). Then, 20–60 μg of protein were resolved on 10–12% SDS-PAGE gels and transferred onto PVDF membranes. Blots were preincubated with PBS containing 5% skim milk before incubation with primary antibody against the target protein for 1–3 h at room temperature or overnight at 4 °C. After incubation, the blot was washed four times with PBS containing 0.1% Tween 20 (PBS-T) and then incubated with PBS-T containing 1:10,000-diluted HRP-conjugated secondary antibodies for 1 h at room temperature. After additional washing with PBS-T, immune complexes were visualized using the ECL system (Pierce). Blots were stripped and re-probed anti-β-actin antibody as described above. Western blot analyses were performed using antibodies specific to HuR (Santa Cruz), caspase-3, caspase-7, and c-Myc (Cell Signaling) as suggested by the manufacturer's protocol.
RNA Extraction and Quantitative Real-time PCR (qPCR)
Total RNA was prepared from tongue cancer tissues and HNSCC cell lines using the RNeasy mini kit (Qiagen). qPCR for all mRNA targets was performed using an Applied Biosystems Stepone Plus system with the TaqMan® One-Step RT-PCR kit (Applied Biosystems). Primer sequences are provided in supplemental Table S1.
HuR RNA-binding Protein-IP-ChIP Analysis
HuR RNA-binding Protein-IP-ChIP was performed as previously described (26). Briefly, cell lysates were prepared from exponentially growing UM74B cells with and without hypoxia treatment. Equal amounts of protein were used (100–300 μg). HuR monoclonal antibody 3A2 (Santa Cruz) or isotype control IgG (Sigma) were precoated onto protein A/G-Sepharose beads and extensively washed using NT2 buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm MgCl2, and 0.05% Nonidet P-40 (Nonidet P-40)). Lysates were preabsorbed with IgG (30 μg) and then removed by the addition of protein A/G-Sepharose beads. Individual pulldown assays were performed at 4 °C for 1–2 h to minimize potential reassortment of mRNAs. For RNA analysis, the beads were incubated with 1 ml of NT2 buffer containing 20 units of RNase-free DNase I (15 min, 30 °C), washed twice with 1 ml of NT2 buffer, and further incubated in 1 ml of NT2 buffer containing 0.1% SDS and 0.5 mg/ml proteinase K (15 min, 55 °C) to digest the proteins bound to the beads. RNA was extracted using phenol and chloroform and precipitated in the presence of glycogen. For analysis of individual mRNAs, the RNA isolated from the IP was subjected to reverse transcription (RT) using random hexamers and SuperScriptII reverse transcriptase (Invitrogen). Amplification and quantification of the PCR products were performed using the Applied Biosystems Stepone Plus system (Applied Biosystems) and Power SYBR Green PCR Master Mix (Applied Biosystems). Input GAPDH mRNA was used as a loading control. Each reaction was carried out in triplicate, and three independent experiments were performed.
Statistics
The data are expressed as the mean ± S.D. Two-sample t tests with equal variances were used to assess differences between means. Results with p values less than 0.05 were considered significant.
RESULTS
Oral Cancer Tissues Exhibit Cleavage of HuR
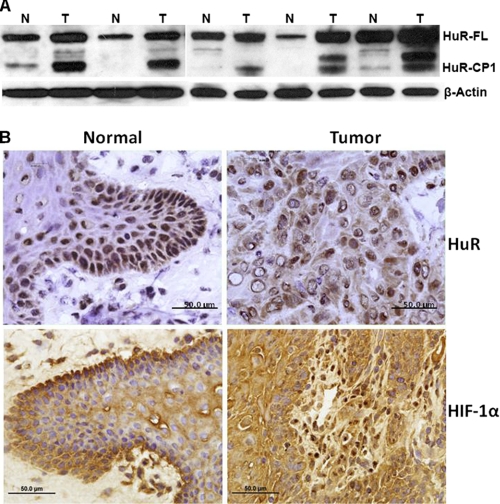
It is well known that hypoxic tumor microenvironment may influence proteins that alter mammalian gene expression patterns (33). To test how hypoxia induces HuR cleavage, tongue tumor tissues from patients were used for Western blot and immunohistochemistry analysis. Data show both overexpression and cleavage of HuR in tongue tumor tissue samples (Fig. 1A). In particular, HuR-CP1 was significantly overexpressed in tumor tissues compared with normal adjacent tissues, and more than 20–30% of total HuR was cleaved compared with normal adjacent tissues. Surprisingly, we did see additional protein bands below the HuR-CP1 and predict that they are either additional cleavage products of HuR-CP1 or degradation products of HuR-CP1. We are currently exploring these additional protein products. In addition, the HuR antibody (HuR-3A2- Santa Cruz) was raised against the epitope, which is present in RRM1 (RNA recognition motif-1) of the HuR protein (34). The cleavage amino acid aspartate 226 is present at the C-terminal region of the protein. Therefore, our experiments describe only the HuR-CP1 and not the HuR-CP2 (contains RRM3 and portion of HNS region; described in Mazroui et al. (15). Next, to determine whether tissues were hypoxic, we stained the tissues with antibodies for HuR and the hypoxic marker HIF-1α (Fig. 1B). Strong expression of HIF-1α positively correlates with hypoxia. Cytoplasmic expression of HuR revealed that a stress-induced nuclear-cytoplasmic translocation occurred in the cancer tissues. Because hypoxia is a strong promoter of apoptosis, the data suggest a correlation between HuR cleavage and apoptosis in oral cancer tissues. Hence, we used oral cancer cell lines to study the relationship between hypoxia-mediated apoptosis and HuR cleavage.
FIGURE 1.
Expression and cleavage of HuR in oral cancer tissues. A, total protein (50 μg) from the tongue tumor (T) and adjacent normal (N) tissues used for Western blot analysis is shown. The blots were probed for HuR and β-actin (used as loading control). HuR-FL indicates the 36-kDa full-length (FL) HuR; HuR-CP1 is a 24-kDa protein. B, tissue sections from representative tumors were subjected to immunohistochemistry using a primary monoclonal antibody to HuR and HIF-1α followed by peroxidase-conjugated goat anti-mouse secondary antibody. Expression of all proteins was relatively high in tumors (brown), where HuR and HIF-1α were significantly expressed in the cytoplasm. The scale bar denotes 50 μm in diameter.
Hypoxia Induces Apoptosis and HuR Cleavage in Oral Cancer Cells
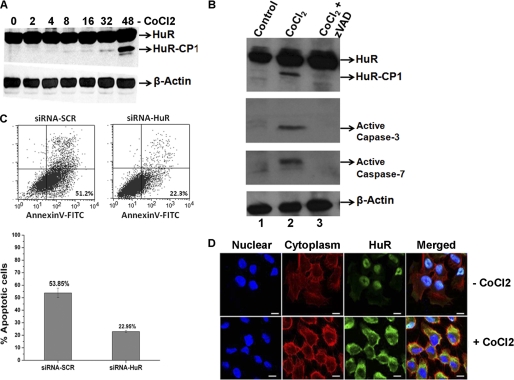
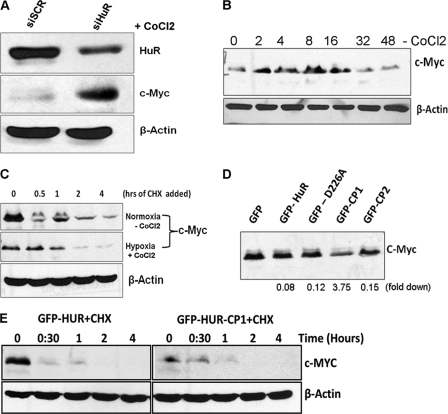
To understand the post-translational modification and nuclear-to-cytoplasmic translocation of HuR during hypoxia, the oral cancer cell line UM74B was subjected to oxygen deprivation by treatment with the hypoxia mimetic compound CoCl2 (250 μm). As shown in Fig. 2A, HuR cleavage in UM74B oral cancer cells begins after 16 h of hypoxic treatment, and it is extensively cleaved at 48 h (Fig. 2A). This observation suggests that acute treatment with CoCl2 (<8 h) does not induce HuR cleavage in UM74B cells, but chronic treatment (>8 h) induces HuR cleavage. Because caspases are known to induce cleavage of HuR (15, 27), we investigated whether this event in UM74B cells is dependent on caspase activity. We saw increased activated caspases-3 and -7 with CoCl2-induced hypoxia (48 h) by Western blot analysis. Caspase proteins activity and HuR cleavage were both abolished by the addition of the caspase inhibitor benzyloxycarbonyl-Val-Ala-Asp (zVAD, Fig. 2B). Next, to test whether HuR-depleted cells underwent apoptosis during CoCl2 treatment, we quantified the percentage of annexin V/propidium iodide-positive cells using flow cytometry. As shown in Fig. 2C, HuR depletion during CoCl2 treatment significantly decreased the number of annexin V-positive cells (from 53.85 to 23.95%; Fig. 2C, compare siRNA-SCR and siRNA-HuR dot plots and bar graph). Thus, we concluded that depleting HuR significantly reduces apoptosis. Together, these findings indicate that introduction of hypoxic stress conditions induces activation of caspases-7 and -3, promoting apoptosis and cleavage of HuR in oral cancer cells.
FIGURE 2.
Expression and cleavage of HuR during CoCl2-induced hypoxia. A, total protein (50 μg) from UM74B oral cancer cells was grown under 250 μm CoCl2 used for Western blot analysis. Cells were treated and harvested at the indicated time points to identify HuR cleavage. The blots were probed for HuR and β-actin (used as loading control). B, active caspases are required for the appearance of the 24-kDa HuR fragment. Cells were incubated with DMSO (first lane 1), CoCl2 (second lane), and CoCl2 + benzyloxycarbonyl-Val-Ala-Asp (zVAD; third lane) followed by Western blotting for HuR, performed as described above and probed with antibodies to active caspases-3 and -7. β-Actin serves as a loading control. C, UM74B cells treated with siRNA are described under “Experimental Procedures” in the presence of 250 μm CoCl2 for 36 h were analyzed by staining with annexin V-FITC and propidium iodide by flow cytometry. The percentage of apoptotic cells (top boxes) upon CoCl2 treatment was determined for HuR (siRNA-HuR) or control siRNA (siRNA-SCR)-treated HeLa cells. The values were normalized to control untreated cells. The graph represents the number of apoptotic cells after treatment as described in C. Values are the means ± S.E. (error bars) from three independent experiments. D, immunofluorescence detection of HuR in UM74B cells, either untreated or treated with CoCl2 as indicated. Distribution of cytoplasmic HuR (merged panel) was observed after 24 h. Blue indicates DAPI staining used to detect nuclei, red indicates β-actin staining to detect cytoplasm, and green indicates HuR immunofluorescence. The scale bar denotes 5 μm.
It has previously been shown that HuR is exported to the cytoplasm under conditions of stress such as UV treatment (36), serum starvation (37), heat shock (34), and staurosporine administration (15). We, therefore, speculated that CoCl2 treatment, mimicking hypoxic stress, would influence HuR translocation. To ascertain the CoCl2-induced export of HuR in UM74B cells, we observed HuR translocation by immunofluorescence microscopy. Under untreated conditions, HuR was localized in the nucleus and/or the perinuclear region of UM74B cells, whereas within 24 h of CoCl2 treatment, cells exported HuR to the cytoplasm (Fig. 2D). Of note, we observed no cytoplasmic HuR during acute stress (less than 8 h of CoCl2 treatment; data not shown). But after chronic stress (24 h of treatment), HuR was localized completely to the cytoplasm. It has been reported that during staurosporine-induced lethal stress, HuR is transported and accumulates in the cytoplasm undergo cleavage by caspases (15). Altogether, these studies indicate that during the hypoxic stress HuR is exported to the cytoplasm and cleaved by caspases.
CoCl2-induced Hypoxia Affects Stability of ARE-containing mRNA
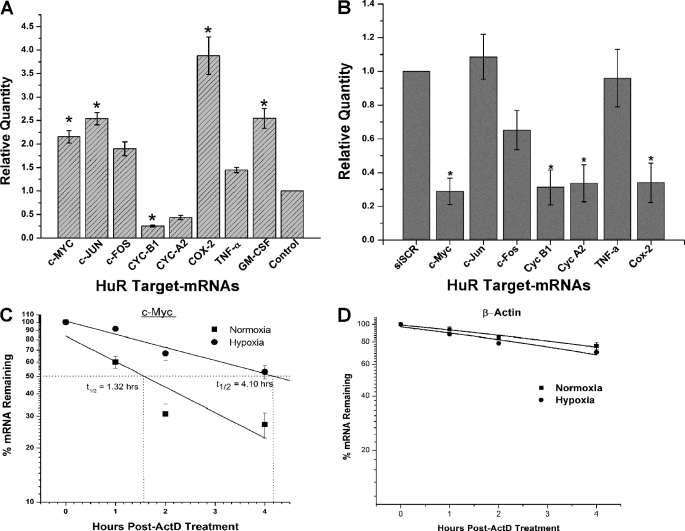
Hypoxia-induced post-transcriptional control plays a critical role in gene expression when cells undergo oxygen deprivation. The transcription factor HIF-1α plays a major role in the transcriptional activation of hypoxia-inducible genes (38). Interestingly, HuR has been shown to be associated with the 3′-UTR of HIF-1α mRNA and promotes its translation during hypoxia (18). To investigate whether CoCl2-induced hypoxia affects other ARE-containing mRNAs, we analyzed known HuR ARE target transcripts (39) using qPCR to examine alterations in their abundance in response to CoCl2 treatment. We observed increased mRNAs encoding c-Myc (p < 0.05), c-Jun (p < 0.05), c-Fos, TNFα, GM-CSF (p < 0.05), and Cox-2 (p < 0.05) and repression of cyclins A2 and B1 (p < 0.05) during CoCl2-induced hypoxic stress (Fig. 3A) when compared with untreated UM74B cells. To confirm that ARE mRNA quantity is directly controlled by HuR under hypoxic conditions, we analyzed their expression after knockdown of HuR using RT-qPCR. As shown in Fig. 3B, under CoCl2-induced hypoxia, ARE-mRNAs such as c-Myc, cyclin B1, cyclin A2, and Cox-2 (p < 0.05) are reduced in HuR-knockdown cells compared with control siRNA-transfected cells. We did not see significant changes in c-Jun, c-Fos, and TNFα ARE mRNAs. This effect may be due to other RNA-binding proteins possibly controlling these transcripts. Interestingly, both cyclin B1 and cyclin A2 mRNA levels were significantly down-regulated in both CoCl2-treated cells with HuR (Fig. 3A) and HuR knockdown cells (Fig. 3B). This down-regulation of cyclins may possibly be due to cell cycle arrest in both experimental conditions. We are currently studying the regulation of these mRNAs during hypoxic stress. Next, we examined the half-life (t½) of the ARE mRNA c-Myc during hypoxia. The quantities of c-Myc mRNA in untreated and treated CoCl2 UM74B cells were measured by qPCR at 0, 30, 60, and 120 min after actinomycin D (ActD) treatment and calculation of the c-myc t½ (Fig. 3C). In untreated UM74B cells, c-myc mRNA the t½ was 1.32 h, whereas in CoCl2-treated hypoxic cells, c-myc t½ increased to 4.10 h. Control mRNA β-actin did not significantly change with CoCl2 treatment (Fig. 3D). These data indicate that hypoxia increases the t½ of c-myc mRNA. In other words, the c-Myc mRNA is stabilized under CoCl2-induced hypoxia, which potentially mimics the conditions in oral cancer progression.
FIGURE 3.
CoCl2-induced hypoxia regulates AU-rich-containing mRNAs. A, shown is the relative quantity of ARE mRNAs measured by RT-qPCR comparing normoxia versus hypoxia after 36 h of CoCl2 treatment, and β-actin serves as a loading control. B, the relative amounts of ARE mRNAs expressed in UM74B cells transfected with HuR siRNA or control siRNA after CoCl2 treatment were measured by RT-qPCR. The values are normalized using a factor calculated from β-actin gene expression. C and D, the decay rates of the c-Myc mRNA and β-actin as indicated were assessed in UM74B cells by RT-qPCR after transcription inhibition with actinomycin D. Error bars denote the S.E. of three sets of experiments. *, p < 0.05; n = 3.
HuR-CP1 Binds ARE of c-Myc mRNA during Hypoxia
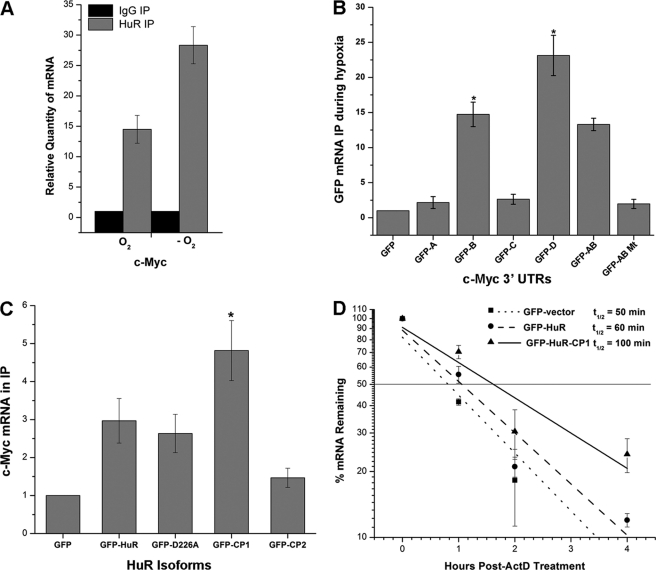
To determine whether the observed changes in c-myc mRNA during hypoxia were due to direct interaction with HuR, we performed ribonucleoprotein (RNP) IP on control and CoCl2-treated UM74B cells using a monoclonal antibody against HuR. HuR-interacting mRNA was then analyzed by RT-qPCR. Consistent with Fig. 3A, we observed a 2-fold enrichment of HuR-bound c-myc mRNA (p < 0.05) (Figs. 4A and supplemental Fig. S1) in CoCl2-treated hypoxic cells compared with untreated cells. Other targets such as c-fos and Cox-2 mRNAs were also more strongly associated with HuR IP samples during hypoxic cells compared with normoxic cells (data not shown). Next, to confirm that during CoCl2-induced hypoxia HuR binds to the c-myc mRNA, additional GFP-based reporter constructs of c-myc 3′-UTR (supplemental Fig. S2, A–D (26)) were tested. UM74B cells were transiently transfected with all the c-myc 3′-UTR reporter constructs, and extracts were prepared 24 h after CoCl2-induced hypoxic stress. The extracts were subjected to RNP IP with HuR antibody followed by qPCR analysis. qPCR carried out with GFP primers revealed that fragments B and D (ARE motifs and U-rich regions), which contained a sequence motif shown previously to interact with HuR (26), significantly associated (15- and 25-fold, respectively compared with GFP vector alone) with HuR during CoCl2 treatment compared with untreated cells (Fig. 4B). But at the same time the ARE mutant of c-myc 3′-UTR regions A and B did not interact with HuR (last bar on the graph). Together, these findings demonstrate that hypoxia-induced HuR associates with the ARE-containing c-myc 3′-UTR regions B and D, leading to increased mRNA stability.
FIGURE 4.
HuR associates with c-myc mRNA during CoCl2-induced hypoxic stress. A, UM74B cells were grown with and without treatment of CoCl2 (O2 indicates no treatment, and −O2 represents CoCl2 treatment) and subjected to RNP IP followed by RT-qPCR analysis to measure the relative quantity of c-myc mRNA in HuR IP compared with control IgG IP. GAPDH serves as a loading control. B, shown are plasmids expressing chimeric RNAs spanning all the four c-myc 3′-UTR segments (A–D) combination plasmid vectors, AB and AB mutant (“Experimental Procedures”; see supplemental Fig. S2). 48 h after transfection, binding of HuR to each chimeric RNA was tested by RNP IP followed by GFP mRNA detection by RT-qPCR. C, forty-eight hours after transfection with a control plasmid, GFP or a plasmid overexpressing full-length HuR, HuR-D226A, HuR-CP1, and HuR-CP2 (“Experimental Procedures,” supplemental Fig. S3) c-Myc was measured after RNP IP using GFP antibody (loading control GAPDH from input lysates). D, treatment (48 h) with plasmids expressing GFP, GFP-HuR, and GFP-HuR-CP1 followed by the decay rates of c-myc was assessed in UM74B cells by RT-qPCR after transcription inhibition with actinomycin D (ActD). Values were normalized using a factor calculated from GAPDH gene expression. Error bars denote the S.E. of three sets of experiments (*, p < 0.05, n = 3).
To determine the role of HuR cleavage products in c-myc mRNA stability during CoCl2-induced hypoxia, we expressed GFP-tagged HuR, GFP-HuRD226A (non-cleavable HuR), GFP-HuR-CP1, and GFP-HuR-CP2 (see supplemental Fig. S3). We then performed RNP IP using an anti-GFP antibody followed by RT-qPCR analysis. As expected, under hypoxia, HuR-CP1 significantly associated with c-Myc mRNA (p < 0.05) as compared with the other isoforms of HuR (Fig. 4C). Next, to validate the changes we observed in c-myc mRNA half-life, we performed an actinomycin D pulse-chase experiment on UM74B cells transfected with either GFP-HuR or GFP-HuR-CP1 under CoCl2-induced hypoxia. As expected, we observed that excess HuR-CP1 results in an increase in the t½ of c-myc mRNA (Fig. 4D) to 100 min compared with 60 min with the full-length HuR. We observed no significant changes in c-myc mRNA stability with other HuR-isoforms such as HuR-D226A and HuR-CP2 (data not shown).
HuR-CP1 Represses c-Myc Protein Expression during CoCl2-induced Hypoxic Stress
In contrast to the effect of HuR on c-myc mRNA stability, it has been reported that siRNA silencing of HuR resulted in increased levels of c-Myc protein under non-stress conditions, suggesting that HuR could block c-myc translation (26) under normoxic conditions. To test whether HuR influences c-Myc translation under hypoxic conditions, we used siRNA knockdown of HuR and studied c-Myc protein expression. Consistent with the above observation, we observed that knockdown of HuR increases c-Myc protein in oral cancer cells under CoCl2 induced hypoxia (Fig. 5A). This observation suggests that HuR could act as a possible translational repressor for c-Myc during hypoxic conditions. Next, we wished to determine if the effect of c-Myc is due to translational repression or degradation of protein. Under hypoxic conditions, we found that c-Myc expression shows a biphasic profile (Fig. 5B) with Western blot analysis. However, during 32 and 48 h of CoCl2 treatment, c-Myc protein was found to be returned as its control level (0-h time point), suggesting that under chronic hypoxic stress, c-Myc protein was not induced in comparison with that under acute-stress conditions. In agreement with this observation, it has been reported that hypoxia decreased c-Myc protein in Beas-2B cells after 24 h of treatment (40). It has been reported that hypoxia-induced proteasomal degradation of c-Myc protects the cells from c-Myc-mediated apoptosis (41). Hence, we aimed to determine whether c-Myc protein is destabilized in hypoxic cells without knockdown of HuR. c-Myc protein t½ was measured in UM74B cells in both untreated and CoCl2-treated cells after treatment with the protein synthesis inhibitor cycloheximide (CHX). We found that the c-Myc protein stability between normoxic and hypoxic cells was not significantly different after adding CHX (Fig. 5C). These observations suggest that c-Myc protein is possibly controlled at the translational level during hypoxia. Also, quite possibly HuR protein activities differ under normoxic versus hypoxic conditions, and therefore, have different effects on c-Myc expression.
FIGURE 5.
Knockdown of HuR enhances c-Myc protein levels. A, forty-eight hours after transfection of UM74B cells with control (siSCR) siRNA or HuR-directed siRNA (siHuR) as described previously (26) and treated with CoCl2 (36 h), lysates were prepared to assess the levels of c-Myc, HuR, and loading control β-actin by Western blot analysis. B, 48 h after CoCl2 treatment of UM74B cells, lysates were prepared to assess the protein levels of c-Myc and loading control β-actin by Western blot analysis. C, CoCl2 treatment decreased c-Myc protein levels in UM74B cells in a time-dependent manner after treating with CHX at a concentration of 5 μg/ml. D, after transfection (48 h) with the indicated plasmid vectors, the lysates were prepared to assess the protein levels of c-Myc. GFP vector serves as loading control normalized by cell sorting using flow cytometry. The Western blot data are representative of three sets of experiments. E, HuR-CP1 regulates c-Myc expression by reducing translation and independent of protein degradation. UM74B cells transfected with GFP fused with full-length HuR and HuRCP1 were treated with CoCl2. Subsequently, CHX was added at a concentration of 5 μg/ml, and c-Myc levels were studied in a time-dependent manner.
As described earlier, HuR is significantly cleaved after 24 h of CoCl2 treatment (shown in Fig. 2A), suggesting a connection between HuR cleavage and c-Myc protein expression under hypoxic conditions. To address the possible relationship between HuR cleavage and c-Myc levels, we overexpressed HuR cleavage isoforms in UM74B cells under CoCl2 treatment and measured c-Myc expression via Western blot analysis. We found that overexpression of HuR-CP1 decreases c-Myc protein more than 3-fold (Fig. 5D) compared with the other HuR isoforms. Altogether, these data establish that HuR-CP1 inhibits the translation of c-myc mRNA under hypoxic conditions. To address whether the effects of HuR-CP1 occur at the level of protein synthesis or degradation of existing c-Myc protein, we added CHX to cells transfected with full-length versus CP1 products of HuR fused with GFP. We found there was no difference in the level of degradation between the cells expressing either full-length HuR or HuR-CP1, suggesting that the degradation level of the c-Myc protein was unaffected by HuR-CP1 (Fig. 5E). This suggests that the mechanism for reduced c-Myc protein levels through HuR-CP1 (as shown in Fig. 5D) is likely to be through inhibition of protein synthesis rather than increased protein degradation. Thus, under hypoxia HuR-CP1 regulates c-Myc levels by reducing protein expression.
HuR-CP1 Decreases Cell Viability Compared with the Non-cleavage Mutant HuR-D226A
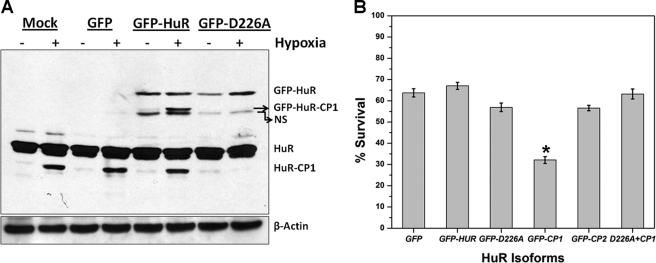
HuR-CP1 has been shown to be involved in the apoptotic pathway and to promote apoptosis (15, 27). The HuR Asp226 amino acid residue is reportedly by the caspases-3 and -7 in HeLa cells (15). Mutation of amino acid Asp226 to alanine abolishes HuR cleavage. Hence, we sought to test cleavage of full-length GFP-HuR and the non-cleavable isoform GFP-HuR-D226A under conditions of CoCl2-induced hypoxia. Our Western blot data revealed that after CoCl2 treatment, cells overexpressing wild type HuR-GFP fusion showed cleavage of both the exogenously expressed GFP-HuR as well as the endogenous HuR protein. In contrast, overexpression of the non-cleavable mutant HuR-D226A showed no cleavage of the exogenous protein GFP-HuR-D226A. Interestingly, it also inhibited the cleavage of endogenous HuR (Fig. 6A). These data indicate that overexpression of the non-cleavable mutant HuR-D226A acts in a dominant-negative fashion, blocking the cleavage of cellular HuR. We decided to determine the functional effects of these results in oral cancer progression. Therefore, we transiently transfected the HuR isoforms in oral cancer UM74B cells under CoCl2-induced hypoxia and measured cell viability using an MTT assay. Among cells expressing different HuR isoforms, HuR-CP1 significantly reduced UM74B cell viability (65%, p < 0.05) (Fig. 6B). Co-transfection of HuR-D226A with HuR-CP1 rescued the impaired cell viability caused by HuR-CP1 (Fig. 6B, last bar), thus exhibiting the functional manifestation of the dominant-negative effects of HuR-D226A. It also validates the role of HuRCP1 in diminishing cell survival under hypoxic conditions, corresponding with its role as a pro-apoptotic molecule (15).
FIGURE 6.
HuR-D226A mutant acts in a dominant-negative manner during stress. A, measurement of HuR in normoxic (no CoCl2) or hypoxic cells (with CoCl2) is shown. UM74B cells were transfected with indicated plasmid vectors (GFP, GFP-HuR, and GFP-D226A) followed by 36 h of hypoxic treatment, and cell extracts (50 μg) were analyzed by Western blotting using antibodies against HuR and the β-actin antibody. All cells were normalized based on GFP counts using flow cytometry. NS represents a nonspecific band. B, shown is the growth rate of representative plasmids transfected after 48 h. Cells were plated at a density of 103 cells per ml and counted at 48 h. All the transfection vectors were normalized by GFP cell sorting using flow cytometry. Triplicate samples were counted at each time point, and the S.D. values were used to plot the error bars (*, p < 0.05, n = 3).
DISCUSSION
Our studies have uncovered a mechanism by which HuR-CP1 stabilizes c-myc mRNA and controls its translation during hypoxic stress in HNSCC. We report that during hypoxic stress in cultured oral cancer cells or in hypoxic human oral cancer tissues, RNP HuR is exported to the cytoplasm, undergoing caspase-mediated cleavage. HuR-CP1 triggers an increase in c-myc mRNA t½ and represses its protein translation. Therefore, we suggest a model in which, during hypoxia, a portion of HuR is cleaved, generating HuR-CP1 that strongly associates with the c-myc ARE containing 3′-UTR regions B and D and promotes c-myc mRNA stability. This association possibly represses c-myc translation and regulates its expression under hypoxic conditions. Previously, the contribution of HuR to c-Myc expression has not been established due to the difficulty of obtaining accurate measurements of c-myc mRNA decay (26). Here, we introduced a stressor such as hypoxia to cells that contributed to HuR modification, and reduction in c-Myc expression was subsequently measured. It is known that stress-induced HuR cleavage promotes apoptosis in HeLa cells and is a key regulator of the pro-apoptotic pathway (15). In addition, it has been shown that HuR-CP1 is associated with import factor TRN2, which allows HuR to accumulate in the cytoplasm and promote myogenesis (27). This result indicated that cytoplasmic accumulation is linked to a promyogenic function of HuR and supported a model whereby, during the transition phase from myoblasts to myotubes, a proportion of HuR is cleaved, generating HuR-CP1. By interfering with the TRN2-mediated import of HuR, this CP1 product helps non-cleaved HuR accumulate in the cytoplasm, thus, promoting myogenesis. It has been proposed that the HuR cleavage event is dependent on the protein kinase RNA. In response to lethal stress, unphosphorylated protein kinase RNA activates the FADD/caspase-8/caspase-3 pathway to trigger HuR cleavage. The resulting HuR-CPs thus formed were capable of promoting apoptosis (42). These observations clearly indicated that HuR-CP1 could be actively engaged in a pro-apoptotic pathway. Our observations impinge on the HuR-CP1 interaction with c-myc regions B and D and its implications in mediating hypoxic stress effects. During normoxic conditions there is no Hur cleavage or repression of c-Myc gene expression. Likewise, HuR-CP1 can modulate target mRNA stability during stress conditions. Hence, we speculate that AU-rich elements such as those present in the c-myc 3′-UTR and stress-induced RNPs like HuR that bind to them are extensively involved in modulating mRNA stability and translation. Further studies are needed to establish whether the cleavage of HuR broadly implicates survival of the cell by interfering with other mRNAs such as the c-Myc.
The tumor microenvironment plays a critical role in determining the therapeutic prognosis of solid tumors (43). Hypoxia, a common characteristic of solid malignances, is an important component of the tumor microenvironment. Hypoxia was suggested to contribute to the aggressive phenotypes of head and neck cancer, characterized by large tumor size, high histologic grade, high proliferation rate, and loco-regional spread (44, 45). Hence, we evaluated the influence of hypoxia in HNSCC cells in the context of HuR associated gene expression. HuR-regulated mRNAs could play a role in the therapeutic response of hypoxic solid tumors. For example, HuR has been shown to impact on a common chemotherapeutic compound gemcitabine, specifically by regulating a key gemcitabine metabolic enzyme, deoxycytidine kinase mRNA (46). It has been shown that hypoxic tumors are enriched for the hypoxic factor HIF-1α and angiogenesis factor VEGF, which are targets of HuR (18, 19). We observed that in hypoxic tumors, HIF-1α and HuR were significantly overexpressed. Additionally, the overexpressed protein was cleaved (Fig. 1A), yielding HuR-CP1. Recently, a growing body of evidence has supported the concept that deregulated c-Myc expression occurs in many human cancers and globally reprograms cells, driving them toward proliferation as well as apoptosis (22–24). Our current findings show that during hypoxia, overexpression of HuR strongly up-regulates c-myc mRNA. It has been reported that c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression (47), with effects apparently opposite to that of HIF-1α. Furthermore, c-Myc has also been shown to activate glucose transporter 1 and glycolytic genes (48), which are transcriptional targets of HIF-1α as well. Notably, c-Myc is responsible for normoxic induction of genes, whereas HIF-1α controls gene expression under hypoxic conditions (49). Thus, both HIF-1α and c-Myc may function coherently to regulate glycolysis in tumor tissues under normal and hypoxic conditions. Testing this hypothesis would involve examining the requirement of c-Myc for increased glycolysis in normoxic conditions. Alternatively, decreased c-Myc expression under hypoxic stress was also reported in other cell lines (40, 50). In fact, our observation of reduced t½ and expression of c-Myc protein during hypoxia confirms this observation.
HuR has been predominantly characterized as a regulatory protein that binds specifically to consensus ARE sequences within the 3′-untranslated regions of multiple genes and enhances mRNA stability. HuR enhances mRNA stability apparently by competing with other ARE-binding proteins, which would otherwise target the mRNA for degradation from the 3′ end (51). In the case of the c-myc mRNA, HuR binds to the 3′-UTR of the c-Myc transcript, providing the opportunity for the translation apparatus to encounter this regulatory protein. Thus, it is reasonable to anticipate a role for HuR in the regulation of the translation efficiency of an mRNA. Indeed, we observed that binding of HuR to its target sequences within the c-Myc 3′-UTR was associated with a decrease in translational efficiency of the c-myc mRNA. This observation was validated by the increase in c-myc mRNA protein upon knockdown of HuR in oral cancer cells. HuR has been implicated in numerous cellular responses against the various types of stress encountered during the course of cell growth. Depending on the stress stimulus, HuR is localized to the cytoplasm and controls mRNA stability of the target genes. Interestingly, overexpression of the cleaved mutant HuR-CP1 significantly decreased cell viability, suggesting a possible role for HuR in modulating cell physiology during hypoxic conditions. A systematic analysis of the potential association of HuR with pro- and anti-apoptotic mRNA transcripts under various stress conditions such as hypoxia is awaited.
The ability of HuR to modulate repression of the c-myc gene allows for appropriate fluctuations in c-Myc expression in response to changes in tumor microenvironment or stress levels. Based on the location of its binding sites and its function in mRNA stability and translational repression, HuR is poised to serve as a critical regulator of gene expression at the protein level (52). HuR also binds to p27kip1 mRNA (35) and functions as a translation repressor in this context as well (9). However, this leaves us with an interesting question regarding the function of HuR in simultaneously inhibiting the expression of both a cell cycle inhibitor as well as a proliferation enhancer. Based on the evidence presented so far, the influence of cleavage of HuR on its apoptotic function likely depends on the magnitude of the stress, timing of the stress response, and proliferative status of the cell. Although we gained some insight into the functions of HuR, a detailed analysis of its role in apoptosis is warranted. This may help design potential therapeutic targets directed to processes in which HuR regulates target mRNAs.
Supplementary Material
Acknowledgments
We thank Sundar Balasubramanian (Dept. of Cardiology, College of Medicine, Medical University of South Carolina, Charleston, SC) for assistance in immunofluorescence. We also thank Kevin Paneerselvam and Saraswathi Ramachandran for technical assistance. We gratefully acknowledge Drs. Myriam Gorospe, Courtney Haycraft, and Jennifer Schnellmann for reviewing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R00DE018165 (to V. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
- HuR
- Hu antigen R
- HNSCC
- head and neck squamous cell carcinoma
- AREs
- AU- and U-rich elements
- CP1 and CP2
- cleavage products 1 and 2
- HIF-1α
- hypoxia-inducible factor 1α
- qPCR
- quantitative real-time PCR
- RNP
- ribonucleoprotein
- CHX
- cycloheximide
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- IP
- immunoprecipitate.
REFERENCES
- 1. Keene J. D. (2007) Nat. Rev. Genet 8, 533–543 [DOI] [PubMed] [Google Scholar]
- 2. Siomi H., Siomi M. C. (2010) Mol. Cell 38, 323–332 [DOI] [PubMed] [Google Scholar]
- 3. Garneau N. L., Wilusz J., Wilusz C. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 113–126 [DOI] [PubMed] [Google Scholar]
- 4. Hasegawa H., Kakuguchi W., Kuroshima T., Kitamura T., Tanaka S., Kitagawa Y., Totsuka Y., Shindoh M., Higashino F. (2009) Br. J. Cancer 100, 1943–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kakuguchi W., Kitamura T., Kuroshima T., Ishikawa M., Kitagawa Y., Totsuka Y., Shindoh M., Higashino F. (2010) Mol. Cancer Res. 8, 520–528 [DOI] [PubMed] [Google Scholar]
- 6. Masuda K., Abdelmohsen K., Gorospe M. (2009) J. Cell Mol. Med. 13, 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brennan C. M., Gallouzi I. E., Steitz J. A. (2000) J. Cell Biol. 151, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen J. L., Lin H. H., Kim K. J., Lin A., Ou J. H., Ann D. K. (2009) Autophagy 5, 244–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kullmann M., Göpfert U., Siewe B., Hengst L. (2002) Genes Dev. 16, 3087–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leandersson K., Riesbeck K., Andersson T. (2006) Nucleic Acids Res. 34, 3988–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Abdelmohsen K., Pullmann R., Jr., Lal A., Kim H. H., Galban S., Yang X., Blethrow J. D., Walker M., Shubert J., Gillespie D. A., Furneaux H., Gorospe M. (2007) Mol. Cell 25, 543–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doller A., Huwiler A., Müller R., Radeke H. H., Pfeilschifter J., Eberhardt W. (2007) Mol. Biol. Cell 18, 2137–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li H., Park S., Kilburn B., Jelinek M. A., Henschen-Edman A., Aswad D. W., Stallcup M. R., Laird-Offringa I. A. (2002) J. Biol. Chem. 277, 44623–44630 [DOI] [PubMed] [Google Scholar]
- 14. Abdelmohsen K., Srikantan S., Yang X., Lal A., Kim H. H., Kuwano Y., Galban S., Becker K. G., Kamara D., de Cabo R., Gorospe M. (2009) EMBO J. 28, 1271–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mazroui R., Di Marco S., Clair E., von Roretz C., Tenenbaum S. A., Keene J. D., Saleh M., Gallouzi I. E. (2008) J. Cell Biol. 180, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lal A., Kawai T., Yang X., Mazan-Mamczarz K., Gorospe M. (2005) EMBO J. 24, 1852–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abdelmohsen K., Lal A., Kim H. H., Gorospe M. (2007) Cell Cycle 6, 1288–1292 [DOI] [PubMed] [Google Scholar]
- 18. Galbán S., Kuwano Y., Pullmann R., Jr., Martindale J. L., Kim H. H., Lal A., Abdelmohsen K., Yang X., Dang Y., Liu J. O., Lewis S. M., Holcik M., Gorospe M. (2008) Mol. Cell. Biol. 28, 93–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Levy N. S., Chung S., Furneaux H., Levy A. P. (1998) J. Biol. Chem. 273, 6417–6423 [DOI] [PubMed] [Google Scholar]
- 20. Evan G. I., Wyllie A. H., Gilbert C. S., Littlewood T. D., Land H., Brooks M., Waters C. M., Penn L. Z., Hancock D. C. (1992) Cell 69, 119–128 [DOI] [PubMed] [Google Scholar]
- 21. Brunelle J. K., Santore M. T., Budinger G. R., Tang Y., Barrett T. A., Zong W. X., Kandel E., Keith B., Simon M. C., Thompson C. B., Hay N., Chandel N. S. (2004) J. Biol. Chem. 279, 4305–4312 [DOI] [PubMed] [Google Scholar]
- 22. Adhikary S., Eilers M. (2005) Nat. Rev. Mol. Cell Biol. 6, 635–645 [DOI] [PubMed] [Google Scholar]
- 23. Cole M. D., McMahon S. B. (1999) Oncogene 18, 2916–2924 [DOI] [PubMed] [Google Scholar]
- 24. Grandori C., Cowley S. M., James L. P., Eisenman R. N. (2000) Annu. Rev. Cell Dev. Biol. 16, 653–699 [DOI] [PubMed] [Google Scholar]
- 25. Lafon I., Carballès F., Brewer G., Poiret M., Morello D. (1998) Oncogene 16, 3413–3421 [DOI] [PubMed] [Google Scholar]
- 26. Kim H. H., Kuwano Y., Srikantan S., Lee E. K., Martindale J. L., Gorospe M. (2009) Genes Dev. 23, 1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beauchamp P., Nassif C., Hillock S., van der Giessen K., von Roretz C., Jasmin B. J., Gallouzi I. E. (2010) Cell Death Differ. 17, 1588–1599 [DOI] [PubMed] [Google Scholar]
- 28. Kuwano Y., Kim H. H., Abdelmohsen K., Pullmann R., Jr., Martindale J. L., Yang X., Gorospe M. (2008) Mol. Cell. Biol. 28, 4562–4575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chung J., Bachelder R. E., Lipscomb E. A., Shaw L. M., Mercurio A. M. (2002) J. Cell Biol. 158, 165–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van de Loosdrecht A. A., Beelen R. H., Ossenkoppele G. J., Broekhoven M. G., Langenhuijsen M. M. (1994) J. Immunol. Methods 174, 311–320 [DOI] [PubMed] [Google Scholar]
- 31. Balasubramanian S., Mani S. K., Kasiganesan H., Baicu C. C., Kuppuswamy D. (2010) PLoS One 5, e11470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 33. Leo C., Giaccia A. J., Denko N. C. (2004) Semin. Radiat. Oncol. 14, 207–214 [DOI] [PubMed] [Google Scholar]
- 34. Gallouzi I. E., Brennan C. M., Stenberg M. G., Swanson M. S., Eversole A., Maizels N., Steitz J. A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 3073–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Millard S. S., Vidal A., Markus M., Koff A. (2000) Mol. Cell. Biol. 20, 5947–5959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fernau N. S., Fugmann D., Leyendecker M., Reimann K., Grether-Beck S., Galban S., Ale-Agha N., Krutmann J., Klotz L. O. (2010) J. Biol. Chem. 285, 3896–3904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Atasoy U., Watson J., Patel D., Keene J. D. (1998) J. Cell Sci. 111, 3145–3156 [DOI] [PubMed] [Google Scholar]
- 38. Rankin E. B., Giaccia A. J. (2008) Cell Death Differ. 15, 678–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. López de Silanes I., Zhan M., Lal A., Yang X., Gorospe M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2987–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li Q., Costa M. (2009) Biochimie 91, 1307–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Corn P. G., Ricci M. S., Scata K. A., Arsham A. M., Simon M. C., Dicker D. T., El-Deiry W. S. (2005) Cancer Biol. Ther. 4, 1285–1294 [DOI] [PubMed] [Google Scholar]
- 42. von Roretz C., Gallouzi I. E. (2010) J. Biol. Chem. 285, 16806–16813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liotta L. A., Kohn E. C. (2001) Nature 411, 375–379 [DOI] [PubMed] [Google Scholar]
- 44. Kurokawa T., Miyamoto M., Kato K., Cho Y., Kawarada Y., Hida Y., Shinohara T., Itoh T., Okushiba S., Kondo S., Katoh H. (2003) Br. J. Cancer 89, 1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Janssen H. L., Haustermans K. M., Balm A. J., Begg A. C. (2005) Head Neck 27, 622–638 [DOI] [PubMed] [Google Scholar]
- 46. Costantino C. L., Witkiewicz A. K., Kuwano Y., Cozzitorto J. A., Kennedy E. P., Dasgupta A., Keen J. C., Yeo C. J., Gorospe M., Brody J. R. (2009) Cancer Res. 69, 4567–4572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Baudino T. A., McKay C., Pendeville-Samain H., Nilsson J. A., Maclean K. H., White E. L., Davis A. C., Ihle J. N., Cleveland J. L. (2002) Genes Dev. 16, 2530–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Osthus R. C., Shim H., Kim S., Li Q., Reddy R., Mukherjee M., Xu Y., Wonsey D., Lee L. A., Dang C. V. (2000) J. Biol. Chem. 275, 21797–21800 [DOI] [PubMed] [Google Scholar]
- 49. Kim J. W., Gao P., Liu Y. C., Semenza G. L., Dang C. V. (2007) Mol. Cell. Biol. 27, 7381–7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang H., Gao P., Fukuda R., Kumar G., Krishnamachary B., Zeller K. I., Dang C. V., Semenza G. L. (2007) Cancer Cell 11, 407–420 [DOI] [PubMed] [Google Scholar]
- 51. Chen C. Y., Gherzi R., Ong S. E., Chan E. L., Raijmakers R., Pruijn G. J., Stoecklin G., Moroni C., Mann M., Karin M. (2001) Cell 107, 451–464 [DOI] [PubMed] [Google Scholar]
- 52. Meng Z., King P. H., Nabors L. B., Jackson N. L., Chen C. Y., Emanuel P. D., Blume S. W. (2005) Nucleic Acids Res. 33, 2962–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.