Abstract
This study explores the changes in expression of microRNA (miRNA) and related genes under simulated microgravity conditions. In comparison with static 1 × g, microgravity has been shown to alter global gene expression patterns and protein levels in cultured cells or animals. miRNA has recently emerged as an important regulator of gene expression, possibly regulating as many as one-third of all human genes. However, very little is known about the effect of altered gravity on miRNA expression. To test the hypothesis that the miRNA expression profile would be altered in zero gravity resulting in altered regulation of gene expression leading to metabolic or functional changes in cells, we cultured TK6 human lymphoblastoid cells in a high aspect ratio vessel (bioreactor) for 72 h either in the rotating condition to model microgravity in space or in the static condition as a control. Expression of several miRNAs was changed significantly in the simulated microgravity condition including miR-150, miR-34a, miR-423-5p, miR-22, miR-141, miR-618, and miR-222. To confirm whether this altered miRNA expression correlates with gene expression and functional changes of the cells, we performed DNA microarray and validated the related genes using quantitative RT-PCR. Expression of several transcription factors including EGR2, ETS1, and c-REL was altered in simulated microgravity conditions. Taken together, the results reported here indicate that simulated microgravity alters the expression of miRNAs and genes in TK6 cells. To our knowledge, this study is the first to report the effects of simulated microgravity on the expression of miRNA and related genes.
Keywords: Biophysics, Cell Biology, Gene Expression, Gene Regulation, MicroRNA, Gene Regulation, Gene Expressions, PCR Array, Simulated Microgravity
Introduction
Living organisms on the surface of the Earth will undoubtedly experience a shock and subsequent adaptation when the gravity environment changes from Earth gravity (1 × g) to microgravity in space. Such changes have already been documented for biological consequences ranging from microbial growth to immune functions in astronauts. Spaceflight provides an isolated environment where microgravity, stress, and radiation can influence immune function and lead to a higher incidence of medical conditions including formation of renal stones (1), impaired autonomic cardiovascular control (2), reduced cellular immune function (3), and bone loss (4). The altered expression of genes that control important cellular functions may also increase the risk for cancer (5). Gene expression profile changes in space have not only been observed in mammalian cells (6, 7) but also in bacterial and other living organisms (8). Microarray analysis of space-flown W138 human fibroblasts showed a modification in expression of tumor necrosis factor (TNF) and interleukin (IL)-related gene families, which are involved in either the regulation of bone density and as such the development of spaceflight osteopenia or in the development of proinflammatory status (9).
To expand our knowledge of life in space and also for the safety of astronauts during spaceflight, it is critical to understand the cellular responses to altered gravity at the molecular level. Molecular changes at the gene level such as gene and protein expressions may compromise cell function and present undesirable effects at the tissue level. Microgravity may influence specific repair pathways for DNA damage induced by elevated levels of oxidative stress during spaceflight or by exposures to space radiation. Recent advancement in the field of molecular biology has revealed that a different class of RNA, the small non-coding miRNA,3 can have a broad effect on gene expression networks mainly by inhibiting the translation process.
miRNAs represent a class of single-stranded non-coding regulatory RNA molecules (∼22 nucleotides) that control gene expressions by inhibiting the translation of mRNA to proteins. Any given miRNA may target up to a thousand different genes, each of which harbors several different miRNA response elements (10). miRNAs play an important role in various cellular processes including development, differentiation, cell growth, morphogenesis, apoptosis, and neurologic disorders (11, 12). Among human diseases, it has been shown that miRNAs are either overexpressed or suppressed in cancer, suggesting that they may play a role as a novel class of oncogenes or tumor suppressor genes. miRNAs are known to regulate gene expressions and play a key role in cellular responses to stress. Although the role of miRNAs in the cellular response to environmental stresses such as radiation exposure (13) has been demonstrated, no reports have been published on expression alterations of miRNA in a simulated microgravity environment. The overall goal of our investigations is to study the effects of simulated microgravity on the expression of miRNA as well as related genes using bioreactors. In the present study, we investigated changes in the miRNA expression profiles of human lymphoblastoid TK6 cells cultured in NASA-developed HARV bioreactors.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Specific primers for Cyclin E2 (CCNE2), high mobility gene A2 (HMGA2), early growth response 2 (EGR2/Krox20), zinc finger protein 145 (ZNF-145), ubiquitin C (UBQTN C), v-ets erythroblastosis virus E26 oncogene homolog 1 (ETS1), c-REL, β-ACTIN, GAPDH; reagents for cDNA synthesis and polymerase chain reaction (PCR); and miRNA isolation and miRNA PCR array kits were purchased from Qiagen (Valencia, CA). Guava ViaCount reagent was purchased from Millipore (Billerica, CA), and TRIzol extraction reagent was from Invitrogen.
Cell Lines and Culture
TK6 human lymphoblastoid cells were purchased from ATCC (Manassas, VA) and maintained in the log phase of cell growth by culturing in RPMI 1640 medium supplemented with 15% fetal bovine serum and 1.0% penicillin/streptomycin (Invitrogen) at 37 °C in 5% CO2, 95% air. For ground-based simulation of microgravity, HARV rotating suspension culture bioreactors (Synthecon, Houston, TX) were used. Actively growing (96–98% viable) TK6 cells were seeded in the bioreactor and rotated at 12 rpm/min. In parallel, cells at the same cell densities were maintained in bioreactors in a normal gravity (static) condition as controls. The bioreactors were kept in an incubator at 37 °C in 5% CO2, 95% air during the culture period.
Cell Viability and Growth
At various time points (24, 48, and 72 h), a small aliquot was taken from both static and rotated cells to check the viability using both trypan blue staining and Guava ViaCount assay. For the trypan blue assay, cells that stained blue were counted as dead, and cell counts were performed using a hemocytometer. For the Guava ViaCount method, cells were stained with ViaCount reagent for 5 min, and viability was assessed by differential permeation of two DNA-binding dyes using a flow cytometer (Millipore). Cell growth was measured using a Beckman counter (Beckman Coulter, Brea, CA).
Total RNA and Small (Micro) RNA Isolation
After being cultured for 72 h in either the static or rotating condition, the total RNA was isolated from three biological replicates by TRIzol extraction (Invitrogen) as described by the manufacturer. The quality and purity of the RNA samples were assessed by an Agilent RNA 6000 Nano kit and an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). MicroRNA was isolated and enriched using an RT2 qPCR miRNA Isolation kit. Briefly, 40 μg of total RNA in 400 μl of RNase-free water was mixed with appropriate ethanol concentrations and passed through a special silica membrane spin column followed by washing and eluting the enriched microRNA. Enriched microRNA was then reverse transcribed into cDNA using an RT2 miRNA First Strand kit (Qiagen).
miRNA Expression PCR Array Analysis
miRNA expression analysis of 352 miRNAs from both static and bioreactor-cultured cells was performed using an RT2 miRNA PCR Array kit (Qiagen) and a Bio-Rad CFX96 real time PCR instrument. The -fold change of miRNA expression was calculated using the provided Excel-based PCR array data analysis program (Qiagen).
RNA Labeling and Microarray Hybridization
Illumina HumanWG-6 V4 BeadChip (Illumina, Inc.) human whole-genome expression arrays were used in this study. Each RNA sample was amplified using the Ambion TotalPrep RNA Amplification kit with biotin-UTP (Enzo) labeling. The Ambion Illumina RNA Amplification kit uses T7 oligo(dT) primer to generate single-stranded cDNA followed by a second strand synthesis to generate double-stranded cDNA, which is then column-purified. In vitro transcription was done to synthesize biotin-labeled cRNA using T7 RNA polymerase. The cRNA was then column-purified. The cRNA was then checked for size and yield using the Bio-Rad Experion system. 1.5 μg of cRNA was hybridized for each array using standard Illumina protocols; streptavidin-Cy3 (GE Healthcare) was used for detection. Slides were scanned on an Illumina Beadstation.
Data Processing and Visualization
Expression values were extracted using BeadStudio v3.3. The data were background-subtracted using the MBCB algorithm (14) and quantile-normalized.
Target Gene Analysis
Seven microRNAs with 2-fold difference and ∼400 genes with substantial changes in the expression were integrated into GeneGo MetaCore pathway analysis software (St. Joseph, MI) to analyze the gene ontology, pathway distribution, and putative gene targets.
Validation of Selected Genes Using Quantitative PCR
cDNA from total RNA used in the miRNA PCR array analysis was synthesized using a SuperScript First-Strand kit (Invitrogen) according to the manufacturer's instructions. Gene expression was measured by quantitative RT-PCR using a SYBR Green kit (Qiagen) and a Bio-Rad CFX96 cycler. cDNA was subjected to PCR using specific primers according to the manufacturer's instructions. RT-negative reactions were run on each plate to confirm the absence of genomic DNA contamination. Relative expression values were calculated by ΔΔCt analysis using the average of two reference genes and normalized to control for -fold changes.
Statistical Analysis
The data from all the assays are presented as mean ± S.D. The statistical analysis was performed using Student's t test, and p < 0.05 was considered statistically significant.
RESULTS
Effect of Simulated Microgravity on Cell Growth and Viability
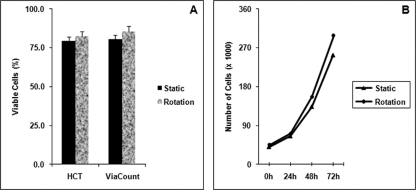
We first determined the effect of simulated microgravity on cell growth and viability every 24 h after the cells were seeded in bioreactors in either the rotating or static condition. In Fig. 1A, we present the cell viability as determined using trypan blue staining and Guava ViaCount methods. Both methods showed no significant differences in the percentage of viable cells between the two cell culture conditions after 72 -h culture in bioreactors. Cell growth was increased after 24 h of culture, and growth rates were similar between both culture conditions. Similar cell growth rates between the rotating and static culture conditions (Fig. 1B) allowed us to rule out the possibility of cell growth as a major contributing factor to the changes in the gene and miRNA expression profiles.
FIGURE 1.
Effect of simulated microgravity on cell viability and growth. TK6 lymphoblastoid cells were cultured in bioreactors either in the rotating (12 rpm/min) or static condition for 72 h at 37 °C. At different time points, cell viability (A) and growth (B) were analyzed using a hemocytometer (HCT), ViaCount staining, and a cell counter (mean ± S.D. from three independent experiments).
Simulated Microgravity Altered Expression of miRNAs
We next analyzed the effect of modeled microgravity on the miRNA expression profile in TK6 cells using an RT2 miRNA PCR Microarray kit. Among 352 miRNAs, the expression of some miRNAs was significantly altered under the simulated microgravity condition. We considered seven overexpressed miRNAs with significant p values (<0.05) as regulated under simulated microgravity condition. Of these, miR-150 (4.7-fold), miR-34a (2.8-fold), miR-423-5p (2.3-fold), miR-22 (2.1-fold), miR-141 (2.2-fold), miR-618 (1.9-fold), and miR-222 (1.9-fold) showed increased expression in the altered simulated microgravity condition (Table 1). Expression of these seven miRNAs in different types of human cancer is shown in Table 2. A list of the expression of 352 miRNAs analyzed is shown in supplemental Table 1.
TABLE 1.
List of altered miRNAs under simulated microgravity condition
miRNA PCR array data of rotated versus static cells are shown. The -fold change of miRNA expression from three independent experiments was calculated using the provided Excel-based PCR array data analysis program. hsa, Homo sapiens.
| miRNA | -Fold change | p value |
|---|---|---|
| hsa-miR-34a | 2.8 | 0.002 |
| hsa-miR-423-5p | 2.3 | 0.002 |
| hsa-miR-22 | 2.1 | 0.01 |
| hsa-miR-141 | 2.2 | 0.02 |
| hsa-miR-618 | 1.9 | 0.02 |
| hsa-miR-222 | 1.9 | 0.02 |
| hsa-miR-150 | 4.7 | 0.05 |
TABLE 2.
Expression of seven simulated microgravity-altered miRNAs in cancer
↑ = Upregulation ↓ = Downregulation.
The expression of altered miRNAs in different types of cancer was taken from miR2Disease, a manually curated database. A brief description of the miRNA-disease relationship, miRNA expression pattern in the disease state, detection method for miRNA expression, experimentally verified miRNA target gene(s), and literature references can be obtained from this database. NSCLC, non-small cell lung cancer; CLL, chronic lymphocytic leukemia; AML, acute myeloid leukemia; HNSCC, head and neck squamous cell carcinoma; EOC, epithelial ovarian cancer; hsa, H. sapiens.
| hsa-miR-34a | CLL (↓), colorectal cancer (↓), glioblastoma (↓), hepatocellular carcinoma (↓), malignant melanoma (↓), neuroblastoma (↓), pancreatic cancer (↓), NSCLC (↓), prostate cancer (↓) |
| hsa-miR-423-5p | Breast cancer (↑), promyelocytic leukemia (↑), gastric cancer (↑) |
| hsa-miR-22 | Acute lymphoblastic leukemia (↓), AML (↑), breast cancer (↑), HNSCC (↓), lung cancer (↓), prostate cancer (↓) |
| hsa-miR-141 | B-cell chronic lymphocytic leukemia (↑), breast cancer (↓), colorectal cancer (↓), EOC (↑), gastric cancer (↓), HNSCC (↓), lung cancer (↑), malignant melanoma (↓), prostate cancer (↓) |
| hsa-miR-618 | None |
| hsa-miR-222 | AML (↑), bladder cancer (↑), breast cancer (↑), gastric cancer (↑), glioblastoma (↑), glioma (↑), hepatocellular carcinoma (↑), malignant melanoma (↑), ovarian cancer (↑), pancreatic cancer (↑), prostate cancer (↓) |
| hsa-miR-150 | CLL (↓), EOC (↓), lung cancer (↑), pancreatic ductal adenocarcinomas (↑), pituitary adenocarcinomas (↑), diffuse large B-cell lymphoma (↓) |
Effect of Simulated Microgravity on Gene Expression
Because miRNAs are known to regulate gene expression, we further checked whether the alteration in expression of miRNAs correlates with the gene expression pattern by performing a DNA microarray. The results revealed that the expression of ∼400 genes was altered substantially with at least 2-fold changes and significant p values (<0.05) under simulated microgravity conditions (data not shown). Among these 400 altered genes, 205 were overexpressed or down-regulated in different types of cancer, and 12 genes including ABL1, AKT1, BCL2L1, BCR (breakpoint cluster region), CTNND1 (cadherin-associated protein), ETS1, LMO1 (rhombotin 1), PRLR (prolactin receptor), PTH (parathyroid hormone), REL, and SKIL (SKI-like oncogene) were oncogenes (supplemental Table 2).
Direct Interaction between Altered miRNA and Genes
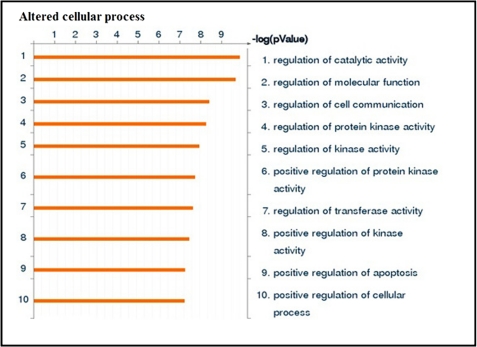
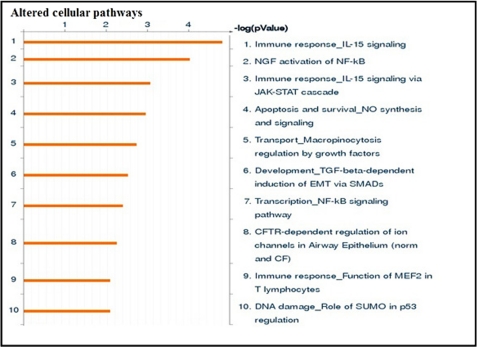
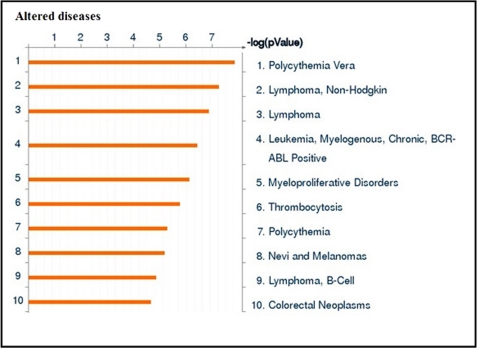
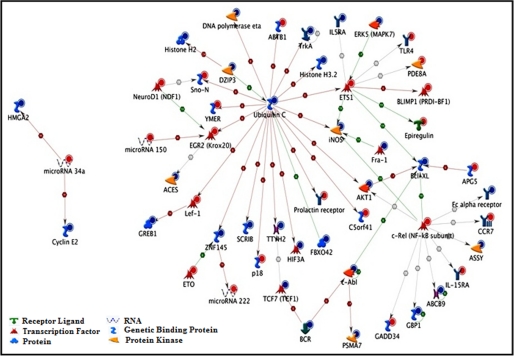
To identify the interaction between regulated miRNAs and genes under simulated microgravity, seven altered miRNAs and 400 genes were loaded into the GeneGo MetaCore pathway analysis database. The involvement of these gene targets in different signaling network and pathways and identification of shared gene ontologies (gene ontology processes) were explored using the systems biology tool MetaCore from GeneGo. Interestingly, the regulation of cell communication and catalytic activity, especially kinase activity; immune response_IL-15 signaling; and NGF activation of NF-κB pathways were significantly altered under the simulated microgravity condition. Diseases related to blood cells including leukemia and lymphoma were also altered under the simulated microgravity condition. The two ontology processes, top seven pathways, and diseases regulated by the altered miRNAs are shown in Figs. 2, 3, and 4. GeneGo MetaCore identified several projected networks with c-REL, ETS1, and UBQTN C as key factors. It also identified four genes, CCNE2, HMGA2, EGR2, and ZNF-145, that showed direct interaction with altered miRNAs (Fig. 5 and Table 3). These seven genes were selected for further validation.
FIGURE 2.
Altered cellular processes under simulated microgravity condition. Seven simulated microgravity-altered miRNAs and 400 genes were loaded into the GeneGo MetaCore analysis software. The altered cellular processes (gene ontology distribution) were ranked based upon significant p value. Bars represent the inverse log of the p value.
FIGURE 3.
Altered cellular pathways under simulated microgravity condition. Seven simulated microgravity-altered miRNAs and 400 genes were loaded into the GeneGo MetaCore analysis software. The altered cellular pathways were ranked based upon significant p value. Bars represent the inverse log of the p value. CF, cystic fibrosis; CFTR, cystic fibrosis transmembrane conductance regulator; norm, normal; SUMO, small ubiquitin-like modifier; EMT, epithelial-mesenchymal transition. SMADs, Caenorhabditis elegans protein SMA-mothers against decapentaplegic (MAD).
FIGURE 4.
Altered diseases under simulated microgravity condition. Seven simulated microgravity-altered miRNAs and 400 genes were loaded into the GeneGo MetaCore analysis software. The altered diseases were ranked based upon significant p value. Bars represent the inverse log of the p value. BCR, breakpoint cluster region.
FIGURE 5.
Direct interaction between simulated microgravity-altered miRNAs and genes. Seven simulated microgravity-altered miRNAs and 400 genes were integrated into GeneGo MetaCore, and the networks were computationally generated on the basis of the evidence stored in the database from published data. The intensity of the node (miRNA or gene) color (blue for down-regulation and red for up-regulation) indicates the expression levels. BCR, breakpoint cluster region. ETO, myeloid transforming gene 8; SCRIB, protein scribble homolog; ACES, angiotensin 1 converting enzyme; DZIP3, DAZ interacting protein 3.
TABLE 3.
List of GeneGo MetaCore-identified altered genes that showed direct interaction with altered miRNAs under simulated microgravity condition
Seven simulated microgravity-altered miRNAs and 400 genes were integrated into GeneGo MetaCore as mentioned in Fig. 5. Student's t test was performed with three independent experiments, and the -fold change value represents a mean value of three individual samples.
| Genes | Up/down | p value | -Fold changes | Related miRNA | Others |
|---|---|---|---|---|---|
| CCNE2 | Down | 0.008 | −2.2 | miR-34a | |
| HMGA2 | Down | 0.004 | −2.2 | miR-34a | |
| EGR2 (Krox20) | Up | 0.01 | 2.1 | miR-150 | |
| ZNF-145 | Down | 0.027 | −2.3 | miR-222 | |
| UBQTN C | Down | 0.025 | −2.3 | Key in direct interaction | |
| ETS1 | Up | 0.023 | 2.2 | Key in direct interaction | |
| c-REL | Up | 0.038 | 2.1 | Key in direct interaction |
Validation of Gene Expression Changes in Modeled Microgravity
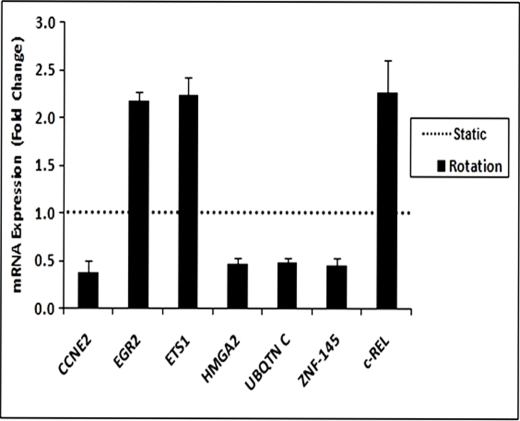
To further validate the effect of simulated microgravity on selected genes, we checked the expression of these genes in bioreactor-cultured TK6 cells using quantitative RT-PCR and specific primers (Table 4). The results showed that expression profiles of these genes were similar to those obtained from the GeneGo MetaCore pathway analysis database. The set of genes whose expression level was significantly altered relative to control static cells comprised CCNE2, HMGA2, EGR2, ZNF-145, UBQTN C, ETS1, and c-REL. Of these, EGR2 (2.2-fold), ETS1 (2.2-fold), and c-REL (2.3-fold) were up-regulated, and CCNE2 (−2.1-fold), HMGA2 (−2.0-fold), ZNF-145 (−2.0-fold), and UBQTN C (−2.0-fold) were down-regulated (Fig. 6).
TABLE 4.
Real time quantitative RT-PCR primer sequences
The list of primer sequences used in quantitative RT-PCR is shown. hsa, H. sapiens.
| Gene | Primer sequences |
|---|---|
| hsa-CCNE2 | 5′-GGGGATCAGTCCTTGCATTA-3′ (forward) |
| 5′-TCAGGCAAAGGTGAAGGATT-3′ (reverse) | |
| hsa-HMGA2 | 5′-GGCAGCAAAAACAAGAGTCC-3′ (forward) |
| 5′-TTGTGGCCATTTCCTAGGTC-3′ (forward) | |
| hsa-EGR2 | 5′-TTCGCCTGTGACTACTGTGG-3′ (forward) |
| 5′-CACTGCTTTTCCGCTCTTTC-3′ (reverse) | |
| hsa-ZNF-145 | 5′-CTATGGGCGAGAGGAGAGTG-3′ (forward) |
| 5′-ACGGAGTAGATGCCCAGATG-3′ (reverse) | |
| hsa-UBQTN C | 5′-CGCACCCTGTCTGACTACAA-3′ (forward) |
| 5′-GCCAGTGAGTGTCTTCACGA-3′ (forward) | |
| hsa-ETS1 | 5′-TGACTACCCCTCGGTCATTC-3′ (forward) |
| 5′-GGGGATCAGTCCTTGCATTA-3′ (forward) | |
| hsa-c-REL | 5′-GGGGTTTCAAGTCAAGCAGA-3′ (forward) |
| 5′-GCTTGAAGAAGGCAGAGGTG-3′ (reverse) |
FIGURE 6.
Expression of altered genes in TK6 cells under simulated microgravity condition. TK6 cells were cultured in a bioreactor as stated in Fig. 1 legend. The expression of altered genes was quantified by quantitative RT-PCR using specific primers. Relative expression values were calculated by ΔΔCt analysis using the average of two reference genes and normalized to control for -fold changes. The values represent mean ± S.D. (n = 3).
DISCUSSION
The major hypothesis of this study is that ground-based simulated microgravity would alter microRNA and gene expression profiles. To study the effect of simulated microgravity on lymphoblastoid cells, we used a specialized HARV bioreactor. Bioreactors have been widely used as a ground model to simulate the cell culture condition of the weightless environment in space. In a bioreactor, cells in growth medium are continuously rotated to be in a free fall condition with minimal shear forces and excellent mass transfer of nutrients and oxygen (15). Altered gene expression profiles were also reported in cells cultured in HARV bioreactors (16, 17). Growth of human activated T-cells in HARV bioreactors resulted in 4–8% of genes having alterations in gene expression with a -fold change greater than 1.5 (18). Clement et al. (19, 20) observed significant alterations in the mRNA expression levels of 139 of 22,000 genes in human liver cells and 162 genes in epidermal keratinocytes when cultured in the modeled microgravity condition. Canova et al. (21) reported a greater yield of mutation in the HPRT gene and formation of micronuclei but a significant reduction of apoptosis in TK6 cells cultured in rotating wall bioreactors after radiation exposure versus those in cells cultured in the static 1 × g condition. In our study, we did not observe significant differences in cell growth or percentage of viable cells between cells cultured in the bioreactor and cells in the control static condition, which would rule out the possibility that the differences in miRNA and RNA expression levels were due to the growth rate. The culture was started at a low concentration. Extended culture time (72 h) allowed the cells to reach steady growth state and adaptation to the altered growth environment before measurements were made. We were interested in the changes between two different culture conditions rather than transient changes from the static to the rotating condition. Our cell growth results were in agreement with the previous study (22). The effect of the rotating culture condition on the cell growth rate appears to be cell type-specific. Mouse satellite cells proliferated normally, but their differentiation was significantly inhibited when cultured in a rotating wall vessel bioreactor (22).
Several forms of cellular stress have already been reported to alter miRNA expression profiles. It has been suggested that the alterations in gene expression from hypoxia treatment of cells are the result of combined effects on transcription, translation, and adjustment mechanisms such as the induction of miRNAs (23). A more commonly studied form of cellular stress is damage from radiation exposure. Radiation effects on miRNA expressions appear to vary according to cell type, radiation dose, and postirradiation time point. In our study, miRNA PCR array and DNA microarray results revealed that expression of seven miRNA and 400 genes was altered substantially under the simulated microgravity compared with the static condition.
Several studies have shown that the members of the miR-34 family are direct targets of p53, and particularly miR-34a is the downstream effector of p53. Some genes like CCNE2 and cyclin-dependent kinase play a vital role in cell signaling pathways that activate downstream transcriptional programs necessary for cell proliferation and differentiation (12, 24). In the present study, bioreactor-cultured cells showed increased miR-34a and decreased CCNE2 and HMGA2 expression, which are consistent with previous reports. Mackley et al. (25) observed that primary fibroblasts isolated from fetal mouse cornea, skin, and tendon upon linear shear stress resulted in decreased expression of CCNE2 mRNA. In a review, Hermeking (26) stated that HMGA2 is a target gene for miR-34a and is involved in inhibition of cell proliferation and senescence. HMGA2 is a non-histone chromatin-binding protein belonging to the HMGA family and is phosphorylated by protein kinases (27, 28). Boo et al. (29) showed that HMGA2 sensitizes cells to genotoxic stresses such as chemotherapy and x-ray irradiation through the modulation of basal and DNA damage-dependent phosphatidylinositol 3-kinase-related protein kinase activation.
miR-150 plays a key role in hematopoiesis, and it is preferentially expressed in mature resting B- and T-cells (but not in their progenitors) and controls their differentiation by blocking the transition from pro-B- to pre-B-cells (30, 31). In our study, in response to miR-150 expression, EGR2 expression was up-regulated under the simulated microgravity compared with the static condition. EGR2 is a highly conserved zinc finger transcription factor and part of the multigene EGR family that also includes EGR1 and EGR3. It has been shown that EGR family members play key roles in monocyte/macrophage cell fate differentiation, regulation of proliferation, and other cellular responses to extracellular stimuli (32–34).
Both miR-22 and miR-222 are highly expressed in acute myeloid lymphoma, the most common acute leukemia in adults (35). Forced miR-22 expression by 12-O-tetradecanoylphorbol-13-acetate (a phorbol ester) in HL-60 leukemia cells resulted in accumulation of cells and inhibition of cell cycle progression and differentiation (36). Aberrant expression of miRNA-221 and miRNA-222 inhibits normal erythropoiesis and blocks erythroleukemic cell growth and differentiation (37). ZNF-145 is a predicted gene target for miR-222 based on the miRDB on-line database for miRNA target prediction and functional annotations. We also noticed decreased expression of ZNF-145 in response to the up-regulation of miR-222 in cells cultured in rotating bioreactors compared with static cells.
miR-423-5p, miR-141, and miR-618 were also up-regulated in the bioreactor-cultured condition compared with the control static condition. miR-423-5p has been reported as a diagnostic predictor or circulating biomarker for heart failure (38). Uhlmann et al. (39) showed that miR-141 is involved in cell cycle progression, and expression of the miR-200a/141 cluster in breast cancer resulted in G1 arrest and decreased CDK6 expression. The miRDB database showed myeloid/lymphoid or mixed lineage leukemia, trithorax homolog (MLLT4) gene as a predicted target of miR-618.
The NF-κB family of transcription factors comprises p65 (RELA), RELB, c-REL, NF-κB1 (p50), and NF-κB2 (p52). c-REL is exclusively expressed in immune cells and plays a role in the regulation of proliferation, adhesion, survival, and immune and inflammatory responses (40, 41). Induction of c-REL is seen in lymphocytes and myeloid cells during an inflammatory response and is highly associated with the development of leukemia and lymphoma. Activation of NF-κB/REL protein involves phosphorylation, ubiquitination, and subsequent degradation of the inhibitory protein IκB, which in turn results in the nuclear translocation of NF-κB/REL proteins (42). In the present study, cells cultured in bioreactors showed decreased UBQTN C and increased c-REL expression compared with the static environment. In our experiment, we cannot rule out the possibility of participation of other UBQTN family members in the degradation of IκB and activation c-REL/NF-κB in rotated cells. Sharma et al. (43) showed that exposure of mice to simulated microgravity using hind limb suspension resulted in increased activation of NF-κB in testis.
ETS1 is a member of the Ets transcription factor family and plays a role in cellular growth, differentiation, organ development, and angiogenesis (44, 45). In the present study, ETS1 expression was up-regulated under simulated microgravity compared with the static condition. Other researchers have also observed the up-regulation of this gene under different types of stress conditions. Nakatsuka et al. (46) showed that controlled levels of shear stress induce the expression of hepatocyte plasminogen activator inhibitor 1 (PAI-1) through cooperative SP1/ETS1 activation of transcription in rat hepatocytes.
In summary, this study showed that simulated microgravity could alter the expression of miRNAs. Several transcription factors including EGR2, ETS1, and c-REL were up-regulated under these conditions. Alteration of miRNA expression under simulated microgravity influenced several genes that are involved in the regulation of the NF-κB-related pathway network. Further study is needed to investigate the role of altered miRNAs in this pathway network in detail. Targeting miRNA will improve our knowledge to fully understand the regulation of gene expression in stress-related conditions like simulated microgravity. This study provides the first evidence for the alteration in miRNA expression under simulated microgravity conditions.
Supplementary Material
This work was supported in part by the University of Houston Institute for Space Systems Operations Program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
- miRNA
- microRNA
- miR
- miRNA
- CCNE2
- Cyclin E2
- EGR
- early growth response
- ETS1
- v-ets erythroblastosis virus E26 oncogene homolog 1
- ZNF-145
- zinc finger protein 145
- HARV
- high aspect ratio vessel
- HMGA
- high mobility gene A
- UBQTN
- ubiquitin.
REFERENCES
- 1. Pietrzyk R. A., Jones J. A., Sams C. F., Whitson P. A. (2007) Aviat. Space Environ. Med. 78, (suppl.) A9–A13 [PubMed] [Google Scholar]
- 2. Baevsky R. M., Baranov V. M., Funtova I. I., Diedrich A., Pashenko A. V., Chernikova A. G., Drescher J., Jordan J., Tank J. (2007) J. Appl. Physiol. 103, 156–161 [DOI] [PubMed] [Google Scholar]
- 3. Leach C. S., Dietlein L. F., Pool S. L., Nicogossian A. E. (1990) Acta Astronaut. 21, 659–666 [DOI] [PubMed] [Google Scholar]
- 4. Vico L., Collet P., Guignandon A., Lafage-Proust M. H., Thomas T., Rehaillia M., Alexandre C. (2000) Lancet 355, 1607–1611 [DOI] [PubMed] [Google Scholar]
- 5. Barr Y. R., Bacal K., Jones J. A., Hamilton D. R. (2007) Aviat. Space Environ. Med. 78, (suppl.) A26–A37 [PubMed] [Google Scholar]
- 6. Hammond T. G., Lewis F. C., Goodwin T. J., Linnehan R. M., Wolf D. A., Hire K. P., Campbell W. C., Benes E., O'Reilly K. C., Globus R. K., Kaysen J. H. (1999) Nat. Med. 5, 359 [DOI] [PubMed] [Google Scholar]
- 7. Hammond T. G., Benes E., O'Reilly K. C., Wolf D. A., Linnehan R. M., Taher A., Kaysen J. H., Allen P. L., Goodwin T. J. (2000) Physiol. Genomics 3, 163–173 [DOI] [PubMed] [Google Scholar]
- 8. Wilson J. W., Ott C. M., Ramamurthy R., Porwollik S., McClelland M., Pierson D. L., Nickerson C. A. (2002) Appl. Environ. Microbiol. 68, 5408–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Semov A., Semova N., Lacelle C., Marcotte R., Petroulakis E., Proestou G., Wang E. (2002) FASEB J. 16, 899–901 [DOI] [PubMed] [Google Scholar]
- 10. Mazière P., Enright A. J. (2007) Drug Discov. Today 12, 452–458 [DOI] [PubMed] [Google Scholar]
- 11. Ambros V. (2004) Nature 431, 350–355 [DOI] [PubMed] [Google Scholar]
- 12. Mendell J. T. (2005) Cell Cycle 4, 1179–1184 [DOI] [PubMed] [Google Scholar]
- 13. Maes O. C., An J., Sarojini H., Wu H., Wang E. (2008) J. Cell. Biochem. 105, 824–834 [DOI] [PubMed] [Google Scholar]
- 14. Ding L. H., Xie Y., Park S., Xiao G., Story M. D. (2008) Nucleic Acids Res. 36, e58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hammond T. G., Hammond J. M. (2001) Am. J. Physiol. Renal Physiol. 281, F12–F25 [DOI] [PubMed] [Google Scholar]
- 16. Cooper D., Pride M. W., Brown E. L., Risin D., Pellis N. R. (2001) In Vitro Cell Dev. Biol. Anim. 37, 63–65 [DOI] [PubMed] [Google Scholar]
- 17. Wilson J. W., Ramamurthy R., Porwollik S., McClelland M., Hammond T., Allen P., Ott C. M., Pierson D. L., Nickerson C. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13807–13812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ward N. E., Pellis N. R., Risin S. A., Risin D. (2006) J. Cell. Biochem. 99, 1187–1202 [DOI] [PubMed] [Google Scholar]
- 19. Clement J. Q., Lacy S. M., Wilson B. L. (2007) J. Gravit. Physiol. 14, P121–122 [PubMed] [Google Scholar]
- 20. Clement J. Q., Lacy S. M., Wilson B. L. (2008) Genomics Proteomics Bioinformatics 6, 8–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Canova S., Fiorasi F., Mognato M., Grifalconi M., Reddi E., Russo A., Celotti L. (2005) Radiat. Res. 163, 191–199 [DOI] [PubMed] [Google Scholar]
- 22. Molnar G., Schroedl N. A., Gonda S. R., Hartzell C. R. (1997) In Vitro Cell Dev. Biol. Anim. 33, 386–391 [DOI] [PubMed] [Google Scholar]
- 23. Rocha S. (2007) Trends Biochem. Sci. 32, 389–397 [DOI] [PubMed] [Google Scholar]
- 24. Sun F., Fu H., Liu Q., Tie Y., Zhu J., Xing R., Sun Z., Zheng X. (2008) FEBS Lett. 582, 1564–1568 [DOI] [PubMed] [Google Scholar]
- 25. Mackley J. R., Ando J., Herzyk P., Winder S. J. (2006) Biochem. J. 396, 307–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hermeking H. (2010) Cell Death Differ. 17, 193–199 [DOI] [PubMed] [Google Scholar]
- 27. Lee Y. S., Dutta A. (2007) Genes Dev. 21, 1025–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nissen M. S., Langan T. A., Reeves R. (1991) J. Biol. Chem. 266, 19945–19952 [PubMed] [Google Scholar]
- 29. Boo L. M., Lin H. H., Chung V., Zhou B., Louie S. G., O'Reilly M. A., Yen Y., Ann D. K. (2005) Cancer Res. 65, 6622–6630 [DOI] [PubMed] [Google Scholar]
- 30. Zhou B., Wang S., Mayr C., Bartel D. P., Lodish H. F. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7080–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xiao C., Calado D. P., Galler G., Thai T. H., Patterson H. C., Wang J., Rajewsky N., Bender T. P., Rajewsky K. (2007) Cell 131, 146–159 [DOI] [PubMed] [Google Scholar]
- 32. Carter J. H., Lefebvre J. M., Wiest D. L., Tourtellotte W. G. (2007) J. Immunol. 178, 6796–6805 [DOI] [PubMed] [Google Scholar]
- 33. Thiel G., Cibelli G. (2002) J. Cell. Physiol. 193, 287–292 [DOI] [PubMed] [Google Scholar]
- 34. Unoki M., Nakamura Y. (2003) Oncogene 22, 2172–2185 [DOI] [PubMed] [Google Scholar]
- 35. Wang Y., Li Z., He C., Wang D., Yuan X., Chen J., Jin J. (2010) Blood Cells Mol. Dis. 44, 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ting Y., Medina D. J., Strair R. K., Schaar D. G. (2010) Biochem. Biophys. Res. Commun. 394, 606–611 [DOI] [PubMed] [Google Scholar]
- 37. Felli N., Fontana L., Pelosi E., Botta R., Bonci D., Facchiano F., Liuzzi F., Lulli V., Morsilli O., Santoro S., Valtieri M., Calin G. A., Liu C. G., Sorrentino A., Croce C. M., Peschle C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18081–18086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tijsen A. J., Creemers E. E., Moerland P. D., de Windt L. J., van der Wal A. C., Kok W. E., Pinto Y. M. (2010) Circ. Res. 106, 1035–1039 [DOI] [PubMed] [Google Scholar]
- 39. Uhlmann S., Zhang J. D., Schwäger A., Mannsperger H., Riazalhosseini Y., Burmester S., Ward A., Korf U., Wiemann S., Sahin O. (2010) Oncogene 29, 4297–4306 [DOI] [PubMed] [Google Scholar]
- 40. Grilli M., Chiu J. J., Lenardo M. J. (1993) Int. Rev. Cytol. 143, 1–62 [DOI] [PubMed] [Google Scholar]
- 41. Grimm S., Baeuerle P. A. (1993) Biochem. J. 290, 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Verma I. M., Stevenson J. K., Schwarz E. M., Van Antwerp D., Miyamoto S. (1995) Genes Dev. 9, 2723–2735 [DOI] [PubMed] [Google Scholar]
- 43. Sharma C. S., Sarkar S., Periyakaruppan A., Ravichandran P., Sadanandan B., Ramesh V., Thomas R., Hall J. C., Wilson B. L., Ramesh G. T. (2008) Mol. Cell. Biochem. 313, 71–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sementchenko V. I., Watson D. K. (2000) Oncogene 19, 6533–6548 [DOI] [PubMed] [Google Scholar]
- 45. Sharrocks A. D. (2001) Nat. Rev. Mol. Cell Biol. 2, 827–837 [DOI] [PubMed] [Google Scholar]
- 46. Nakatsuka H., Sokabe T., Yamamoto K., Sato Y., Hatakeyama K., Kamiya A., Ando J. (2006) Am. J. Physiol. Gastrointest. Liver Physiol. 291, G26–G34 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.