Abstract
Derivation of patient-specific induced pluripotent stem cells (iPSCs) opens a new avenue for future applications of regenerative medicine. However, before iPSCs can be used in a clinical setting, it is critical to validate their in vivo fate following autologous transplantation. Thus far, preclinical studies have been limited to small animals and have yet to be conducted in large animals that are physiologically more similar to humans. In this study, we report the first autologous transplantation of iPSCs in a large animal model through the generation of canine iPSCs (ciPSCs) from the canine adipose stromal cells and canine fibroblasts of adult mongrel dogs. We confirmed pluripotency of ciPSCs using the following techniques: (i) immunostaining and quantitative PCR for the presence of pluripotent and germ layer-specific markers in differentiated ciPSCs; (ii) microarray analysis that demonstrates similar gene expression profiles between ciPSCs and canine embryonic stem cells; (iii) teratoma formation assays; and (iv) karyotyping for genomic stability. Fate of ciPSCs autologously transplanted to the canine heart was tracked in vivo using clinical positron emission tomography, computed tomography, and magnetic resonance imaging. To demonstrate clinical potential of ciPSCs to treat models of injury, we generated endothelial cells (ciPSC-ECs) and used these cells to treat immunodeficient murine models of myocardial infarction and hindlimb ischemia.
Keywords: Embryonic Stem Cell, Epigenetics, Gene Regulation, Induced Pluripotent Stem (iPS) Cell, Stem Cells, Molecular Imaging, Reprogramming
Introduction
In recent years, human embryonic stem cells have gained popularity as a potentially ideal cell candidate for regenerative medicine (1). Clinical use of human embryonic stem cells, however, remains challenging due to concerns about the use of human embryos and the risk of transplant rejection (2). One way to circumvent these issues is to generate induced pluripotent stem cells (iPSCs)4 from adult somatic tissues (3). iPSCs have been generated successfully from a variety of adult cell types through exogenous expression of various transcription factors (3, 4). Numerous preclinical studies in small animals have demonstrated that adult cells can be successfully reprogrammed into iPSCs, differentiated into therapeutic cell types, and transplanted into damaged tissue for repair (5, 6).
Despite promising results in small animal models, further evaluation is needed in large animal models, which are physiologically more similar to humans. Among large animals, the canine model is especially well suited for translational studies of iPSC-based therapies. Unlike the pig, canine embryonic stem cells (cESCs) have already been isolated and characterized and thus provide criteria for pluripotent canine cells (7–9). Although both monkey ESCs and iPSCs have been previously derived, the use of primates remains controversial (7, 10). In this study, we generated ciPSCs resembling cESCs from the somatic skin and fat of adult mongrel dogs. We tested the efficacy of a canine model for preclinical optimization of iPSC delivery to the heart by monitoring stem cell fate using positron emission tomography (PET) reporter gene imaging and iron oxide labeling by magnetic resonance (MR) imaging.
EXPERIMENTAL PROCEDURES
Adipose Cell Harvest and Culture of Canine iPSCs (ciPSCs)
Adipose tissue and skin were resected from three individual 1-year-old mongrel dogs (Marshall Farms, North Rose, NY) for digestion with collagenase II (Invitrogen) to a single cell solution for plating and culture in appropriate mediums. Derived ciPSCs were maintained on mouse embryonic fibroblast (MEF) feeder layers with ciPSC medium.
Lentivirus Production and Transduction
Lentivirus for reprogramming transgenes or Fluc-RFP-HSVttk (firefly luciferase, red fluorescent protein, and herpes simplex truncated thymidine kinase) was collected from the supernatant of transfected 293FT cells and concentrated as described previously (11, 12).
Immunofluorescence and Alkaline Phosphatase Staining
After cells were fixed, permeabilized, and blocked, incubation with primary and secondary antibody was performed. A list of the antibodies used in this study is included in supplemental Table 1. Alkaline phosphatase staining was performed using the Quantitative Alkaline Phosphatase ES Characterization kit (Millipore) according to the manufacturer's instructions.
Quantitative PCR
Following isolation of total RNA and cDNA, quantitative PCR was performed with Taqman Gene Expression Assays using a SteponePlus Realtime PCR System (Applied Biosystems). All primers are listed in supplemental Table 2.
In Vitro Differentiation
ciPSCs were differentiated into embryoid bodies (EBs) in suspension culture for 8 days. EBs were seeded in adherent culture after which spontaneous differentiation into ectodermal, mesodermal, and endodermal lineages was assessed by quantitative PCR.
Teratoma Formation
Approximately 2 million ciPSCs were injected into the dorsal flanks of male SCID mice (n = 6). After 8 weeks, tumors were dissected and fixed with 10% formaldehyde in PBS. Paraffin embedded tissue sections were stained with hematoxylin and eosin (H&E).
Microarray Hybridization and Data Analysis
Total RNA samples were hybridized to Affymetrix GeneChip Canine Genome 2.0 Array and then normalized and annotated by the Affymetrix Expression Console software.
Canine Myocardial Delivery of Transfected Cells
Dogs were anesthetized, tracheally intubated, and mechanically ventilated as described previously (12). Cells were delivered by injection through the fourth and fifth intercostal spaces into three adjacent sites of the anterior left ventricular myocardium.
Clinical PET-Computed Tomography (PET/CT) Imaging
Imaging was performed with a clinical PET-CT scanner (Discovery LightSpeed Plus; GE Medical Systems, Waukesha, WI) as described previously (12).
Clinical MR Imaging
Imaging was performed on a Signa 3.0T Excite HD scanner (GE Healthcare Systems) and eight-element cardiac phased array coil. A T2 weighted GRE sequence was used to image ciPSCs incubated overnight with iron particles. Cine images of the left ventricle in short and long axes were acquired with a steady-state free precession sequence as previously described (13, 14).
Generation of Canine Endothelial Cells (ciPSC-ECs) from ciPSCs
ciPSCs were differentiated into EBs in ultra-low attachment dishes using differentiation medium. After 16 days in culture, EBs were dissociated into single cells and placed in adherent cell culture conditions. Cells were lifted and FACS sorted for CD31. ciPSC-ECs were cultured using EBM-2 (Lonza, Hopkinton, MA).
Generation of Myocardial Infarction and Intramyocardial Delivery of ciPSC-ECs
8–10-week-old SCID Beige mice (Charles Rivers, MA) were anesthetized by inhaled isoflurane (2% to 3%) and intubated and ventilated. A left thoracotomy was performed followed by ligation of the left anterior descending artery for 30 min followed by reperfusion as described previously (15). After 30 min, 1 × 106 ciPSC-ECs stably expressing Fluc-RFP-HSVttk were injected intramyocardially into three sites near the periinfarct zone at 20 ml of total volume (n = 8). Control animals received PBS injection instead (n = 8).
Optical Bioluminescence (BLI) of Cell Survival and Localization
To assess ciPSC-EC survival and engraftment in vivo, BLI was performed on animals receiving 375 mg/kg body weight d-luciferin reporter probe. Living Image 4.0 (Caliper Life Sciences) was used to quantify signal intensity as described previously (15, 16).
Echocardiographic Measurement of Cardiac Function
Animals receiving left anterior descending artery ligation were scanned on days 0, 14, 28, and 42 using a Siemens-Acuson Sequoia C512 system equipped with a multifrequency (8–14 MHz) 15L8 transducer. Fractional shortening was acquired using standard M-mode image acquisitions of left ventricular short axis images as described previously (15, 16).
Hindlimb Ischemia Model
SCID Beige mice were anesthetized with isoflurane (2–3%) after which unilateral hindlimb ischemia was induced by ligating the femoral artery as described previously (17). To test the therapeutic potential of ciPSC-ECs, 1 × 106 cells stably expressing Fluc-RFP-HSVttk were delivered by intramuscular injection into the region of ischemia (n = 5). Control animals received PBS (n = 5).
For more information on procedures, see supplemental Methods.
RESULTS
Derivation of ciPSCs from Adipose Stromal Cells and Fibroblasts
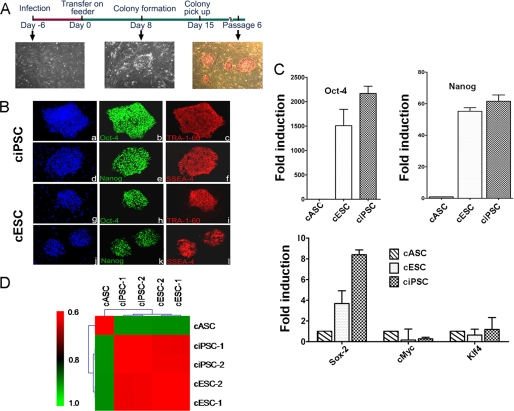
We successfully reprogrammed canine adipose stromal cells (cASCs) and canine fibroblasts (cFibros) into ciPSCs from three individual dogs. We used lentivirus containing human Oct4, Sox2, Klf4, and c-Myc at a 1:1:1:1 ratio without chemical inhibitors. From days 12 to 15 after transduction, we observed clearly recognizable, tightly packed colonies with morphologic appearance similar to cESCs under bright field microscopy (Fig. 1A). As previous studies characterizing cESCs have used TRA-1–60 as a marker of undifferentiation (7), we used TRA-1-60 in conjunction with typical ESC-like morphology to track the progression of the putative ciPSC colonies. In our reprogramming experiments, we consistently observed the appearance of TRA-1-60-positive colonies as early as day 12, following introduction of virus into cells. From days 15 to 18, cESC-like colonies grew large enough for mechanical isolation and were transferred onto MEF feeder layers. Prior to transfer, we recorded the number of ciPSCs that stained positive for TRA-1-60 and had typical ESC-like morphology to calculate reprogramming efficiency. Although the timelines for ciPSC derivation from cASCs and cFibros were nearly identical, cASCs had double the reprogramming efficiency compared with cFibros (1.74% ± 0.09 versus 0.84% ± 0.07, p < 0.05).
FIGURE 1.
Generation of ciPSCs. A, schematic diagram of the generation of ciPSCs. ciPSC colonies can be picked out approximately 12–15 days and are alkaline phosphatase-positive. B, immunofluorescence staining of pluripotent markers. Similar to cESCs, ciPSCs are positive for pluripotent stem cell markers Oct-4 (b and h), Tra-1-60 (c and i), Nanog (e and k), and SSEA-4 (f and l), with nuclear staining by DAPI (a, d, g, and j). C, quantitative PCR analysis of expression of pluripotent stem cell markers Oct-4, Nanog, Sox-2, c-Myc, and Klf-4. Y axis value represents -fold differences (log2) in expression of select genes. D, Pearson correlation analysis for gene expression in cASCs, ciPSCs, and cESCs using transcripts with S.D. >0.2 among all samples (17,895 probes, p < 1.0E-15).
Comprehensive Analyses Confirmed Pluripotency of ciPSCs
Previously we have shown enhanced reprogramming efficiency of human iPSCs from ASCs compared with fibroblasts (11). Hence, we focused first on derivation and subsequent detailed characterization of cASC-derived ciPSCs for this study. Following transfer to fresh feeders, putative ciPSC colonies were characterized by immunostaining and quantitative PCR for pluripotent markers. Similar to cESCs, ciPSCs demonstrated positive staining for alkaline phosphatase, Oct-4, Sox2, Nanog, TRA-1-60, and SSEA-4 (Fig. 1, A and B, and supplemental Fig. 1). RT-PCR of these ciPSCs revealed expression of canine pluripotency genes Oct-4, Sox2, and Nanog at levels comparable with cESCs (Fig. 1C and supplemental Fig. 2). By comparison, cASCs isolated from canine tissue samples revealed very low or no expression of these genes.
Successful reprogramming of ciPSCs was further confirmed by whole genome expression profiling using microarray analysis comparing ciPSCs with cASCs and cESCs. ciPSCs showed a high degree of similarity in their gene expression profile with cESCs and importantly, were distinct from cASCs (Fig. 1D and supplemental Fig. 3). In addition, cFibro-derived ciPSCs were found to be similar to cASC-derived iPSCs and cESCs as well (supplemental Fig. 4).
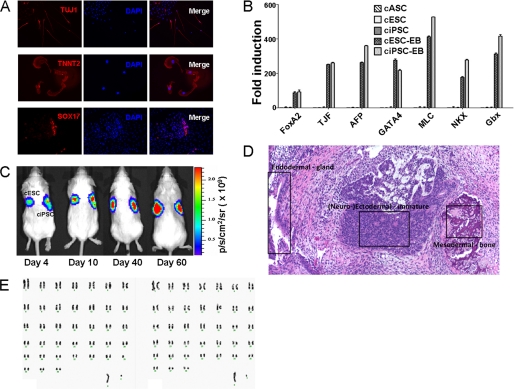
To test pluripotency of ciPSCs derived from cASCs and cFibros, we differentiated ciPSCs in vitro by EB formation. ciPSCs derived from both cASCs as well as cFibros gave rise to cells of all three germ layers as evidenced by immunostaining and quantitative PCR (Fig. 2, A and B, and supplemental Figs. 5 and 6). Teratoma formation assays confirmed potential of ciPSCs to form tumors composed of derivatives from all three germ layers in vivo (Fig. 2, C and D). ciPSCs were found to have a normal karyotype after extended culture for 30 passages with 78 chromosomes and no translocations, indicating genomic stability of clones over time (Fig. 2E).
FIGURE 2.
Confirmation of pluripotency of ciPSCs. A, immunofluorescence staining of differentiated ciPSCs. Differentiated cells are positive for ectodermal cell marker Tuj1, mesodermal cell marker TNNT2, and endodermal cell marker Sox-17. B, quantitative PCR analysis of genes expressed from the three germ layers. Differentiated ciPSCs express genes from all three germ layers, whereas ciPSCs and cESCs do not express these genes at detectable levels. C, longitudinal BLI after injection of ciPSCs and cESCs into SCID mice. BLI shows progressive increase in Fluc activity within teratomas. The left side of animals is control cESCs, and the right side is ciPSCs. D, histological section of a solid, encapsulated tumor removed from the subcutaneous flank of a SCID mouse that had been injected with ciPSCs. The highly differentiated tumor contained a wide range of tissues, composed of endodermal, neuroectodermal, and mesodermal lineages. E, karyotyping of parental cell cASC- and ASC-derived ciPSCs at passage 30. All cells had a normal canine cell karyotype of 2n = 78 (x,y).
In Vivo Tracking of Transplanted Canine iPSCs Using PET and MR Imaging
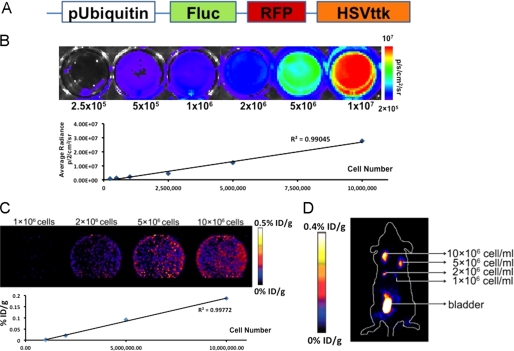
To simulate use of ciPSCs for cell therapy, we next prepared c-ASC-iPSCs for autologous delivery into the same animals from which parental cells were isolated. To visualize anatomic localization and survival of cells after in vivo injection, ciPSCs were stably transduced with a lentivirus carrying a triple-fusion (TF) reporter gene consisting of Fluc-RFP-HSVttk driven by the ubiquitin promoter (Fig. 3A). Following flow cytometry sorting for RFP-positive cells, transfected cells were tested in vitro and in vivo to verify luciferase and HSVttk expression (Fig. 3, B–D). The minimal number of cells required for detection by PET in vivo was calculated for both murine and canine models by injection of varying cell concentration into the flanks of immunodeficient mice and autologous dogs. The minimum detectable cell concentration in the mouse was 1 × 106 cells/ml using microPET (Fig. 3D). For the dog, the signal could be detected from the subcutaneous regions of the animal at a minimum concentration of 5 × 106 cells/ml using a clinical PET/CT scanner. Specifically, the in vivo reporter gene expression for 5 and 10 million ciPSCs was 0.0004 ± 0.0001 and 0.001 ± 0.0003% injected dose/g of tissue (% ID/g), respectively. As typical human trials for cell-based therapies can utilize hundreds of millions of cells (18), PET-based reporter gene imaging modalities at a detection level of 5 million cells appears sensitive enough to enable accurate tracking of ciPSC delivery in vivo.
FIGURE 3.
Molecular imaging of ciPSCs in vitro and in vivo. A, schematic for the TF reporter gene carrying LV-Fluc-RFP-HSVttk driven by the ubiquitin promoter. B, ciPSCs stably expressing the TF reporter gene tested in vitro to verify Fluc expression. A linear correlation was observed between cell number and Fluc activity. C, ciPSCs stably expressing the TF reporter gene tested in vitro to verify HSVttk expression. A linear correlation was observed between cell number and HSVttk activity. The activities were 0.0008 ± 0.0003 (1 × 106), 0.0210 ± 0.0010 (2 × 106), 0.0910 ± 0.0240 (5 × 106), and 0.1860 ± 0.0670% ID/g (1 × 107). D, 1 × 106, 2 × 106, 5 × 106, and 10 × 106 ciPSCs stably expressing the TF reporter gene injected into the subcutaneous flanks of mice. In vivo imaging by microPET was performed 60 min later. The activities were 0.0208 ± 0.0033 (1 × 106), 0.1830 ± 0.0510 (2 × 106), 0.2910 ± 0.1160 (5 × 106), and 0.3770 ± 0.0880% ID/g (10 × 106). The minimal detectable number of ciPSCs was ∼1 × 106.
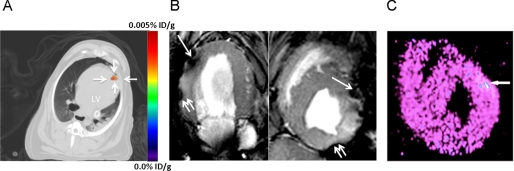
We next tested whether ciPSCs expressing the HSVttk reporter gene could be imaged at an anatomical site where signal attenuation due to tissue thickness is problematic (e.g. myocardium). Due to the therapeutic potential of iPSC therapy for cardiovascular disease and challenge of imaging cell delivery to the heart, we chose the left ventricular myocardial wall as a target. To confirm PET signals in our cardiac model as specific, we labeled ciPSCs with iron oxide particles for co-localization with MR imaging. Following this, 1 × 108 cells were autologously delivered to the hearts of animals from which ciPSCs were originally derived. Fusion imaging of PET/CT demonstrated focal 9-[4-[18F]fluoro-3-(hydroxymethyl)butyl]guanine ([18F]FHBG) radiotracer uptake in the anterior myocardial wall (Fig. 4A and supplemental video). The in vivo reporter gene expression after injection of 100 million cells was 0.0025 ± 0.0010% ID/g of the injected dose/g of myocardial tissue. MR imaging of these cells co-labeled with iron oxide particles confirmed localization of tracer activity in the anterior wall of the myocardium (Fig. 4B). After imaging, canine hearts were explanted immediately after in vivo imaging and cut into 100-mm sections. The location of the cells in the myocardium was further confirmed by ex vivo microPET imaging (Fig. 4C) and histological analysis (supplemental Fig. 7).
FIGURE 4.
Clinical PET-CT and MR imaging of injected cells in autologous canine model. ciPSCs were dual labeled with HSVttk reporter gene and iron oxide particles for PET imaging of cell viability and MR imaging of cell location, respectively. A, representative axial nonenhanced PET-CT fusion image of ciPSCs delivered to the left ventricle acquired 1.5 h after administration of FHBG. A distinct signal (0.0025% ID/g) can be seen at the site of injection close to the left ventricular apex. B, T2 weighted GRE images by MRI showing iron-labeled cells as hypointense signals (dark areas noted by the single arrow) in the anterior apical wall, corresponding to PET imaging. Images are shown in the two-chamber (lower left) and short axis views (lower right). Signal voids along two different tissue interfaces are susceptibility artifacts (double arrows). C, ex vivo analysis of short axis sections of myocardium in the same dog confirming distinct signal in the anterior apical wall, which correspond to the in vivo PET and MR signals. MicroPET imaging was performed 4 h after tracer injection and 2 h after animal sacrifice.
Generation of Endothelial Cells from ciPSCs and Transplantation into Murine Hindlimb Ischemia Model
As undifferentiated iPSCs are not a cell type directly amenable for clinical transplantation, we sought to demonstrate therapeutic applications of these cells through differentiation of ciPSCs into ECs (ciPSC-ECs). Like human and murine ESC-ECs and iPSC-ECs generated in previous studies (15–17, 19, 20), ciPSC-ECs were positive for CD31, capable of DiI-ac-LDL uptake, and formed tubules when cultured on Matrigel in vitro (supplemental Fig. 8A). To assess in vivo therapeutic potential of ciPSC-ECs, we stably transduced ciPSC-ECs with a TF reporter gene carrying RFP, Fluc, and HSVttk driven by the ubiquitin promoter (Fig. 3A) and delivered these cells into a murine model of hindlimb ischemia through intramuscular injection. BLI demonstrated engraftment of ciPSC-ECs in the ischemic murine hindlimb for 14 days following ligation of the femoral artery (supplemental Fig. 8B). Laser Doppler imaging of blood perfusion in ischemic hindlimbs demonstrated significantly improved revascularization in mice receiving ciPSC-ECs compared with mice receiving PBS only at day 7 (ischemic leg/control leg ratio; ciPSC-EC: 0.31 ± 0.09 versus PBS: 0.28 ± 0.13; p < 0.05) and day 14 (ischemic leg/control leg ratio; ciPSC-EC: 0.35 ± 0.08 versus PBS: 0.30 ± 0.17; p < 0.05) (supplemental Fig. 8, C and D).
Transplantation of ciPSC-ECs for Murine Myocardial Injury Model
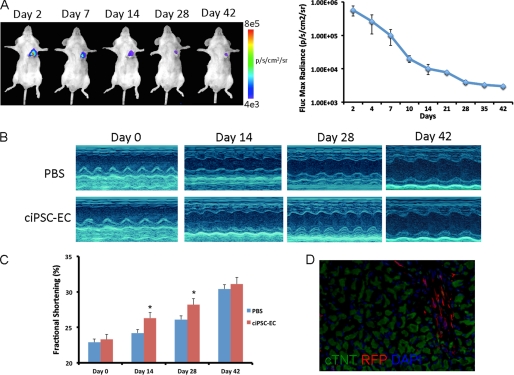
Previous studies have demonstrated potential of iPSC- and ESC-derived endothelial cells to improve systolic function following myocardial infarction through revascularization (15, 16). Hence, we next delivered ciPSC-ECs stably transduced with TF reporter gene into the border zone of infarcted murine hearts. Cardiac engraftment of ciPSC-ECs was monitored noninvasively by BLI for 42 days after cell delivery. Engraftment of ciPSC-ECs was still detectable at 42 days after infarction (Fig. 5A). Measurement of systolic function by echocardiography revealed improvement in cardiac contractility for animals receiving ciPSC-ECs compared with animals receiving PBS at days 14 and 28 after myocardial infarction. However, as in the murine hindlimb model, BLI demonstrated progressive death of donor cells following transplantation into the ischemic heart. Hence, significant differences in systolic function between the two groups of animals were not observed at week 6 (Fig. 5, B and C). Histologic confirmation of ciPSC-ECs in the heart was performed using RFP immunofluorescence at day 42 following cell delivery (Fig. 5D).
FIGURE 5.
Delivery of ciPSC-ECs into murine model of myocardial infarction. A, representative BLI images of an animal receiving 1 × 106 ciPSC-ECs demonstrate strong cell localization to the heart at day 2. Progressive decrease in signal is observed over the next 6 weeks, but persistent cell engraftment is still observed at day 42. Quantitative analysis of BLI signal in animals receiving ciPSC-ECs is expressed in photons/s per cm2 per sr. B, representative M-mode echocardiographic views of infarcted hearts receiving ciPSC-ECS (n = 8) or PBS (n = 8). C, quantification of fractional shortening reveals significant improvement in systolic function of animals receiving ciPSC-ECs at days 14 and 28 (p < 0.05) after myocardial infarction, but not at day 42 compared with animals receiving PBS. D, immunofluorescence staining reveals engraftment of RFP cells in the murine heart 42 days after injection.
DISCUSSION
Previous studies have shown that iPSCs can be derived from small animals such as mice and rats, demonstrating proof of principle for the therapeutic potential of pluripotent stem cell-based regenerative therapies (21, 22). However, these models have several limitations for extrapolation to human patients due to differences in animal physiology and size. Development of large animal models is likely necessary for the creation of clinical criteria for iPSC-based human trials in the future. Unfortunately, efforts to establish allogeneic or autologous cell transplant models in large animals have been hampered by the difficulty in isolation of stable ESC lines from farm animal species and the inability to optimize cell culture conditions (23). The reasons for these reported difficulties in derivation of ESCs from large animals are not clear but likely stem from the inability to isolate cells from the inner cell mass at the appropriate stage of embryonic development (23). Thus, the generation of stable iPSC lines from somatic tissue of large animals is a novel approach by which pluripotent stem cell studies may be established.
Several studies have demonstrated the feasibility of deriving iPSCs from larger animals, including pigs and monkeys (10, 24). More recently, the derivation of ciPSCs from canine embryonic fibroblasts and testicular tissue has been described (25, 26). However, the ciPSCs derived in these preliminary studies were not shown capable of teratoma formation nor were compared with cESCs in terms of comprehensive analysis for markers of pluripotency. Derivation of ciPSCs that comprehensively resemble cESCs is thus still required.
Generation and maintenance of ciPSCs had several noticeable differences between that of murine and human iPSCs. First, both murine and human somatic cells have been shown to be capable of induction to a pluripotent state using feeder free systems (11). By comparison, in the generation of ciPSCs, we were unable to propagate undifferentiated cells without a feeder layer of MEFs. Withdrawal of MEFs resulted in cell differentiation and loss of colony morphology. Second, both leukemia inhibitory factor (LIF) and basic FGF were necessary for the derivation and maintenance of ciPSCs. This stands in contrast to murine ESCs and iPSCs, which require LIF only (27), and human ESCs and iPSCs, which require basic FGF only (23). Without the addition of both LIF and basic FGF, ciPSCs lost ESC-like colony morphology as well as expression of Oct-4 and TRA-1-60. Third, directed differentiation of ciPSCs into therapeutic cell populations may be different from methods employed for human counterparts. For example, to differentiate ciPSC-ECs from ciPSCs, we found that 4 to 10 times the amount of VEGF was required compared with differentiation of human ESCs and iPSCs (19, 20). In addition, efforts to obtain sufficient numbers of iPSC-derived cardiac cells using published protocols for human cells have proven difficult (28, 29). Hence, further studies will be needed to understand differences in the maintenance and differentiation of ciPSCs into specific cell types.
In summary, we have successfully generated 12 ciPSC lines from three adult dogs. Our culture system for ciPSCs may provide additional clues to help establishing other large animal ESC lines like pig and sheep for which culture conditions have yet to be determined. Unlike the pig and sheep for which ESC lines are not available, isolation and culture of cESCs have been documented (7–9), thus allowing us to verify pluripotency and self-renewal properties of ciPSCs derived in this study. Importantly, we have used a noninvasive multimodality imaging approach to monitor autologous iPSC transplantation in a large animal model of cardiac delivery. Both the PET reporter gene and the MR iron oxide labeling techniques we employed allow for accurate localization of cells and are directly applicable to human patients (30). Furthermore, we have generated functional endothelial cells from ciPSCs and demonstrated the therapeutic potential of these cells in murine models of hindlimb ischemia and myocardial infarction. Validating preclinical iPSC imaging in large animal models can be an arduous process and will be different for every cell type and targeted tissue. Undoubtedly, further endeavors will be needed to optimize further both the imaging protocols and iPSC biology synergistically to allow successful translation of pluripotent stem cell therapies to patients in the future.
Supplementary Material
Acknowledgments
We thank Andrew J. Connolly for assistance with histological analysis as well as the Stanford Functional Genomics Facility for assistance with microarrays.
This work was supported, in whole or in part, by National Institutes of Health Grants DP2OD004437, HL089027, HL099117, and EB009689 (to J. C. W.), HL100490 (to M. T. L.), and HL099776 (to R. C. R.). This work was also supported by the Mallinckrodt Foundation (to J. C. W.), the Howard Hughes Medical Institute (to A. S. L. and K. H. N.), the Radiological Society of North America (to A.S.L. and K. H. N.), the Stanford Bio-X Program (to A. S. L.), the California Institute for Regenerative Medicine (CIRM) T1-00001, CIRM RL1-00662-1, and and the Oak Foundation and the Hagey Laboratory for Pediatric Regenerative Medicine (to M. T. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–8, Tables 1 and 2, a Video, Methods, and additional references.
- iPSC
- induced pluripotent stem cell
- BLI
- bioluminescence imaging
- cASC
- canine adipose stromal cell
- cESC
- canine embryonic stem cell
- cFibro
- canine fibroblast
- ciPSC
- canine iPSC
- CT
- computed tomography
- EB
- embryoid body
- [18F]FHBG
- 9-[4-[18F]fluoro-3-(hydroxymethyl)butyl]guanine
- Fluc
- firefly luciferase
- HSVttk
- herpes simplex truncated thymidine kinase
- LIF
- leukemia inhibitory factor
- LV
- lentivirus
- MEF
- mouse embryonic fibroblast
- MR imaging
- magnetic resonance imaging
- PET
- positron emission tomography
- RFP
- red fluorescent protein
- TF
- triple-fusion.
REFERENCES
- 1. Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. (1998) Science 282, 1145–1147 [DOI] [PubMed] [Google Scholar]
- 2. Drukker M., Katz G., Urbach A., Schuldiner M., Markel G., Itskovitz-Eldor J., Reubinoff B., Mandelboim O., Benvenisty N. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 9864–9869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 4. Park I. H., Zhao R., West J. A., Yabuuchi A., Huo H., Ince T. A., Lerou P. H., Lensch M. W., Daley G. Q. (2008) Nature 451, 141–146 [DOI] [PubMed] [Google Scholar]
- 5. Alipio Z., Liao W., Roemer E. J., Waner M., Fink L. M., Ward D. C., Ma Y. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 13426–13431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hanna J., Wernig M., Markoulaki S., Sun C. W., Meissner A., Cassady J. P., Beard C., Brambrink T., Wu L. C., Townes T. M., Jaenisch R. (2007) Science 318, 1920–1923 [DOI] [PubMed] [Google Scholar]
- 7. Wilcox J. T., Semple E., Gartley C., Brisson B. A., Perrault S. D., Villagómez D. A., Tayade C., Becker S., Lanza R., Betts D. H. (2009) Stem Cells Dev. 18, 1167–1178 [DOI] [PubMed] [Google Scholar]
- 8. Hayes B., Fagerlie S. R., Ramakrishnan A., Baran S., Harkey M., Graf L., Bar M., Bendoraite A., Tewari M., Torok-Storb B. (2008) Stem Cells 26, 465–473 [DOI] [PubMed] [Google Scholar]
- 9. Vaags A. K., Rosic-Kablar S., Gartley C. J., Zheng Y. Z., Chesney A., Villagómez D. A., Kruth S. A., Hough M. R. (2009) Stem Cells 27, 329–340 [DOI] [PubMed] [Google Scholar]
- 10. Liu H., Zhu F., Yong J., Zhang P., Hou P., Li H., Jiang W., Cai J., Liu M., Cui K., Qu X., Xiang T., Lu D., Chi X., Gao G., Ji W., Ding M., Deng H. (2008) Cell Stem Cell 3, 587–590 [DOI] [PubMed] [Google Scholar]
- 11. Sun N., Panetta N. J., Gupta D. M., Wilson K. D., Lee A., Jia F., Hu S., Cherry A. M., Robbins R. C., Longaker M. T., Wu J. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 15720–15725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Willmann J. K., Paulmurugan R., Rodriguez-Porcel M., Stein W., Brinton T. J., Connolly A. J., Nielsen C. H., Lutz A. M., Lyons J., Ikeno F., Suzuki Y., Rosenberg J., Chen I. Y., Wu J. C., Yeung A. C., Yock P., Robbins R. C., Gambhir S. S. (2009) Radiology 252, 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li Z., Lee A., Huang M., Chun H., Chung J., Chu P., Hoyt G., Yang P., Rosenberg J., Robbins R. C., Wu J. C. (2009) J. Am. Coll. Cardiol 53, 1229–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Z., Suzuki Y., Huang M., Cao F., Xie X., Connolly A. J., Yang P. C., Wu J. C. (2008) Stem Cells 26, 864–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Z., Wilson K. D., Smith B., Kraft D. L., Jia F., Huang M., Xie X., Robbins R. C., Gambhir S. S., Weissman I. L., Wu J. C. (2009) PLoS One 4, e8443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li Z., Wu J. C., Sheikh A. Y., Kraft D., Cao F., Xie X., Patel M., Gambhir S. S., Robbins R. C., Cooke J. P., Wu J. C. (2007) Circulation 116, I46–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang N. F., Niiyama H., De A., Gambhir S. S., Cooke J. P. (2009) J. Vis. Exp. 23, 1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wollert K. C., Meyer G. P., Lotz J., Ringes-Lichtenberg S., Lippolt P., Breidenbach C., Fichtner S., Korte T., Hornig B., Messinger D., Arseniev L., Hertenstein B., Ganser A., Drexler H. (2004) Lancet 364, 141–148 [DOI] [PubMed] [Google Scholar]
- 19. Li Z., Hu S., Ghosh Z., Han Z., Wu J. C. (2011) Stem Cells Dev., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Narsinh K. H., Sun N., Sanchez-Freire V., Lee A. S., Almeida P., Hu S., Jan T., Wilson K. D., Leong D., Rosenberg J., Yao M., Robbins R. C., Wu J. C. (2011) J. Clin. Invest. 121, 1217–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 22. Liao J., Cui C., Chen S., Ren J., Chen J., Gao Y., Li H., Jia N., Cheng L., Xiao H., Xiao L. (2009) Cell Stem Cell 4, 11–15 [DOI] [PubMed] [Google Scholar]
- 23. Keefer C. L., Pant D., Blomberg L., Talbot N. C. (2007) Anim. Reprod. Sci. 98, 147–168 [DOI] [PubMed] [Google Scholar]
- 24. Ezashi T., Telugu B. P., Alexenko A. P., Sachdev S., Sinha S., Roberts R. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10993–10998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shimada H., Nakada A., Hashimoto Y., Shigeno K., Shionoya Y., Nakamura T. (2010) Mol. Reprod. Dev. 77, 2 [DOI] [PubMed] [Google Scholar]
- 26. Luo J., Suhr S., Chang E. A., Wang K., Ross P. J., Nelson L., Venta P., Knott J., Cibelli J. B. (2011) Stem Cells Dev., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ogawa K., Nishinakamura R., Iwamatsu Y., Shimosato D., Niwa H. (2006) Biochem. Biophys. Res. Commun. 343, 159–166 [DOI] [PubMed] [Google Scholar]
- 28. Laflamme M. A., Chen K. Y., Naumova A. V., Muskheli V., Fugate J. A., Dupras S. K., Reinecke H., Xu C., Hassanipour M., Police S., O'Sullivan C., Collins L., Chen Y., Minami E., Gill E. A., Ueno S., Yuan C., Gold J., Murry C. E. (2007) Nat. Biotechnol. 25, 1015–1024 [DOI] [PubMed] [Google Scholar]
- 29. Yang L., Soonpaa M. H., Adler E. D., Roepke T. K., Kattman S. J., Kennedy M., Henckaerts E., Bonham K., Abbott G. W., Linden R. M., Field L. J., Keller G. M. (2008) Nature 453, 524–528 [DOI] [PubMed] [Google Scholar]
- 30. Zhu J., Zhou L., XingWu F. (2006) N. Engl. J. Med. 355, 2376–2378 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.