Abstract
The Fanconi anemia (FA) pathway is essential for the repair of DNA interstrand cross-links. At the heart of this pathway is the monoubiquitination of the FANCI-FANCD2 (ID) complex by the multiprotein “core complex” containing the E3 ubiquitin ligase FANCL. Vertebrate organisms have the eight-protein core complex, whereas invertebrates apparently do not. We report here the structure of the central domain of human FANCL in comparison with the recently solved Drosophila melanogaster FANCL. Our data represent the first structural detail into the catalytic core of the human system and reveal that the central fold of FANCL is conserved between species. However, there are macromolecular differences between the FANCL proteins that may account for the apparent distinctions in core complex requirements between the vertebrate and invertebrate FA pathways. In addition, we characterize the binding of human FANCL with its partners, Ube2t, FANCD2, and FANCI. Mutational analysis reveals which residues are required for substrate binding, and we also show the domain required for E2 binding.
Keywords: Crystal Structure, Crystallography, DNA Damage, E3 Ubiquitin Ligase, Protein Structure
Introduction
Fanconi anemia (FA)4 is an X-linked or autosomal recessive disorder with a range of clinical presentations including bone marrow failure and high incidence of cancer (1). Mutations in one of at least 14 genes associated with FA lead to dysfunction in DNA interstrand cross-link repair, rendering patients susceptible to DNA-damaging agents (2). Of the FA genes, eight of the gene products form the FA core complex (FANCA, -B, -C, -E, -F, -G, -L, and -M) (3–6). The role of the complex, in conjunction with the E2 ubiquitin-conjugating (UBC) enzyme Ube2T (7), is to monoubiquitinate each of the components of the FANCI-FANCD2 (ID) complex (8–11). Mutation in any of the genes coding for the core complex results in the loss of this monoubiquitination event. The FA pathway is at least partially present in all eukaryal organisms, with yeast having only a FANCM homologue (12) but all higher organisms possessing at least FANCD2. However, in the case of the core complex, vertebrates have the full complement, conserved from zebrafish to humans (13), whereas the invertebrates vary in their FA composition; Caenorhabditis elegans have no obvious core complex (14) and the slime mold Dictyostelium discoideum has a FANCE homologue (15), whereas Drosophila melanogaster has FANCM and FANCL homologues (16).
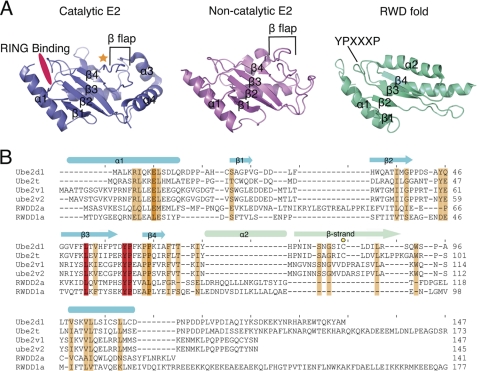
FANCL is the E3 ubiquitin ligase of the core complex (17). It was originally predicted from primary sequence to be a WD40-propeller fold, with a RING domain at the C terminus. A recent study also suggested the presence in human FANCL of an N-terminal RWD domain (18). The RWD classification is a submember of the UBC superfamily (19, 20). The UBC superfamily has a core four-stranded β-meander, flanked by an N-terminal helix and one or two C-terminal helices. In the catalytic E2s, this meander has a C-terminal β-flap containing the catalytic cysteine (Fig. 1A). Most E2 folds contain a YPXXXP motif between strands 3 and 4 that forms a triple turn motif also predicted to be characteristic of the RWD-like fold (18, 20). In the RWD fold, the β-flap assumes a helical conformation (19, 20) (Fig. 1A).
FIGURE 1.
Structural evolution of the UBC family folds. A, the UBC fold of a catalytic E2 (Ube2T, PDB code 1YH1) shown in blue, of a non-catalytic E2 (MMS2, PDB code 1J74 (39)) shown in pink, and the RWD fold (RWDD1a, PDB code 2EBM). A gold star shows the catalytic cysteine, and a hot pink patch shows the RING domain-binding region. The β-flap indicative to a UBC fold is labeled, as is the YPXXXP motif. B, a structural alignment of UBC folds for both catalytic E2s (Ube2D1 and Ube2T) and non-catalytic E2s (Ube2V1 and Ube2V2) and for the RWD folds (RWDD1a and RWDD2a). A scale of red-orange-white shows conservation, with red being 100% conserved and white being 0%. A gold circle labels the catalytic cysteine. The secondary structure is shown above in blue, with the main difference between the UBC fold (a β-strand) and the RWD fold (α2) shown in light green. The most conserved region is the YPXXXP motif.
We recently reported the structure of full-length FANCL from D. melanogaster (21). DmFANCL comprises three domains, an N-terminal E2-like fold (ELF), a double RWD (DRWD) domain, and a C-terminal RING domain. The DmFANCL DRWD domain harbors no YPXXXP motifs, yet each lobe contains a helix in place of the β-flap, giving rise to the DRWD terminology.
Conservation in the core between the Drosophila and the vertebrate FANCLs suggests that they would be broadly similar in structure. However, the sequence identity between human and DmFANCL is low, at 21% (21). Of the three domains, the DRWD domain, particularly the N-terminal half, has the least sequence identity, at 9%. Given that the requirements for a core complex are apparently different between species and that in humans, the loss of any member of the complex results in failure to ubiquitinate the FANCI-FANCD2 complex, we set out to solve the structure of the human FANCL protein. We report here the structure of the central domain of human FANCL, representing ∼50% of the protein. We show that it is globally similar to the DmDRWD domain but that there are macromolecular differences between the proteins that may account for the biological differences in the systems. In addition, we demonstrate that the central human FANCL domain is dispensable for interaction with Ube2T but is essential to bind substrates FANCD2 and FANCI.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Previous efforts to structurally characterize human FANCL had proved unsuccessful, but the understanding of the domain architecture of DmFANCL allowed us to design new constructs. We synthesized a codon-optimized version of the human FANCL coding sequence, using GeneArt. We then subcloned the central domain (residues 109–294) and the RING domain (residues 289–375) into a vector containing an N-terminal His-Smt3 tag using restriction-free cloning (22). Human Ube2T cDNA was purchased as an I.M.A.G.E. clone (Geneservice) and cloned into the same vector as described above. Proteins were expressed in Escherichia coli BL21 cells (Invitrogen) in lysogeny broth medium supplemented with antibiotics and with 0.5 mm ZnCl2 in the case of the RING domain. Cells were cultured at 37 °C to an A600 of 0.6 nm, induced with 0.25 mm isopropyl-1-thio-β-d-galactopyranoside (0.5 mm isopropyl-1-thio-β-d-galactopyranoside in the case of Ube2T), and cultured overnight at 16 °C. Cells were harvested by centrifugation and lysed by sonication on ice (10-s bursts followed by 30 s on ice, repeated four times) in 100 mm Tris, pH 8.0, 500 mm NaCl, 20 mm imidazole, 250 μm tris(carboxyethyl)phosphine (10 μm ZnCl2 added when purifying the RING domain). Sonicated samples were subjected high-speed centrifugation at 4 °C to remove cell debris. Clarified supernatants were applied to an Ni2+-nitrilotriacetic acid affinity resin (Invitrogen) equilibrated with sonication buffer. Fusion proteins were cleaved overnight at 4 °C with Smt3 protease, Ulp1, at a w/w of 1:30 Ulp1:His-Smt3-protein. Flow-through was collected, concentrated, and applied to a Superdex 200 or Superdex 75 column for size-exclusion chromatography. Purified proteins were concentrated to 20 mg/ml (central domain), 8 mg/ml (RING), and 15 mg/ml (Ube2T), flash-frozen in liquid nitrogen, and stored at −80 °C. Human central domain mutants were generated using the Stratagene site-directed mutagenesis method with Pfu Turbo DNA polymerase (Stratagene) and verified by sequencing. The central domain mutants were prepped as the wild type (WT). Human His-FANCD2 was a kind gift from M. R. Hodskinson and K. J. Patel and prepared as described (23). Xenopus laevis FLAG-FANCI was a kind gift from P. Knipscheer and J. C. Walter and prepared as described (24).
Crystallization and Structure Determination
Human FANCL central domain crystals were grown by hanging drop vapor diffusion at 4 °C, in space group C2, with cell dimensions a = 147.6 Å, b = 102.5 Å, c = 65.8 Å, α = 90, β = 94.09, γ = 90. Solvent-content estimates suggested two monomers per asymmetric unit, with a solvent content of 79%. Crystallization conditions were: 0.2 m l-proline, 0.1 m trisodium citrate, pH 5.5, 2% PEG 3350. Crystals were harvested and cryocooled after protection with 30% glycerol. Data were collected at the DIAMOND synchrotron light source beamline I03, at 0.98 Å wavelength. Both a low-resolution pass and a high-resolution pass were collected (3.5 and 1.75 Å, respectively). The two datasets were processed separately using Mosflm (25) and then merged using XDS and scaled using XSCALE (26, 27). The last 55 images from the high-resolution data set were omitted from data processing due to radiation damage to the crystal, which accounts for 75% completeness in the outer shell (2.05–2.0 Å, overall completeness 79%). The data were phased using the DmFANCL DRWD domain, residues 109–292, as a search model (Protein Data Bank (PDB) code 3K1L (21)), with the program PHASER (28). The model was rebuilt manually through iterative cycles of refinement using COOT (29)for building and PHENIX (30) for refinement. Omit maps were generated across regions of the protein to assess model bias. The structure was refined to 2.0 Å resolution. The asymmetric unit contains two molecules, A and B. A is complete and in a single conformation throughout and is used for the analyses reported. Copy B, however, has a region of disorder where residues Cys-174–Ser-182 in loop 6 appear in the electron density maps in multiple conformations. To reflect this movement/disorder, these atoms have been assigned varying occupancies in the PDB file as judged manually in the final 2Fo − Fc and Fo − Fc maps. Along with the high resolution of the data and the fact that chain B has crystal contacts, the non-crystallographic symmetry was not used for some of the loop regions (a total of 80 residues left out of the non-crystallographic symmetry). The final model possesses excellent stereochemistry with all residues in the favored regions of the Ramachandran diagram (98.7%) on the basis of an analysis with MolProbity (31). Table 1 summarizes the data collection and refinement statistics.
TABLE 1.
Data collection and refinement statistics
Highest resolution shell is given in parentheses.
| Data collection | |
| Wavelength (Å) | 0.98 |
| Space group | C121 |
| Cell dimensions (Å) | a = 147.6, b = 102.5, c = 65.8 |
| Cell dimensions (o) | α = 90, β = 94.09 γ = 90 |
| Resolution range (Å) | 45–2.0 (2.05–2.0) |
| No. observations | 153,360 |
| No. unique reflections | 103,454 |
| Completeness (%) | 79% (75%) |
| Multiplicity | 1.5 (1.5) |
| I/σ | 8.91 (2.03) |
| Rmeas | 7.2% (59.1%) |
| Refinement | |
| Number of reflections used | 98,143 |
| Number of protein atoms | 3031 |
| Number of solvent atoms | |
| Water | 413 |
| Na+ ion | 2 |
| PEG | 103 |
| Proline | 16 |
| Mean B values (Å2) | |
| Protein | 39.5 |
| Water | 54.5 |
| Na+ ion | 35.0 |
| PEG | 62.8 |
| Proline | 81.3 |
| r.m.s.a deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.0 |
| Final Rwork | 0.183 |
| Final Rfree | 0.207 |
| Ramachandran regions | |
| Favored | 98.7% |
| Outliers | 0.0% |
a r.m.s., root mean square.
Structural Analyses
All structural analyses were carried out on Chain A of the model. Figures were produced using PyMOL (32), and structure-based alignments were produced using MegaAlign and manual adjustments. To produce the electrostatic surfaces, programs pdb2pqr (33) and apbs (34) were used in conjunction with PyMOL.
Thioester Charge Assay
2 μm E2, His-UbcH7 (Boston Biochem), or human central domain was charged with an excess (50 μm) of HA-Ub (Caltag Medsystems) using 150 nm E1 (Boston Biochem). A total reaction volume of 10 μl was made up with assay buffer containing 50 mm Tris, pH 8, 250 mm NaCl, 2 mm dithiothreitol (DTT), 5 mm MgCl, and 5 mm ATP. Reactions were left for 30 min at room temperature. Reactions were then terminated by the addition of 40 μl of 2× non-reducing SDS buffer. 20 μl of each reaction then had 1 μl of 1 m β-mercaptoethanol added to reduce the thioester bond. 4 μl of each sample was loaded on SDS-PAGE gels and visualized using Western blots. Antibodies used were anti-His (GE Healthcare), anti-HA, and anti-FANCL raised against recombinant human FANCL central domain (Pettingill Technology Ltd.).
In Vitro Pulldown Assays
In a total reaction volume of 1 ml containing assay buffer: 0.5 m NaCl, 0.1 m Tris, pH 8, and 250 μm tris(carboxyethyl)phosphine, human His-FANCD2 (225 nm) or X. laevis FLAG-FANCI (225 nm) was added with an excess of human FANCL central domain or mutants at 454 nm. Reactions were left to bind for 1 h at room temperature. 100 μl of Ni2+-nitrilotriacetic acid-agarose (Qiagen) or 100 μl of anti-FLAG affinity gel (Sigma) was equilibrated in assay buffer, added to the 1-ml reaction, and left on a roller at 4 °C for at least 1 h. Samples were washed with 10 ml of assay buffer. Agarose/affinity gel was then resuspended in 100 μl of assay buffer, 50 μl of 2× SDS buffer was added, and the sample was boiled. 9 μl of the samples was loaded onto SDS page gel, subjected to Western blotting, and probed with the appropriate antibodies. Anti-FLAG antibody was purchased from Abcam.
Analytical Gel Filtration
Initial establishment of interactions between the human central domain (109–294), and human RING domain (289–375) with Ube2T was carried out by analytical gel filtration. Interaction buffer contained 100 mm Tris, pH 8, 250 μm tris(carboxyethyl)phosphine, 10 μm ZnCl2 with 100 mm NaCl. An increased amount of NaCl to 500 mm was used for central domain interactions as the protein is unstable in lower amounts of NaCl. Samples were incubated on ice for 1 h with a total volume of 500 μl. Samples were then loaded onto a Superdex 75 column (equilibrated with the sample buffer), and 0.5-ml fractions were collected for analysis on 12% SDS-PAGE gels.
Isothermal Titration Calorimetry (ITC)
Kinetic information of Ube2T binding interactions with FANCL domains was established using the iTC200 microcalorimeter (MicroCal, Northampton, MA). Sample buffers were the same as interaction buffers used for analytical gel filtration. Experiments were carried out at 8 °C. The syringe contained Ube2T between 650 and 750 μm that injected a total volume of 40 μl by 2.5-μl injections. The cell contained a total volume of 205 μl of either RING (70 μm) or central domain (65 μm). Cell concentrations were adjusted for a 1:1 stoichiometric interaction, and MicroCal Origin software version 7.0 was used to determine dissociation constants (Kd). All measurements were repeated at least twice.
RESULTS
The Structure of Human FANCL Central Domain
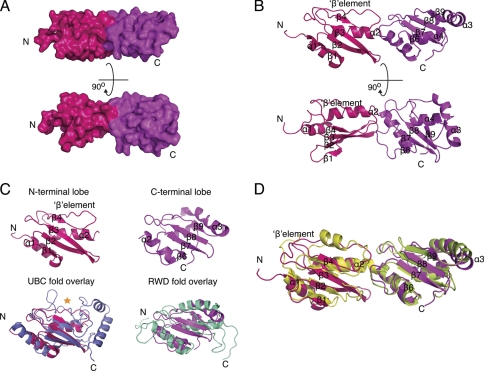
We have determined the structure of the predicted human DRWD domain (residues 109–294) to 2.0 Å resolution. Overall, the human FANCL central domain has a fused bilobal shape with dimensions: 70 × 25 × 20 Å, consisting of four α-helices and nine β-strands (Fig. 2A). Each lobe contains a four-stranded β-meander followed by a β-element in the N-terminal lobe and an α-helix in the C-terminal lobe. The lobes are fused by a kinked helix that runs from one lobe to the other. In addition, there is an α-helix N-terminal to the first β-meander and a C-terminal α-helix (Fig. 2B).
FIGURE 2.
Structure of human FANCL central domain. A, surface representation of the human central domain, with hot pink representing the N-terminal lobe and magenta representing the C-terminal lobe. B, secondary structural representation of the human central domain. C, structural comparison of the human N-terminal lobe, shown in hot pink overlaid with the UBC fold of Ube2T shown in blue with its catalytic cysteine highlighted by a gold star. A comparison of the human C-terminal lobe is shown in magenta overlaid with the RWD fold of GCN2, shown in cyan. D, a secondary structure comparison of the human central domain shown in hot pink and magenta with the DmDRWD domain overlaid shown in yellow and green.
Global Comparison with DmFANCL
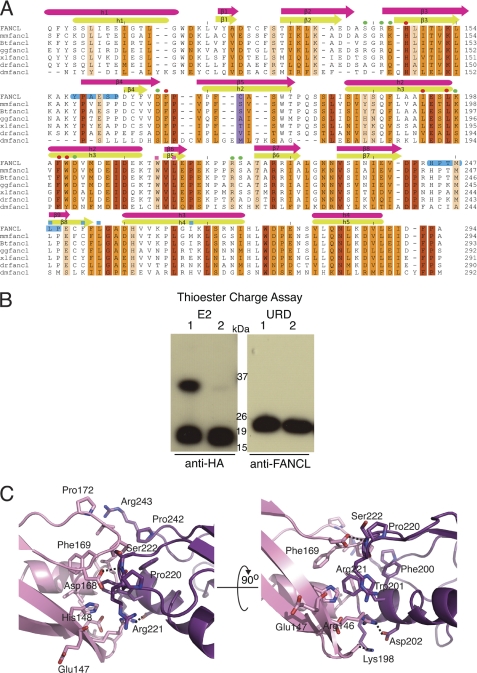
A comparison of known structures using the DALI server unsurprisingly reveals DmFANCL (PDB code 3K1L) as the closest structural homologue. However, a DALI comparison using only the N-terminal lobe (residues 109–194) or C-terminal lobe (residues 195–294) of the human central domain lists E2 UBC enzymes as the most structurally similar for both, with significant RWD fold hits only for the C-terminal lobe (Fig. 2C). This is due to the single biggest difference between the DmDRWD and the human central domain being the change of secondary structure element in the N-terminal lobe (Fig. 2D). In the DmDRWD, both lobes contain a helix after the four-stranded β-meander, indicative of an RWD fold, even in the absence of the triple turn YPXXXP motif. The human central domain, however, harbors the YPXXXP motif in the N-terminal lobe and a related HPXXXP motif in the C-terminal lobe (Fig. 3A). However, the human N-terminal lobe has a β-element in the position normally occupied by a helix in RWD folds. This puts the N-terminal lobe of the human central domain in an almost intermediary position between an E2 fold with a β-flap in the catalytic position and an α-helix in the non-catalytic RWD-like folds (Fig. 2C). Therefore the human central domain is not a DRWD but in fact a UBC-RWD domain. For the remainder of this study, the human central FANCL domain will be referred to as the UBC-RWD domain (URD). Structure-based sequence alignment with other FANCL proteins reveals that this element swap is likely to be in all the vertebrate FANCLs (Fig. 3A). Therefore we refer to this as a vertebrate-specific feature, and it represents a key difference between the vertebrate and invertebrate FANCLs.
FIGURE 3.
Structural comparison of human and Drosophila central domains. A, a structural alignment of the central domains across species. A scale of red-orange-white shows conservation, with red being 100% conserved and white being 0%. Secondary structure is shown above with hot pink for the human central domain and green for the DmDRWD domain. The YPXXXP and HPXXXP motifs are highlighted in blue. The cysteine found in the differing β-strand region of the human central domain is highlighted in lilac. Residues involved in the lobe interactions have a green circle above them, and those that are found in the hydrophobic groove of the lobe interacting site have red circles above them. Residues mutated to alanine to determine the region for substrate binding have a square above them, with blue squares representing patch 2 and pink squares representing patch 4. mm, Mus musculus; Bt, Bos taurus; gg, Gallus gallus; xl, X. laevis; dr, Danio rerio. B, thioester charge assay to determine whether the human β-element cysteine thioester links ubiquitin in a manner analogous to catalytic E2s. Lanes 1 and 2 for each panel correspond to the sample in non-reducing buffer and reducing buffer to determine a thioester linkage, respectively. It is clear that that the URD cannot form a thioester link to ubiquitin. C, a close-up view of the interactions between the human N-terminal (pink) and C-terminal (mauve) central domain lobes. Hydrogen bonds are depicted as black dashed lines.
Intriguingly, the human β-element in the N-terminal lobe contains a cysteine (Fig. 3A). Superposition of the human N-terminal lobe with Ube2T, with a root mean square deviation of 2 Å, reveals that the cysteine in the URD β-element lies structurally in the same position as the catalytic cysteine of Ube2T (Fig. 2C). We wondered whether this cysteine was capable of forming a thioester bond with ubiquitin as in the case of the catalytic E2s. To test whether the cysteine in the URD is catalytic, we carried out a thioester charge assay. The thioester assay revealed that the URD is unable to form a thioester linkage between its cysteine and the activated C terminus of ubiquitin (Fig. 3B).
The interacting residues between the N- and C-terminal lobes of the URD and DmDRWD are not conserved (Fig. 3, A and C). However the residues that make up the hydrophobic core between the two lobes are highly conserved (Fig. 3A). A structural alignment of the human and Drosophila domains shows an overall conservation of 50%, with only 13% of residues absolutely conserved (Fig. 3A). There are also differences in conservation between the two lobes, with the C-terminal lobe being more conserved than the N-terminal lobe (30 and 20%, respectively).
Surface Composition Reveals Extensive Hydrophobic Exposure
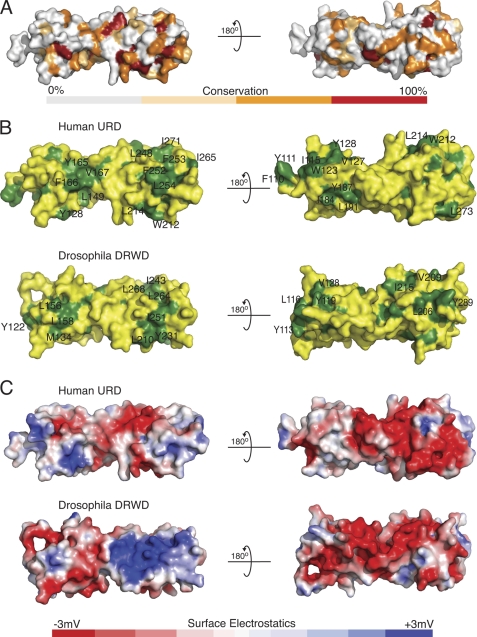
It is likely that differences in a core complex requirement for the vertebrate system arise from the need to stabilize FANCL. We reasoned that there would likely be differences in the surface conservation and surface chemical composition between DmFANCL and human FANCL (HsFANCL). To assess this, we generated surface maps of the URD and compared it with the DmDRWD domain (Fig. 4).
FIGURE 4.
Surface comparisons of the human URD and the Drosophila DRWD domains. A, a surface conservation comparison between the human URD and the Drosophila DRWD. The surface shown here is of the human URD. B, a comparison of surface hydrophobic elements shown in green, for both the human and the Drosophila domains. C, a comparison of surface electrostatics for both the human and Drosophila domains, with blue for basic charge and red for acidic.
The surface conservation between DmDRWD and the URD is ∼32%. The surface is more conserved on the C-terminal lobe at 20% as compared with the N-terminal lobe at 12% (Figs. 3A and 4A).
Comparison of solvent-exposed hydrophobic residues between the URD and the DmDRWD reveals additional surface hydrophobic residues on the URD. A small hydrophobic patch resides on the N-terminal lobe of the URD, between the last β-strand of the four-stranded β-meander (β4) and the β-element (residues Leu-149, Tyr-165, Phe-166, and Val-167, Fig. 4B). Additionally a patch on the URD lies between α-helix 1 and the N-terminal region of the large kinked helix, corresponding to residues Leu-121, Ala-140, Ile-184, Tyr-187, and Leu-191 (Fig. 4B). Interestingly, a reasonably conserved region resides on the N-terminal region corresponding to residues Tyr-112, Tyr-113, and Leu-116 in DmDRWD, and more extensively in humans, Ile-115, Trp-123, Leu-126, Val-127, Tyr-128, Ala-129, and Phe-133 (Fig. 4B). On the C-terminal domain of the DmDRWD is a hydrophobic groove between the last β-strand of the four-stranded β-meander and α-helix 3 that follows (residues Ile-243, Met-246, Leu-248, Leu-264, Leu-268, and Trp-271). In the human URD, there is also a hydrophobic groove formed by residues Phe-253, Val-260, and Leu-268, but it is extended by comparison with the DmFANCL to include Leu-248, Phe-252, Leu-254, Ile-265, and Ile-271 (Fig. 4B). In the URD, there is an additional small hydrophobic patch that consists of Trp-212, Val-213, and Leu-214. In Drosophila DRWD, the corresponding residues are His-208, Val-209, and Leu-210. This patch in the Drosophila DRWD is partly occluded by the DRWD-RING loop in the full-length FANCL structure.
A comparison of the electrostatic potential of the surfaces of the URD and the DmDRWD reveals distinct differences between the species (Fig. 4C). Interestingly, there is a large positive patch on the DmDRWD, which is absent in the URD. Overall, the URD has a greater negative charge then the DmDRWD, which wraps around the URD like a ring. However, on the backside of the central FANCL domain in both human and Drosophila, there is a similar overall negative charge.
The Human RING Domain Binds Ube2T
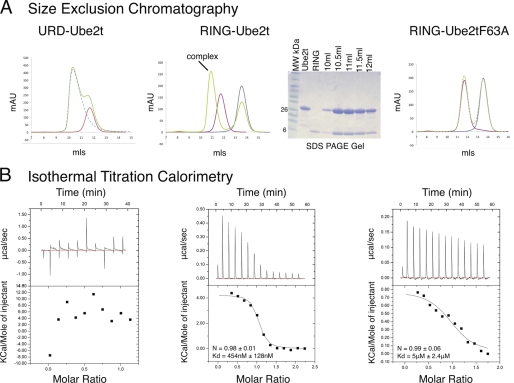
To assess whether the URD plays any role in Ube2T binding, we carried out size-exclusion chromatography and ITC. Initial results from the size-exclusion chromatography showed no change in migration that would correspond to a complex formation between the URD and Ube2T (Fig. 5A). To further assess the interaction, we employed ITC, which again showed no interaction between the URD and Ube2T (Fig. 5B). We then tested the purified RING domain to see whether it was sufficient for Ube2T binding. We initially observed a 1:1 stoichiometric interaction between the RING and Ube2T as judged by the migration of the complex during size-exclusion chromatography (Fig. 5A). To ascertain the affinity of RING for Ube2T, we carried out ITC. After titration of Ube2T into RING, the data were fitted with a stoichiometry of 1:1 as seen in the size-exclusion chromatography, resulting in an equilibrium binding dissociation constant (Kd) of 454 nm (Fig. 5B). Based on the structure of UbcH7 bound to the RING E3 c-cbl (35), we mutated Phe-63 in Ube2T, predicted to be interacting with the RING, to alanine. No complex shift was observed between Ube2T-F63A and RING using analytical gel filtration (Fig. 5A). Further analysis using ITC revealed that mutant Ube2T-F63A was able to bind the RING domain, but with an 11-fold decrease in binding affinity (Kd = 5 μm, Fig. 5B).
FIGURE 5.
RING domain is necessary and sufficient for E2 binding. A, size-exclusion chromatograms showing molecular mass shifts if complex formation has occurred. Green lines on the chromatogram represent the proteins allowed to form complexes, and red lines show Ube2T or Ube2T-F63A. From the size-exclusion chromatography experiments, it is clear that complex formation only occurs between RING and Ube2T. The SDS-PAGE gel also show that both the RING and the Ube2T proteins are present in the shifted peak. mAU, milliabsorbance units; MW, molecular mass markers. B, ITC experiments of the interactions between URD-Ube2T, RING-Ube2T, and RING-Ube2T-F63A.
The Human Central Domain Binds FANCI and FANCD2
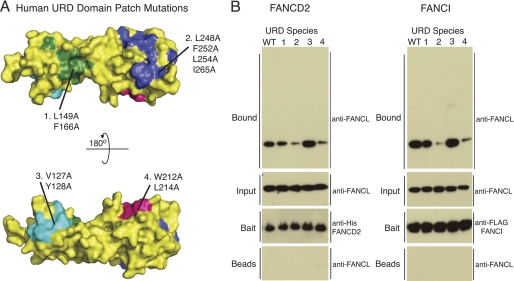
In the Drosophila system, pulldown studies indicated that the ELF domain was not required for substrate binding (21). Given the role of the RING domain in E2 binding, we hypothesized that the URD domain would be sufficient for substrate binding. To test this, we performed in vitro pulldown experiments with FANCI, FANCD2, and the URD (Fig. 6). As expected, the URD was pulled down by both substrates. Our structural analysis reveals several hydrophobic patches exposed on the surface of the URD (Fig. 4B). To establish which patch(es) may be required for substrate binding, we generated the following mutants: L149A/F166A; L248A/F252A/L254A/I265A; V127A/Y128A; and W212A/L214A (Fig. 6A). All mutants were purified to homogeneity as wild type URD and subjected to pulldown arrays with FANCI and FANCD2. Both substrates showed reduced binding with mutants L248A/F252A/L254A/I265A and W212A/L214A as compared with wild type, indicating that the exposed hydrophobic patches on the C-terminal lobe are required for substrate binding. In contrast, mutations in the patches on the N-terminal lobe behaved as wild type (Fig. 6B).
FIGURE 6.
URD domain binds substrates FANCD2 and FANCI. A, structural representation of the surface hydrophobic patch mutants of the URD domain. B, pulldown of substrates His-FANCD2 and FLAG-FANCI with WT URD and the patch mutants of the URD, with the mutations indicated in A. URD input and URD incubated with beads alone control are shown using anti-FANCL antibody. The pulldown shows either His-FANCD2 with anti-His or FLAG-FANCI with anti-FLAG and the URD species that have been pulled down with anti-FANCL antibody.
DISCUSSION
The structure of the human central FANCL domain (URD) adopts the same overall topology as the Drosophila DRWD domain, confirming that the FANCL proteins belong to the UBC and RING superfamilies and are not WD40 proteins as predicted (17). The human central domain does differ from the DmFANCL as it does not have a DRWD but contains a UBC fold fused to an RWD fold (URD). Gene duplication resulting in fused domains is not a novel occurrence in protein topology. However, the fused consecutive UBC superfamily folds, seen in both the URD and the DmDRWD, are unique and appear to be specific to FANCL.
In the context of the full-length human FANCL, there may be subtle differences in domain associations. Structure-based sequence alignments of vertebrate FANCL proteins to Drosophila FANCL reveal that the loops connecting ELF-URD (or ELF-DRWD) and URD-RING (or DRWD-RING) are divergent between species (21). The ELF-URD loop is conserved in length, but not composition, and in the Drosophila structure, there are no interactions between the ELF and any other part of the protein. One possible explanation for the existence of a β-element in place of the helix in human FANCL URD may be to allow a more intimate association between the URD and ELF domains. The URD-RING loop in human FANCL is 7 residues shorter than in DmFANCL. We show a high affinity of human RING for the E2, Ube2T, which may reflect a different arrangement of the URD and RING domains in the human protein. Further structural analyses of the relationship between the human URD and RING domains and other members of the FANCL family will be required to tease out these differences.
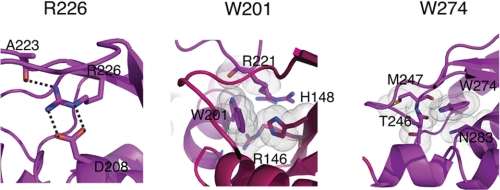
Although we have shown that the URD domain is not required for Ube2T binding, the URD domain appears to be essential for the function of FANCL (36). It has been previously shown in the Drosophila system that the ELF domain is not required to bind substrates FANCD2 and FANCI (21). In this analysis, we show that the human URD domain is sufficient for binding substrates. Additionally, we also reveal the molecular determinants of this binding (Fig. 6). In particular, the residues Trp-212, Leu-214, Leu-248, and Leu-254 are conserved across species (Fig. 3A). Previous mutational work based on the predicted structure of FANCL indicated that the then uncharacterized URD domain was essential for core complex assembly (36), with point mutants R226E, W201A, and W274A (human numbering, numbered as 275 in Ref. 36) abolishing core complex assembly. Our structure of human FANCL URD provides a molecular rationale for these observations and indicates that loss of core complex assembly is in fact due to structural instability (Fig. 7). As predicted from the Drosophila FANCL structure (21), Arg-226 forms buried salt bridges with Asp-208 and the main-chain oxygen of Ala-223 (Fig. 7). Mutation of this residue to the oppositely charged glutamate therefore destabilizes these salt bridges and consequently the surrounding fold. Trp-201 is right in the middle of the interface connecting the N- and C-lobes of the URD domain (Fig. 7). A W201A mutation would therefore impact the folding of the lobes as this residue is clearly seen in the structure to be stacking against and making multiple contacts with residues His-148, Arg-146, and Arg-221. Therefore, although the hydrophobic nature of the residue is maintained, the contacts in the core are not (Fig. 7). Finally, Trp-274 is completely conserved between all the FANCL species, including Drosophila (21). In our URD structure, Trp-274 forms a hydrogen bond between the side-chain nitrogen of the tryptophan and the main-chain oxygen of Thr-246. Mutation of this Trp-274 to alanine removes this hydrogen bond as well as multiple van der Waals contacts with residues Met-247 and Asn-283 (Fig. 7). We can conclude that these particular mutations destabilize the URD domain, and we can speculate, based on the extent of surface-exposed hydrophobic patches on the URD domain, that this domain is very likely to be involved in binding one or more of the core complex proteins. The presence of such patches certainly suggests a number of protein-protein interaction sites. Interestingly, a recent study implicates the URD domain in (PCNA) proliferating cell nuclear antigen binding, providing a potential link between the translesion synthesis and FA pathways (37). Coupled with our observation that the URD binds FANCI and FANCD2, it appears that the URD domain of FANCL is responsible for several interactions. Finally, there has been speculation that phosphorylation of FANCI, shown to trigger the FA pathway in the chicken system, may enhance the ability of FANCL to bind FANCI (38). For this to be the case, we would anticipate the existence of a basic patch on FANCL to mediate interaction with a cluster of negatively charged phosphate ions. From our surface electrostatic analysis of the URD domain, there is no such patch, yet intriguingly, there is in the Drosophila protein. Further structural studies of FANCL in complex with substrates and other binding partners will be required to determine the functions of these surfaces.
FIGURE 7.
Structural explanation for FANCL mutations. Left panel, close-up of the salt-bridge formed between Arg-226 and Asp-208. The hydrogen bonding is depicted as black dashed lines. Middle panel, close-up view of the contribution of Trp-201 to the hydrophobic core of the DRWD N-/C-terminal interface. Van der Waals contacts are represented by gray dotted clouds. Right panel, close-up view of the completely conserved Trp-274 and its contacts in the URD core. Hydrogen bonding is depicted as black dashed lines, and van der Waals contacts are depicted as gray dotted clouds.
Acknowledgments
We thank Michael Way for discussions and criticisms on the manuscript and members of the Protein Structure and Function laboratory for suggestions and support. We also thank K. J. Patel for the plasmid for HIS-FANCD2 and P. Knipscheer and J. C. Walter for the FLAG-FANCI plasmid. We thank the protein production facility for generating the viruses for the insect cell protein expression.
This work was supported by a grant from Cancer Research UK.
The atomic coordinates and structure factors (code 3ZQS) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- FA
- Fanconi anemia
- DmFANCL
- D. melanogaster FANCL
- Ub
- ubiquitin
- UBC
- ubiquitin-conjugating
- URD
- UBC-RWD domain
- DRWD
- double RWD
- DmDRWD
- D. melanogaster DRWD
- ELF
- E2-like fold
- ITC
- isothermal titration calorimetry
- RWD
- found in RING, WD-containing proteins, DEXD helicases.
REFERENCES
- 1. Alter B. P. (1996) Am. J. Hematol. 53, 99–110 [DOI] [PubMed] [Google Scholar]
- 2. German J., Schonberg S., Caskie S., Warburton D., Falk C., Ray J. H. (1987) Blood 69, 1637–1641 [PubMed] [Google Scholar]
- 3. Garcia-Higuera I., Kuang Y., Denham J., D'Andrea A. D. (2000) Blood 96, 3224–3230 [PubMed] [Google Scholar]
- 4. Garcia-Higuera I., D'Andrea A. D. (1999) Blood 93, 1430–1432 [PubMed] [Google Scholar]
- 5. Medhurst A. L., Huber P. A., Waisfisz Q., de Winter J. P., Mathew C. G. (2001) Hum. Mol. Genet. 10, 423–429 [DOI] [PubMed] [Google Scholar]
- 6. de Winter J. P., Rooimans M. A., van Der Weel L., van Berkel C. G., Alon N., Bosnoyan-Collins L., de Groot J., Zhi Y., Waisfisz Q., Pronk J. C., Arwert F., Mathew C. G., Scheper R. J., Hoatlin M. E., Buchwald M., Joenje H. (2000) Nat. Genet. 24, 15–16 [DOI] [PubMed] [Google Scholar]
- 7. Machida Y. J., Machida Y., Chen Y., Gurtan A. M., Kupfer G. M., D'Andrea A. D., Dutta A. (2006) Mol. Cell 23, 589–596 [DOI] [PubMed] [Google Scholar]
- 8. Timmers C., Taniguchi T., Hejna J., Reifsteck C., Lucas L., Bruun D., Thayer M., Cox B., Olson S., D'Andrea A. D., Moses R., Grompe M. (2001) Mol. Cell 7, 241–248 [DOI] [PubMed] [Google Scholar]
- 9. Sims A. E., Spiteri E., Sims R. J., 3rd, Arita A. G., Lach F. P., Landers T., Wurm M., Freund M., Neveling K., Hanenberg H., Auerbach A. D., Huang T. T. (2007) Nat. Struct. Mol. Biol. 14, 564–567 [DOI] [PubMed] [Google Scholar]
- 10. Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M. S., Timmers C., Hejna J., Grompe M., D'Andrea A. D. (2001) Mol. Cell 7, 249–262 [DOI] [PubMed] [Google Scholar]
- 11. Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E. R., 3rd, Hurov K. E., Luo J., Ballif B. A., Gygi S. P., Hofmann K., D'Andrea A. D., Elledge S. J. (2007) Cell 129, 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mosedale G., Niedzwiedz W., Alpi A., Perrina F., Pereira-Leal J. B., Johnson M., Langevin F., Pace P., Patel K. J. (2005) Nat. Struct. Mol. Biol. 12, 763–771 [DOI] [PubMed] [Google Scholar]
- 13. Titus T. A., Selvig D. R., Qin B., Wilson C., Starks A. M., Roe B. A., Postlethwait J. H. (2006) Gene 371, 211–223 [DOI] [PubMed] [Google Scholar]
- 14. McVey M. (2010) Environ. Mol. Mutagen. 51, 646–658 [DOI] [PubMed] [Google Scholar]
- 15. Zhang X. Y., Langenick J., Traynor D., Babu M. M., Kay R. R., Patel K. J. (2009) PLoS Genet. 5, e1000645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marek L. R., Bale A. E. (2006) DNA Repair 5, 1317–1326 [DOI] [PubMed] [Google Scholar]
- 17. Meetei A. R., de Winter J. P., Medhurst A. L., Wallisch M., Waisfisz Q., van de Vrugt H. J., Oostra A. B., Yan Z., Ling C., Bishop C. E., Hoatlin M. E., Joenje H., Wang W. (2003) Nat. Genet. 35, 165–170 [DOI] [PubMed] [Google Scholar]
- 18. Alpi A. F., Pace P. E., Babu M. M., Patel K. J. (2008) Mol. Cell 32, 767–777 [DOI] [PubMed] [Google Scholar]
- 19. Burroughs A. M., Jaffee M., Iyer L. M., Aravind L. (2008) J. Struct. Biol. 162, 205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nameki N., Yoneyama M., Koshiba S., Tochio N., Inoue M., Seki E., Matsuda T., Tomo Y., Harada T., Saito K., Kobayashi N., Yabuki T., Aoki M., Nunokawa E., Matsuda N., Sakagami N., Terada T., Shirouzu M., Yoshida M., Hirota H., Osanai T., Tanaka A., Arakawa T., Carninci P., Kawai J., Hayashizaki Y., Kinoshita K., Güntert P., Kigawa T., Yokoyama S. (2004) Protein Sci. 13, 2089–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cole A. R., Lewis L. P., Walden H. (2010) Nat. Struct. Mol. Biol. 17, 294–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pace P., Mosedale G., Hodskinson M. R., Rosado I. V., Sivasubramaniam M., Patel K. J. (2010) Science 329, 219–223 [DOI] [PubMed] [Google Scholar]
- 23. Knipscheer P., Räschle M., Smogorzewska A., Enoiu M., Ho T. V., Schärer O. D., Elledge S. J., Walter and J. C. (2009) Science 326, 1698–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van den Ent F., Löwe J. (2006) J. Biochem. Biophys. Methods 67, 67–74 [DOI] [PubMed] [Google Scholar]
- 25. Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 26. Kabsch W. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kabsch W. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 31. Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) Nucleic Acids Res. 32, W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 33. Dolinsky T. J., Nielsen J. E., McCammon J. A., Baker N. A. (2004) Nucleic Acids Res. 32, W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Unni S., Huang Y., Hanson R. M., Tobias M., Krishnan S., Li W. W., Nielsen J. E., Baker N. A. (2011) J. Comput. Chem. 32, 1488–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zheng N., Wang P., Jeffrey P. D., Pavletich N. P. (2000) Cell 102, 533–539 [DOI] [PubMed] [Google Scholar]
- 36. Gurtan A. M., Stuckert P., D'Andrea A. D. (2006) J. Biol. Chem. 281, 10896–10905 [DOI] [PubMed] [Google Scholar]
- 37. Geng L., Huntoon C. J., Karnitz L. M. (2010) J. Cell Biol. 191, 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ishiai M., Kitao H., Smogorzewska A., Tomida J., Kinomura A., Uchida E., Saberi A., Kinoshita E., Kinoshita-Kikuta E., Koike T., Tashiro S., Elledge S. J., Takata M. (2008) Nat. Struct. Mol. Biol. 15, 1138–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moraes T. F., Edwards R. A., McKenna S., Pastushok L., Xiao W., Glover J. N., Ellison M. J. (2001) Nat. Struct. Biol. 8, 669–673 [DOI] [PubMed] [Google Scholar]