Abstract
Iridoids are one of the most widely distributed secondary metabolites in higher plants. They are pharmacologically active principles in various medicinal plants and key intermediates in the biosynthesis of monoterpenoid indole alkaloids as well as quinoline alkaloids. Although most iridoids are present as 1-O-glucosides, the glucosylation step in the biosynthetic pathway has remained obscure. We isolated a cDNA coding for UDP-glucose:iridoid glucosyltransferase (UGT85A24) from Gardenia jasminoides. UGT85A24 preferentially glucosylated the 1-O-hydroxyl group of 7-deoxyloganetin and genipin but exhibited only weak activity toward loganetin and no activity toward 7-deoxyloganetic acid. This suggests that, in the biosynthetic pathway of geniposide, a major iridoid compound in G. jasminoides, glucosylation occurs after methylation of 7-deoxyloganetic acid. UGT85A24 showed negligible activity toward any acceptor substrates other than iridoid aglycones. Thus, UGT85A24 has a remarkable specificity for iridoid aglycones. The mRNA level of UGT85A24 overlaps with the marked increase in genipin glucosylation activity in the methyl jasmonate-treated cell cultures of G. jasminoides and is related to iridoid accumulation in G. jasminoides fruits.
Keywords: Enzyme Catalysis, Glycoconjugate, Glycosylation, Metabolism, Plant, Biosynthesis, Iridoid, Natural Product, Secondary Metabolism
Introduction
Iridoids are cyclopenta[c]pyran monoterpenoids distributed in numerous plant families, usually as glucosides (1). In plants, iridoids function as important defense compounds in plant-herbivore interactions (2). They play an important biogenetic role linking terpenes and alkaloids (3). Iridoids exhibit a wide range of pharmacological activities, such as cardiovascular, anti-hepatotoxic, choleretic, anti-inflammatory, immunomodulatory, and purgative activities, and are present in various folk medicinal plants as bioactive principles (4, 5).
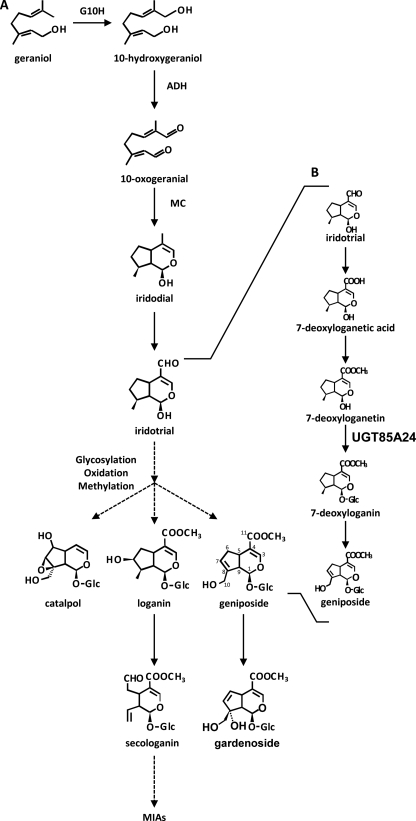
In vivo tracer studies have shown that the iridane skeleton is formed by the cyclization of 10-oxogeranial (10-oxoneral) to yield iridodial, which is subsequently oxidized to iridotrial. Iridotrial is converted to a series of iridoid compounds through various reactions, including oxidation, methylation, and glycosylation as shown in Fig. 1 (6, 7). However, few studies have investigated the biochemical and molecular biological characterization of the iridoid biosynthetic pathway. In particular, the 1-O-glucosylation step in the iridoid biosynthetic pathway has been poorly characterized, although most iridoids are present as 1-O-glucosides in nature. Glucosyltransferase activity toward loganetin, 7-deoxyloganetin, and 7-deoxyloganetic acid was detected in the crude extract of Lonicera japonica (8). However, genes for enzymes relevant to the glucosylation step in the iridoid biosynthetic pathway have yet to be characterized.
FIGURE 1.
A, iridoid biosynthetic pathway. B, proposed biosynthetic pathway from iridotrial to geniposide based on the present results. G10H, geraniol 10-hydroxylase; ADH, acyclic monoterpene primary alcohol dehydrogenase; MC, monoterpene cyclase; MIAs, monoterpenoid indole alkaloids.
Glucosylation steps in plant secondary metabolism are catalyzed by family 1 plant secondary product glycosyltransferases (PSPGs)2 that attach a sugar molecule from a UDP-sugar to a low-molecular-weight acceptor substrate (9–11). PSPGs are defined by the presence of a 44-amino acid C-terminal signature motif designated the PSPG box (12). Recent investigations on the three-dimensional structures of PSPGs by x-ray crystallography revealed that the PSPG box functions as a sugar donor-binding pocket (13–15). Homology-based cloning using the conserved amino acid sequence within the PSPG box has been shown to be an efficient tool for isolating cDNAs encoding PSPGs (16, 17).
Gardenia jasminoides (Rubiaceae) fruits accumulate iridoid compounds, such as geniposide and gardenoside (18), and the dried fruits have been used as a crude drug in traditional Chinese medicine. Cultured G. jasminoides cells have been used to investigate iridoid biosynthesis because they produce small amounts of iridoids even after dedifferentiation (19–21). Thus, suspension cultures of G. jasminoides are suitable as sources for cDNA isolation and the molecular characterization of glucosyltransferases that catalyze the 1-O-glucosylation step in the iridoid biosynthetic pathway. Geniposide is a major iridoid compound present in G. jasminoides fruits, and genipin, an aglycone of geniposide, is the only iridoid aglycone commercially available in relatively large amounts. Homology-based cloning using the conserved amino acid sequence within the PSPG box combined with screening of the glucosylation activity of recombinant enzymes from candidate cDNA clones with genipin as a glucose-accepting substrate, instead of the intrinsic substrates such as 7-deoxyloganetic acid and 7-deoxyloganetin, which are difficult to obtain, led to the isolation of a cDNA encoding an iridoid-specific glucosyltransferase for the first time. Here, we describe the molecular cloning and characterization of a UDP-glucose:iridoid glucosyltransferase (UGT85A24) from G. jasminoides in culture.
EXPERIMENTAL PROCEDURES
Plant Materials
Cell suspension cultures of G. jasminoides and Catharanthus roseus were originally obtained from seedlings of each plant and maintained in LS medium (22) supplemented with 3% sucrose, 1 μm 2,4-dichlorophenoxyacetic acid, and 1 μm kinetin. The cells were cultured on a rotary shaker (100 rpm) at 25 °C in the dark and subcultured at 2-week intervals. Methyl jasmonate was dissolved in dimethyl sulfoxide and aseptically added to the cultures at 3 days after cell inoculation through membrane filters at a final concentration of 250 μm.
Chemicals
The reagent 7-deoxyloganin tetraacetate was kindly provided by Professor K. Inoue (Yokohama College of Pharmacy). 7-Deoxyloganin and 7-deoxyloganic acid were prepared from 7-deoxyloganin tetraacetate according to the method described previously (23). Loganetin, 7-deoxyloganetin, and 7-deoxyloganetic acid were enzymatically prepared from loganin, 7-deoxyloganin, and 7-deoxyloganic acid, respectively, as follows. A 20-mg aliquot of each glucoside was dissolved in 10 ml of 50 mm phosphate-citrate buffer (pH 4.8) containing 50 units/ml almond β-glucosidase (Sigma) and incubated for 3 h at 37 °C. After centrifugation, the reaction mixture was extracted three times with ethyl acetate. The ethyl acetate fraction was concentrated under reduced pressure to yield each aglycone. The purity of each aglycone thus obtained was estimated by quantitative 1H NMR analysis (24). Geniposide, genipin, and loganin were obtained from Wako Pure Chemicals (Osaka, Japan). Gardenoside was a generous gift from Tsumura & Co. (Tokyo, Japan). All other chemicals were commercial reagent-grade quality.
Biotransformation of Genipin
Genipin (25 μmol) was dissolved in Me2SO and added to the cultures through membrane filters. The cells were collected by vacuum filtration at an appropriate time, immediately frozen in liquid nitrogen, and stored at −70 °C until used.
The frozen cells (∼0.5 g) were extracted with 1 ml of methanol by sonication, and the methanol extracts were subjected to HPLC analysis on a reversed-phase column (COSMOSIL 5C18-AR-II, 4.6 × 150 mm; Nacalai Tesque, Kyoto, Japan). The eluate was monitored using a photodiode array detector. The following gradient elution program was used at a flow rate of 1.0 ml/min: 0–15 min, 15–35% methanol; 15–17 min, 35–100% methanol; and 17–20 min, 100% methanol. The amounts of products formed were calculated on the basis of calibration curves prepared using the respective genipin glucosides.
Isolation and Identification of Glucosylation Products of Genipin
To identify the glucosylation products, the methanol extract was concentrated to dryness in vacuo. The residue was suspended in water and extracted first with ethyl acetate and then with water-saturated butanol. The butanol extract was applied to a silica gel TLC plate, which was developed with chloroform/methanol (3:1). The bands corresponding to two glycosylation products were scraped and extracted with methanol. The products thus obtained were analyzed by 1H and 13C NMR spectroscopies using a JNM α-500 spectrometer (JEOL, Tokyo).
PCR Cloning of Glycosyltransferase cDNAs
Total RNA was isolated from G. jasminoides cells using an RNeasy plant minikit (Qiagen, Hilden, Germany). RT-PCR was performed using a CapFishing full-length cDNA premix kit (Seegene, Rockville, MD). Two degenerate primers, UGT2mFw (TT(T/C)(T/C/G)TI(A/T)(G/C)ICA(T/C)TG(T/C)GGITGGAA) and PSPG2Fw (TG(T/C)GGITGGAA(T/C)TCI(A/G)(T/C)I(T/C)TIGA), were designed on the basis of amino acid sequences F(L/V)(T/S)HCGWN and CGWNS(T/V)LE, respectively, in the conserved PSPG box of plant glucosyltransferases (12, 16). A 5-μl aliquot of the cDNA was used as a template for PCR amplification in a 50-μl reaction mixture containing 1 μm primer UGT2mFw, 0.2 μm 3′-RACE primer from the CapFishing full-length cDNA premix kit, and 25 μl SeeAmp TaqPlus Master Mix (Seegene). A portion of the first PCR product was used as the template for nested PCR using the PSPG2Fw and 3′-RACE primers. The first and nested PCRs were performed under the following conditions: denaturation at 94 °C for 3 min; 35 cycles at 94 °C for 30 s, 45 °C for 1 min, and 72 °C for 1 min; and a final extension step at 72 °C for 5 min. Amplified fragments of ∼500 bp were recovered from an agarose gel and subcloned into the pT7Blue-2 (Novagen, Madison, WI) or pBluescript II SK(−) (Stratagene, La Jolla, CA) vector. Randomly selected cloned inserts were sequenced using a BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA) with an ABI PRISM 3100 genetic analyzer (Applied Biosystems).
The 5′-ends of these cDNAs were obtained using the gene-specific primers shown in supplemental Table S1 and the 5′-RACE primer from the CapFishing full-length cDNA premix kit. The full-length cDNA clones were amplified and sequenced using the 5′- and 3′-sequences as specific primers (supplemental Table S2).
Heterologous Expression and Purification of Recombinant Proteins
The full-length cDNAs were amplified by PCR with KOD FX DNA polymerase (Toyobo, Tokyo). The nucleotide sequences of the PCR primers containing appropriate restriction sites are shown in supplemental Table S3. The PCR products were subcloned into the pT7Blue-2 or pBluescript II SK(−) vector to confirm the sequence and then into the pQE-30 vector (Qiagen) after digestion with appropriate restriction enzymes to create N-terminal fusion proteins with a His6 tag. Escherichia coli strain JM109 was used as the host for expression. Transformed cells were cultured at 37 °C until the absorbance at 600 nm reached 0.6. After incubation overnight at 18 °C, the cells were harvested. The recombinant protein was affinity-purified on a nickel-nitrilotriacetic acid-agarose matrix (Qiagen) according to the manufacturer's instructions. Protein content in the enzyme preparations was estimated using the Bradford method (25).
Recombinant Enzyme Assay
The standard reaction mixture (total volume of 50 μl) contained 50 mm Tris-HCl (pH 8.0), 5 mm UDP-sugar (UDP-glucose, UDP-galactose, or UDP-glucuronic acid), 1 mm acceptor substrates, and the enzyme preparation. The reaction was performed at 30 °C and then terminated by the addition of 100 μl of methanol. After centrifugation at 12,000 × g for 5 min, the reaction products were analyzed by HPLC, and elution was monitored using the photodiode array detector.
To separate the substrates and their glucosides, the following gradient elution programs were used: for iridoid glucosides, 30–60% methanol for 0–16 min and 60% methanol for 16–19 min; for flavonoids, coumarins, and various phenolic glucosides, 15–52% acetonitrile for 0–26 min, 52–100% acetonitrile for 26–29 min, and 100% acetonitrile for 29–33 min; and for crocetin derivatives, 20–40% methanol for 0–20 min, 40–100% methanol for 20–25 min, and 100% methanol for 25–27.5 min. For trans-zeatin and its glucosides, the HPLC conditions were as described by Hou and co-workers (26).
To determine the kinetic parameters, enzyme assays were performed in triplicate at each substrate concentration with 10–25 μg of the purified enzyme at 30 °C for 10 min. The substrate concentrations used were 0.1–10 mm sugar acceptors with UDP-glucose at 5 mm for acceptor kinetics and 0.05–2.5 mm UDP-glucose with 7-deoxyloganetin at 0.5 mm for donor kinetics. The initial velocity data were visualized by Lineweaver-Burk plots, and kinetic parameters were calculated based on linear regression analysis using Excel 2007 (Microsoft Japan, Tokyo).
Determination of Genipin Glucosyltransferase Activity in Cultured Cells
G. jasminoides cells frozen in liquid nitrogen were ground to a fine powder using a mortar and pestle. The powdered cells were homogenized in 3 volumes of extraction buffer (50 mm Tris-HCl (pH 7.5), 5 mm EDTA, 0.5 mm phenylmethylsulfonyl fluoride, and 5% (w/v) polyvinylpolypyrrolidone). The homogenate was filtered through two layers of Miracloth (Merck) and centrifuged at 12,000 × g for 20 min at 4 °C. The supernatant was desalted on a PD-10 column (GE Healthcare), which was pre-equilibrated with 50 mm Tris-HCl (pH 7.5). To determine enzymatic activity, a 100-μl reaction mixture containing 5 mm UDP-glucose, 1 mm genipin, and the enzyme preparation was used. The reaction was performed at 30 °C for 30 min and then terminated in liquid nitrogen. After centrifugation at 12,000 × g for 5 min, the reaction products were analyzed by HPLC as described above.
Analysis of Gene Expression by RNA Gel Blot Analysis and RT-PCR
Total RNA was prepared from cultured cells and organs of G. jasminoides using the RNeasy plant minikit and a Fruit-mate (Takara, Shiga, Japan) for RNA purification. For RNA gel blot analysis, RNA (7 μg) was separated by formamide-containing 1% agarose gel electrophoresis and then blotted onto a nylon membrane (Hybond N+, GE Healthcare). The membrane was hybridized with a digoxigenin-labeled probe of full-length G. jasminoides (Gj) UGT2 cDNA and detected using a chemiluminescent reagent. The specificity of the probe for GjUGT2 cDNA was confirmed by Southern blotting of the probe with the cDNA inserts of GjUGT1–GjUGT13 (supplemental Fig. S2).
First-strand cDNAs for RT-PCR were synthesized from 0.5 μg of total RNA using SuperScript III RNase H− reverse transcriptase (Invitrogen). The specific primers GjUGT2_Fw3 (GATGCCCTCAATGGCTTGCCT) and GjUGT2_Rv3 (TCTTGGCTCTCTCAGTCTCTTGC) were used for GjUGT2 amplification using AmpliTaq Gold PCR Master Mix (Applied Biosystems). The specific annealing of the primer set to GjUGT2 cDNA was confirmed by PCR using cDNA clones of GjUGT1–GjUGT13 as templates. The 60 S acidic ribosomal protein P0 (RPP0C) primer pair was used as an internal control. PCR was performed under the following conditions: precycling at 94 °C for 7 min, followed by 32 cycles of 30 s at 94 °C, 30 s at 60 °C, and 30 s at 72 °C and then 5 min at 72 °C. The PCR products were separated on a 1.5% agarose gel and stained with ethidium bromide, followed by fluorescent signal detection. Analysis of signal intensity was performed using ImageJ software. The RT-PCR products were extracted from the gel using a Wizard SV gel and PCR clean-up system (Promega, Madison, WI), subcloned into T-Vector pMD20 (Takara Biotechnology, Otsu, Japan), and sequenced using a BigDye terminator cycle sequencing kit with an ABI PRISM 3100 genetic analyzer.
Quantitative Determination of Genipin Glucosides
Plant tissues of G. jasminoides were ground in liquid nitrogen to a fine powder using a mortar and pestle, and the powdered tissues (0.5 g) were extracted with 1 ml of 70% (v/v) methanol by sonication. The methanol extracts were subjected to HPLC separation. The following gradient elution program was used: 0–40 min, 10–50% methanol; 40–42 min, 50–80% methanol; and 42–47 min, 80% methanol. The amounts of genipin glucosides formed were calculated on the basis of a calibration curve prepared using their respective standards.
RESULTS
Genipin Glucosylation by Cultured G. jasminoides Cells
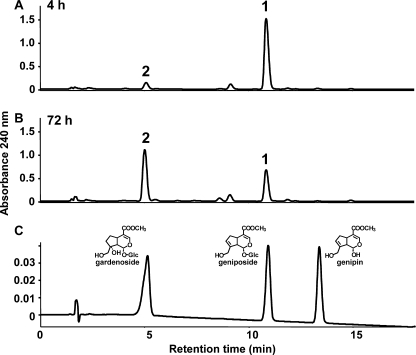
Genipin was added to cell suspension cultures of G. jasminoides at a final concentration of 0.25 mm at 7 days after cell inoculation. The cells were collected at 4, 8, 24, 48, and 72 h after the addition of genipin. Two potentially glycosylated products (1 and 2) were detected when the methanol extract of G. jasminoides cells supplemented with genipin was analyzed by HPLC as shown in Fig. 2. These products were isolated by preparative TLC and identified as geniposide (1) and gardenoside (2) on the basis of their 1H and 13C NMR data (27, 28).
FIGURE 2.
HPLC profiles of the biotransformation products of genipin by cell suspension cultures of G. jasminoides. Genipin was added to cell suspension cultures of G. jasminoides and incubated for 4 h (A) and 72 h (B). The methanol extracts of the collected cells were subjected to HPLC analysis. C, HPLC profile of a mixture comprising standards for genipin, geniposide, and gardenoside.
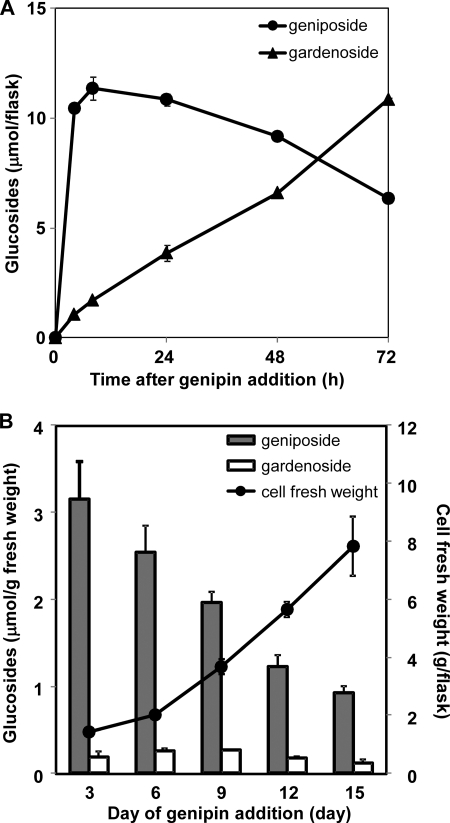
Time course changes in genipin glucosylation in G. jasminoides cell cultures were examined (Fig. 3A). Immediately after addition, genipin was rapidly converted to geniposide. The geniposide content reached a maximum level at 8 h after the addition of genipin and then gradually decreased. In contrast, gardenoside formation gradually increased until 72 h after the addition of genipin. About 70% of the exogenously added genipin was transformed into its glucosides after 3 days. The glucosylation activity of cultured G. jasminoides cells was examined during the growth cycle of the cell suspension cultures (Fig. 3B). Cultures at 3, 6, 9, 12, and 15 days after cell inoculation were incubated with genipin for 4 h before cell collection. The amount of genipin glucosides (geniposide plus gardenoside) per gram of cells (fresh weight) was highest when genipin was added to the cultures on day 3 (during the early growth phase) and then decreased during the later stages of the growth cycle. The addition of genipin did not affect the growth G. jasminoides cell suspension cultures.
FIGURE 3.
Genipin glucosylation by cell suspension cultures of G. jasminoides. A, time course changes in the accumulation of glucosylation products in cultured G. jasminoides cells. G. jasminoides cell cultures were supplemented with genipin (25 μmol/flask). The cells were collected at 4, 8, 24, 48, and 72 h after the addition of genipin, and the amounts of the glucosides formed from genipin were estimated by HPLC. B, glucosylation activity of G. jasminoides cell cultures. Genipin (25 μmol/flask) was added to the cell cultures at 3, 6, 9, 12, and 15 days after cell inoculation, and glucoside formation was estimated after the cells were cultured for 1 additional day. ●, biomass fresh weight per flask at 3, 6, 9, 12, and 15 days after cell inoculation. Each circle and bar represent an average value with S.D. from triplicate measurements.
Cloning of Glycosyltransferase cDNAs from Cultured G. jasminoides Cells
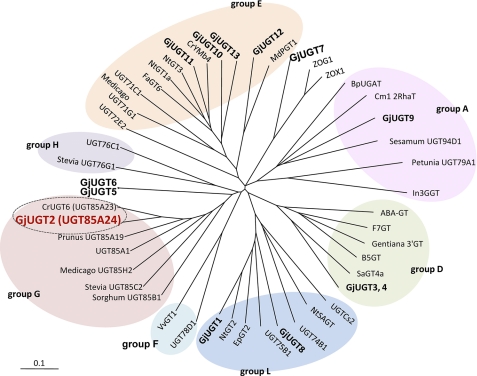
On the basis of the results of our biotransformation experiments, we used G. jasminoides cells collected during the early growth phase for isolating cDNAs encoding iridoid glucosyltransferases. Total RNA was prepared from the cells and used as the template for RT-PCR designed on the basis of the conserved amino acid sequence within the PSPG box. Thirteen partial cDNA fragments with their deduced amino acid sequences similar to the C-terminal sequences of various PSPGs in the database were obtained. Using these partial cDNA fragments, we obtained 13 nearly full-length cDNAs by 5′-RACE and designated them as GjUGT1–GjUGT13. The nucleotide sequences of GjUGT1–GjUGT13 are provided in the DDBJ/GenBankTM/EBI DNA Database under the accession numbers shown in the legend for Fig. 4. Phylogenetic analysis based on the deduced amino acid sequences indicated that, of the 13 PSPGs, four (GjUGT10–GjUGT13) belonged to group E, two (GjUGT3 and GjUGT4) to group D, two (GjUGT1 and GjUGT8) to group L, and one (GjUGT2) to group G (Fig. 4). Four other PSPGs were distinctly separated from the abovementioned PSPGs.
FIGURE 4.
Non-rooted molecular phylogenetic tree of family 1 PSPGs. The PSPG clones isolated in this investigation are shown in boldface, and GjUGT2 (UGT84A24) is shown in red. The tree was generated by the neighbor-joining method following multiple alignments by the ClustalW algorithm. The bar indicates 0.1 amino acid substitution/site. The nucleotide sequences of GjUGT1–GjUGT13 have been submitted to the DDBJ/GenBankTM/EBI DNA Database under accession numbers AB555731–AB555743, respectively. For the names and DDBJ/GenBankTM/EBI accession numbers of other PSPGs, see supplemental Table S4.
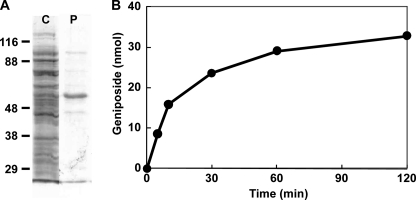
The ORFs of the 13 cDNAs were expressed in E. coli as N-terminal fusion proteins with His6 tags. SDS-PAGE analysis confirmed that all recombinant proteins were successfully obtained in the soluble fraction from the bacterial cells, although the expression levels were variable to some extent. The crude enzyme extracts corresponding to GjUGT1–GjUGT13 were used for glucosyl transfer assays with genipin as an acceptor substrate in the presence of UDP-glucose. The crude enzyme containing recombinant GjUGT2 converted genipin to geniposide. A small amount of the second product, presumably genipin 10-O-glucoside, was detected by prolonged incubation, whereas gardenoside was not formed even after prolonged incubation. Genipin glucosylation catalyzed by recombinant GjUGT2 purified nearly to homogeneity by nickel affinity chromatography proceeded linearly over a 10-min period, and then the rate gradually decreased under the incubation conditions used (Fig. 5). The GjUGT2 cDNA contains an ORF corresponding to a protein (UGT85A24) of 481 amino acids with a predicted molecular mass of 53.6 kDa. It shares 66% amino acid identity with UGT85A9 from Prunus dulcis (29). The yield of the purified UGT85A24 protein from the bacterial culture was 2.5 mg/liter. Neither the other recombinant proteins nor the crude enzyme preparation from E. coli harboring the control vector produced glucosylation products from genipin.
FIGURE 5.
Analysis of the glucosyltransferase activity of recombinant UGT85A24 expressed in E. coli. A, results from SDS-PAGE analysis of the crude protein from E. coli JM109 harboring pQE-30-GjUGT2 (left lane: C) and the recombinant enzyme (UGT85A24) purified using a nickel-nitrilotriacetic acid resin column (right: P). B, time course changes in genipin glucosylation by incubation with recombinant UGT85A24.
Functional Characterization of UGT85A24
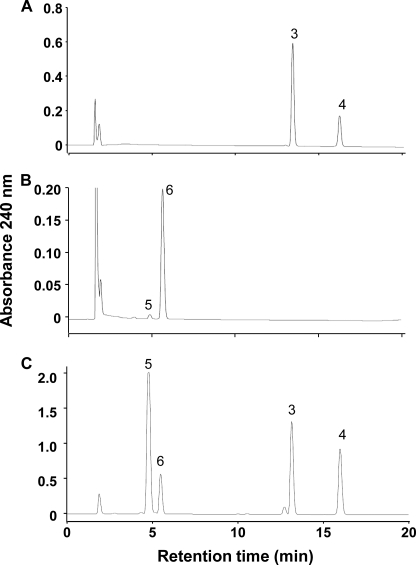
The glucosyl acceptor specificity of UGT85A24 was examined using various substrates. The chemical structures of the compounds used are shown in supplemental Fig. S1. HPLC analysis revealed the formation of a product with a retention time and UV spectrum identical to those of 7-deoxyloganin (7-deoxyloganetin 1-O-glucoside, 3) when 7-deoxyloganetin (4) was used as a substrate (Fig. 6A). Loganin (loganetin 1-O-glucoside, 5) was also produced from loganetin (6), but the yield was extremely low compared with that from 7-deoxyloganin (Fig. 6B). No product was formed from 7-deoxyloganetic acid. UGT85A24 exhibited little glucosyltransferase activity toward several phenolic compounds, including 4-nitrophenol, curcumin, flavonoids, coumarins, crocetin, and phytohormones such as trans-zeatin and indoleacetic acid. As for the glycosyl donor specificity, UGT85A24 exhibited no activity toward either UDP-galactose or UDP-glucuronic acid.
FIGURE 6.
A and B, glucosylation of 7-deoxyloganetin and loganetin, respectively, by UGT85A24. Each iridoid aglycone was incubated with UGT85A24 for 60 min, and the assay mixture was subjected to HPLC analysis. C, HPLC profile of a mixture comprising standards. 3, 7-deoxyloganin; 4, 7-deoxyloganetin; 5, loganin; 6, loganetin.
The kinetic parameters of UGT85A24 for iridoid aglycones using UDP-glucose as the sugar donor substrate were determined using the affinity-purified protein (Table 1). The apparent Km values for genipin and 7-deoxyloganetin were 8.8 and 0.61 mm, respectively. The kcat/Km ratio for 7-deoxyloganetin was 2-fold higher than that for genipin. The kinetic parameters for UDP-glucose were also determined using 7-deoxyloganetin as the sugar acceptor (Table 1).
TABLE 1.
Kinetic parameters of recombinant UGT85A24 for iridoid aglycones and UDP-glucose
Data represent means ± S.D. from triplicate measurements. NA, no activity; ND, cannot be determined because only trace amounts of the product were formed (detection limit, 0.1 nmol of product/50 μl of reaction mixture).
| Km | kcat | kcat/Km | |
|---|---|---|---|
| mm | s−1 | mm−1s−1 | |
| Genipin | 8.82 ± 0.09 | 1.04 ± 0.04 | 0.12 ± 0.004 |
| 7-Deoxyloganetin | 0.61 ± 0.08 | 0.13 ± 0.04 | 0.21 ± 0.033 |
| 7-Deoxyloganetic acid | NA | NA | NA |
| Loganetin | ND | ND | ND |
| UDP-glucose | 0.17 ± 0.03 | 0.16 ± 0.02 | 1.00 ± 0.21 |
Expression of the UGT85A24 Gene in G. jasminoides
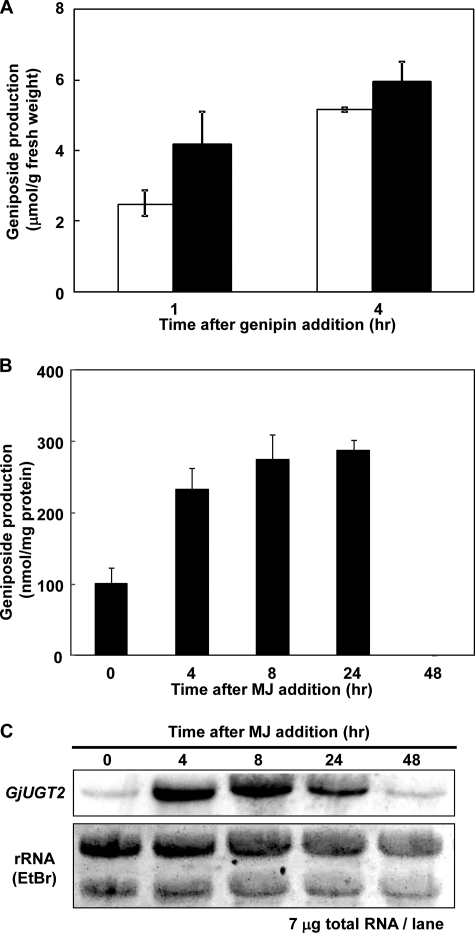
The relationship between the enzymatic activity of genipin glucosyltransferase and the mRNA level of UGT85A24 in cultured G. jasminoides cells was examined. The genipin glucosylation activity of the cultured cells (Fig. 7A) and that of the crude enzyme extract from the cultured cells (Fig. 7B) were markedly increased by adding methyl jasmonate to the cell cultures. The mRNA level of UGT85A24 in the cells was also strongly up-regulated, and the pattern of up-regulation correlated well with the pattern of increased enzymatic activity (Fig. 7C).
FIGURE 7.
MJ-induced activity of genipin glucosylation in G. jasminoides. A, the suspension cultures of G. jasminoides were supplemented with methyl jasmonate at a final concentration of 250 μm 3 days after cell inoculation, and genipin was exogenously added to the cells 24 h after methyl jasmonate addition. The geniposide contents of the cells were estimated 1 and 2 days after genipin addition. Open bars, geniposide formation in mock (Me2SO)-treated cells; closed bars, geniposide formation in methyl jasmonate-treated cells. B, genipin glucosylation activity in the crude enzyme from methyl jasmonate (MJ)-treated cells. Each bar represents an average value with S.D. from triplicate measurements. C, Northern blot analysis of mRNA levels of UGT85A24. Methyl jasmonate was added to the cells as described above, and the crude enzyme and total RNA were prepared 0, 4, 8, 24, and 48 h after methyl jasmonate addition.
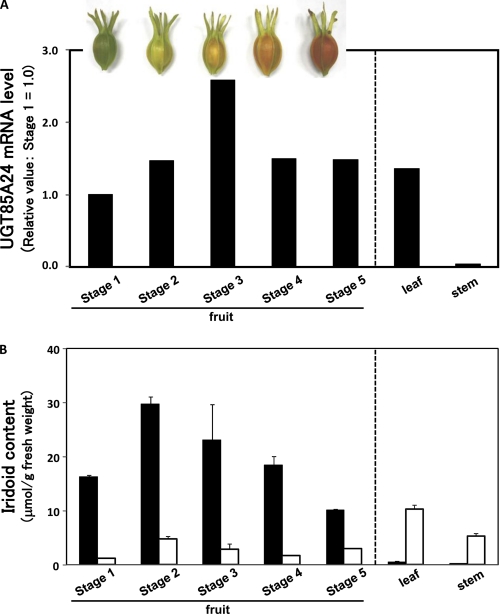
The expression levels of UGT85A24 mRNA in various G. jasminoides organs were examined by RT-PCR (Fig. 8A). To examine the specificity of the PCR primers for UGT85A24, the PCR products were subcloned into T-Vector pMD20. Twenty clones were randomly selected and sequenced. All of the clones exhibited the same sequence with UGT85A24 cDNA, indicating that the PCR primers annealed specifically to UGT85A24 cDNA. UGT85A24 mRNA was detected at similar levels in all organs examined except the stems, in which the expression level was very low. The mRNA level gradually increased during the early stage of fruit ripening, whereas it remained low in the mature fruits. Geniposide and gardenoside contents in G. jasminoides organs were quantitatively determined (Fig. 8B). Geniposide was found to be a predominant iridoid in G. jasminoides fruits, whereas gardenoside was found to be a major iridoid compound in other organs. Iridoid contents in the fruits gradually increased in the early stages of development and gradually decreased afterward. Iridoid content was somewhat lower in the vegetative tissues than in the flowers and fruits. A strict correlation between the mRNA level and iridoid content in the plant organs was not exhibited, except that both the mRNA level of UGT85A24 and the iridoid content were low in the stems compared with those in the other organs.
FIGURE 8.
Temporal and tissue distribution of UGT85A24 mRNA (A) and iridoid glucosides (B) in G. jasminoides. A, fruits, leaves, and stems were collected from a single G. jasminoides plant on the same day, powdered in liquid nitrogen, and stored until used. The fruits were classified into five maturation stages based on the red coloration. Total RNA and methanol extract were prepared from the tissues and used for RT-PCR analysis and HPLC analysis, respectively. B, closed and open bars represent contents of geniposide and gardenoside, respectively, and represent average values with S.D. from five replicates.
DISCUSSION
Cultured G. jasminoides cells efficiently glucosylated exogenously added genipin to geniposide and converted geniposide to gardenoside. We also examined genipin glucosylation using cell suspension cultures of C. roseus, which produces no geniposide in planta. Only ∼3% of the initially supplemented genipin was converted to glucoside in the cell suspension cultures, and no further transformation of geniposide to gardenoside was observed. Therefore, the cellular activity of genipin glucosylation is specific to G. jasminoides, which has biosynthetic activity for geniposide-related iridoid glucosides. The glucosylation activity in the G. jasminoides cells was increased by the addition of methyl jasmonate to the cultures, which is consistent with the possible roles of iridoid glucosides as defense compounds against herbivores (2).
Homology-based cloning using a characteristic motif conserved in the C-terminal part of glycosyltransferases involved in plant secondary metabolism (PSPGs) led to isolation of 13 cDNA clones (GjUGT1–GjUGT13) encoding novel PSPGs from cultured G. jasminoides cells. Biochemical analysis revealed that GjUGT2 encodes a glucosyltransferase (UGT85A24) that catalyzes 1-O-glucosylation of iridoid aglycones, such as genipin and 7-deoxyloganetin, but does not exhibit glucosyltransferase activity toward any other substrates examined that have unique chemical structures. The deduced amino acid sequence of UGT85A24 showed high identity to that of UGT85A21 from Lycium barbarum (78% identity), UGT85A19 from P. dulcis (66%), and UGT85A1 from Arabidopsis thaliana (62%). Molecular phylogenetic analysis revealed that UGT85A24 belongs to group G of PSPGs with other UGT85 family enzymes. PSPGs of this group exhibit glucosyltransferase activity toward various acceptor substrates. For example, UGT85A19 from P. dulcis is a mandelonitrile glucosyltransferase (29), and UGT85A1 from A. thaliana exhibits glucosyltransferase activity toward zeatin (26). UGT85A24 exhibits glucosyltransferase activity toward neither mandelonitrile nor zeatin. Recently, we isolated a cDNA clone encoding an iridoid glucosyltransferase (UGT85A23) from C. roseus. The deduced amino acid sequence of UGT85A23 is 88% identical to that of UGT85A24. UDP-glucose:iridoid glucosyltransferases may form a subgroup within the group G PSPGs as shown in Fig. 4.
UGT85A24 is apparently responsible for iridoid glucosylation in cultured G. jasminoides cells because 1) marked enhancement of genipin glucosylation activity by adding methyl jasmonate to G. jasminoides cell cultures was consistent with the strong up-regulation of UGT85A24 mRNA expression, and 2) G. jasminoides cells efficiently glucosylated exogenously added 7-deoxyloganetin to glucoside but poorly glucosylated 7-deoxyloganetic acid (data not shown), which is consistent with the substrate preferentiality of UGT85A24. UGT85A24 mRNA expression was also detected in various G. jasminoides organs. The gradual increase in the mRNA level during the early phase of fruit ripening correlates with an increase in iridoid accumulation during the fruit development. Furthermore, the relatively low level of iridoid accumulation (geniposide plus gardenoside) in the vegetative tissues is consistent with the distribution pattern of UGT85A24 mRNA in the plant organs. Thus, UGT85A24 may at least partially limit the iridoid accumulation in planta.
Iridoid biosynthesis has been extensively studied using cell cultures of C. roseus and L. japonica because the biosynthesis of secologanin, a major iridoid metabolite in these species, has been considered a bottleneck for monoterpenoid indole alkaloid production in C. roseus cell cultures (7). Tracer experiments and enzymatic studies revealed that the iridane skeleton is formed by the cyclization of 10-oxogeranial biosynthesized from geraniol through 10-hydroxygeraniol. Conversion of the cyclization product iridodial to loganin and then to secologanin involves several steps, including 1-O-glucosylation of the iridane skeleton (Fig. 1). However, the glucosylation step has remained elusive so far. Kinetic analysis of UGT85A24 indicated that the Km value for 7-deoxyloganetin is ∼13-fold lower than that for genipin, whereas the kcat value for genipin is ∼8 times higher than that for 7-deoxyloganetin. Thus, the catalytic efficiency (kcat/Km) is ∼2-fold higher for 7-deoxyloganetin than for genipin. This result suggests that 7-deoxyloganetin is the preferential sugar acceptor for UGT85A24 and probably an endogenous substrate of the glucosylation step in the biosynthetic pathways of geniposide and gardenoside in G. jasminoides, although genipin is a good substrate because of its structural similarity to 7-deoxyloganetin. In G. jasminoides, 7-deoxyloganetic acid is converted to 7-deoxyloganetin, which is then glucosylated to 7-deoxyloganin. Although the Km value of UGT85A24 for 7-deoxyloganetin (∼0.6 mm) seems to be higher compared with the values of, for example, flavonoid glucosyltransferases (30, 31), it is in the same range of the Km values for terpenoid glucosyltransferases so far reported (32, 33).
Hydroxylation of 7-deoxyloganin at C-10 results in the formation of geniposide as shown in Fig. 1B. Yamamoto and co-workers (8) examined the kinetic parameters of the partially purified glucosyltransferase preparation from L. japonica cells and noted that the preparation had the highest affinity for 7-deoxyloganetic acid compared with 7-deoxyloganetin and loganetin, although the specific activity was the highest for loganetin. On the basis of their result, they proposed that 7-deoxyloganetic acid is a sugar-accepting substrate in the iridoid biosynthetic pathway leading to loganin. Glucosylation of iridoid precursors may occur in different steps between loganin biosynthesis in L. japonica and geniposide biosynthesis in G. jasminoides. However, the partially purified enzyme preparation from L. japonica cells may be a mixture of glucosyltransferases, which makes the estimated the kinetic parameters difficult to discern.
In conclusion, we isolated a novel UDP-glucose:7-deoxyloganetin glucosyltransferase, UGT85A24 from G. jasminoides. Our results shed light not only on the glucosylation step in iridoid biosynthesis in G. jasminoides but also on the isolation of various iridoid-specific glucosyltransferases from diverse iridoid- and indole alkaloid-producing plant species, leading to the engineering of biosynthetic pathways producing iridoids and/or monoterpenoid indole alkaloids.
Supplementary Material
Acknowledgment
We thank Professor K. Inoue for the generous gift of 7-deoxyloganin tetraacetate.
This work was supported by a grant-in-aid for scientific research and a grant-in-aid for young scientists from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1–S3.
- PSPG
- plant secondary product glycosyltransferase
- RACE
- rapid amplification of cDNA ends
- Gj
- G. jasminoides.
REFERENCES
- 1. Dinda B., Debnath S., Harigaya Y. (2007) Chem. Pharm. Bull. 55, 159–222 [DOI] [PubMed] [Google Scholar]
- 2. Harvey J. A., van Nouhuys S., Biere A. (2005) J. Chem. Ecol. 31, 287–302 [DOI] [PubMed] [Google Scholar]
- 3. El-Sayed M., Verpoorte R. (2007) Phytochem. Rev. 6, 277–305 [Google Scholar]
- 4. Ghisalberti E. L. (1998) Phytomedicine 5, 147–163 [DOI] [PubMed] [Google Scholar]
- 5. Dinda B., Debnath S., Harigaya Y. (2007) Chem. Pharm. Bull. 55, 689–728 [DOI] [PubMed] [Google Scholar]
- 6. Meijer A. H., Verpoorte R., Hoge J. H. (1993) J. Plant Res. 3, 145–164 [Google Scholar]
- 7. Oudin A., Courtois M., Rideau M, Clastre M. (2007) Phytochem. Rev. 6, 259–276 [Google Scholar]
- 8. Yamamoto H., Sha M., Kitamura K., Yamaguchi Y., Katano N., Inoue K. (2002) Plant Biotechnol. 19, 295–301 [Google Scholar]
- 9. Vogt T., Jones P. (2000) Trends Plant Sci. 5, 380–386 [DOI] [PubMed] [Google Scholar]
- 10. Lim E. K., Bowles D. J. (2004) EMBO J. 23, 2915–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gachon C. M., Langlois-Meurinne M., Saindrenan P. (2005) Trends Plant Sci. 10, 542–549 [DOI] [PubMed] [Google Scholar]
- 12. Hughes J., Hughes M. A. (1994) DNA Seq. 5, 41–49 [DOI] [PubMed] [Google Scholar]
- 13. Offen W., Martinez-Fleites C., Yang M., Kiat-Lim E., Davis B. G., Tarling C. A., Ford C. M., Bowles D. J., Davies G. J. (2006) EMBO J. 25, 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li L., Modolo L. V., Escamilla-Trevino L. L., Achnine L., Dixon R. A., Wang X. (2007) J. Mol. Biol. 370, 951–963 [DOI] [PubMed] [Google Scholar]
- 15. Brazier-Hicks M., Offen W. A., Gershater M. C., Revett T. J., Lim E. K., Bowles D. J., Davies G. J., Edwards R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20238–20243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Noguchi A., Fukui Y., Iuchi-Okada A., Kakutani S., Satake H., Iwashita T., Nakao M., Umezawa T., Ono E. (2008) Plant J. 54, 415–427 [DOI] [PubMed] [Google Scholar]
- 17. Masada S., Terasaka K., Oguchi Y., Okazaki S., Mizushima T., Mizukami H. (2009) Plant Cell Physiol. 50, 1401–1415 [DOI] [PubMed] [Google Scholar]
- 18. Yu Y., Xie Z. L., Gao H., Ma W. W., Dai Y., Wang Y., Zhong Y., Yao X. S. (2009) J. Nat. Prod. 72, 1459–1464 [DOI] [PubMed] [Google Scholar]
- 19. Ueda S., Kobayashi K., Muramatsu T., Inouye H. (1981) Planta Med. 41, 186–191 [DOI] [PubMed] [Google Scholar]
- 20. Uesato S., Ueda S., Kobayashi K., Inouye H. (1983) Chem. Pharm. Bull. 31, 4185–4188 [Google Scholar]
- 21. Uesato S., Ueda S., Kobayashi K., Miyauchi M., Inouye H. (1984) Tetrahedron Lett. 25, 573–576 [Google Scholar]
- 22. Linsmaier E. M., Skoog F. (1965) Physiol. Plant. 18, 100–127 [Google Scholar]
- 23. Inoue K., Ono M., Nakajima H., Fujie I., Inouye H., Fujita T. (1992) Heterocycles 33, 673–695 [Google Scholar]
- 24. Hasada K., Yoshida T., Yamazaki T., Sugimoto N., Nishimura T., Nagatsu A., Mizukami H. (2010) J. Nat. Med. 64, 161–166 [DOI] [PubMed] [Google Scholar]
- 25. Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 26. Hou B., Lim E. K., Higgins G. S., Bowles D. J. (2004) J. Biol. Chem. 279, 47822–47832 [DOI] [PubMed] [Google Scholar]
- 27. Chaudhuri R. K., Afifi-Yazar F. U., Sticher O. (1980) Tetrahedron 36, 2317–2326 [Google Scholar]
- 28. Ono M., Ueno M., Masuoka C., Ikeda T., Nohara T. (2005) Chem. Pharm. Bull. 53, 1342–1344 [DOI] [PubMed] [Google Scholar]
- 29. Franks T. K., Yadollahi A., Wirthensohn M. G., Guerin J. R., Kaiser B. K., Sedgley M., Ford C. M. (2008) Funct. Plant Biol. 35, 236–246 [DOI] [PubMed] [Google Scholar]
- 30. Nakatsuka T., Sato K., Takahashi H., Yamamura S., Nishihara M. (2008) J. Exp. Bot. 59, 1241–1252 [DOI] [PubMed] [Google Scholar]
- 31. Noguchi A., Horikawa M., Fukui Y., Fukuchi-Mizutani M., Iuchi-Okada A., Ishiguro M., Kiso Y., Nakayama T., Ono E. (2009) Plant Cell 21, 1556–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Achnine L., Huhman D. V., Farag M. A., Sumner L. W., Blount J. W., Dixon R. A. (2005) Plant J. 41, 875–887 [DOI] [PubMed] [Google Scholar]
- 33. Naoumkina M. A., Modolo L. V., Huhman D. V., Urbanczyk-Wochniak E., Tang Y., Sumner L. W., Dixon R. A. (2010) Plant Cell 22, 850–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.