Abstract
In eukaryotic cells, maintenance of cellular ATP stores depends mainly on mitochondrial oxidative phosphorylation (OXPHOS), which in turn requires sufficient cellular oxygenation. The crucial role of proper oxygenation for cellular viability is reflected by involvement of several mechanisms, which sense hypoxia and regulate activities of respiratory complexes according to available oxygen concentrations. Here, we focus on mouse nitric oxide-associated protein 1 (mNOA1), which has been identified as an important component of the machinery that adjusts OXPHOS activity to oxygen concentrations. mNOA1 is an evolutionary conserved GTP-binding protein that is involved in the regulation of mitochondrial protein translation and respiration. We found that mNOA1 is located mostly in the mitochondrial matrix from where it interacts with several high molecular mass complexes, most notably with the complex IV of the respiratory chain and the prohibitin complex. Knock-down of mNOA1 impaired enzyme activity I+III, resulting in oxidative stress and eventually cell death. mNOA1 is transcriptionally regulated in an oxygen-sensitive manner. We propose that oxygen-dependent regulation of mNOA1 is instrumental to adjusting OXPHOS activity to oxygen availability, thereby controlling mitochondrial metabolism.
Keywords: Bioenergetics, Cell Death, Hypoxia, Mitochondria, Oxidative Stress
Introduction
Eukaryotic cells mainly depend on mitochondrial oxidative phosphorylation with oxygen as the terminal electron acceptor for maintenance of cellular ATP stores. Because oxygen is essential for cellular viability, mechanisms have evolved to sense hypoxia (1), to redistribute intracellular oxygen (2), or to deactivate mitochondrial respiration under conditions when partial pressures of oxygen reach critically low levels (3). Molecular mechanisms underlying the mitochondrial response to decreased oxygen levels include changes of the mitochondrial shape and supramolecular respiratory chain organization but are not yet fully understood (for review, see Ref. 4). Ideas about the regulation of the mitochondrial respiratory chain, which is composed of four complexes that couple electron shuttling with proton pumping and the F1F0-ATPase, have been influenced by two different models, the fluid state and the solid state model. According to the fluid state model complexes I–IV exist as single units that mediate electron transfer by lateral diffusion induced random collisions (5). In contrast, the solid state model assumes that respiratory complexes form high molecular mass supercomplexes of different stoichiometries in a reversible and dynamic manner thereby improving electron shuttling by spatial proximity (6–8).
Many factors that modulate mitochondrial structure or function contain GTPase domains (for recent review, see Ref. 9). GTPase domain-containing factors are involved in the regulation of the mitochondrial network affecting bioenergetics in a bidirectional manner (10). GTP concentrations within mitochondria depend primarily on ATP levels, although it has been suggested that GTP-binding proteins might help to establish local GTP gradients to facilitate fast adaptive response within mitochondria. Moreover, mitodynamin GTPases modulate the organization of the mitochondrial network under hypoxia.
We reasoned that GTPase-dependent regulatory circuits might also be involved in the adjustment of respiratory chain complex organization/activity in response to altered oxygen levels. Guided by the recent description of the mitochondrial proteome (11), we focused on poorly known mitochondrial GTPases. Of these, the mouse homolog of Arabidopsis thaliana nitric oxide-associated protein 1, mNOA1,3 stood out as a potential factor, which might regulate adaptive responses to changes in oxygen concentration, because mNOA1 is known to control respiratory activity and cell death (12, 13) as well as the assembly or stability of ribosomes although its mode of action is enigmatic (14–16).
Here, we describe a comprehensive biochemical analysis of mNOA1. We found that mNOA1 interacts with and stabilizes mitochondrial respiratory complexes, which has a direct impact on mitochondrial enzyme activities. Loss of mNOA1 destabilizes respiratory supercomplexes which leads to oxidative stress originating from the respiratory chain, activation of apoptosis, and cell death. Our findings define mNOA1 as a crucial regulator of mitochondrial activity, which links oxygen availability to mitochondrial respiration.
EXPERIMENTAL PROCEDURES
Plasmid Construction
mNOA1 (NM_019836) constructs were expressed using pcDNA5/TO or pcDNA3.1+ vectors (Invitrogen). U6 + 2 tetO Stuffer (17) was used for mNOA1 knock-down with the shRNA target sequence (5′-3′): ggttcacagttgtggcttccaactt.
Cell Culture
C2C12, HEK 293, HeLa, NIH 3T3 and 143B.TK-K7 cells were grown under standard conditions. Transfections were performed using FuGENE HD (Roche Diagnostics) or Lipofectamine 2000 (Invitrogen). The stable isotope labeling with amino acids (SILAC) experiments and subsequent mass spectrometric analysis were performed as described previously (18). Briefly, C-terminally FLAG-tagged mNOA1 was expressed in C2C12 myoblasts cultivated in medium containing [13C6]lysine. Interaction partners of mNOA1 were recovered using a FLAG-antibody and compared with FLAG-vector transfected C2C12 cells cultivated in standard tissue culture medium. The ratio of heavy to light amino acids (H/L) distinguishes specific interaction partners of mNOA1 from nonspecific binders. C2C12 cells were treated with 5 μg/ml actinomycin D or 50 μg/ml cycloheximide to estimate RNA and protein stability, respectively. Cellular viability was assessed by cultivating C2C12 in the presence of 10 mm N-acetylcysteine or 1 μm cyclosporin A (CsA) for 24 h. Cells were harvested in 0.4% trypan blue solution (Sigma-Aldrich), and living cells were counted in a Neubauer hemocytometer. Hypoxia (0.5% O2, 5% CO2, 94.5% N2) was applied in a COY box (Toepffer Laborsysteme GmbH, Göppingen, Germany).
Confocal Laser Scanning Microscopy
Mitochondrial compartments were labeled using MitoTracker Red CMXRos and Deep Red 633 (Molecular Probes) (inner membrane), pEGFP-OMP25-Fr (19) (outer membrane) or Mito-EGFP (matrix) and imaged as described (20). For induction of mega-mitochondria, cells were incubated with lactic acid-acidified medium (pH 6.5) or with valinomycin (21).
Mitochondrial Fractionation
C57BL/6 heart mitochondria were isolated by tissue homogenization and differential centrifugation. The sucrose gradient fractionation and blue native-PAGE (BN-PAGE) of digitonin-solubilized samples were performed as described (22, 23).
Immunoblotting and Immunoprecipitation (IP)
Immunoreactive bands were visualized and analyzed using the VersaDoc Imaging system (Bio-Rad). Antibodies were obtained from AbFrontier (Seoul, Korea) anti-Trx-2 (LF-PA0012); AnaSpec (San Jose, CA) anti-GFP (53882); Eurogentec (Köln, Germany) anti-mNOA1 raised against the peptide CYRMFKRQRRLQEDAT; Invitrogen anti-complex I 30-kDa subunit (A-21343), anti-complex IV subunit IV (A-21348) Total OxPhos Complex kit (458099); Mitosciences (Eugene, OR) anti-cytochrome c (MSA06), anti-VDAC/porin (MSA03), anti-ATPase β subunit (MS503); New England Biolabs (Frankfurt, Germany) anti-pan-actin (NEB 4968), anti-caspase-3 (NEB 9662), anti-PARP (NEB 9542); Novus Biologicals (Littleton, CO) anti-REA (PHB2) (NB100-1809); Sigma-Aldrich anti-FLAG M2. For Trx-2 redox analysis lysates were incubated with 15 mm AMS (Molecular Probes). The NE-PER kit (Pierce, Thermo Scientific) was used for C2C12 fractionation, and the FLAG-tagged Protein Immunoprecipitation kit (Sigma-Aldrich) was used for IP.
Enzymatic Activity and ATP Determination
The rotenone-sensitive NADH:CoQ1 oxidoreductase (I), NADH:cytochrome-c oxidoreductase (I+III), and citrate synthase activities were measured spectrophotometrically using an Amersham Biosciences UV/visible Ultraspec 3000 pro (GE Healthcare) as described (24). ATP was determined using the ATP lite-M assay (Packard Biosciences, Groningen, The Netherlands).
FACS Analysis
JC-1 (Molecular Probes) and MitoSOX Red (Molecular Probes) were used to measure mitochondrial membrane potential (ΔΨm) and superoxide production, respectively, in C2C12. Cells were analyzed on a BD LSR II flow cytometer.
Reverse Transcription-PCR
mRNA was isolated using TRIzol (Invitrogen). RT-PCR was performed using the H-Minus First Strand cDNA Synthesis kit (Fermentas, St. Leon-Roth, Germany) and Absolute SYBR Green Fluorescein premix (ABgene) in a real-time PCR system (iQ5; Bio-Rad). Primer sequences (5′-3′) were mNOA1 forward, cctatttgcaacccgactcc and reverse, gtcataaaaccagtgggcgtc; β-actin forward, gtgggccgccctaggcacca and reverse, gttggccttagggttcaggggg.
Statistical Analysis
All data are presented as mean ± S.D. of at least three independent experiments. Statistical significance was assessed using the Student's t test with a p value smaller than 0.05 being considered significant.
RESULTS
Submitochondrial Localization of NOA1
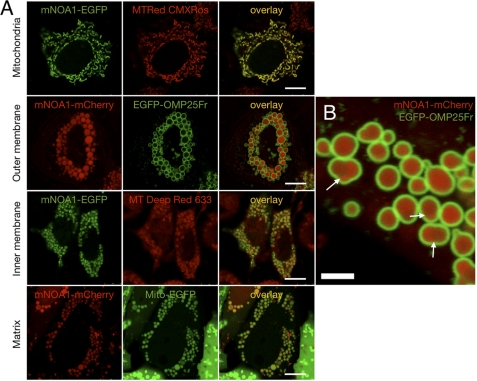
To obtain a better idea about the function of mNOA1 in the regulation of mitochondrial activities, we first determined its precise submitochondrial localization. Fluorescent mNOA1 fusion proteins were constructed and transfected into different human and mouse cell lines followed by confocal microscopy analysis. Analysis of normal mitochondria does not provide a resolution that permits assignment of fluorescent proteins to a specific submitochondrial compartment. Therefore, we induced megamitochondria using two different methods. This approach, together with the use of specific fluorescent markers that label mitochondrial compartments, allowed us to localize the bulk of mNOA1 protein to the mitochondrial matrix (Fig. 1). This result was further substantiated by transmitochondrial scans (supplemental Fig. 1) and time lapse experiments uncovering a rapid movement of mNOA1 within the mitochondrial matrix (supplemental Movie 1).
FIGURE 1.
mNOA1 resides within the mitochondrial inner membrane. A, co-localization of fluorescent fusion proteins of mNOA1 with mitochondrial dyes is shown. mNOA1 co-localizes with the mitochondrial matrix marker (Mito-EGFP) but not with markers for outer membrane (EGFP-OMP25-Fr) or inner membrane (MitoTracker Deep Red 633) in megamitochondria. Cell lines used were HeLa, NIH 3T3, and 143B.TK-K7. The scale bars, 10 μm. B, mitochondrial matrix localization of mNOA1-mCherry in HeLa with valinomycin-induced megamitochondria. Outer membrane is stained by EGFP-OMP25-Fr. Note that matrices are separated by unfused inner membranes (marked by arrows). The white bar represents 2.5 μm.
NOA1 Co-migrates with Respiratory Complexes and Forms Dimers or Multimers
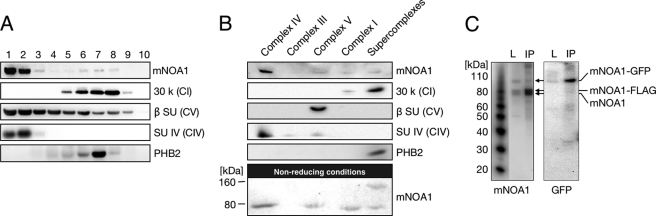
We next investigated whether mNOA1 might interact with complexes of the respiratory chain. Digitonin-solubilized mouse heart mitochondria were fractionated by sucrose gradient centrifugation and probed with a rabbit antibody, which we had raised against mNOA1. The highest concentrations of mNOA1 were detected in fractions containing complex IV. Interestingly, significant amounts of mNOA1 were also found in higher molecular mass fractions, which included complex I and prohibitin 2 (PHB2) (Fig. 2A). Both complex I of the respiratory chain and the PHB complex are estimated to have a molecular mass of approximately 1 MDa (25, 26) and may be involved in respiratory supercomplex formation (6).
FIGURE 2.
mNOA1 co-migrates with high molecular mass complexes and forms dimers or multimers. A and B, solubilized whole heart mitochondria were separated by sucrose gradient centrifugation (A) or by BN-PAGE (B). Sucrose gradient fractions and isolated respiratory chain complexes were analyzed by Western blotting under denaturing and reducing conditions. BN-PAGE isolated complexes were prepared under denaturing but nonreducing conditions to detect mNOA1 dimers or multimers. C, C2C12 myoblasts were co-transfected with vectors coding for mNOA1-FLAG and mNOA1-EGFP. FLAG-IP and Western blot analysis against mNOA1 revealed three bands, and against GFP revealed one band. These monomeric protein bands are indicative of the formation of mNOA1 dimers or multimers.
Because the resolution of sucrose gradient centrifugation is limited, we turned to BN-PAGE, which allows better separation of different complexes. BN-PAGE of digitonin-solubilized mitochondria identified four individual respiratory complexes and five distinct supercomplexes, each representing a supercomplex of different composition (6, 8). Western blot analysis of separated respiratory complexes and the total supercomplex fraction revealed that mNOA1 predominantly co-segregated with complex IV and only to a lesser extent with the complexes I, V, and pooled supercomplexes (Fig. 2B). Interestingly, we also observed an additional 160-kDa band, which reacted with the mNOA1 antibody in the pooled supercomplexes fraction when samples were prepared under nonreducing conditions (Fig. 2B). We reasoned that mNOA1, which shows a molecular mass of 80 kDa in SDS-PAGE, might form dimers or multimers when getting contact to supercomplexes. To prove this assumption, mNOA1 FLAG- and EGFP-tagged were co-expressed in C2C12 myoblasts and subjected to FLAG immunoprecipitation and Western blot analysis using anti-mNOA1 and anti-EGFP antibodies, respectively (Fig. 2C). Results from both experimental approaches indicate that mNOA1 proteins interact with each other to form dimers or multimers.
NOA1 Interacts Specifically with Multiple Mitochondrial Complexes
To characterize further the interaction partners of mNOA1 we employed IP and mass spectrometry combined with SILAC (27) (supplemental Fig. 2). SILAC is based on incorporation of either unlabeled or heavy isotope-labeled amino acids into proteins of cultured cells and allows relative quantification of protein concentrations. Only interaction partners that bind with high affinity and specificity will show a significant ratio of heavy amino acid-labeled proteins compared with light amino acid-labeled proteins (H/L) when IPs with bait constructs (heavy) are compared with IPs with control constructs (light). False positive interaction partners will be precipitated to the same extent under both conditions and hence will show only a low (H/L) ratio (28). In the current case, we transfected C2C12 myoblasts with FLAG-tagged mNOA1 and FLAG control vector followed by FLAG IP and mass spectrometry analysis. Protein interactions were considered specific when the H/L ratio exceeded 3.0, the p value was <0.05, and at least three unique peptides were detected. Our analysis revealed that mNOA1 interacts with several proteins that belong to subunits of respiratory complexes (including components of complex IV), which confirmed and extended the results of our fractionation experiments. In addition, we also found specific interactions with several members of the PHB complex (supplemental Table 1).
NOA1 Enhances Mitochondrial Enzyme Activities in a GTPase-dependent Manner
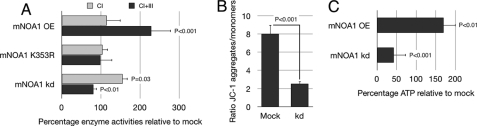
Because complex IV acts as a regulator for the supramolecular organization of the respiratory chain and affects the stability of individual complexes (29–31), we investigated the effects of mNOA1 on mitochondrial enzyme activities in C2C12 myoblasts using overexpression or knock-down approaches. We also generated a mutant form of mNOA1, which contained an amino acid substitution (K353R) in the P-loop sequence (13, 32), to disclose the potential necessity of an intact GTPase domain for mNOA1 function. Transfection of mNOA1 resulted in a strong enhancement of complex I+III activity as measured by cytochrome c reduction using NADH as a substrate (corresponding to complex I+III activity). In contrast, the activity of isolated complex I was unaltered when coenzyme Q1 was used as electron acceptor to monitor NADH oxidation (corresponding to complex I activity) (24) (Fig. 3A). The mNOA1-K353R P-loop variant failed to induce activation of complexes I+III, suggesting that the GTPase domain is critical for mNOA1-mediated activation of respiratory complexes (Fig. 3A). shRNA mediated knock-down of mNOA1 confirmed its critical role in the regulation of respiratory complexes because we observed a drop of complex I+III activity of ∼20% after transfection of shRNAs directed against mNOA1 whereas the activity of complex I increased concomitantly (Fig. 3A). The reduced complex I+III activity suggested that the knock-down of mNOA1 might also lead to a decreased mitochondrial membrane potential (ΔΨm). In fact, we measured a reduced ΔΨm using the cationic, lipophilic dye JC-1 (Fig. 3B) together with a decline in cellular ATP levels of ∼50% (Fig. 3C). Interestingly, we also observed an increased cellular content of ATP after overexpression of mNOA1, reinforcing our hypothesis that a dynamic change in the mitochondrial concentration of mNOA1 directly affects oxidative phosphorylation activity and cellular energy production.
FIGURE 3.
mNOA1 modulates mitochondrial enzyme activities in C2C12 myoblasts. A, enzymatic activities of complexes I and I+III upon overexpression (OE) of wild-type mNOA1, a K353R P-loop variant, or transient mNOA1 knock-down (kd) normalized to the mitochondrial marker enzyme citrate synthase and depicted as percentage. B, mitochondrial membrane potential after transient knock-down of mNOA1 using JC-1. Data represent the ratio of JC-1 aggregates to monomers. C, cellular ATP levels after transient knock-down or overexpression of mNOA1. Data are given as mean ± S.D. (error bars).
Loss of NOA1 Specifically Destabilizes Complex IV and Supercomplexes
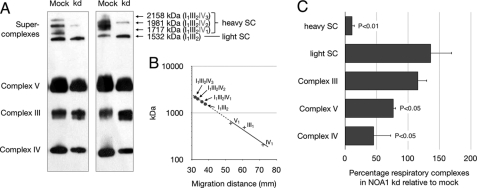
The finding that mNOA1 favors activity of complex I+III whereas its knock-down stimulates the isolated complex I raised the question whether mNOA1 might stabilize respiratory supercomplexes. Therefore, we knocked down mNOA1 in C2C12 myoblasts and studied formation of respiratory complexes by Western blot analysis. Reduction of mNOA1 protein caused a massive loss of complex IV and some supercomplexes (Fig. 4A). Analysis of the molecular mass of supercomplexes revealed that specifically those supercomplexes, which contain complex IV at different stoichiometries, disappeared after loss of mNOA1. In contrast, complex III and supercomplexes that do not contain complex IV were unchanged (Fig. 4, B and C). These findings corresponded nicely to our enzyme assays, which also indicated reduced respiratory activity.
FIGURE 4.
mNOA1 knock-down (kd) reduces stability of mitochondrial heavy supercomplexes and complex IV. A, Western blot analysis of digitonin-solubilized transfected cells separated on a BN-PAGE with the Total OxPhos Complex kit is shown. B, to estimate protein mass of supercomplexes, known molecular masses of complex IV, III, and V and the corresponding migration distance were used. +, protein complexes used for calibration; ●, analyzed supercomplexes. Supercomplex composition according to the calculated molecular mass as described by Wittig et al. (47). C, quantitative analysis of respiratory complex levels is shown. Data are given as mean ± S.D. (error bars).
Loss of NOA1 Causes Oxidative Stress and Cell Death in C2C12 Myoblasts
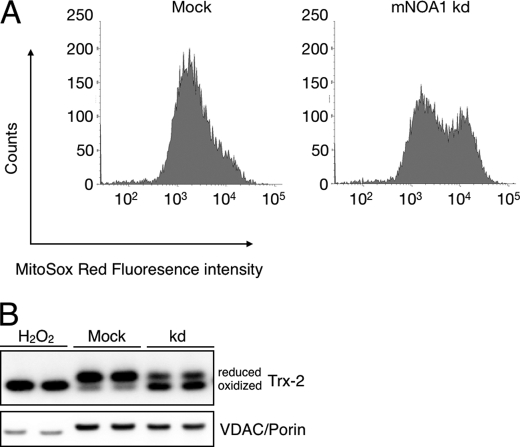
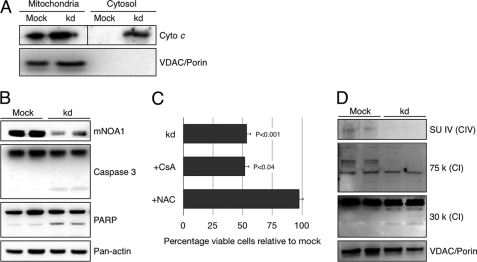
The reduction of complex I+III activity in the presence of enhanced complex I activity suggested that loss of mNOA1 causes severe oxidative stress due to the imbalance of enzyme activities. To investigate this scenario we analyzed the generation of superoxide anions after mNOA1 knock-down in C2C12 myoblasts using the MitoSOX Red fluoroprobe, which selectively detects superoxide in mitochondria of live cells. We observed a clear increase in MitoSOX Red fluorescence in mNOA1 knock-down but not in control cells (Fig. 5A). Further support for our hypothesis that loss of mNOA1 leads to increased oxidative stress came from a redox-sensitive Western blot analysis, which revealed a shift of the reduced to the oxidized form of mitochondrial antioxidant thioredoxin-2 (Fig. 5B). The increase of oxidative stress after knock-down of mNOA1 was accompanied by the release of mitochondrial cytochrome c (Fig. 6A) and activation of caspase-3 and PARP (Fig. 6B). We also detected a marked decrease in cellular viability, which was restored by addition of the antioxidant N-acetylcysteine but not CsA, an inhibitor of the mitochondrial permeability transition pore (mPTP) (Fig. 6C). We concluded that the impairment of cellular viability was caused by respiratory dysfunction due to instability of complex IV in the absence of mNOA1 (Fig. 6D). This conclusion was also supported by the occurrence of cleavage products of the complex I subunits 30 k and 75 k (Fig. 6D), which indicates activation of caspase-3 (33).
FIGURE 5.
mNOA1 knock-down (kd) causes mitochondrial oxidative stress in C2C12 myoblasts. A, FACS histograms of mitochondrially produced superoxide using MitoSOX Red. B, representative Western blots of thioredoxin-2 as a sensitive marker for oxidative stress normalized to VDAC/porin. H2O2-treated samples served as positive control.
FIGURE 6.
mNOA1 knock-down (kd) activates the mitochondrial apoptotic pathway in C2C12 myoblasts. A, cytochrome c release upon knock-down of mNOA1. VDAC/porin served as loading control. B, representative Western blots of mNOA1, caspase-3 and PARP as markers for apoptotic pathway activation. Pan-actin served as loading control. C, cell survival upon mNOA1 knock-down in the presence of 1 μm CsA or 10 mm N-acetylcysteine (NAC) for 24 h. Data are given as mean ± S.D. (error bars). D, Western blot analysis of subunit IV of complex IV as well as subunits 30 k and 75 k of complex I. VDAC/porin served as loading control.
NOA1 Expression Depends on Oxygen Availability
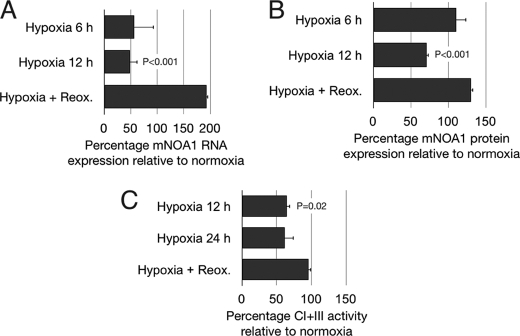
We reasoned that the expression of mNOA1 should closely follow oxygen availability if mNOA1 is indeed instrumental for complex IV-related oxygen sensing. To test this concept we investigated the expression of mNOA1 in C2C12 myoblasts, which were cultivated under hypoxic conditions (0.5% O2) for up to 48 h. Hypoxia resulted in a reduction of mNOA1 mRNA and protein levels by ∼50% compared with normoxia. Reoxygenation for 2 h reversed the hypoxic effect (Fig. 7, A and B). Consistent with the changes in mNOA1 protein and mRNA levels, we also measured a reduction of normalized mitochondrial enzyme activity I+III under hypoxia and a restoration of complex I+III activity to control levels after reoxygenation (Fig. 7C). Oxygen-dependent regulation of protein activities might occur on the transcriptional level or on the protein level by control of protein stability. Determination of the half-life of mNOA mRNA and protein under normoxia using inhibitors of RNA (actinomycin D) and protein synthesis (cycloheximide) revealed that the mNOA1 protein is more stable (half-life ≤12 h) than its mRNA (half-life ≤4 h) (data not shown), which indicates that mNOA1 transcriptional regulation is more dynamic compared with its protein stability.
FIGURE 7.
mNOA1 expression is regulated by the concentration of oxygen. A, mNOA1 mRNA expression levels normalized to β-actin in C2C12 myoblasts under normoxia, after 6-h or 12-h hypoxia or after 24-h hypoxia and 2-h reoxygenation (Reox.). B, mNOA1 protein levels normalized to pan-actin in C2C12 myoblasts after 6-h or 12-h hypoxia or after 24-h hypoxia and 2-h reoxygenation. C, mitochondrial enzyme activity I+III normalized to the mitochondrial marker enzyme citrate synthase in C2C12 myoblasts after 12-h or 24-h hypoxia or after 24-h hypoxia and 2-h reoxygenation. Data are given as mean ± S.D. (error bars).
DISCUSSION
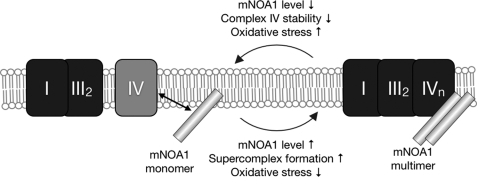
In this study we describe the interaction of mNOA1 with several high molecular mass complexes most notably with the complex IV of the respiratory chain and the PHB complex, which conferred oxygen-dependent regulation of mitochondrial metabolism. We showed that absence of mNOA1 impaired enzyme activity I+III, resulting in oxidative stress and eventually cell death. The most prominent effect on the protein level of respiratory complexes is based on the stabilizing effect of mNOA1 on complex IV and respiratory supercomplexes containing complex IV (Figs. 4A and 8).
FIGURE 8.
Model of the role of mNOA1 in the control of mitochondrial respiration. Expression of mNOA1 on the transcriptional level is controlled by oxygen. NOA1 interacts with complex IV of the respiratory chain. By stabilization of complex IV it favors formation or stabilization of respiratory supercomplexes. Oxidative stress is minimal. Loss of mNOA1 destabilizes complex IV and supercomplexes. Oxidative stress is increased.
Interaction of mNOA1 with different respiratory complexes was demonstrated by two different means: (i) fractionation and (ii) immunoprecipitation. (i) Sucrose gradient centrifugation and BN-PAGE revealed co-migration of mNOA1 with the complex IV of the respiratory chain and, to a lesser degree, with respiratory supercomplexes. It is important to emphasize that the interaction of complex IV with supercomplexes is easily lost during fractionation (34), which might lead to a systematic underestimation of the importance of mNOA1 for formation and/or maintenance of supercomplexes. (ii) The combined IP-mass spectrometry approach yielded a comprehensive array of interaction partners belonging to respiratory complexes and the PHB complex. This does not mean that mNOA1 necessarily interacts directly with all of these proteins. Instead, it seems more likely that mNOA1 forms contact only with some integral components of the respective complexes. Additional interaction studies using isolated components will be necessary to distinguish between direct and indirect interactions and to map protein domains that are bound by mNOA1. At present, it is also possible that mNOA1 interacts simultaneously with multiple proteins within the respiratory supercomplex thereby facilitating assembly and/or upholding this complex structure. In this context it was also interesting to note that mNOA1 forms dimers or multimers that appear only in the fraction of respiratory supercomplexes. mNOA1 contains a predicted leucine zipper motif (13), which is likely responsible for the observed dimerization.
Complex IV is the final respiratory complex and known to act as oxygen acceptor and sensor (35). Interestingly, complex IV stabilizes complex I indirectly (29–31), which explains why mechanisms that alter the function or stability of complex IV exert effects on other respiratory complexes. The PHB complex, on the other hand, acts as a mitochondrial chaperone and assembly factor for respiratory complexes (26, 36) and is indispensable for mitochondrial translation (37). Only recently it became clear that processes such as fission-fusion-controlled changes in mitochondrial activity partially rely on the PHB complex (38, 39). We would therefore like to propose that mNOA1 acts as an adaptor to bring together complex IV and the PHB complex to assemble or stabilize the complex IV. This in turn favors a shift toward assembly and/or stabilization of respiratory supercomplexes.
In support of this hypothesis, we were able to demonstrate that mNOA1 is instrumental for optimal enzyme activity I+III. Knock-down of mNOA1 and disruption of its GTPase domain reduced complex I+III activity, which is in agreement to findings from mNOA1 homologs (13, 15). Surprisingly, reduction of complex I+III activity after inhibition of mNOA1 was associated with increased activity of complex I. The resulting imbalance of respiratory complex activities nicely explains the induction of oxidative stress and reduced cellular viability after depletion of mNOA1.
It might seem puzzling that the reduction of cellular viability induced by the knock-down of mNOA1 was rescued by N-acetylcysteine but not by the mPTP inhibitor CsA. However, there is experimental evidence of a mPTP-independent, CsA-insensitive, release of cytochrome c and induction of the caspase-induced cytochrome c feedback loop (40–42). Moreover, loss of the mNOA1 interaction partner PHB2 does also initiate caspase-dependent apoptotic cell death (38, 43), which further strengthens the existence of mPTP-independent release of cytochrome c.
Our data support the conclusion that mNOA1 modulates enzyme activities by facilitating supercomplex formation. This mechanism might become particularly important when oxygen reaches critically low levels. It is obvious that mitochondrial respiration needs to be tuned down under hypoxic conditions. The reduced expression of mNOA1 at low oxygen levels might achieve this adjustment by favoring dissolution of the respiratory supercomplex thereby reducing its activity and oxygen consumption. The relatively long half-life of mNOA1, however, suggests that this mechanism might be important only for a long term adaptation to hypoxic conditions. Clearly, for an immediate reduction of respiratory activity other mechanisms prevail, which do not rely on mNOA1 (44).
Induction of megamitochondria, which increases the spatial resolution substantially, allowed us to localize the greater part of mNOA1 to the mitochondrial matrix compartment although the techniques applied do not rule out that mNOA1 acquires a different localization under specific conditions. The presence of mNOA1 in the mitochondrial matrix fits also to its described functions in mitochondrial translation (14–16). It has been claimed that NOA1 is unable to rescue a plant mutant (for review, see Ref. 45). Based on our localization studies it is probably necessary to reevaluate these results critically because the authors used a mitochondrial targeting sequence derived from the farnesyl-diphosphate synthase isoform 1L of A. thaliana. This targeting sequence was previously used to functionally complement a lack of inner membrane proteins but not matrix proteins (46). Nevertheless, it remains possible that the function of mNOA1 in plants differs from its mammalian homologs although homologs of NOA1 are evolutionary conserved across species.
Taken together, our data disclose an intimate relationship among mitochondrial enzyme activities, mNOA1 levels, and oxygen availability. The repression of the transcriptional activity of mNOA1 by hypoxia is intriguing and indicates a pivotal oxygen-dependent role of mNOA1 in the regulation of the respiratory chain, in which mNOA1 might act as a scaffolding protein or stabilizer.
Supplementary Material
Acknowledgments
We thank Dr. F. Czauderna for providing the plasmid U6 native tetO; Dr. J. E. Walker for providing the 75-kDa subunit of complex I antibody; and Dr. S. Goffart, Dr. J. Pohjoismaki, and Dr. M. Wheeler for critical comments and valuable discussions on the manuscript.
This work was supported by the Max-Planck-Society, the Deutsche Forschungsgemeinschaft Grant Br1416, the Excellence Initiative “Cardiopulmonary System,” and the University of Giessen-Marburg Lung Center (to T. B.), and the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 815 (Redox-Proteomics), Project Z1 (to I. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1, Figs. 1 and 2, and Movie 1.
- mNOA1
- mouse nitric oxide-associated protein 1
- AMS
- 4-acetoamido-4′-maleimidylstilbene-2,2′-disulfonic acid
- BN-PAGE
- blue native-PAGE
- CsA
- cyclosporin A
- H/L
- heavy/light
- mPTP
- mitochondrial permeability transition pore
- PHB
- prohibitin
- SILAC
- stable isotope labeling with amino acids
- VDAC
- voltage-dependent anion channel.
REFERENCES
- 1. Wang G. L., Semenza G. L. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 4304–4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hagen T., Taylor C. T., Lam F., Moncada S. (2003) Science 302, 1975–1978 [DOI] [PubMed] [Google Scholar]
- 3. Galkin A., Abramov A. Y., Frakich N., Duchen M. R., Moncada S. (2009) J. Biol. Chem. 284, 36055–36061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Solaini G., Baracca A., Lenaz G., Sgarbi G. (2010) Biochim. Biophys. Acta 1797, 1171–1177 [DOI] [PubMed] [Google Scholar]
- 5. Hackenbrock C. R., Chazotte B., Gupte S. S. (1986) J. Bioenerg. Biomembr. 18, 331–368 [DOI] [PubMed] [Google Scholar]
- 6. Acín-Pérez R., Fernández-Silva P., Peleato M. L., Pérez-Martos A., Enriquez J. A. (2008) Mol. Cell 32, 529–539 [DOI] [PubMed] [Google Scholar]
- 7. Schägger H. (2002) Biochim. Biophys. Acta 1555, 154–159 [DOI] [PubMed] [Google Scholar]
- 8. Schägger H., Pfeiffer K. (2001) J. Biol. Chem. 276, 37861–37867 [DOI] [PubMed] [Google Scholar]
- 9. Jezek P., Plecitá-Hlavatá L. (2009) Int. J. Biochem. Cell Biol. 41, 1790–1804 [DOI] [PubMed] [Google Scholar]
- 10. Benard G., Bellance N., James D., Parrone P., Fernandez H., Letellier T., Rossignol R. (2007) J. Cell Sci. 120, 838–848 [DOI] [PubMed] [Google Scholar]
- 11. Pagliarini D. J., Calvo S. E., Chang B., Sheth S. A., Vafai S. B., Ong S. E., Walford G. A., Sugiana C., Boneh A., Chen W. K., Hill D. E., Vidal M., Evans J. G., Thorburn D. R., Carr S. A., Mootha V. K. (2008) Cell 134, 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parihar M. S., Parihar A., Chen Z., Nazarewicz R., Ghafourifar P. (2008) Biochim. Biophys. Acta 1780, 921–926 [DOI] [PubMed] [Google Scholar]
- 13. Tang T., Zheng B., Chen S. H., Murphy A. N., Kudlicka K., Zhou H., Farquhar M. G. (2009) J. Biol. Chem. 284, 5414–5424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kolanczyk M., Pech M., Zemojtel T., Yamamoto H., Mikula I., Calvaruso M. A., van den Brand M., Richter R., Fischer B., Ritz A., Kossler N., Thurisch B., Spoerle R., Smeitink J., Kornak U., Chan D., Vingron M., Martasek P., Lightowlers R. N., Nijtmans L., Schuelke M., Nierhaus K. H., Mundlos S. (2011) Mol. Biol. Cell 22, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moreau M., Lee G. I., Wang Y., Crane B. R., Klessig D. F. (2008) J. Biol. Chem. 283, 32957–32967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sudhamsu J., Lee G. I., Klessig D. F., Crane B. R. (2008) J. Biol. Chem. 283, 32968–32976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Czauderna F., Santel A., Hinz M., Fechtner M., Durieux B., Fisch G., Leenders F., Arnold W., Giese K., Klippel A., Kaufmann J. (2003) Nucleic Acids Res. 31, e127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Krüger M., Kratchmarova I., Blagoev B., Tseng Y. H., Kahn C. R., Mann M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2451–2456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nemoto Y., De Camilli P. (1999) EMBO J. 18, 2991–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kukat A., Kukat C., Brocher J., Schäfer I., Krohne G., Trounce I. A., Villani G., Seibel P. (2008) Nucleic Acids Res. 36, e44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Malka F., Guillery O., Cifuentes-Diaz C., Guillou E., Belenguer P., Lombès A., Rojo M. (2005) EMBO Rep. 6, 853–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taylor S. W., Warnock D. E., Glenn G. M., Zhang B., Fahy E., Gaucher S. P., Capaldi R. A., Gibson B. W., Ghosh S. S. (2002) J. Proteome Res. 1, 451–458 [DOI] [PubMed] [Google Scholar]
- 23. Wittig I., Braun H. P., Schägger H. (2006) Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 24. Gellerich F. N., Deschauer M., Chen Y., Müller T., Neudecker S., Zierz S. (2002) Biochim. Biophys. Acta 1556, 41–52 [DOI] [PubMed] [Google Scholar]
- 25. Antonicka H., Ogilvie I., Taivassalo T., Anitori R. P., Haller R. G., Vissing J., Kennaway N. G., Shoubridge E. A. (2003) J. Biol. Chem. 278, 43081–43088 [DOI] [PubMed] [Google Scholar]
- 26. Nijtmans L. G., de Jong L., Artal Sanz M., Coates P. J., Berden J. A., Back J. W., Muijsers A. O., van der Spek H., Grivell L. A. (2000) EMBO J. 19, 2444–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 28. Blagoev B., Kratchmarova I., Ong S. E., Nielsen M., Foster L. J., Mann M. (2003) Nat. Biotechnol. 21, 315–318 [DOI] [PubMed] [Google Scholar]
- 29. Diaz F., Fukui H., Garcia S., Moraes C. T. (2006) Mol. Cell. Biol. 26, 4872–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oswald C., Krause-Buchholz U., Rödel G. (2009) J. Mol. Biol. 389, 470–479 [DOI] [PubMed] [Google Scholar]
- 31. Stroh A., Anderka O., Pfeiffer K., Yagi T., Finel M., Ludwig B., Schägger H. (2004) J. Biol. Chem. 279, 5000–5007 [DOI] [PubMed] [Google Scholar]
- 32. Lu C., Wang A., Wang L., Dorsch M., Ocain T. D., Xu Y. (2005) Biochem. Biophys. Res. Commun. 331, 1114–1119 [DOI] [PubMed] [Google Scholar]
- 33. Ricci J. E., Muñoz-Pinedo C., Fitzgerald P., Bailly-Maitre B., Perkins G. A., Yadava N., Scheffler I. E., Ellisman M. H., Green D. R. (2004) Cell 117, 773–786 [DOI] [PubMed] [Google Scholar]
- 34. Schägger H., Pfeiffer K. (2000) EMBO J. 19, 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Castello P. R., David P. S., McClure T., Crook Z., Poyton R. O. (2006) Cell Metab. 3, 277–287 [DOI] [PubMed] [Google Scholar]
- 36. Nijtmans L. G., Artal S. M., Grivell L. A., Coates P. J. (2002) Cell Mol. Life Sci. 59, 143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Týc J., Faktorová D., Kriegová E., Jirkù M., Vávrová Z., Maslov D. A., Lukes J. (2010) Int. J. Parasitol. 40, 73–83 [DOI] [PubMed] [Google Scholar]
- 38. Merkwirth C., Dargazanli S., Tatsuta T., Geimer S., Löwer B., Wunderlich F. T., von Kleist-Retzow J. C., Waisman A., Westermann B., Langer T. (2008) Genes Dev. 22, 476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Twig G., Elorza A., Molina A. J., Mohamed H., Wikstrom J. D., Walzer G., Stiles L., Haigh S. E., Katz S., Las G., Alroy J., Wu M., Py B. F., Yuan J., Deeney J. T., Corkey B. E., Shirihai O. S. (2008) EMBO J. 27, 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen Q., Gong B., Almasan A. (2000) Cell Death Differ. 7, 227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schild L., Keilhoff G., Augustin W., Reiser G., Striggow F. (2001) FASEB J. 15, 565–567 [DOI] [PubMed] [Google Scholar]
- 42. Shimizu S., Tsujimoto Y. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 577–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kasashima K., Ohta E., Kagawa Y., Endo H. (2006) J. Biol. Chem. 281, 36401–36410 [DOI] [PubMed] [Google Scholar]
- 44. Acin-Perez R., Gatti D. L., Bai Y., Manfredi G. (2011) Cell Metab. 13, 712–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gas E., Flores-Pérez U., Sauret-Güeto S., Rodríguez-Concepción M. (2009) Plant Cell 21, 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cunillera N., Boronat A., Ferrer A. (1997) J. Biol. Chem. 272, 15381–15388 [DOI] [PubMed] [Google Scholar]
- 47. Wittig I., Beckhaus T., Wumaier Z., Karas M., Schägger H. (2010) Mol. Cell Proteomics 9, 2149–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.