Abstract
Understanding the component stoichiometry of the T cell antigen receptor (TCR) triggering apparatus is essential for building realistic models of signal initiation. Recent studies suggesting that the TCR and other signaling-associated proteins are preclustered on resting T cells relied on measurements of the behavior of membrane proteins at interfaces with functionalized glass surfaces. Using fluorescence recovery after photobleaching, we show that, compared with the apical surface, the mobility of TCRs is significantly reduced at Jurkat T cell/glass interfaces, in a signaling-sensitive manner. Using two biophysical approaches that mitigate these effects, bioluminescence resonance energy transfer and two-color coincidence detection microscopy, we show that, within the uncertainty of the methods, the membrane components of the TCR triggering apparatus, i.e. the TCR complex, MHC molecules, CD4/Lck and CD45, are exclusively monovalent or monomeric in human T cell lines, implying that TCR triggering depends only on the kinetics of TCR/pMHC interactions. These analyses also showed that constraining proteins to two dimensions at the cell surface greatly enhances random interactions versus those between the membrane and the cytoplasm. Simulations of TCR-pMHC complex formation based on these findings suggest how unclustered TCR triggering-associated proteins might nevertheless be capable of generating complex signaling outputs via the differential recruitment of cytosolic effectors to the cell membrane.
Keywords: Cell Surface, Membrane Proteins, Protein Structure, Receptors, Signal Transduction, Single Molecule Biophysics, T Cell Receptor, Bioluminescence Resonance Energy Transfer, Protein Stoichiometry
Introduction
T lymphocytes play crucial roles in adaptive immune responses, where their surface interactions with antigen-presenting cells are critical for the detection and subsequent elimination of pathogens. Many of these receptor-ligand pairs have no intrinsic enzymatic activity, as is the case for the T cell antigen receptor (TCR),4 which not only has to rely on extrinsic tyrosine kinases to propagate signaling but also must do so while discriminating between ligands of differing quality. The ligands for the TCR are peptides presented in the context of the MHC molecule (pMHC) by the antigen-presenting cells. Discrimination by the TCR is intimately linked to the longevity of the TCR/pMHC interaction, where the decision to trigger can be influenced by small changes in the binding kinetics. The TCR must initiate differential signaling pathways based on these differences, from titrating positive and negative selection during thymocyte development to causing a potent immune response in the periphery.
These diverse mechanisms must all be intrinsic to the structure and organization of the antigen receptor triggering apparatus and the enzymatic processes initiated by receptor engagement (1). It is unfortunate, therefore, that the structure and organization of the receptor triggering apparatus are so mired in controversy. The TCR itself has been the most contentious and illustrates the extent to which inferences about the stoichiometry of the proteins influence thinking about mechanism (2, 3). We previously attempted to resolve this issue for the TCR using a single-molecule in situ approach that examined TCRs diffusing at the apical surface of T cells resting on a glass surface, which strongly suggested that the TCR is monovalent (4). Very recently, however, high resolution measurements of the behavior of proteins at the cell/glass interface suggested that the TCR is instead preclustered in groups of 7–25 molecules in resting cells (5).
The organization of the other components of the triggering apparatus, i.e. CD4/Lck, CD45 and MHC molecules (1), is also contentious. In the case of the co-receptor CD4, although initial analysis of the extracellular region limited any oligomerization to a very low affinity interaction (6), functional significance has been attributed to homodimeric interactions of the membrane-proximal domain observed in crystals of its extracellular region (7). CD45 has no apparent ligand, but there has been much interest in the possibility that it too is regulated by oligomerization. An initial structure of a tyrosine phosphatase domain revealed a homodimer in the lattice (8) and suggested a general mechanism of phosphatase inhibition (9). More recently it was proposed that CD45 is regulated by glycosylation-controlled dimerization of its extracellular region (10). Finally, there has been speculation that MHC class II forms functional dimers of dimers, based principally on the first crystal structure of HLA-DR (11, 12). However, other evidence points to there being no higher level of organization above the MHC heterodimer (discussed in Ref. 13), and a role for its oligomerization in T cell activation is unproven (12).
Here we readdress the stoichiometry of the TCR (4, 14) and extend the analysis to other membrane components of the TCR triggering apparatus, i.e. to CD4/Lck, CD45 and MHC class II. We present evidence that contact with a functionalized glass surface alters the behavior of the TCR, complicating measurements at this interface. We show that the components of the TCR triggering apparatus are all largely if not completely monovalent or monomeric and that these membrane-bound molecules participate in unexpectedly high levels of nonspecific association within the membrane due to an increase in their effective concentration, in marked contrast to membrane and cytosolic proteins, whose encounters are likely to be much less frequent. Because the TCR requires recruitment of a cytoplasmic tyrosine kinase to the membrane, we speculate that these rate differences could affect the mode and tempo of signaling by this receptor.
EXPERIMENTAL PROCEDURES
Cell Culture
HEK-293T cells used in the BRET experiments were grown in DMEM (Sigma) supplemented with 10% FBS (Sigma), 2 mm glutamine (Sigma), and antibiotics (Sigma) and passaged using trypsin (Sigma). The Jurkat, J.RT3, J45, and PM1 T cell lines and THP-1 monocyte cell line were grown in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS, 10 mm HEPES (Sigma), 1 mm sodium pyruvate (Invitrogen) and antibiotics.
Vector Construction and Transfection
Oligonucleotide primers and cloning strategies used in this study can be found in the supplemental Experimental Procedures. Transient transfection of HEK-293T was performed using GeneJuice (Novagen) according to the manufacturer's protocol. For the TCRβ BRET assay, VSV-G pseudotyped virus produced from transiently transfected HEK-293T cells was used to infect 5 × 105 J.RT3 cells with the TCRβLuc vector for 16 h. Infected cells were recovered, stained for surface expression of CD3ϵ, and cell sorted by using a MoFlo cell sorter, forming the J.Luc line. These cells were then infected in an analogous manner with the TCRβGFP or CD3ϵGFP vectors, and the cell sorter was used to define four subpopulations with increasing GFP expression.
FRAP Microscopy
FRAP was performed as described by Ayoub et al. (15). Approximately 1 × 106 cells were labeled at 4 °C for at least 30 min with saturating UCHT1 Alexa Fluor 488-labeled Fab. Cells were washed and added to glass-bottomed dishes for imaging on a Zeiss LSM510 at 37 °C in PBS, using a 40 × 1.2 NA lens with the 488-nm line of an argon laser operating at 6.1 A. A circular region ∼2 μm in diameter was bleached using 100% laser intensity for 2.4 s. The aperture of the pinhole was adjusted to obtain slices of <2 μm depth. Images were acquired postbleaching every second, with bleaching and background corrected for. To obtain the asymptotic fluorescence values (F∞), each recovery curve was fitted to Equation 1 in Ref. 16.
Bioluminescence Resonance Energy Transfer
The BRET assay was performed essentially as described (17), except for CD45Ex, which was transfected for 48 h to give as high a level of wild-type expression as possible. For the CD4/Lck co-expression experiments, a constant amount (0.5 μg) of the Lck-mCherry (or MyrNck1-mCherry) vector was used in combination with varying ratios of CD4Luc to CD4GFP (total DNA of 0.5 μg), and expression of the mCherry-tagged proteins was confirmed by fluorescence microscopy. For the J.Luc cell lines expressing either TCRβGFP or CD3ϵGFP, 5 × 106 cells of each subpopulation were pelleted, resuspended to 200 μl in PBS, and the BRETeff values measured as for the normal assay.
Two-color Coincidence Detection Analysis
The principles and experimental setup of TCCD have been described previously (4). Antibodies to CD2 (OKT11), CD3 (UCHT1), CD45 (GAP8.3), and HLA-DR (L243) were purified from hybridoma supernatant using protein A or G. Anti-CD4 (Q4120) antibody was obtained from the Centre for AIDS reagents, National Institute for Biological Standards and Control, UK. All antibodies were fragmented to Fabs using immobilized papain and labeled with either Alexa Fluor 488 or 647.
Statistical Analysis
The TCCD dataset for CD2 was used to define a 95% confidence interval to assign monomeric interactions for the TCCD data, defined as the mean ± 1.96 × S.E. This equation was also used to calculate the confidence interval at each time point of the FRAP datasets. When required, measurement error was appropriately propagated.
RESULTS
Methodology
We have previously made use of two techniques, BRET (17) and TCCD (4), to probe membrane protein stoichiometry in situ. BRET detects the physical proximity of fluorophore-tagged proteins by measuring the efficiency of energy transfer (BRETeff) from a donor (luciferase, Luc) to acceptor (green fluorescent protein, GFP) and is highly dependent on donor/acceptor separation, effectively limited to distances of 3–10 nm. By varying the ratio of the acceptor-tagged proteins to those of the donor-fused versions, it is possible to derive additional information about whether physical proximity is due to a random or a specific oligomeric interaction, at near physiological levels of protein expression. BRET-based stoichiometric analysis does require the use of nonnative reporter constructs in heterologous cells. Complementing this approach, therefore, is TCCD, which permits the organization of native proteins to be studied. TCCD uses monovalent antibody fragments (Fabs) labeled with either of two spectrally distinct fluorophores to detect proteins of interest. By focusing two overlapping lasers at the cell surface, it is possible to resolve fluorescence bursts corresponding to single molecules passing through the confocal volume. The spatio-temporal coincidence (i.e. co-localization in time) of the bursts from each fluorophore is used to detect associated proteins. The level of interaction, expressed as the association quotient value, Q, is the fraction of all detected fluorophores exhibiting coincidence, after correction for random events (4). Importantly, the measurement of Q does not rely on any physical process and so is only length-limited by the size of the confocal region, ∼500 nm. The analysis software used to process the data optimizes the threshold above which fluorescence bursts are detected. This alters the effective detection region so that the rate of detected bursts falls within the single molecule range, allowing calculation of Q (4, 18). This results in the detection of comparable numbers of red and blue events in the experiments. The coincident event rates, the rates of red and blue events, and the total numbers of cells and events analyzed in the present experiments are given in Table 1. The absolute number of labeled proteins per cell, measured by fluorescence-activated cell sorting, was above 10,000/cell in all experiments (data not shown).
TABLE 1.
Association quotients for T cell surface proteins labeled at 37 °C with fluorescently labeled Fab fragments, together with the significant event rate (the rate of coincident events above the rate due to random diffusion), the rate of red and blue events, and the numbers of cells and events analyzed
| Cells | Association quotient | Coincidence rate | Red event rate | Blue event rate | No. of cells analyzed | No. of files analyzed |
|---|---|---|---|---|---|---|
| s−1 | s−1 | s−1 | ||||
| Jurkats | ||||||
| CD2/CD2 | 5.5 ± 2.1 | 0.33 | 10.4 | 7.7 | 22 | 56 |
| CD4/CD4 | 3.6 ± 1.8 | 0.31 | 8.6 | 7.6 | 28 | 80 |
| CD45/CD45 | 4.0 ± 1.5 | 0.31 | 9.2 | 8.3 | 15 | 78 |
| PM1 | ||||||
| CD4/CD4 | 1.9 ± 2.6 | 0.32 | 8.0 | 9.0 | 29 | 68 |
| CD45/CD45 | 8.2 ± 3.8 | 0.45 | 9.0 | 8.5 | 32 | 80 |
| CD4/CD45 | 5.7 ± 2.9 | 0.36 | 9.6 | 7.4 | 33 | 74 |
| CD3/CD3 | 28.4 ± 4.3 | 0.31 | 7.6 | 7.6 | 102 | 172 |
| THP | ||||||
| MHCII/MHCII | 5.2 ± 2.2 | 0.27 | 8.3 | 6.6 | 43 | 79 |
Establishing the True “Resting” State of the T Cell Surface
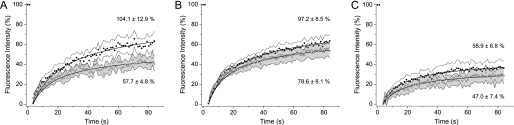
Recent experiments have suggested that proteins involved in triggering are preclustered prior to activation (5). This evidence derives from resolving proteins at the plasma membrane in close apposition to a supporting glass slide or bilayer. We wanted to determine whether there was any significant difference between collecting stoichiometric data from the apical (upper) or basal (lower) T cell membranes. To do this, we performed a FRAP experiment on both surfaces of the same cell attached to a polylysine-coated coverslip, as previously used by Lillemeier et al. (5). Wild-type Jurkat T cells, and CD45low J45 and Lck-deficient J.CaM1.6 cells were each labeled with a fluorophore-conjugated Fab against a component of the TCR (CD3ϵ) and allowed to settle for at least 5 min onto the coated glass surface. Following photobleaching, we observed a striking difference between the apical and basal regions for the wild-type Jurkat cells (p = 0.002), with much lower recovery found at the basal surface (Fig. 1A). The fraction of mobile TCR measured for the apical surface is ∼100%, in excellent agreement with previous measurements (19), whereas it is reduced to ∼58% on the basal surface. In contrast, for the J45 and J.CaM1.6 cells, which cannot be physiologically activated (20, 21), we observed no significant differences (p = 0.118, 0.244, respectively) in the mobility of the TCR complex at the two surfaces, suggesting that there is at most only a modest effect of the glass surface on TCR dynamics in the absence of triggering (Fig. 1, B and C). Given that the “preclustered” TCR complexes previously observed were essentially static (5), these results suggest that the functionalized surface may have induced some weak triggering of the TCR, with at least a quarter of the labeled TCRs likely becoming clustered compared with the equivalent region of the J45 or J.CaM1.6 cells. It also means that to determine the organization of endogenous proteins on the T cell surface in a basal state it is essential to perform experiments on the apical cell surface, as in the case of the TCCD experiments described here and elsewhere (4).
FIGURE 1.
Nonspecific T cell binding to a glass surface leads to changes in TCR diffusion. FRAP analysis of Alexa Fluor 488-labeled TCR (CD3ϵ) is presented. Mean recovery curves are shown with data bounded by 95% confidence intervals at each point, with the apical surface in solid circles and basal in open circles for Jurkat cells [n = 20] (A), J45 cells [n = 32] (B) and J.CaM1.6 cells [n = 20] (C). The mean ± S.E. of asymptotic fluorescence recovery for each dataset are also shown.
The TCR Triggering Apparatus Is Composed of Monomeric and/or Monovalent Proteins
TCR
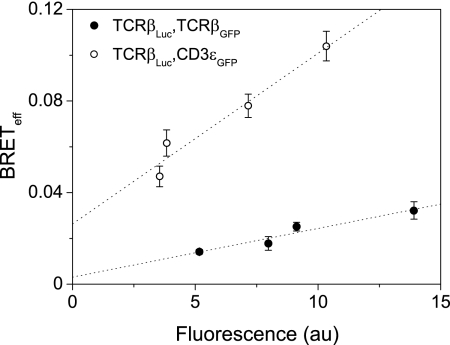
We have previously shown that the murine TCR complex is monovalent using TCCD analysis (4). Despite demonstrating that the antibody fragment used to detect the antigen receptor did not affect TCR triggering, the caveat still remained that Fab binding interfered with the basal stoichiometry of the complex. We used BRET-based analysis, which localizes the label to the cytoplasmic domain, to circumvent this possibility. The complete TCR complex cannot readily be transfected into cells, which prevents systematic variation of the acceptor/donor ratios required for conventional BRET analysis, necessitating an alternative approach. The J.RT3 cell line is a derivative of the Jurkat T cell that lacks TCRβ expression, resulting in the receptor being absent from the cell surface (22). Using viral gene transduction, we stably expressed native TCRβ fused to luciferase (TCRβLuc), producing the J.Luc cell line, which restores expression of the TCR (supplemental Fig. 1). We reasoned that if the TCR complex was multivalent then more than one TCRβ chain must be present in each complex. Therefore, by co-expressing a GFP-tagged version of TCRβ (TCRβGFP) in the J.Luc cells, we would expect to see significant energy transfer between the two fluorophores (analogous to “type 2” BRET experiments) (17).
Cells expressing both TCRβLuc and TCRβGFP were sorted into four subpopulations with increasing GFP. The assay was then carried out on these sublines and the BRETeff values plotted against fluorescence (Fig. 2). The values were low and tended to zero when extrapolated to minimal levels of GFP expression, consistent with the observed energy transfer being the result of random interactions within the membrane rather than oligomerization of the TCRβ chains. To confirm this, CD3ϵGFP was expressed as the acceptor in J.Luc cells. At the low levels of expression sorted for in each population, it is very unlikely that both chains in the minimal complex were GFP-tagged, because CD3ϵGFP must compete with native CD3ϵ for incorporation into the TCR. Because CD3ϵ is an obligate component of the minimal receptor complex, we expected these constructs to define the maximal energy transfer between TCR chains within the complex. BRETeff for CD3ϵGFP-expressing J.Luc cells was indeed substantially higher than that measured for J.Luc cells expressing TCRβGFP and did not extrapolate to zero at low expression (Fig. 2). In BRET experiments, it is not possible to distinguish between surface and intracellular signals, but our assumption is that the contribution of the intracellular signals should be equivalent between the TCRβLuc/TCRβGFP and the TCRβLuc/CD3ϵGFP experiments. We conclude from these experiments that the TCR contains only a single TCRβ chain, or that only a fraction of the minimal complexes form oligomers. The ratio between the TCR/TCR and TCR/CD3ϵ datasets of BRETeff extrapolated to zero expression gives an estimate of this fraction, which would correspond to 0.10 ± 0.11. Oligomer formation is, however, very unlikely given the results of the TCCD analysis of the murine TCR (4).
FIGURE 2.
The Jurkat TCR complex contains only one β-chain. Jurkat cells co-expressing TCRβ fused to luciferase or GFP show low energy transfer, as detected by BRET (filled circles). Expression of CD3ϵGFP with TCRβLuc gave significant BRET that does not extrapolate to zero at low acceptor levels, as expected for a specific association (open circles). GFP fluorescence was measured by flow cytometry.
MHC Class II
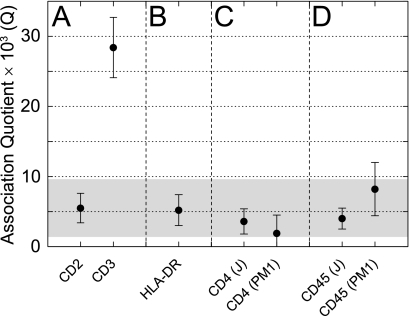
A monovalent TCR is likely to bind a monovalent ligand. Because the correct expression of MHC molecules is intimately linked with the process of peptide loading, heterologous expression of the heterodimer is unlikely to mimic the physiological assembly or localization of the protein, making TCCD the most appropriate assay for MHC oligomerization. To define the dynamic range of the technique for human cells and to establish a positive control for our subsequent analyses, we first confirmed that we could detect two CD3ϵ chains within each TCR complex of the PM1 human T cell line (a HUT78 derivative; Fig. 3A). We then used the adhesion receptor, CD2, which is known to be monomeric (17), to define the value of Q expected for nonassociated plasma membrane proteins. Human CD2 expressed by PM1 cells gave a nonzero value for Q that was significantly lower than that obtained for CD3ϵ (Fig. 3A) and equivalent to that observed previously for monomeric control proteins (4). This demonstrated that monomers and dimers present on human cells are readily distinguishable.
FIGURE 3.
The endogenous membrane proteins of the triggering apparatus are monomeric at the cell surface. A, Fabs against the known monomer CD2 define the expected Q value from TCCD for noninteracting proteins within the membrane. The presence of two CD3ϵ chains per TCR complex is confirmed by TCCD and constitutes the positive control. B, HLA-DR expressed on THP-1 monocyte cells showed no evidence for a dimer of dimers. C, the CD4 co-receptor on two independent T cell lines is monomeric by TCCD. D, CD45 is also shown to be monomeric on both of these cell types. The shaded part of the graph is a 95% confidence interval based on the CD2 dataset that defines the most likely Q values for monomeric proteins.
The monocyte line THP-1 was used to determine the stoichiometry of the MHC class II molecule, HLA-DR. Generally, MHC expression on antigen-presenting cells is extremely high and outside the regime of single-molecule detection required for TCCD. However, THP-1 cells express a low level of HLA-DR prior to differentiation to macrophages (23), allowing TCCD analysis. We found no evidence for the formation of dimers of dimers at the antigen-presenting cell surface (Fig. 3B), with the level of association consistent with HLA-DR, and presumably all MHC class II molecules being monovalent as in the case of their class I counterparts (13).
CD4/Lck
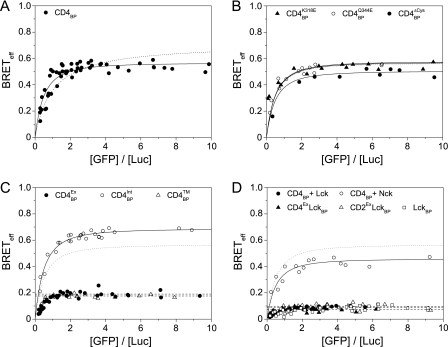
We used both BRET and TCCD to determine whether CD4 oligomerizes at the T cell surface. Full-length CD4 was genetically fused to either Luc or GFP and expressed as a “BRET pair” in HEK-293T cells at physiologically meaningful levels (i.e. in the range of 103-105 molecules/cell; supplemental Fig. 2), as described previously (17). The BRETeff values measured for CD4 were high and dependent on the acceptor/donor ratio, indicative of an oligomeric interaction (Fig. 4A). BRETeff for CD4 was comparable with that for CD80, a membrane protein where the monomeric and dimeric forms are in equilibrium (17). Because the dissociation constant for CD80 homodimerization is ∼ 35 μm (24), it suggested factors other than the putative membrane-proximal domain interaction (Kd > 1 mm) (7) were likely to contribute to the CD4 association. Furthermore, the acceptor/donor ratio dependence of the BRETeff values could be readily fitted as an oligomeric association (n ≥ 3) but not as a dimeric one (Fig. 4A). To test whether the membrane-proximal domain interaction seen in sCD4 crystals (7) was responsible for the oligomerization observed, we mutated the two residues, CD4K318E and CD4Q344E, believed to disrupt it (25). These mutations had no effect on BRETeff (Fig. 4B), implying that CD4 does not oligomerize via the interface observed in the crystals. We also excluded the unpaired cysteines in the intracellular region by mutating them to serine (CD4ΔCys) and repeating the assay (Fig. 4B).
FIGURE 4.
CD4 co-receptor is monomeric in the presence of Lck kinase. A, expression of CD4 as a BRET pair in HEK-293T cells shows oligomerization (fitted solid line) that does not fit as well to a dimer model (dotted line). B, mutation of residues thought to disrupt CD4 oligomerization or of the intracellular cysteines had a negligible effect on BRETeff values; the fit to wild type CD4 (from A) is shown for comparison in this and all subsequent panels (dotted line). C, truncations of the co-receptor isolated the intracellular region of CD4 as the driving force for oligomerization. D, co-expression of Lck with CD4 leads to the complete and specific disruption of oligomerization to levels equivalent to the monomeric controls and Lck itself.
To identify the region of CD4 causing the observed oligomerization, a series of truncations of the protein was generated: CD4Ex lacked the intracellular region, CD4Int lacked the extracellular region, and CD4TM consisted only of the transmembrane segment of the protein. The CD4Ex and CD4TM constructs gave BRETeff profiles that were indistinguishable from monomeric proteins (17), indicating that the self-association of CD4 is driven solely by its intracellular region (Fig. 4C). What role could this region have for CD4 oligomerization? Unlike many type I membrane proteins, this region has defined tertiary structure when bound to Lck, including an amphipathic α-helix (26) and a cysteine-utilizing Zn2+ ion “clasp” (27). Because we had already excluded a role for the cysteine residues (Fig. 4B), the amphipathic α-helix seemed the most likely candidate. Co-expression of Lck with the CD4 BRET pair essentially abolished oligomerization (Fig. 4D), consistent with the helix driving assembly of the oligomers. A control protein, myristoylated Nck1 had no effect, further implicating specific helical interactions (Fig. 4D). The low BRETeff observed in these experiments is due to the co-transfection of the unlabeled proteins; the acceptor/donor ratio dependence of the data is unaffected, as expected. A chimeric molecule consisting of CD4Ex fused to Lck (CD4ExLck), which has previously been shown to restore T cell function in CD4−/− mice (28), yielded BRET data characteristic of monomers that were indistinguishable from those for an analogous CD2ExLck negative control (Fig. 4D). Lck expressed as a BRET pair in the absence of CD4 exhibited no evidence of oligomerization, as expected, and was essentially equivalent to CD4ExLck (Fig. 4D). In TCCD experiments on endogenous CD4 using both Jurkat and PM1 T cell lines, the Q values obtained were equivalent to that obtained for the monomeric control, CD2 (Fig. 3C). This strongly suggests that CD4 is invariably bound to Lck at the cell surface.
CD45
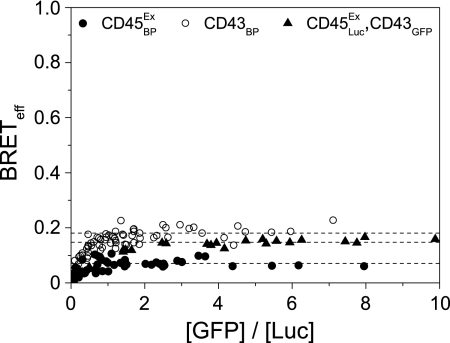
In TCCD experiments with a pan-CD45 Fab, we found no evidence of CD45 oligomerization on either PM1 or Jurkat T cells (Fig. 3D). Both of these cell lines predominantly express CD45RO (29, 30), the isoform proposed previously to dimerize (10, 31). Because the dimerization of this, the smallest of the isoforms, could have been obfuscated by the larger isoforms, we used BRET to determine whether the extracellular domain of CD45RO self-associates by truncating the protein sequence at the start of the cytoplasmic region, removing the phosphatase domains (giving CD45Ex). This also minimized the possibility that these large domains could impede the association of GFP and Luc fluorophores, as observed for multipass transmembrane proteins (17). CD45 is very difficult to express but by adding a histidine tag to the N terminus of the mature protein, it was possible to transiently express CD45 to the levels observed on T cell lines (supplemental Fig. 3). The BRETeff values obtained for CD45Ex were very low and showed no dependence on the acceptor/donor ratio (Fig. 5). To demonstrate that the high level of glycosylation of CD45 was not affecting the analysis, the similarly large and heavily glycosylated protein, CD43, was examined. CD43 exhibited the same behavior as known monomers and gave BRETeff values higher than that for CD45 (Fig. 5). Expressing CD45ExLuc and CD43GFP as a BRET pair gave values intermediate between the two (Fig. 5), suggesting that the lower BRETeff values of CD45Ex may result from a larger hydrodynamic radius. Full-length CD45RO BRET constructs gave BRETeff values even lower than those observed for CD45Ex (data not shown). These observations suggest that, in resting cells, CD45 is very unlikely to be regulated via dimeric associations of its extracellular region.
FIGURE 5.
The extracellular domain of CD45RO is monomeric. The BRET assay was used to determine the level of CD45 self-association in HEK-293T cells. No specific oligomerization of the phosphatase could be detected compared with appropriate controls. All data were fitted as monomers (dashed lines).
Within-membrane Interactions Are Greatly Enhanced Compared with Those with Cytoplasmic Proteins
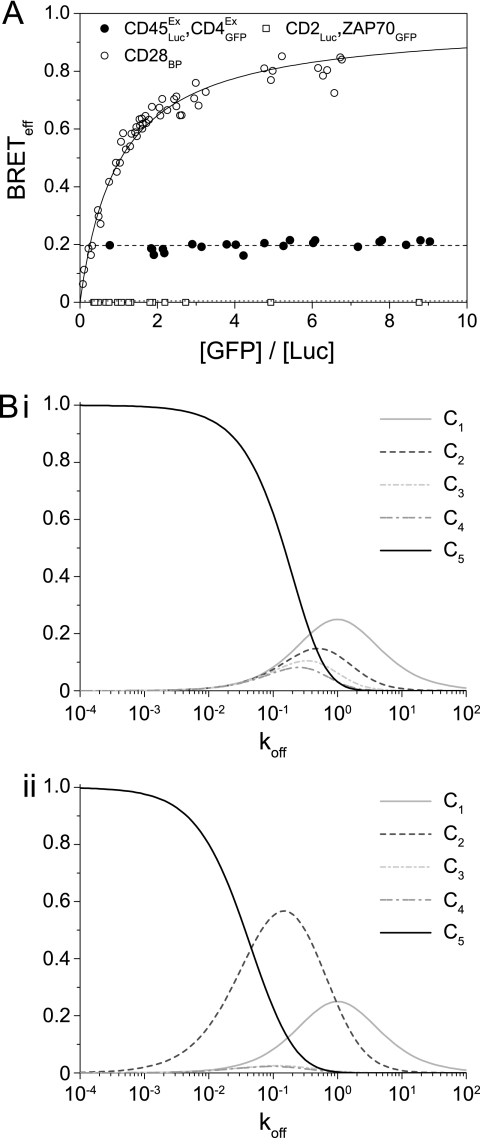
As noted previously, even randomly interacting monomeric membrane proteins such as CD2 and CD86 give very significant nonzero BRETeff values (17). Illustrating the effect for proteins of interest in the context of receptor triggering, the values obtained for the CD45ExLuc/CD4ExGFP BRET pair were a considerable fraction of that measured for the homodimer CD28 (Fig. 6A). TCCD analysis of the interaction of the native proteins gave Q values similar to those for monomer control proteins (Q = 5.7 ± 2.9), arguing against the specificity of interaction and functional association between these proteins that was claimed previously (31). Given that BRET efficiency is a measure of proximity, this significant “background” energy transfer can be explained if membrane confinement effectively concentrates proteins, recasting the plasma membrane as an organelle that facilitates molecular interactions. To examine the scale of this concentrating effect we determined the efficiency of energy transfer when membrane-bound CD2Luc is co-expressed with a cytosolic GFP-tagged protein, ZAP70 (1), and found it to be undetectable (Fig. 6A). Membrane confinement by acylation has similar effects (32), which are very unlikely to be the result of anisotropic effects from cell surface localization (33). To determine whether this concentrating effect would have implications for the rate of reaction of differentially located proteins, we ran a stochastic simulation using SmolDyn (34), which was evaluated at length scales on the order of nanometers. The rate that molecules either diffusing in solution, or confined to a cell surface, reacted with a membrane receptor was used to estimate the advantage of membrane localization. We found the rate to be higher for membrane localization at all concentrations and that it increased disproportionately within the range of cellular protein expression (supplemental Fig. 4).
FIGURE 6.
Interactions within the membrane are highly favored compared with those with molecules from the cytosol. A, the BRETeff values for a CD45/CD4 pair are nonspecific but significant when compared with those of the covalent homodimer CD28. Co-expression of the monomer CD2 with cytosolic GFP-tagged ZAP70 gives undetectable energy transfer. B, data from the molecular simulation are shown, with the relative proportions of the various complexes [C] with the original (i) or revised (ii) KP model, as the koff for the TCR/pMHC interaction is varied.
Implications for Differential Signaling
To explore the likely implications of differences in reaction rates for membrane versus cytosolic proteins, we incorporated these findings into the Kinetic Proofreading (KP) model as applied to antigen receptors (35). The original KP model explains how apparently small changes in the strength of pMHC binding to the TCR could lead to appropriate activation only in the presence of non-self-antigen. The model proposed that serial TCR-modifying events of equal duration could compound the relatively small differences in koff values of different peptides (in complex with MHC) into large differences in formation of signaling-competent TCR complexes. Based on our observations, we introduced a “slow” step in the serial pathway to simulate ZAP70 recruitment and activation at the phosphorylated antigen receptor (supplemental Fig. 5). We then compared the outcomes of the original and revised models based on their response to changes in koff value of the TCR/pMHC interaction. For the original model, variation of koff shifted the majority of TCR-pMHC complexes [C] from the inactive [C1] to the fully active [C5] state in effectively a single step (Fig. 6Bi). In marked contrast, the revised version shows an accumulation of an intermediate state [C2] at koff values ranging from 1 to 0.01 (Fig. 6Bii). The length of time required to form [C3] meant that many TCR-pMHC complexes dissociate before becoming the fully modified (and therefore activated) form and suggests that ZAP70 recruitment and activation would be intrinsically dependent on the strength of the complex. Accumulation of [C2] occurs even when the slow step is only 2-fold lower than the other modification steps.
DISCUSSION
We set out to establish the “ground state” of the TCR triggering apparatus using a pair of complementary, minimally perturbative in situ methods for characterizing the structural organization of cell surface proteins. Although BRET requires heterologous protein expression, the luminescence detection mode allows molecular interactions to be studied at near native expression levels (17) so that the observations are relevant to the behavior of the native proteins, which can be studied directly using the TCCD method (4). The TCCD measurements presented here show with 95% confidence that all analyzed proteins are monomeric. When combined with the orthogonal BRET method for which, in the case of the analysis of the TCR, the upper limit on the fraction of oligomers is estimated to be 10%, these two techniques allow us to be confident that these assignments represent the state of the proteins at the cell surface.
Critically, our working definition of the ground state of the plasma membrane ex vivo, i.e. as one that is not in close apposition to a rigid surface, likely provides the clearest view of the resting organization of the cell surface. Exactly how surface contact leads to the apparently TCR triggering-dependent changes in receptor diffusion detected in our FRAP experiments remains to be determined. We suggest that the TCR preclusters observed at a cell/glass interface by Lillemeier et al. likely correspond to very early precursors of the microclusters observed by others (36, 37). Whether or not the TCR is preclustered in this way in vivo because of contacts with the surfaces of other cells also remains to be established. What does seem clear is that the TCR does not spontaneously oligomerize. Although we have not directly tested the diffusive mobility of the proteins studied here besides the TCR, they have been shown elsewhere to have insignificant immobile fractions, and for CD45 changes in mobility have been linked to cellular activation (38).
The data indicate that exclusively monomeric or monovalent membrane proteins are likely to drive receptor triggering in T cells, offering a simplified structural framework for understanding receptor triggering. A monovalent TCR complex and pMHC restricts the types of models able to account for the initial steps of TCR triggering. At least for the initial contact between antigen receptor and ligand, these entities likely form a 1:1 stoichiometric complex. We have also used single-molecule tracking techniques to characterize individual TCR complexes at the cell surface, which also suggested that the TCR is monovalent (14). TCR triggering is thus likely to be dependent only on the kinetics of the TCR/pMHC interaction, where downstream signaling is a consequence of extended complex formation. We acknowledge, as we have previously (4), that others (3) have observed apparently multivalent forms of the TCR on paraformaldehyde-fixed cells labeled with antibodies and polyvalent gold beads. We note, however, that even very high levels of chemical fixatives cannot be relied upon to prevent antibody-dependent receptor aggregation following fixation (39).
Our BRET and TCCD analyses imply that CD4 is also monovalent when complexed with Lck and that there is very little if any free CD4 capable of oligomerizing when Lck is present. CD4 is almost undetectable at the surface of Lck-deficient Jurkat cells, but appears upon exogenous expression of the kinase (40). If the co-receptor must associate with Lck to reach the location where it is active, it seems likely that CD4 is only ever functionally monovalent. The putative oligomerization motif seems to reside in the intracellular amphipathic α-helix, which is known to interact with the HIV-1 proteins Vpu (41) and Nef (42). The finding that the K318E and Q344E mutations interfere with T cell activation suggests that the D4 domain of CD4 may nevertheless be functionally important. One possibility is that D4 associates with the TCR complex during MHC class II engagement (43).
Although the notion that the intracellular domains of CD45 and CD45-related receptor-type protein phosphatases dimerize is gradually losing favor (44, 45), there has been much interest in the possibility that the activity of these molecules is modulated by the oligomerization state of their extracellular regions. This has largely been driven by attempts to explain CD45 isoform diversity, where dimerization is thought to be most prevalent for the smaller forms of the protein (10). We find, however, that even the smallest isoform has no propensity to dimerize, regardless of how we test for it. The simplest view is that, in resting cells, CD45 is a highly abundant, constitutively active but largely nonspecific phosphatase that both counteracts and modulates the activities of kinases present at the cell surface (46).
Although its constituent components are relatively simple, the configuration of the receptor triggering apparatus nevertheless permits multiple signaling outcomes. An unexpected observation that emerged from our BRET experiments is the extent to which membrane confinement of proteins increased their effective concentration compared with that of cytosolic proteins. As noted previously (47), for reaction-limited interactions the protein concentrating effects of membrane confinement most likely translate into enhanced association rates, by as much as 100–1000-fold. Such concentrating effects might be further compounded by the “corralling” action of the actin cytoskeleton (14, 48). We believe that these differences in reaction rates at the membrane could have quantifiable effects on TCR triggering. Of the initial mediators of TCR signaling, almost all, including Lck and LAT, are membrane-bound, and only the essential Syk-family kinase, ZAP70, is cytosolic. Our reconfiguration of the KP model with a slower step suggests that the reduced rate of accumulation of cytoplasmic effectors could make ZAP70 recruitment to the phosphorylated TCR complex and its subsequent activation a rate-limiting step in receptor signaling that may potentiate a minimally triggered antigen receptor.
Is there any evidence that the triggering apparatus may function in this way? ZAP70 recruitment is relatively slow and delayed (49), and a membrane-bound form of the kinase (in the presence of Lck) causes activation in the absence of stimulation (50). Similarly, the artificially induced, transient recruitment of ZAP70 to the cell surface is sufficient to activate T cells (51). The intermediate state described in the revised KP model might correspond to one induced by altered peptide ligands. These low affinity TCR ligands induce anergic T cell responses where ZAP70 is not activated (52, 53), because incomplete TCR triggering primarily yields the monophosphorylated pp21 form of CD3ζ that is incapable of recruiting ZAP70 (54). The monophosphorylated immunoreceptor tyrosine-based activation motifs may allow other, presumably membrane-associated proteins to initiate distinct signals leading to anergy, as recently observed in B cells (55). Exactly how such effects are translated into distinct functional outcomes remains to be elucidated.
Supplementary Material
Acknowledgments
We thank Professor Anton van der Merwe for a critical reading of the manuscript and Kristina Ganzinger for help with the FRAP analysis. The reagent ARP318 was obtained from the Programme EVA Centre for AIDS Reagents, National Institute for Biological Standards and Control United Kingdom, and was donated by Dr. Q Sattentau.
This work was supported by the Wellcome Trust, the Biotechnology and Biological Sciences and Research Council, the United Kingdom Medical Research Council, and the Fundação para a Ciência e a Tecnologia of Portugal.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and Experimental Procedures.
- TCR
- T cell antigen receptor
- BRET
- bioluminescence resonance energy transfer
- FRAP
- fluorescence recovery after photobleaching
- KP
- kinetic proofreading
- Luc
- luciferase
- pMHC
- peptide-major histocompatibility complex
- TCCD
- two-color coincidence detection.
REFERENCES
- 1. Smith-Garvin J. E., Koretzky G. A., Jordan M. S. (2009) Annu. Rev. Immunol. 27, 591–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Salzmann M., Bachmann M. F. (1998) Mol. Immunol. 35, 271–277 [DOI] [PubMed] [Google Scholar]
- 3. Schamel W. W., Arechaga I., Risueño R. M., van Santen H. M., Cabezas P., Risco C., Valpuesta J. M., Alarcón B. (2005) J. Exp. Med. 202, 493–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. James J. R., White S. S., Clarke R. W., Johansen A. M., Dunne P. D., Sleep D. L., Fitzgerald W. J., Davis S. J., Klenerman D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 17662–17667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lillemeier B. F., Mörtelmaier M. A., Forstner M. B., Huppa J. B., Groves J. T., Davis M. M. (2010) Nat. Immunol. 11, 90–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Davis S. J., Ward H. A., Puklavec M. J., Willis A. C., Williams A. F., Barclay A. N. (1990) J. Biol. Chem. 265, 10410–10418 [PubMed] [Google Scholar]
- 7. Wu H., Kwong P. D., Hendrickson W. A. (1997) Nature 387, 527–530 [DOI] [PubMed] [Google Scholar]
- 8. Bilwes A. M., den Hertog J., Hunter T., Noel J. P. (1996) Nature 382, 555–559 [DOI] [PubMed] [Google Scholar]
- 9. Hoffmann K. M., Tonks N. K., Barford D. (1997) J. Biol. Chem. 272, 27505–27508 [DOI] [PubMed] [Google Scholar]
- 10. Xu Z., Weiss A. (2002) Nat. Immunol. 3, 764–771 [DOI] [PubMed] [Google Scholar]
- 11. Brown J. H., Jardetzky T. S., Gorga J. C., Stern L. J., Urban R. G., Strominger J. L., Wiley D. C. (1993) Nature 364, 33–39 [DOI] [PubMed] [Google Scholar]
- 12. Hayball J. D., Lake R. A. (2005) Mol. Cell. Biochem. 273, 1–9 [DOI] [PubMed] [Google Scholar]
- 13. Choudhuri K., van der Merwe P. A. (2007) Semin. Immunol. 19, 255–261 [DOI] [PubMed] [Google Scholar]
- 14. Dunne P. D., Fernandes R. A., McColl J., Yoon J. W., James J. R., Davis S. J., Klenerman D. (2009) Biophys. J. 97, L5–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ayoub N., Jeyasekharan A. D., Bernal J. A., Venkitaraman A. R. (2008) Nature 453, 682–686 [DOI] [PubMed] [Google Scholar]
- 16. Dushek O., Coombs D. (2008) J. Biochem. Biophys. Methods 70, 1224–1231 [DOI] [PubMed] [Google Scholar]
- 17. James J. R., Oliveira M. I., Carmo A. M., Iaboni A., Davis S. J. (2006) Nat. Methods 3, 1001–1006 [DOI] [PubMed] [Google Scholar]
- 18. Clarke R. W., Orte A., Klenerman D. (2007) Anal. Chem. 79, 2771–2777 [DOI] [PubMed] [Google Scholar]
- 19. Dushek O., Mueller S., Soubies S., Depoil D., Caramalho I., Coombs D., Valitutti S. (2008) PLoS One 3, e3913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koretzky G. A., Picus J., Schultz T., Weiss A. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 2037–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Straus D. B., Weiss A. (1992) Cell 70, 585–593 [DOI] [PubMed] [Google Scholar]
- 22. Ohashi P. S., Mak T. W., Van den Elsen P., Yanagi Y., Yoshikai Y., Calman A. F., Terhorst C., Stobo J. D., Weiss A. (1985) Nature 316, 606–609 [DOI] [PubMed] [Google Scholar]
- 23. Yunis J. J., Band H., Bonneville F., Yunis E. J. (1989) Blood 73, 931–937 [PubMed] [Google Scholar]
- 24. Ikemizu S., Gilbert R. J., Fennelly J. A., Collins A. V., Harlos K., Jones E. Y., Stuart D. I., Davis S. J. (2000) Immunity 12, 51–60 [DOI] [PubMed] [Google Scholar]
- 25. Moldovan M. C., Yachou A., Lévesque K., Wu H., Hendrickson W. A., Cohen E. A., Sékaly R. P. (2002) J. Immunol. 169, 6261–6268 [DOI] [PubMed] [Google Scholar]
- 26. Wray V., Mertins D., Kiess M., Henklein P., Trowitzsch-Kienast W., Schubert U. (1998) Biochemistry 37, 8527–8538 [DOI] [PubMed] [Google Scholar]
- 27. Kim P. W., Sun Z. Y., Blacklow S. C., Wagner G., Eck M. J. (2003) Science 301, 1725–1728 [DOI] [PubMed] [Google Scholar]
- 28. Glaichenhaus N., Shastri N., Littman D. R., Turner J. M. (1991) Cell 64, 511–520 [DOI] [PubMed] [Google Scholar]
- 29. Lusso P., Cocchi F., Balotta C., Markham P. D., Louie A., Farci P., Pal R., Gallo R. C., Reitz M. S., Jr. (1995) J. Virol. 69, 3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rothstein D. M., Saito H., Streuli M., Schlossman S. F., Morimoto C. (1992) J. Biol. Chem. 267, 7139–7147 [PubMed] [Google Scholar]
- 31. Dornan S., Sebestyen Z., Gamble J., Nagy P., Bodnar A., Alldridge L., Doe S., Holmes N., Goff L. K., Beverley P., Szollosi J., Alexander D. R. (2002) J. Biol. Chem. 277, 1912–1918 [DOI] [PubMed] [Google Scholar]
- 32. Tran T. M., Jorgensen R., Clark R. B. (2007) Biochemistry 46, 14438–14449 [DOI] [PubMed] [Google Scholar]
- 33. Corry B., Jayatilaka D., Martinac B., Rigby P. (2006) Biophys. J. 91, 1032–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Andrews S. S., Bray D. (2004) Phys. Biol. 1, 137–151 [DOI] [PubMed] [Google Scholar]
- 35. McKeithan T. W. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 5042–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Varma R., Campi G., Yokosuka T., Saito T., Dustin M. L. (2006) Immunity 25, 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yokosuka T., Sakata-Sogawa K., Kobayashi W., Hiroshima M., Hashimoto-Tane A., Tokunaga M., Dustin M. L., Saito T. (2005) Nat. Immunol. 6, 1253–1262 [DOI] [PubMed] [Google Scholar]
- 38. Cairo C. W., Das R., Albohy A., Baca Q. J., Pradhan D., Morrow J. S., Coombs D., Golan D. E. (2010) J. Biol. Chem. 285, 11392–11401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tanaka K. A., Suzuki K. G., Shirai Y. M., Shibutani S. T., Miyahara M. S., Tsuboi H., Yahara M., Yoshimura A., Mayor S., Fujiwara T. K., Kusumi A. (2010) Nat. Methods 7, 865–866 [DOI] [PubMed] [Google Scholar]
- 40. Yousefi S., Ma X. Z., Singla R., Zhou Y. C., Sakac D., Bali M., Liu Y., Sahai B. M., Branch D. R. (2003) Clin. Exp. Immunol. 133, 78–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tiganos E., Yao X. J., Friborg J., Daniel N., Cohen E. A. (1997) J. Virol. 71, 4452–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gratton S., Yao X. J., Venkatesan S., Cohen E. A., Sékaly R. P. (1996) J. Immunol. 157, 3305–3311 [PubMed] [Google Scholar]
- 43. Vignali D. A., Vignali K. M. (1999) J. Immunol. 162, 1431–1439 [PubMed] [Google Scholar]
- 44. Barr A. J., Ugochukwu E., Lee W. H., King O. N., Filippakopoulos P., Alfano I., Savitsky P., Burgess-Brown N. A., Müller S., Knapp S. (2009) Cell 136, 352–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nam H. J., Poy F., Saito H., Frederick C. A. (2005) J. Exp. Med. 201, 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Davis S. J., van der Merwe P. A. (2006) Nat. Immunol. 7, 803–809 [DOI] [PubMed] [Google Scholar]
- 47. Haugh J. M., Lauffenburger D. A. (1997) Biophys. J. 72, 2014–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Douglass A. D., Vale R. D. (2005) Cell 121, 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sloan-Lancaster J., Presley J., Ellenberg J., Yamazaki T., Lippincott-Schwartz J., Samelson L. E. (1998) J. Cell Biol. 143, 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kolanus W., Romeo C., Seed B. (1993) Cell 74, 171–183 [DOI] [PubMed] [Google Scholar]
- 51. Graef I. A., Holsinger L. J., Diver S., Schreiber S. L., Crabtree G. R. (1997) EMBO J. 16, 5618–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Irie A., Chen Y. Z., Tsukamoto H., Jotsuka T., Masuda M., Nishimura Y. (2003) Eur. J. Immunol. 33, 1497–1507 [DOI] [PubMed] [Google Scholar]
- 53. Sloan-Lancaster J., Shaw A. S., Rothbard J. B., Allen P. M. (1994) Cell 79, 913–922 [DOI] [PubMed] [Google Scholar]
- 54. Deindl S., Kadlecek T. A., Brdicka T., Cao X., Weiss A., Kuriyan J. (2007) Cell 129, 735–746 [DOI] [PubMed] [Google Scholar]
- 55. Cambier J. C., Getahun A. (2010) Immunol. Lett. 128, 6–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.