Abstract
Members of the caspase family of cysteine proteases coordinate the highly disparate processes of apoptosis and inflammation. However, although hundreds of substrates for the apoptosis effector caspases (caspase-3 and caspase-7) have been identified, only two confirmed substrates for the key inflammatory protease (caspase-1) are known. Whether this reflects intrinsic differences in the substrate specificity of inflammatory versus apoptotic caspases or their relative abundance in vivo is unknown. To address this issue, we have compared the specificity of caspases-1, -3, and -7 toward peptide and protein substrates. Contrary to expectation, caspase-1 displayed concentration-dependent promiscuity toward a variety of substrates, suggesting that caspase-1 specificity is maintained by restricting its abundance. Although endogenous concentrations of caspase-1 were found to be similar to caspase-3, processed caspase-1 was found to be much more labile, with a half-life of ∼9 min. This contrasted sharply with the active forms of caspase-3 and caspase-7, which exhibited half-lives of 8 and 11 h, respectively. We propose that the high degree of substrate specificity displayed by caspase-1 is maintained through rapid spontaneous inactivation of this protease.
Keywords: Apoptosis, Caspase, Enzyme Inactivation, Inflammation, Protease, Caspase Substrates, Caspase-1, Cell Death
Introduction
The caspase family of cysteine proteases plays a conserved role in apoptosis and inflammation (1–3). Although the apoptosis-associated effector caspases (caspases-3 and -7) have been extensively characterized, less is known concerning the mechanism of action of the inflammatory caspases. Caspases-1, -4, and -5 comprise the human inflammatory caspases and these proteases do not appear to play any significant role in apoptosis (1, 2). Instead, caspases-1, -4, and -5 play central roles in inflammatory processes and are essential for the production of the mature forms of certain inflammatory cytokines in response to infectious agents as well as sterile injury (4, 5). To date, over 600 substrates for the apoptosis-associated effector caspases have been identified through a combination of global proteomic and single protein analyses (3, 6). In sharp contrast, only two confirmed substrates for the inflammatory caspases have been identified (interleukin-1β and interleukin-18), which suggests that these proteases are more specific than the apoptotic caspases by more than two orders of magnitude.
The inflammatory caspases are present as dormant precursor enzymes in many cell types but become activated in response to proinflammatory stimuli such as bacterial LPS, uric acid crystals, and several other proinflammatory agents (4, 7). Although the precise mechanism of activation of the inflammatory caspases is still under active investigation, these caspases become activated upon recruitment to a variety of related caspase-activating platforms called inflammasomes (4). Distinct inflammasome complexes, built upon different members of the NALP family of scaffold proteins in tandem with the ASC/PYCARD adaptor molecule, assemble in response to distinct proinflammatory stimuli (8–11).
Because of the paucity of substrates that have been identified for caspase-1, it is often assumed that inflammatory caspases are much less promiscuous than the apoptosis effector caspases. However, because this issue has not been examined formally, it remains unclear whether caspase-1 is indeed more stringent in substrate selection than caspases-3 and -7, thereby explaining why so few substrates have been found for this protease. To address this question, we have compared the specificity of caspases-1, -3, and -7 toward a diverse panel of peptide and protein substrates.
Here, we show that caspase-1 exhibits concentration-dependent promiscuity toward synthetic as well as natural substrates, suggesting that the specificity of this protease is maintained by restricting its availability within the cell. However, concentrations of endogenous procaspase-1 were found to be similar to that of procaspases-3 and -7 in a number of commonly used cell lines, suggesting that caspase-1 specificity is not maintained by regulating the amount of available zymogen. Instead, we have found that caspase-1 is much more labile than caspases-3 or -7 and is rapidly inactivated upon formation of the mature enzyme, with a half-life of ∼9 min at 37 °C. In contrast, caspases-3 and -7 were found to be much more stable enzymes, maintaining essentially full activity for periods of 8–11 h at 37 °C. Collectively, these data suggest that caspase-1 specificity is maintained through rapid destabilization of this protease within minutes of activation, thereby restricting the activity of this enzyme toward highly preferred substrates such as IL-1β and IL-18.
EXPERIMENTAL PROCEDURES
Materials
Antibodies specific to caspase-1 (N-terminal and p10-specific) were purchased from Santa Cruz Biotechnology (catalog nos. sc-622 and sc-515). Anti-caspase-4 and caspase-5 were purchased from MBL (catalog nos. M029-3 and M060-3). Caspase-2, caspase-3, caspase-6, caspase-7, BID, gelsolin, RhoGDI, ROCKI, and XIAP antibodies were purchased from BD Biosciences (catalog nos. 611022, 610323, 556581, 610813, 550365, 610413, 556511, 611136, and 610763). Caspase-8- and caspase-9-specific antibodies were obtained from Apotech (catalog no. APO-20A-071-C050) and Oncogene Research Products (catalog no. AM47), respectively. Anti-active caspase-3 and anti-PAK2 were purchased from Cell Signaling (catalog nos. 96615 and 2608). Anti-co-chaperone p23 was from Affinity Bioreagents (catalog no. MA3-414) and anti-IL-1β was from R&D Systems (catalog no. MAB201). All small peptide substrates and inhibitors (YVAD-CHO, ZVAD-fmk,2 DEVD-AMC, IETD-AMC, WEHD-AMC, YVAD-AMC and ZVAD-AMC) were purchased from Bachem, and biotin-VAD was from ICN.
Expression and Purification of Recombinant Caspases and Substrates
Caspase-1 p30 (lacking N-terminal prodomain) was cloned into pET21a. pET23b caspase-3 and caspase-7 constructs were kindly provided by Dr. Guy Salvesen (Burnham Institute, La Jolla, CA). Sequences encoding interleukin-1β and RhoGDI were cloned into pET45b. Plasmids encoding polyhistidine-tagged proteins were transformed into Escherichia coli BL21/DE3/pLysS and bacteria were induced to express the recombinant proteins in the presence of isopropyl 1-thio-β-d-galactopyranoside. The zymogen forms of caspase-3 and -7 were expressed by restricting the induction time to prevent spontaneous autoprocessing of these proteases. His-tagged fusion proteins were subsequently purified using nickel-nitrilotriacetic acid-agarose beads according to standard procedures.
Active Site Titration of Caspases
The active site concentration of recombinant caspases was determined by the method described by Stennicke and Salvesen (12). Briefly, recombinant caspases were co-incubated for 30 min with the irreversible caspase inhibitor Z-VAD-fmk over a range of concentrations (from 0 to 800 nm). Because of the instability of caspase-1 at 37 °C, incubations with Z-VAD-fmk were performed at 4 °C. Enzymatic activity of enzyme-inhibitor complexes was then measured through hydrolysis of the fluorogenic substrates; WEHD-AMC for caspase-1 and DEVD-AMC for caspases-3 and -7. The rate of substrate hydrolysis was then plotted against the inhibitor concentration to determine active site concentrations. The concentration of recombinant caspase-1, -3, and -7 in the active site titrations depicted in supplemental Fig. S1 were 56, 87, and 74 nm, respectively.
Fluorimetry Assays
Recombinant caspases or THP-1 cell-free extracts were diluted to a final volume of 30 μl in the appropriate buffer containing 50 μm substrate. Fluorescence was then measured over time in an automated fluorimeter at wavelengths of 430 nm (excitation) and 535 nm (emission).
Cell-free Reactions
Cell-free extract was generated from exponentially growing healthy THP-1 or Jurkat cells as described previously (13). Briefly, cells (5 × 108) were harvested by centrifugation at 800 × g into a Dounce-type homogenizer. Cells were then incubated for 15 min in three volumes of ice-cold cell extract buffer (20 mm Hepes, pH 7.5, 10 mm KCl, 1.5 mm MgCl2, 1 mm EDTA, 1 mm EGTA, 1 mm DTT, 100 μm PMSF, 10 μg/ml leupeptin, and 2 μg/ml aprotinin), followed by homogenization using a B-type pestle to the point of cellular but not organellar rupture (i.e. mitochondria remained intact). Lysates were then clarified by centrifugation at 15,000 × g for 20 min to remove nuclei, mitochondria, and other cellular debris. In the case of THP-1 cell-free extracts, cells were prestimulated for 5 h with 1 μg/ml of LPS to promote IL-1β synthesis and inflammasome formation. Extracts were then aliquoted and frozen at −70 °C prior to use. For in vitro activation of inflammatory caspases, THP-1 cell-free extracts were simply incubated at 37 °C. To provoke apoptosome-dependent caspase activation, cytochrome c (bovine heart) and dATP were added to final concentrations of 50 μg/ml and 1 mm, respectively. For Jurkat cell-free experiments, extracts were incubated with recombinant caspases for 2 h at 37 °C. Recombinant IL-1β (10 ng per reaction) was added to cell-free extracts as a positive control for caspase-1 activity.
Biotin-VAD Capture of Active Caspases from THP-1 Cell-free Extracts
Inflammasome or apoptosome activation in THP-1 cell-free extracts was initiated as described above. At each time point (0, 30, 60, and 120 min) 50 μl of cell-free extract was sampled and 5 μl of biotin-VAD was added to a final concentration of 10 μm. Samples were then incubated with biotin-VAD for 30 min at 37 °C to permit labeling of active caspase. Following labeling, samples were diluted to 250 μl with PBS and 30 μl of streptavidin beads were added. Biotin-VAD labeled caspases were captured under constant rotation for 3 h at 4 °C before beads were isolated, resuspended in SDS-PAGE loading buffer, and boiled for 7 min. Captured caspases were analyzed by SDS-PAGE, followed by immunoblotting onto nitrocellulose. Blots were then probed for the full length and processed forms of caspase-1 or caspase-3.
Cross-linking of Recombinant Caspases
Cross-linking was carried out using bis[sulfosuccinimidyl]suberate (BS3) or Bis[maleimido]hexane (BMH, both from Pierce) in 50 mm phosphate buffer at the appropriate pH (8.0 and 6.8, respectively). Cross-linking was achieved by incubation on ice for 2 h with 1–2 mm BMH in the presence of 5 mm EDTA. Cross-linked proteins were run on 12% SDS-PAGE gels and visualized by staining with Commassie Blue.
Chymotrypsin Susceptibility Analysis
Samples of recombinant caspase-1 or caspase-7 (0.5 μg per sample) were preincubated at 4 or 37 °C for 2 h. Caspases were then further incubated for 30 min at 37 °C in the presence or absence of chymotrypsin (from 0.25 to 2 μg/ml). Chymotrypsin-mediated degradation of caspases was then evaluated by SDS-PAGE and Commassie Blue staining.
Two-dimensional Electrophoresis
Approximately 450 μg of proteins were solubilized to a final volume of 350 μl in isoelectric focusing sample buffer (8 m urea, 4% CHAPS, 0.05% SDS, 100 mm DTT, 0.2% w/v Bio-Lyte ampholytesTM, 0.02% w/v bromphenol blue). Immobilized pH gradient strips were actively rehydrated at 50 V in the presence of protein samples overnight. Samples were then focused on a Bio-Rad isoelectric focusing cell under the following conditions: (i) a linear voltage ramp to 500 V over 1 h, (ii) 5 h at a constant 500 V to facilitate desalting, (iii) linear voltage ramp to 3500 V over 5 h, and (iv) 15 h at a constant 3500 V. Following isoelectric focusing, the immobilized pH gradient strips were prepared for second dimension SDS-PAGE by a 10-min incubation in reducing buffer (2% (w/v) DTT in 6 m urea, 375 mm Tris-HCl, pH 8.8, 2% SDS, 20% glycerol), followed by a 10-min incubation in alkylating buffer (2.5% (w/v) iodoacetimide in 6 m Urea, 375 mm Tris-HCl, pH 8.8, 2% SDS, 20% glycerol). Strips were then mounted on 12% SDS-PAGE gels using 1–2 ml of easymelt agarose (Bio-Rad) and electrophoresed at 37.5 mA per gel in a Bio-Rad Protean IIxi electrophoresis cell (Bio-Rad). Proteins were visualized using a mass spectrometry-compatible silver staining protocol (14).
RESULTS
Inflammatory and Apoptotic Caspase Activation Events Can Be Separated in THP-1 Cell-free Extracts
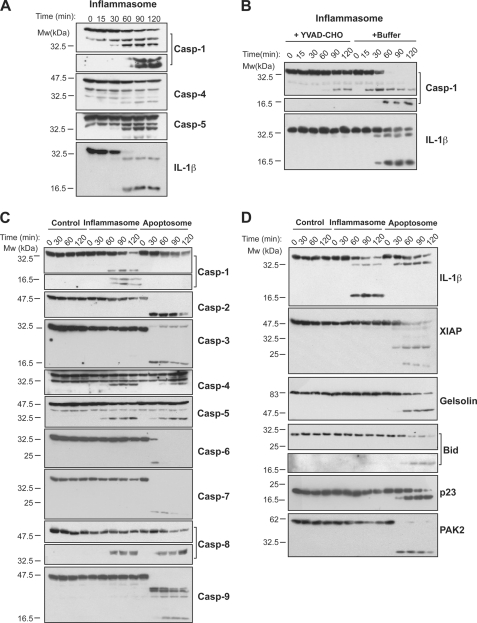
To compare the specificities of apoptotic versus inflammatory caspases, we initially employed a cell-free system based upon concentrated homogenates of LPS-treated cells of the human macrophage cell line, THP-1. This system has been used previously to identify IL-1β as a substrate for caspase-1 (15, 16) and also to explore the composition of the human inflammasome (4). THP-1 cells express all of the inflammatory caspases, which undergo spontaneous activation, in an ASC-dependent manner, upon incubation of cell-free lysates of THP-1 cells at 37 °C (4). Under these conditions, caspases-1, -4, and -5 underwent proteolytic processing to their mature forms and the caspase-1 substrate IL-1β was also cleaved to its biologically active p17 form (Fig. 1A). All of these events were readily suppressed by addition of the tetrapeptide caspase inhibitor, Ac-YVAD-CHO, to the cell-free extracts, and this was used as a control condition in subsequent experiments (Fig. 1B).
FIGURE 1.
Inflammatory versus apoptotic caspase activation in THP-1 cell-free extracts. A, spontaneous processing of inflammatory caspases and IL-1β in THP-1 cell-free extracts incubated at 37 °C. B, caspase-1 and IL-1β processing in THP-1 cell-free extracts in the presence or absence of the caspase inhibitor YVAD-CHO (5 μm). C, processing of endogenous caspases in THP-1 cell-free extracts under control (10 μm YVAD-CHO), inflammasome (37 °C incubation) or apoptosome (50 μg/μl cytochrome c, 1 mm dATP) activating conditions. D, processing of endogenous caspase substrates in THP-1 cell-free extracts under control, inflammasome-activating, and apoptosome-activating conditions. Results are representative of at least three independent experiments.
Of note, under conditions where the inflammatory caspases were processed to their mature forms, the apoptosis-associated caspases (caspases-2, -3, -6, -7, and -9) failed to display any significant processing (Fig. 1C). Consistent with this, we failed to detect proteolytic processing of any of the known substrates for the apoptotic caspases we examined under these conditions (Fig. 1D). However, all of the apoptotic caspases were readily activated upon addition of the co-factors for apoptosome assembly, cytochrome c and dATP, to the same cell-free extracts (Fig. 1C). Under these conditions, multiple apoptotic caspase substrates (XIAP, gelsolin, Bid, co-chaperone p23, and PAK2) were readily cleaved (Fig. 1D). Interestingly, under conditions that permitted assembly and activation of apoptosomes, both caspase-1 and IL-1β processing were suppressed, suggesting that apoptotic and inflammatory caspase activation events may be mutually antagonistic (Fig. 1, C and D).
Apoptotic Caspases Appear more Promiscuous than Inflammatory Caspases
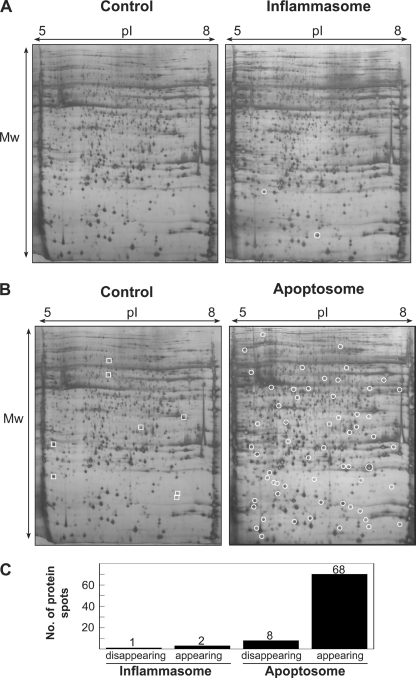
Having established conditions where we could selectively activate inflammatory or apoptotic caspases in the same setting, we then analyzed THP-1 cell-free extracts under these conditions using two-dimensional SDS-PAGE. As Fig. 2 illustrates, whereas numerous alterations to the protein spot patterns were readily detected after incubation of cell-free extracts under conditions permissive for apoptotic caspase activation (i.e. in the presence of cytochrome c and dATP), few alterations were detected under conditions permissive for inflammatory caspase activation. Such analyses resolved ∼2000 of the most abundant proteins present in THP-1 cell-free extracts and strongly suggest that the inflammatory caspases cleave considerably fewer substrates than the apoptosis-associated caspases (Fig. 2C). These observations are entirely consistent with the paucity of inflammatory caspase substrates, and conversely with the abundance of apoptotic caspase substrates, that have been identified to date. Of the few proteins that did become altered upon activation of the inflammatory caspases, we succeeded in identifying only one of these by MALDI mass spectrometry (due to low protein abundance), and this substrate was identified as IL-1β. However, multiple apoptotic caspase substrates were readily identified under conditions of apoptosome activation, and many of these are known substrates for the cell death caspases (supplemental Table S1).
FIGURE 2.
Global proteomic analysis of inflammatory versus apoptotic caspase-dependent substrate proteolysis. A, two-dimensional gel analysis of THP-1 cell-free extracts incubated for 2 h under control (10 μm YVAD-CHO) and inflammasome-activating conditions (37 °C). New protein spots resulting from inflammatory caspase activation are marked with circles. B, two-dimensional gel analysis of THP-1 cell-free extracts incubated for 2 h under control (10 μm YVAD-CHO) or apoptosome-activating conditions (50 μg/ml cytochrome c, 1 mm dATP, 37 °C). Protein spots that disappeared upon activation of the apoptosome are marked with squares, whereas protein spots that appeared upon apoptosome activation are marked by circles. C, summary of the number of two-dimensional gel protein spots that appeared/disappeared as a result of inflammatory or apoptotic caspase activation in THP-1 cell-free extracts. Results are representative of at least three independent experiments.
Specificity of Caspases -1, -3, and -7 toward Synthetic Peptide Substrates
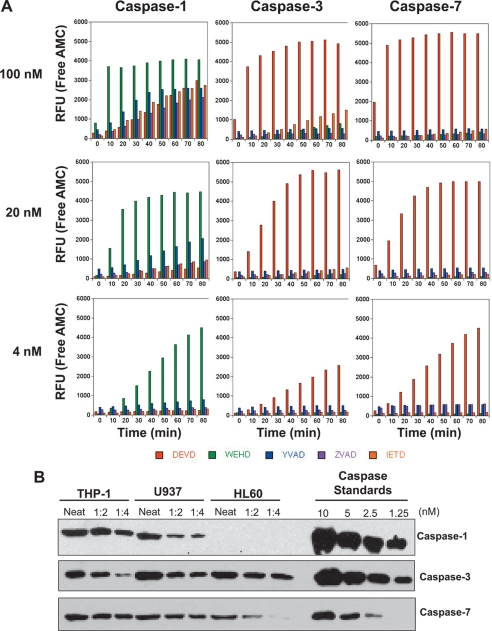
The above observations suggested that inflammatory caspases are much more specific than the apoptotic caspases, as few proteins were cleaved in THP-1 cell-free extracts upon activation of the former but not the latter. However, a major problem with this conclusion is that the relative abundance of each of these proteases in THP-1 cells is not known. Previous studies have suggested that endogenous concentrations of caspases -3 and -7 are in the range of 100–200 nm (17, 18), but the concentration of endogenous caspase-1 is not known. Therefore, a trivial explanation for our observations is that there is simply much less caspase-1 than caspase-3/-7 in THP-1 cells. To address this issue, we expressed each of these proteases in bacteria and active site titrated each enzyme against synthetic peptide substrates to determine the precise molarity of each recombinant protease (supplemental Fig. S1). This enabled us to directly compare the activities of normalized amounts of each caspase against a panel of tetrapeptide substrates. Surprisingly, this analysis gave us a very different result to the cell-free experiments described above and suggested that caspase-1 was considerably more promiscuous than either caspase-3 or caspase-7 at a concentration of 100 nm, with caspase-1 specificity toward WEHD maintained only at very low nanomolar concentrations (Fig. 3A). By comparison, caspases -3 and -7 maintained their specificity for DEVD over the same concentration range (Fig. 3A).
FIGURE 3.
Caspase-1 exhibits greater promiscuity toward synthetic substrates than caspases-3 or -7. A, hydrolysis of synthetic tetrapeptide substrates (DEVD-AMC, WEHD-AMC, YVAD-AMC, ZVAD-AMC, and IETD-AMC) by recombinant caspase-1, -3, or -7 at the indicated enzyme concentrations. B, Relative amounts of endogenous procaspase-1, -3, and -7 in the myelomonocytic cell lines, THP-1, U937, and HL-60. Cell lysates were generated from identical numbers of cells and were ran in a dilution series, as indicated, along with known amounts of the indicated recombinant procaspases. Caspases were detected by SDS-PAGE followed by immunoblotting. Results are representative of at least three independent experiments. RFU, relative fluorescence units.
Concentrations of Endogenous Caspase-1, -3, and -7 Are Similar
As mentioned earlier, one reason for the high substrate specificity of caspase-1 seen upon activation of the endogenous protein was that lower amounts of this protease may be present in THP-1 cells, as compared with caspases -3 and -7. To explore this issue, we carried out quantitative immunoblotting, by comparison with known amounts of recombinant caspases, to determine the relative concentrations of caspases-1, -3, and -7 in THP-1, U937, and HL60 cells. Surprisingly, we found that endogenous amounts of caspase-1 were comparable with those of caspase-3, with caspase-7 being somewhat more abundant than either of the latter caspases (Fig. 3B). Thus, the specificity of endogenous caspase-1 observed in THP-1 cells does not appear to be related to a severely reduced abundance of this protease relative to caspase-3.
Caspase-1 Displays Promiscuity toward Natural Protein Substrates
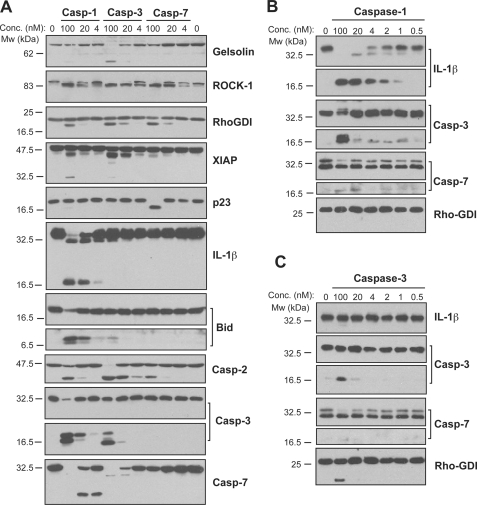
To explore whether the promiscuity displayed by recombinant caspase-1 was related to the use of synthetic (i.e. peptide-based) rather than natural substrates, we repeated our analyses, this time using Jurkat cell-free extracts (which lack endogenous caspase-1) as a source of natural protein substrates (Fig. 4A). Because Jurkat cells do not express endogenous IL-1β, we added recombinant pro-IL-1β to these extracts to serve as a positive control for caspase-1 activity.
FIGURE 4.
Caspase-1 exhibits greater promiscuity toward natural substrates than caspases-3 or -7. A, processing of substrate proteins in Jurkat cell-free extracts after addition of the indicated amounts of recombinant caspase (Casp)-1, -3, and -7 followed by incubation for 2 h at 37 °C. Recombinant caspases were active site titrated (see supplemental Fig. S1) as described under “Experimental Procedures.” Recombinant IL-1β (10 ng per reaction) was added to cell-free extracts as a positive control for caspase-1 activity. Substrate proteolysis was detected by SDS-PAGE followed by immunoblotting. B, caspase-1-mediated processing of the purified recombinant proteins over the indicated concentration (Conc.) range. The indicated proteins were incubated with recombinant active caspase-1 for 2 h at 37 °C. C, caspase-3-mediated processing of the purified recombinant proteins over the indicated concentration range. The indicated proteins were incubated with recombinant active caspase-3 for 2 h at 37 °C. Results are representative of at least three independent experiments.
In agreement with the results of the synthetic tetra-peptide experiments (Fig. 3A), we again observed that concentrations of caspase-1 in excess of 20 nm resulted in proteolysis of multiple proteins that are not normally cleaved upon activation of the inflammasome (Fig. 4A). When added to cell-free extracts at 100 nm, caspase-1 readily cleaved multiple substrates that are cleaved by caspases-3 and -7 during apoptosis (Fig. 4A). Moreover, caspases-3 and -7 failed to cleave the caspase-1 substrate, IL-1β, under conditions where these proteases displayed activity toward their known substrates (Fig. 4A).
To confirm these observations in the absence of any confounding elements (particularly other proteases) present within Jurkat cell-free extracts, we also tested caspase-1 activity against purified recombinant protein substrates. Caspase-1 was highly active toward proIL-1β, as processed cytokine was detectable following incubation with caspase-1 at concentrations as low as 1 nm (Fig. 4B). This suggests that very low intracellular concentrations of active caspase-1 may be sufficient to serve its primary physiological role of cytokine activation. However, caspase-1 was also able to process the proforms of caspase-3 and caspase-7 at concentrations of 100 nm in a manner similar to that of caspase-3 (Fig. 4, B and C). Once again, these data argue that caspase-1 becomes more promiscuous at concentrations approaching 100 nm, which are well within the physiological range.
Caspase-1 Is Much Less Stable Than Caspases-3 and -7
Because caspase-1 loses its marked preference for WEHD/IL-1β at concentrations approaching 100 nm, it is likely that a mechanism exists to limit the availability of active caspase-1 within cells. Otherwise, the consequence of untrammeled caspase-1 activation is likely to be apoptosis or necrosis of the host cell due to proteolysis of numerous substrates. One way of limiting the activity of a protease in vivo is through rapid destabilization. For example, lysis of host plasma membranes by components of the complement system is prevented by factors that immediately dissociate the complement proteases C3bBb and C4b2a (19). Thus, we explored whether a limit on the concentration of active caspase-1 available within a cell might be achieved through rapidly inactivating this protease.
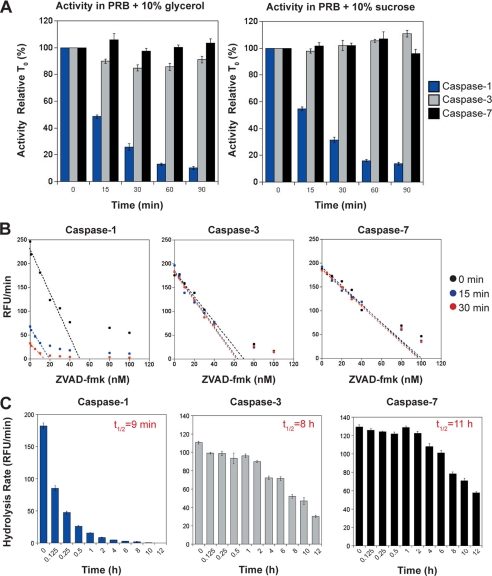
To explore this possibility, we measured the activity of recombinant caspase-1 after incubation for varying periods of time at 37 °C. As Fig. 5A illustrates, we observed a dramatic loss of caspase-1 activity within 15–30 min of incubation at 37 °C, which was independent of buffer composition. In contrast, caspases-3 and -7 failed to display any discernable loss of activity even upon 90 min of incubation under the same conditions (Fig. 5A). We also carried out more quantitative active site titration measurements on all three caspases after incubation periods of up to 2 h at 37 °C, with identical results (Fig. 5B). Further experiments revealed that caspase-1 has an approximate half-life of 9 min at 37 °C (Fig. 5C). By comparison, caspases-3 and -7 displayed half-lives of 8 and 11 h, respectively, under the same conditions (Fig. 5C). Thus, active caspase-1 is ∼50 times less stable that either caspases -3 or -7 under identical conditions.
FIGURE 5.
Processed caspase-1 is highly unstable compared with caspases-3 and -7. A, stability of caspase-1, -3, and -7 (all at 10 nm) at 37 °C under two different buffer conditions. Based on previous optimization studies, protease reaction buffer (PRB) was made to an optimal pH of 7.5 with the inclusion of two alternative stabilizers 10% glycerol or 10% sucrose, as indicated. B, active site titration of caspase-1, -3, and -7 following incubation at 37 °C for 0, 15, and 30 min. Line of best fit extrapolates the concentration of active enzyme remaining under each condition. The estimated concentration of active sites for caspases-3 and -7 was consistent under all conditions (for caspase-3, 0 min, 69 nm; 15 min, 67 nm; 30 min, 62 nm; for caspase-7, 0 min, 99 nm; 15 min, 98 nm; 30 min, 98 nm). However, the measured concentration of caspase-1 dropped by 63% (from 51 to 19 nm) after 15 min of preincubation at 37 °C, and by 73% (from 51 to 14 nm) after 30 min of preincubation 37 °C. C, long term stability of caspase-1, -3, and -7 (at 10 nm initial concentrations). Half-lives of caspase-1 (9 min), caspase-3 (8 h), and caspase-7 (11 h) were estimated by measuring the residual enzyme activity, by comparison with the initial input amounts, after incubation for the indicated times at 37 °C. Results shown represent the mean ± S.E. of three independent experiments. RFU, relative fluorescence units.
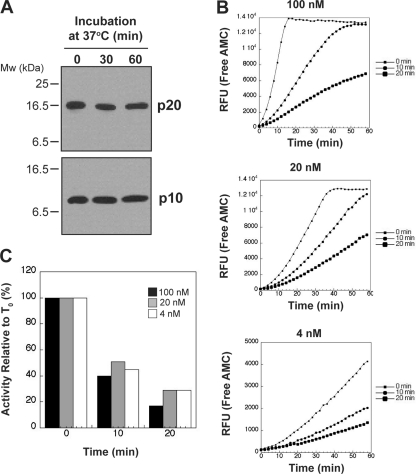
Previous studies on recombinant caspase-1 have noted that at concentrations greater than 2 μm, this protease may inactivate through autolytic digestion within the p10 subunit (20, 21). To examine whether this occurs at the concentrations used here, we incubated 100 nm caspase-1 at 37 °C over 60 min followed by immunoblotting for both the p20 and p10 subunits. As shown in Fig. 6A, we did not observe any evidence for autolytic digestion of caspase-1 at 100 nm. In addition, we also examined whether the instability of caspase-1 was concentration-dependent. We found that, irrespective of initial enzyme concentration (100, 20 or 4 nm), caspase-1 lost activity at a similar rate upon incubation for 10 or 20 min at 37 °C. This argues that activity loss was concentration-independent under the conditions used in our experiments (Fig. 6, B and C).
FIGURE 6.
Instability of caspase-1 at 37 °C is not dependent on autolysis or enzyme concentration. A, caspase-1 (100 nm) was incubated at 37 °C for 0, 30, or 60 min followed by SDS-PAGE/immunoblot analysis to determine whether the p20 or p10 subunits underwent autodigestion. B, caspase-1 (100, 20, or 4 nm) was incubated for 0, 10, or 20 min at 37 °C, followed by assessment of enzyme activity by WEHD-AMC hydrolysis. C, percent loss of caspase-1 activity, relative to time zero (T0), upon incubation for 10 or 20 min at 37 °C at the indicated initial concentrations.
Caspase-1 Instability Is Associated with Loss of Quaternary Structure
Unlike the apoptotic effector caspases which form homodimers as inactive zymogens, proximity induced dimerization appears to be the key activating event for caspase-1 (22). Thus, we examined the possibility that the rapid loss of caspase-1 activity was due to instability of the quaternary structure of the active enzyme, resulting in loss of caspase-1 heterodimers/heterotetramers.
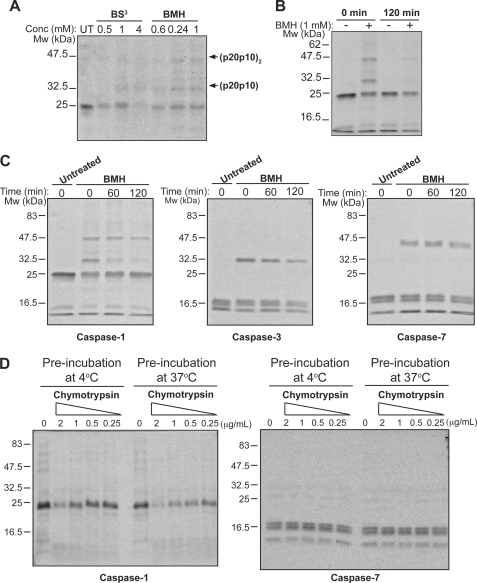
To explore this issue, we used homo-bifunctional cross-linking agents to covalently link heterodimeric (p20p10) and homodimeric (p20p10)2 caspase-1 oligomers before and after incubation at 37 °C, to determine whether a change in these oligomeric forms could be detected. Two different cross-linking reagents (BS3 and BMH) were tested to establish conditions for optimal caspase-1 cross-linking. As seen in Fig. 7A, we found that BMH efficiently cross-linked caspase-1 as indicated by the appearance of two oligomeric forms upon incubation with this cross-linker. Based upon previous studies, these bands most likely represent (p10p20) heterodimers and (p10p20)2 homodimers (Fig. 7A), as indicated. However, caspase-1 that was preincubated for 2 h at 37 °C, and therefore inactivated, displayed a clear reduction in the pattern of oligomers observed upon covalent cross-linking (Fig. 7, B and C). In contrast, little change was observed for the covalent complexes detected under the same conditions for either caspase-3 or caspase-7 (Fig. 7C).
FIGURE 7.
Caspase-1 inactivation is associated with loss of quaternary structure. A, recombinant caspase-1 was treated for 2 h at 4 °C with the indicated amounts of BS3 or BMH cross-linking reagents. Cross-linking with BMH resulted in a pattern of two up-shifted complexes of ∼32 and 47 kDa. In line with previous studies, we interpreted these complexes to represent (p20p10) heterodimers and (p20p10)2 homodimers, respectively. B, cross-linking of recombinant processed caspase-1 after preincubation at 37 °C for the indicated times. C, cross-linking of recombinant processed caspase-1, -3, and -7 after preincubation at 37 °C for the indicated times. D, susceptibility of caspase-1 versus caspase-7 to chymotrypsin-mediated proteolysis after incubation for 2 h at 4 °C versus 37 °C. Following preincubation at 4 or 37 °C, caspases were incubated for a further 30 min at 37 °C in the presence of the indicated concentrations of chymotrypsin. Results are representative of at least three independent experiments.
Digestive proteases, such as chymotrypsin, are useful probes for protein structure and can reveal changes in protein conformation and higher order structure, as these factors influence the rate as well as extent of substrate hydrolysis. Monomerization of caspase-1 is likely to result in the exposure of previously hidden chymotrypsin cleavage sites and increase susceptibility to proteolysis. To test this, we compared the susceptibility of caspase-1 to chymotrypsin treatment, before and after incubation for 2 h at 37 °C. As Fig. 7D illustrates, incubation at 37 °C for 2 h increased the susceptibility of caspase-1 to digestion by chymotrypsin (Fig. 7D). In contrast, caspase-7 remained resistant to chymotrypsin under the same conditions (Fig. 7D). Once again, these data are consistent with a change in the structural configuration of caspase-1 upon incubation at 37 °C, which correlates with a drastic loss in activity of this enzyme.
Endogenous Caspase-1 Is Rapidly Inactivated in THP-1 Cell-free Extracts
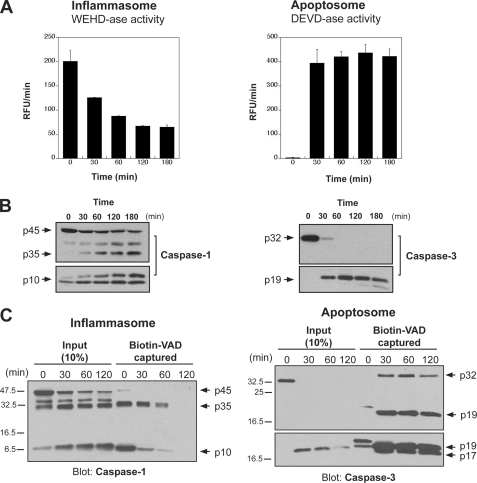
Although the above experiments strongly suggested that caspase-1 is much more labile than either caspases -3 or -7, it was possible that this could be an artifact of using recombinant caspases. To control for this possibility, we also repeated our activity measurements in the THP-1 cell-free system as described earlier, where inflammatory and apoptotic caspases can be differentially activated. As shown in Fig. 8A, measurement of caspase activity under either inflammasome or apoptosome activating conditions revealed two very different scenarios. Although apoptosome-associated caspase activity, once triggered, accumulated and remained high, inflammasome-associated activity was not only comparatively much weaker but also short-lived. This was despite the fact that, as measured by Western blot, processed caspase-1 continued to accumulate (Fig. 8B).
FIGURE 8.
Time-dependent loss of caspase activity occurs upon activation of inflammasomes, but not apoptosomes, in THP cell-free extracts. A, following activation of inflammasomes (by incubation at 37 °C) or apoptosomes (by addition of 50 μg/ml cytochrome c, 1 mm dATP, 37 °C) in THP-1 cell-free extracts, caspase activity was measured at the indicated time points by hydrolysis of WEHD-AMC or DEVD-AMC. B, using the same approach employed above in A, samples were collected at the indicated times to examine caspase-1 or caspase-3 processing by SDS-PAGE followed by immunoblot analysis. C, following activation of inflammasomes or apoptosomes as described in A, samples (50 μl) were taken at the indicated time points and were incubated for 30 min at 37 °C with biotin-VAD (10 μm) to label active caspases. Biotin-VAD-labeled caspase was then captured using streptavidin beads followed by immunoblotting for the appropriate caspase to determine labeling efficiency. Input amounts represent 10% of the total amount of caspase available for capture within the cell-free extracts.
Additionally, we used biotin-VAD-fmk as an affinity ligand to capture active endogenous caspase-1 after different periods of incubation of THP-1 extracts at 37 °C. As Fig. 8C shows, using this approach we observed a sharp time-dependent decline in the quantity of active caspase-1 captured using biotin-VAD-fmk, despite the processed protein being readily detectable in the cell-free extracts. By contrast, using a similar approach to capture endogenous caspase-3 under conditions permissible for activation of apoptosomes in the same THP-1 extracts revealed robust capture at all time points examined (Fig. 8C).
Collectively, these data suggest that caspase-1 is not an especially specific protease, but rather, that this caspase displays promiscuity toward multiple substrates if the concentration of active enzyme exceeds a threshold of ∼50–100 nm. To minimize this likelihood, active caspase-1 displays a rapid loss of proteolytic activity with a half-life of ∼9 min. By comparison, caspases-3 and -7 are highly stable proteases with half-lives of almost two orders of magnitude longer than caspase-1. On the basis of these observations, we propose that caspase-1 substrate specificity is maintained through rapid spontaneous inactivation of this enzyme.
DISCUSSION
Here, we have shown that the key inflammatory caspase, caspase-1, processed a very limited set of substrates upon activation in THP-1 cell-free extracts under conditions where IL-1β was fully processed. In contrast, the apoptosis effector caspases, caspase-3 and -7, cleaved numerous substrates upon activation in the same setting. Although these observations suggest that caspase-1 has a much more restricted substrate range than either caspases-3 or -7, direct comparison of the substrate specificities of all three caspases revealed that caspase-1 was actually more promiscuous toward synthetic as well as natural substrates at concentrations approaching 100 nm. However, although this concentration appears to be within the physiological range of caspase-1 present in macrophages, we also found that caspase-1 underwent spontaneous inactivation ∼50-times more rapidly than either caspase-3 or -7. This leads us to propose that the highly restricted substrate repertoire of caspase-1 is maintained by mechanisms, including rapid spontaneous inactivation of this protease, which operate to limit the amount of caspase-1 that accumulates in response to stimuli capable of activating the inflammasome. Whereas caspase-3 and -7 exhibited half-lives of 8 and 11 h, respectively, at 37 °C, the half-life of active caspase-1 was 9 min under the same conditions. Our study also suggests that structural instability of caspase-1 is linked to a rapid loss of quaternary structure, which disrupts the active site as a result of disassociation of higher order forms of this protease. This is consistent with previous observations concerning caspase-1 stability (20, 21), which suggest that two mechanisms underpin loss of caspase-1 activity. At high concentrations (>2 μm), autolytic proteolysis of the p10 subunit leads to rapid inactivation of this protease. However, it is debatable whether such concentrations would ever be achieved under physiological conditions. At concentrations significantly lower than this, dissociation of the enzyme subunits was found to be the primary factor (20). At the enzyme concentrations used in our study, we found no evidence for caspase-1 autolysis but instead found evidence of structural instability that led to very rapid inactivation of this protease, in sharp contrast to the marked stability of caspase-3 and -7.
The scarcity of validated caspase-1 substrates, when compared with caspase-3/-7 substrates (6), has often been attributed to the specificity of the substrate-binding pocket of caspase-1. Combinatorial peptide library screens have identified WEHD (and to a lesser extent YVAD) as preferred substrates for inflammatory caspases, as compared with DEVD for caspase-3 and -7 (23, 24). However, equating substrate preference with substrate specificity is highly problematic as these factors are greatly influenced by enzyme as well as substrate abundance. Many proteases exhibit substrate specificity that can be lost upon increasing the abundance of enzyme, or alternatively, by adding a less preferred substrate in great abundance. For these reasons, there are serious problems associated with equating the hydrolysis of synthetic substrates in a complex protein mixture with activity of a specific enzyme. As we have shown here, although caspase-1 displayed a marked preference for WEHD at 4–20 nm, this caspase is very capable of hydrolyzing DEVD, IETD, and ZVAD at enzyme concentrations of 100 nm. Similar observations have also been made for the apoptotic caspases where LEHDase activity, for example, is frequently assumed to be indicative of caspase-9 activity. However, caspases-3 and -7 are very capable of hydrolyzing this peptide at physiological concentrations (25, 26).
Although there are only two confirmed substrates for caspase-1 (IL-1β and IL-18) it is generally believed that the range of caspase-1 activity goes beyond the activation of these two cytokines. For example, caspase-1 activity is also thought to be required for the nonclassical secretion pathway and the release of molecules such as IL-1α that are not themselves direct substrates for caspase-1 (27). A number of additional caspase-1 substrates have also been proposed from targeted single protein studies, as well as through large-scale screens. These include MyD88 adapter like protein (Mal), Parkin, and several others (28–33). Indeed, aside from IL-1β and IL-18, more than one hundred additional caspase-1 substrates have been proposed. However, a critical factor in most of these studies is that the concentration of caspase-1 used to demonstrate substrate processing was typically at least 50–100 fold higher than that required for robust IL-1β processing. For example, in three reported caspase-1 screens, an active enzyme concentration of 400 nm or greater was employed (29–31). Due to the very high promiscuity observed with caspase-1 at concentrations of 100 nm or greater, the present study casts considerable doubt on the physiological relevance of such observations, as it is highly unlikely that such concentrations of active caspase-1 could be achieved in vivo. Thus, investigators should be aware of the high likelihood of false-positives when using arbitrarily selected, non-physiological, concentrations of enzyme for such studies.
As we have shown, concentrations of caspase-1 approaching 100 nm or greater are capable of cleaving numerous substrates normally cleaved during apoptosis, as opposed to inflammation. Thus, rapid activation of the total complement of caspase-1 within a cell might be expected to result in apoptosis or even necrosis. Indeed, hyperactivation of the inflammasome has been suggested to drive caspase-1-dependent necrosis or pyroptosis (34). Why this does not occur in most instances may be linked to factors such as the level of inflammasome activation, which could set a threshold on caspase-1 activity. However, under more standard conditions of inflammasome activation, accumulation of the processed form of caspase-1 is typically observed in the absence of proteolysis of ‘apoptotic’ substrates. This strongly suggests that factors such as enzyme stability have an important role in limiting the amount of active caspase-1 present within a cell at any one time.
In conclusion, we have shown that the major inflammatory caspase, caspase-1, is a relatively promiscuous enzyme with the potential to process numerous substrates that are not typically processed during inflammation. However, to counteract this propensity, the specificity of caspase-1 appears to be maintained through rapid spontaneous inactivation of this protease.
Supplementary Material
This work was supported by Strategic Research Cluster and Principal Investigator grants from the Science Foundation Ireland (07/SRC/B1144 and 08/IN.1/B2031) and The Wellcome Trust United Kingdom (082749).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
- Z
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone
- BS3
- bis[sulfosuccinimidyl]suberate
- or BMH
- Bis[maleimido]hexane
- ASC
- apoptosis-associated Speck-like protein containing a caspase recruitment domain.
REFERENCES
- 1. Creagh E. M., Conroy H., Martin S. J. (2003) Immunol. Rev. 193, 10–21 [DOI] [PubMed] [Google Scholar]
- 2. Martinon F., Tschopp J. (2004) Cell 117, 561–574 [DOI] [PubMed] [Google Scholar]
- 3. Taylor R. C., Cullen S. P., Martin S. J. (2008) Nat. Rev. Mol. Cell Biol. 9, 231–41 [DOI] [PubMed] [Google Scholar]
- 4. Martinon F., Burns K., Tschopp J. (2002) Mol. Cell 10, 417–426 [DOI] [PubMed] [Google Scholar]
- 5. Li H., Ambade A., Re F. (2009) J. Immunol. 183, 1528–1532 [DOI] [PubMed] [Google Scholar]
- 6. Lüthi A. U., Martin S. J. (2007) Cell Death Differ. 14, 641–50 [DOI] [PubMed] [Google Scholar]
- 7. Martinon F., Pétrilli V., Mayor A., Tardivel A., Tschopp J. (2006) Nature 440, 237–241 [DOI] [PubMed] [Google Scholar]
- 8. Agostini L., Martinon F., Burns K., McDermott M. F., Hawkins P. N., Tschopp J. (2004) Immunity 20, 319–325 [DOI] [PubMed] [Google Scholar]
- 9. Wang L., Manji G. A., Grenier J. M., Al-Garawi A., Merriam S., Lora J. M., Geddes B. J., Briskin M., DiStefano P. S., Bertin J. (2002) J. Biol. Chem. 277, 29874–29880 [DOI] [PubMed] [Google Scholar]
- 10. Mariathasan S., Newton K., Monack D. M., Vucic D., French D. M., Lee W. P., Roose-Girma M., Erickson S., Dixit V. M. (2004) Nature 430, 213–218 [DOI] [PubMed] [Google Scholar]
- 11. Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D. R., Latz E., Fitzgerald K. A. (2009) Nature 458, 514–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stennicke HR, Salvesen GS. (1999) Methods 17, 313–9 [DOI] [PubMed] [Google Scholar]
- 13. Lüthi A. U., Cullen S. P., McNeela E. A., Duriez P. J., Afonina I. S., Sheridan C., Brumatti G., Taylor R. C., Kersse K., Vandenabeele P., Lavelle E. C., Martin S. J. (2009) Immunity 31, 84–98 [DOI] [PubMed] [Google Scholar]
- 14. Mortz E., Krogh T. N., Vorum H., Grog A. (2001) Proteomics 11, 1359–63 [DOI] [PubMed] [Google Scholar]
- 15. Thornberry N. A., Bull H. G., Calaycay J. R., Chapman K. T., Howard A. D., Kostura M. J., Miller D. K., Molineaux S. M., Weidner J. R., Aunins J. (1992) Nature 356, 768–774 [DOI] [PubMed] [Google Scholar]
- 16. Yamin T. T., Ayala J. M., Miller D. K. (1996) J. Biol. Chem. 271, 13273–82 [DOI] [PubMed] [Google Scholar]
- 17. Rehm M., Huber H. J., Dussmann H., Prehn J. H. (2006) EMBO J. 25, 4338–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walsh J. G., Cullen S. P., Sheridan C., Lüthi A. U., Gerner C., Martin S. J. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12815–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brodbeck W. G., Liu D., Sperry J., Mold C., Medof M. E. (1996) J. Immunol. 156, 2528–2533 [PubMed] [Google Scholar]
- 20. Talanian R. V., Dang L. C., Ferenz C. R., Hackett M. C., Mankovich J. A., Welch J. P., Wong W. W., Brady K. D. (1996) J. Biol. Chem. 271, 21853–21858 [DOI] [PubMed] [Google Scholar]
- 21. Dang L. C., Talanian R. V., Banach D., Hackett M. C., Gilmore J. L., Hays S. J., Mankovich J. A., Brady K. D. (1996) Biochemistry 35, 14910–14916 [DOI] [PubMed] [Google Scholar]
- 22. Gu Y., Wu J., Faucheu C., Lalanne J. L., Diu A., Livingston D. J., Su M. S. (1995) EMBO J. 14, 1923–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thornberry N. A., Rano T. A., Peterson E. P., Rasper D. M., Timkey T., Garcia-Calvo M., Houtzager V. M., Nordstrom P. A., Roy S., Vaillancourt J. P., Chapman K. T., Nicholson D. W. (1997) J. Biol. Chem. 272, 17907–17911 [DOI] [PubMed] [Google Scholar]
- 24. Talanian R. V., Quinlan C., Trautz S., Hackett M. C., Mankovich J. A., Banach D., Ghayur T., Brady K. D., Wong W. W. (1997) J. Biol. Chem. 272, 9677–9682 [DOI] [PubMed] [Google Scholar]
- 25. Pereira N. A., Song Z. (2008) Biochem. Biophys. Res. Commun. 377, 873–877 [DOI] [PubMed] [Google Scholar]
- 26. McStay G. P., Salvesen G. S., Green D. R. (2008) Cell Death Differ. 15, 322–331 [DOI] [PubMed] [Google Scholar]
- 27. Keller M., Rüegg A., Werner S., Beer H. D. (2008) Cell 132, 818–831 [DOI] [PubMed] [Google Scholar]
- 28. Kahns S., Kalai M., Jakobsen L. D., Clark B. F., Vandenabeele P., Jensen P. H. (2003) J. Biol. Chem. 278, 23376–23380 [DOI] [PubMed] [Google Scholar]
- 29. Shao W., Yeretssian G., Doiron K., Hussain S. N., Saleh M. (2007) J. Biol. Chem. 282, 36321–36329 [DOI] [PubMed] [Google Scholar]
- 30. Lamkanfi M., Kanneganti T. D., Van Damme P., Vanden Berghe T., Vanoverberghe I., Vandekerckhove J., Vandenabeele P., Gevaert K., Núñez G. (2008) Mol. Cell Proteomics 7, 2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Agard N. J., Maltby D., Wells J. A. (2010) Mol. Cell. Proteomics 9, 880–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miggin S. M., Pålsson-McDermott E., Dunne A., Jefferies C., Pinteaux E., Banahan K., Murphy C., Moynagh P., Yamamoto M., Akira S., Rothwell N., Golenbock D., Fitzgerald K. A., O'Neill L. A. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3372–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weigert A., Cremer S., Schmidt M. V., von Knethen A., Angioni C., Geisslinger G., Brüne B. (2010) Blood 115, 3531–3540 [DOI] [PubMed] [Google Scholar]
- 34. Bergsbaken T., Fink S. L., Cookson B. T. (2009) Nat. Rev. Microbiol. 7, 99–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.