Abstract
LJM11, an abundant salivary protein from the sand fly Lutzomyia longipalpis, belongs to the insect “yellow” family of proteins. In this study, we immunized mice with 17 plasmids encoding L. longiplapis salivary proteins and demonstrated that LJM11 confers protective immunity against Leishmania major infection. This protection correlates with a strong induction of a delayed type hypersensitivity (DTH) response following exposure to L. longipalpis saliva. Additionally, splenocytes of exposed mice produce IFN-γ upon stimulation with LJM11, demonstrating the systemic induction of Th1 immunity by this protein. In contrast to LJM11, LJM111, another yellow protein from L. longipalpis saliva, does not produce a DTH response in these mice, suggesting that structural or functional features specific to LJM11 are important for the induction of a robust DTH response. To examine these features, we used calorimetric analysis to probe a possible ligand binding function for the salivary yellow proteins. LJM11, LJM111, and LJM17 all acted as high affinity binders of prohemostatic and proinflammatory biogenic amines, particularly serotonin, catecholamines, and histamine. We also determined the crystal structure of LJM11, revealing a six-bladed β-propeller fold with a single ligand binding pocket located in the central part of the propeller structure on one face of the molecule. A hypothetical model of LJM11 suggests a positive electrostatic potential on the face containing entry to the ligand binding pocket, whereas LJM111 is negative to neutral over its entire surface. This may be the reason for differences in antigenicity between the two proteins.
Keywords: Antigen, Isothermal Titration Calorimetry, Ligand-binding Protein, Protein Structure, Serotonin, X-ray Crystallography, Beta Propeller, Catecholamine
Introduction
The salivary secretions of blood-feeding insects contain a rich mixture of proteins that act to prevent host hemostatic and inflammatory responses (1, 2). Individual proteins in the saliva inhibit the essential processes of coagulation, platelet activation, vasoconstriction, inflammation, and mast cell function (2). Additionally, certain of these salivary proteins are immunogenic, conferring protection against cutaneous and visceral leishmaniasis in several animal models following exposure to sand fly bites or immunization with a salivary component (3–9). In most of these studies, protection correlates with a delayed type hypersensitivity (DTH)3 response to saliva or to a protective salivary molecule in the host skin that alters the outcome of infection (10, 11). These studies have established the potential of salivary proteins as components of vaccines against leishmaniasis.
A major protein family in the saliva of the sand fly genera Lutzomyia and Phlebotomus is related by sequence to the “yellow” protein of Drosophila melanogaster and the major royal jelly proteins (MRJPs) of bees. Immunization with LJM11, a yellow protein from saliva of Lutzomyia longipalpis, protects hamsters against visceral leishmaniasis up to 2 months postinfection (4). This partial protection was associated with the induction at the site of challenge of a DTH response composed mainly of a mononuclear infiltrate of macrophages and lymphocytes (4). Furthermore, humans and dogs living in an area where L. longipalpis is prevalent produce specific antibodies to LJM11 (12), demonstrating its broad immunogenicity.
The function of yellow family members from sand fly saliva, including LJM11, has not been established. Understanding the physiological significance and structure of these molecules is relevant to their potential as vaccine candidates. One known function of saliva is the removal of small molecule mediators of hemostasis and inflammation using high affinity ligand-binding proteins that have been referred to as kratagonists (2, 13–17). Among these mediators are the biogenic amines, including serotonin, norepinephrine, epinephrine, and histamine, that induce responses including vasoconstriction, platelet activation, increases in vascular permeability, itching, and pain (18, 19). Because blood feeding has evolved independently in many taxa, different families of proteins perform this function in different taxonomic groups, making it difficult to predict which proteins are kratagonists. In Hemiptera and ticks, members of the lipocalin protein family bind biogenic amines, whereas in the mosquito, insect odorant-binding protein family members perform this role (13, 14, 16, 20). The high expression level of yellow proteins in sand fly saliva suggests that they may serve a kratagonist function because stoichiometric amounts of binding proteins are required for efficient removal of small molecule effectors, such as biogenic amines.
In this study, we demonstrate the DTH-inducing and protective nature of LJM11 against Leishmania infection in mice. We also describe the biogenic amine-binding function and the three-dimensional crystal structure of LJM11 and compare it with LJM111, another yellow protein from the saliva of L. longipalpis that is non-immunogenic. This is the first structural characterization of a member from the large yellow/MRJP family of proteins as well as the first structure of a salivary protein from a vector sand fly.
EXPERIMENTAL PROCEDURES
Sand Flies
L. longipalpis sand flies were reared at the Laboratory of Malaria and Vector Research (NIAID, National Institutes of Health). Salivary glands were dissected from adult sand flies 4–7 days after emergence. Salivary glands were stored in 10 mm sodium phosphate, pH 7.4, 150 mm NaCl at −70 °C until needed.
Mice
6–8-week-old C57BL/6 female mice were obtained from Charles River Laboratories (Germantown, MD) and maintained under pathogen-free conditions. All animal procedures were approved by the NIAID animal care and use committee (Protocol LMVR4E) following the National Institutes of Health guidelines for animal housing and care.
Parasites
Leishmania major clone V1 (MHOM/IL/80/Friedlin) was cultured in complete 199 medium (Invitrogen) as published elsewhere (7). Infective stage metacyclic promastigotes of L. major were isolated from stationary cultures (4–5 days old) by negative selection using peanut agglutinin (Vector Laboratories). 500 metacyclic promastigotes with or without 0.5 pairs of L. longipalpis salivary gland homogenate (SGH) in 10 μl of PBS buffer were inoculated intradermally into the left ear dermis using a 29-gauge needle. The evolution of the lesion was monitored by measuring the ear thickness using a vernier caliper (Mitutoyo Corp.).
Expression and Purification of Recombinant Proteins
For crystallography and binding studies, LJM11, LJM111, and LJM17 were produced as recombinant proteins in Escherichia coli. The cDNA for each mature polypeptide was cloned into the expression vector, pET-17b, and expressed in the BL21(DE3)pLysS strain. The proteins were refolded as described previously for other salivary proteins (13) and purified using gel filtration and ion exchange chromatography. Mutation of Thr-328 and Asn-343 to alanine was performed using PCR (21). The quality of the protein preparations was assessed using gel filtration chromatography of purified proteins on Superdex 75 (supplemental Fig. S4). LJM11 used in vaccination experiments (referred to as LJM11HEK) was expressed in HEK 293 cells, and the supernatant was collected at 72 h as described previously (12). Expressed protein was purified by HPLC (DIONEX) using a HITRAP chelating HP column (GE Healthcare) charged with Ni2SO4 (0.1 m). Proteins were eluted using an imidazole gradient, dialyzed against PBS, and stored at −70 °C (12).
Cloning of DNA Plasmids Encoding L. longipalpis Salivary Proteins
Full-length cDNA coding for the proteins of the L. longipalpis saliva were cloned into a VR2001 vector as described elsewhere (22), and plasmids were purified using the Endofree Plasmid Megaprep kit following the manufacturer's instructions (Qiagen).
Immunization
Anesthetized mice (Isoflurane) were immunized three times in the right ear, intradermally, every 15 days with a volume of 10 μl containing 5 μg of the plasmid of interest. 15 days after the third immunization, mice were challenged in the left ear either with SGH (0.5 salivary gland pair/10 μl) or SGH plus L. major parasites (0.5 salivary gland pair with 500 parasites/10 μl).
Measurements of the DTH Response and Ear Lesion
DTH response was measured as an increase in the ear thickness 48 h after the challenge with SGH. Mice challenged with L. major parasites were followed weekly, and ear thickness measurements were used as an indication of lesion size. Ear measurements were performed using a Vernier caliper (Mitutoyo Corp.) from the dorsal to the ventral side of the ear.
ELISA
ELISA plates (Immulon4-Thermo) were coated overnight at 4 °C with 50 μl of a solution of carbonate-bicarbonate buffer, pH 9.6, containing 1 salivary gland pair/ml of L. longipalpis. After washing with PBS-Tween (0.05%) and blocking with PBS-Tween (0.05%) plus 4% BSA for 2 h at room temperature, sera from immunized mice (1:50) were incubated for 1 h at 37 °C. After washing, plates were incubated with alkaline phosphatase-conjugated anti-mouse IgG (Promega) antibody (1:1000). The plate was revealed using alkaline phosphate substrate (Sigma-Aldrich). Absorbance was recorded at 405 nm.
Measurements of Parasite Loads
Parasite load was determined using a limiting dilution assay as described elsewhere (23). Briefly, ear tissue was excised and homogenized in RPMI medium. The homogenate was serially diluted on Schneider medium (Invitrogen) supplemented with 20% heat-inactivated fetal bovine serum (HyClone), 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm l-glutamine, and 40 mm Hepes and seeded in 96-well plates containing biphasic blood agar (Novy-Nicolle-McNeal). The number of viable parasites was determined from the highest dilution at which promastigotes could be found after 21 days of culture at 23 °C.
Generation of Bone Marrow-derived Dendritic Cells (BMDCs)
Bone marrow cells were removed from the femurs and tibias of mice and cultured in RPMI medium supplemented with 10% heat-inactivated fetal bovine serum (HyClone), 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm l-glutamine, 40 mm Hepes, 5 × 10−5 m 2-mercaptoethanol, and 20 ng/ml GM-CSF (Prepotech). On days 3 and 6, complete medium was added containing 10 ng/ml GM-CSF. BMDCs were pulsed with 10 μg/ml LJM11HEK or one pair of L. longipalpis SGH for 24 h and then activated with 100 ng/ml LPS (Sigma-Aldrich) for 6 h. Dendritic cells were harvested, washed once, and adjusted to 106cells/ml.
Stimulation of Splenocytes with LJM11HEK
Splenocytes were harvested from C57BL/6 mice exposed to uninfected bites of L. longipalpis, and cell suspension was prepared by pressing spleen cells through a nylon strainer. Cells were collected, washed twice, and resuspended in RPMI medium supplemented with 10% heat-inactivated fetal bovine serum (HyClone), 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm l-glutamine, 40 mm Hepes, and 5 × 10−5 m 2-mercaptoethanol. Splenocytes (5 × 106/ml) were incubated for 72 h with 106/ml dendritic cells previously pulsed with LJM11HEK or SGH. Supernatants were collected and tested by ELISA for IFN-γ following the manufacturer's protocol (BD Biosciences).
Crystallization and Data Collection
LJM11 was crystallized using the hanging drop vapor diffusion method from 0.1 m sodium citrate, pH 5.4, 10% PEG 6000, 6 mm NiCl2. The crystals were frozen for data collection in the crystallization buffer described above containing 20–25% glycerol. To prepare a selenomethionine derivative of LJM11, the BL834(DE3)pLysS cells were transformed with the expression construct and grown in SelenoMet medium (Molecular Dimensions). The protein was folded and purified as described above. A complex of LJM11 and serotonin was prepared by soaking crystals for ∼5 min in cryoprotectant solution containing 2 mm serotonin. LJM11 was also cocrystallized with serotonin. In this case, the ligand was added to the protein at a 1.1:1 ratio of ligand to protein prior to preparation of the crystallization drops.
Data collection was performed at beamlines 19-ID of the Structural Biology Center and 22-ID at the Southeast Regional Collaborative Access Team, Advanced Photon Source, Argonne National Laboratory. All of the crystals obtained belong to the space group P6222 and contain two monomers in the asymmetric unit (Table 2). Four data sets were collected from a ligand-free selenomethionine crystal (2.75 Å resolution), a ligand-free crystal (2.93 Å resolution), a serotonin-soaked crystal (2.52 Å resolution), and a serotonin-complexed co-crystal (2.90 Å resolution).
TABLE 2.
Data collection, phasing, and refinement statistics for LJM11 and its serotonin complex
| Selenomethionine | Ligand-free | Serotonin | |
|---|---|---|---|
| Resolution (Å) | 40.0-2.75 | 25.0-2.93 | 50.0-2.52 |
| Beamline | 19-ID | 19-ID | 22-ID |
| Wavelength (Å) | 0.97915 | 0.97937 | 1.0000 |
| Completeness (total/high resolution shell) | 99.3/96.7 | 99.6/99.4 | 99.3/91.1 |
| Average redundancy (total/high resolution shell) | 12.5/12.9 | 14.0/14.6 | 23.0/4.5 |
| Rmerge (total/high resolution shell) | 9.4/71.1 | 9.5/64.7 | 8.1/67.9 |
| I/σI (total/high resolution shell) | 51.3/7.4 | 13.3/7.6 | 11.4/3.2 |
| Observed reflections | 653,588 | 331,743 | 826,286 |
| Unique reflections | 28,420 | 23,723 | 35,867 |
| Space group | P6222 | P6222 | P6222 |
| Unit cell dimensions (Å) | |||
| a | 120.36 | 120.38 | 120.01 |
| b | 120.36 | 120.38 | 120.01 |
| c | 248.93 | 248.12 | 244.75 |
| α, β, γ (degrees) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Phasing statistics | |||
| No. of selenium sites | 15 | ||
| FOM (MLphare) | 0.86 | ||
| Root mean square deviations | |||
| Bond lengths (Å) | 0.011 | 0.014 | 0.011 |
| Bond angles (degrees) | 1.40 | 1.60 | 1.46 |
| Ramachandran plot (favored/allowed) | 85.0/100 | 82.8/100 | 85.3/100 |
| Mean B value for all atoms | 49.46 | 69.84 | 55.95 |
| Rcryst/Rfree | 19.7/26.0 | 19.2/24.4 | 22.0/27.0 |
Structure Solution and Refinement
The structure of LJM11 was determined from the selenomethionine derivative using single wavelength anomalous dispersion methods with data collected at the selenium absorption edge (Table 2). The data were indexed, integrated, and scaled using HKL-3000 (24). Initial phases were obtained using the structure determination features of HKL-3000. This includes the location of selenium sites using SHELX-D and phasing of the reflection data using SHELX-E and MLphare (25–27). A portion of the model was built using Buccaneer (28), and the remainder was built manually using Coot (29). During the rebuilding phase, the model was refined using REFMAC (27) with non-crystallographic symmetry restraints being applied. At the end of the refinement, the non-crystallographic symmetry restraints were relaxed, and a TLS model was applied, utilizing a single TLS group per molecule of LJM11 (27). The structures of ligand-free LJM11 (3Q6T), its selenomethionine derivative (3Q6P) and the serotonin complex after ligand soaking (3Q6K) have been deposited with the Protein Data Bank. A homology model of LJM111 was produced using the Swiss Model Web Server in the project mode. Pairwise alignments of LJM111 with LJM11 were adjusted manually and were then submitted for modeling using the LJM11 structure as a template.
Smooth Muscle Bioassay
Smooth muscle bioassays were performed as described previously (20). Guinea pig ileum contractions in response to histamine were measured isotonically. A modified Tyrode solution (with 10 mm HEPES buffer, pH 7.4) was used for the ileum. All solutions were oxygenated by bubbling air through the bath.
Isothermal Titration Calorimetry
Salivary yellow proteins were prepared for isothermal titration calorimetry (ITC) experiments by dialysis against 20 mm Tris-HCl, 0.15 m NaCl, pH 8.0, for 1 h. Binding experiments were performed on a MicroCal VP-ITC instrument. Aliquots of lipid stock solutions in ethanol or methyl acetate were dried under a stream of nitrogen in a glass vial and then dissolved in 20 mm Tris-HCl, 0.15 m NaCl, pH 8.0, and sonicated for 8 min. Biogenic amine ligands were dissolved in 20 mm Tris-HCl, 0.15 m NaCl, pH 8.0. Assays were performed at 30 °C, and the injection enthalpies were analyzed by fitting to a single-site binding model, after subtraction of the heats of dilution, using the MicroCal Origin software.
Sequence Analysis
A sequence motif containing residues having side chains lying within 5 Å of the bound serotonin ligand in the binding pocket of the LJM11-serotonin complex were identified. The motif was then used to search the non-redundant GenBankTM protein data base with the sequence pattern search program Seedtop from the NCBI Blast package.
Statistical Analysis
Statistical evaluation of the means of experimental groups was done using one-way analysis of variance followed by the Tukey-Kramer post-test. The disease burden was calculated as the area under the lesion curve for each mouse (31). Data from parasite numbers were log-transformed before conducting statistical tests. Significance was determined as p < 0.05. All statistical tests and graphs were done using GraphPad version 5 (GraphPad Software Inc.).
RESULTS
Screening for DTH-inducing Salivary Proteins from L. longipalpis in Mice
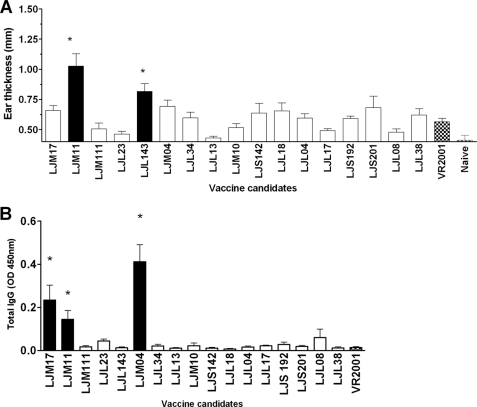
Cell-mediated immune responses to whole sand fly saliva, defined by a DTH response, are largely protective against Leishmania infection (5, 7, 9, 32). We selected 17 abundant salivary proteins (supplemental Table 1) from 35 previously identified secreted L. longipalpis salivary proteins (33) to search for the DTH-inducing molecules. Of these, only LJM11 and LJL143 exhibited a statistically significant DTH response defined by a significant increase in ear thickness over VR2001 (empty vector)-vaccinated mice or naive controls 48 h after challenge with SGH (Fig. 1A). Fig. 1B shows levels of antibodies specific to salivary proteins before challenge with SGH. Vaccination with LJM17, LJM11, and LJM04 induced antibody levels that were significantly higher than VR2001-vaccinated mice or naive controls. Notably, LJM17 and LJM111 yellow proteins, closely related to LJM11, induced only antibodies (LJM17) or, in the case of LJM111, did not elicit a DTH response or antibody production (Fig. 1, A–C).
FIGURE 1.
Salivary proteins LJM11 and LJL143 from L. longipalpis produce a DTH response in C57BL/6 mice. A, mice (n = 5) were injected three times at 2-week intervals in the right ear with 10 μl containing 5 μg of one of 17 L. longipalpis plasmids. Animals were challenged 2 weeks later in the left ear with 0.5 pair of L. longipalpis salivary gland homogenate. DTH was assessed by measuring ear thickness at 48 h. B, mice serum was obtained just before challenge and tested for levels of anti-saliva-specific antibodies by ELISA. Results are expressed as the mean ± S.D. (error bars). *, statistical significance (p < 0.05) relative to naive and empty vector-injected mice. Data represent three independent experiments.
Vaccination with DNA Plasmid Coding for LJM11 Protects Mice against L. major Infection
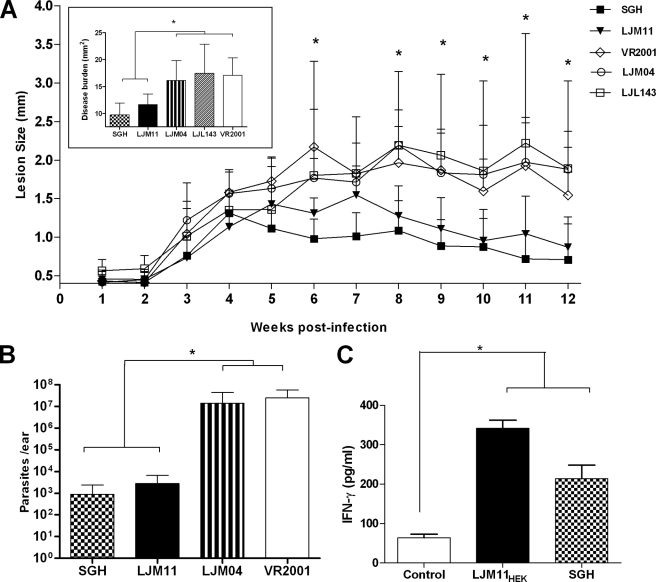
To test the protective potential of DTH-inducing salivary candidates against Leishmania infection, we challenged mice vaccinated with LJM11 and LJL143, the strongest inducers of cellular immunity to saliva (Fig. 1A). In addition, we tested LJM04, a protein that did not induce a DTH response but produced antibodies (Fig. 1B). Following challenge with 500 metacyclics of L. major together with SGH, the VR2001-, LJL143-, and LJM04-vaccinated groups developed ulcerative lesions as early as 3 weeks postinfection (Fig. 2A). In contrast, mice vaccinated with LJM11 or whole SGH controlled lesion development throughout the follow-up period (Fig. 2A). Additionally, lesions in VR2001-, LJL143-, and LJM04-vaccinated mice remained ulcerated, whereas those of SGH- or LJM11-vaccinated mice showed only a small scarred area. Considering disease burden as a whole, it was significantly reduced in LJM11- or SGH-vaccinated animals compared with LJM04-, LJL143-, or VR2001-vaccinated mice (Fig. 2A, inset). Parasite load 12 weeks postinfection corroborated the pathology showing that mice vaccinated with LJM11 or SGH controlled parasite growth showing a 3.6 log reduction in parasite numbers (p < 0.001) compared with LJM04- or VR2001-vaccinated mice (Fig. 2B).
FIGURE 2.
Immunization with LJM11 protects mice from cutaneous leishmaniasis. A, mice (n = 5) were immunized three times every 2 weeks in the right ear with 10 μl containing 5 μg of LJM11, LJM04, or LJL143 plasmids. Two weeks after the last immunization, mice were challenged with 500 L. major parasites and 0.5 pair of L. longipalpis SGH. Disease burden was calculated as the mean of the area under the lesion curve for each mouse (inset). B, parasite load was assessed at 12 weeks postinfection using a limiting dilution assay. C, interferon-γ production by splenocytes from mice exposed to bites of L. longipalpis sand flies following stimulation with BMDCs pulsed with LJM11HEK or SGH and activated with LPS. The control group received BMDCs activated with LPS. Results are expressed as the mean ± S.D. (error bars). *, statistical significance (p < 0.05). Data represent two independent experiments.
Spleen Cells from Mice Exposed to Bites of L. longipalpis Produce IFN-γ upon Stimulation with Recombinant LJM11
We tested whether exposure to bites of L. longipalpis, where mice receive a physiological dose of the native LJM11, could induce a specific immune response to recombinant LJM11 produced in HEK 293 cells (LJM11HEK). Splenocytes from exposed mice produced significantly higher IFN-γ upon stimulation with BMDCs primed with LJM11HEK or SGH compared with cells primed with BMDCs alone (Fig. 2C). These data confirm that LJM11HEK can be recognized by an immune response mounted against the native protein injected by the fly and demonstrate that immunity to LJM11 is Th1-biased and systemic.
The Yellow Salivary Proteins LJM11, LJM111, and LJM17 Are Kratagonists Specific for Biogenic Amines
The biological function of the sand fly salivary proteins in the yellow family has not been previously elucidated. We hypothesized that, like some other highly expressed proteins in the saliva of blood-feeding arthropods, LJM11 and other yellow proteins may bind physiologically active small molecule ligands, such as the prohemostatic and proinflammatory biogenic amines or eicosanoids. Biogenic amines, including serotonin, norepinephrine, and histamine as well as the eicosanoid leukotriene E4 were evaluated as potential ligands in binding assays using ITC. For these experiments, LJM11, LJM111, and LJM17 were expressed in E. coli and refolded (Table 1 and supplemental Fig. S1). All three proteins bound various biogenic amine compounds at a single site (n ∼ 1), in an enthalpy (ΔH)-driven manner. The eicosanoid compound leukotriene E4 did not bind, indicating that none of the proteins has a lipid binding function (data not shown). LJM11 bound serotonin with higher affinity than the catecholamines epinephrine and norepinephrine, as indicated by the value of the dissociation constant, Kd (Table 1 and supplemental Fig. S1). Conversely, LJM111, the variant most closely related to LJM11, bound serotonin with lower affinity than LJM11 (37-fold), whereas norepinephrine and epinephrine were bound extremely tightly by this protein (Table 1).
TABLE 1.
Thermodynamic parameters for the binding of biogenic amines with LJM11, LJM111, and LJM17
S.E. values were calculated from regression residuals.
| Protein | Ligand | ΔH | Kda | nb | TΔS |
|---|---|---|---|---|---|
| kcal/mol | nm | ||||
| LJM11 | Norepinephrine | −23.3 ± 0.2 | 63 ± 4.3 | 0.7 | −13.3 |
| Epinephrine | −8.2 ± 0.2 | 454 ± 80 | 1.3 | 0.6 | |
| Serotonin | −15.8 ± 0.2 | 4.3 ± 1.1 | 1.0 | −4.2 | |
| Dopamine | −19.2 ± 0.1 | 12 ± 1 | 1.0 | −8.2 | |
| Octopamine | −16.3 ± 0.3 | 217 ± 19 | 1.1 | −7.1 | |
| Histamine | >1000 | ||||
| LJM111 | Norepinephrine | −24.0 ± 0.3 | ≤2.5 | 1.0 | |
| Epinephrine | −23.3 ± 0.2 | ≤2.5 | 0.9 | ||
| Serotonin | −9.6 ± 0.4 | 161 ± 37 | 0.9 | −0.2 | |
| Dopamine | −22.7 ± 0.3 | 7.1 ± 1.6 | 1.0 | −11.0 | |
| Octopamine | −18.0 ± 0.5 | 194 ± 38 | 1.3 | −8.7 | |
| Histamine | >1000 | ||||
| LJM17 | Norepinephrine | −15.4 ± 1.0 | 280 ± 58 | 0.7 | −6.4 |
| Epinephrine | >1000 | ||||
| Serotonin | −18.0 ± 0.1 | ≤2.5 | 1.0 | ||
| Dopamine | −15.2 ± 0.3 | 44 ± 8 | 0.9 | −5.0 | |
| Octopamine | −15.9 ± 0.7 | 296 ± 41 | 0.9 | −6.8 | |
| Histamine | −18.9 ± 0.1 | 141 ± 8 | 0.9 | −9.3 |
a A value of ≤2.5 indicates that a protein concentration of 2.5 μm was used, and the measured Kd value was less than or equal to 2 nm (43). Kd values of >1000 indicate weak but detectable binding at concentrations of at least 10 μm protein and 100 μm ligand.
b Values indicate the stoichiometry of ligand binding.
LJM17 is somewhat divergent in sequence from LJM11 (47% amino acid identity) and LJM111 (42% amino acid identity). Like LJM11, LJM17 bound serotonin with very high affinity and norepinephrine with considerably lower affinity. In this case, no detectable binding of epinephrine was observed (Table 1).
Dopamine and octopamine are unlikely physiological targets for the salivary yellow proteins because they do not play a major role in hemostasis or inflammation. Nevertheless, they bound with relatively high affinity to the salivary yellow proteins (Table 1), presumably due to their structural similarity to norepinephrine and epinephrine. Interestingly, the differences in dopamine affinity between the proteins are relatively small, whereas the differences in norepinephrine and epinephrine binding are large. This suggests that the secondary hydroxyl group of norepinephrine and epinephrine is an important determinant of high affinity binding in LJM111. The affinity for octopamine is lower than for dopamine and was not significantly greater in LJM111 than LJM11, despite the presence of a secondary hydroxyl group in this compound (Table 1). This suggests that the ortho arrangement of phenolic hydroxyl groups on the ligand aromatic ring is also important for high affinity catecholoamine binding in LJM111.
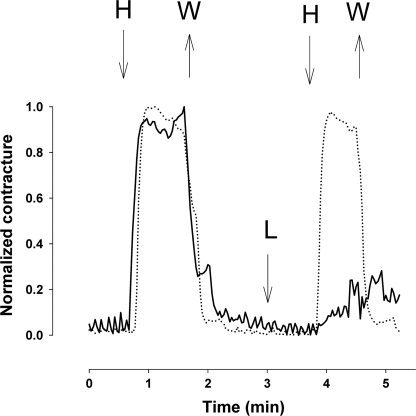
Histamine was a poor ligand for LJM11 and LJM111, producing affinity constants greater than 1 μm, suggesting that this mast cell product is not a target ligand for either of these proteins (Table 1). Conversely, LJM17 binds histamine with a dissociation constant of less than 150 nm, suggesting that it might effectively sequester this compound during insect feeding. To determine if this affinity is sufficient to prevent the interaction of histamine with its receptor, we tested the ability of LJM17 to inhibit the contraction of a physiological preparation of guinea pig ilieum by histamine. The addition of 5.7 μm LJM17 to a solution bathing an ex vivo ileum preparation abrogated the contraction induced by 0.5 μm histamine (Fig. 3).
FIGURE 3.
Effect of LJM17 on the contraction of guinea pig ileum induced by histamine (0.5 μm). In the first experiment (dotted line) guinea pig ileum was contracted for 1 min with histamine (H), followed by replacement of the saline solution with one lacking histamine (W), resulting in relaxation of the preparation. A second administration of histamine and replacement of the bath were then performed to demonstrate reproducibility of the contraction. In a second experiment (solid line), histamine was added to the same preparation and removed as above, resulting in a response very similar to the first addition. In this case, LJM17 was then added to a concentration of 5.7 μm (L) prior to the second pulse of histamine (0.5 mm; H). The response to histamine was almost completely eliminated in the presence of LJM17.
The Crystal Structure of LJM11
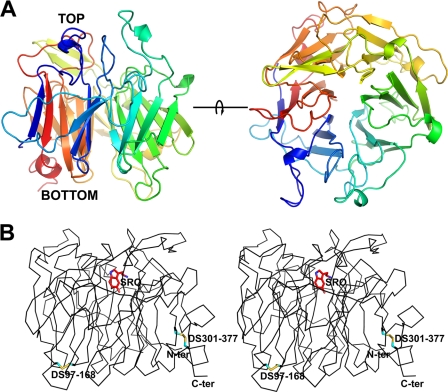
In order to fully understand the biological function of LJM11, we crystallized the E. coli-produced recombinant protein and determined its three-dimensional structure (Table 2 and Fig. 4). LJM11 has a six-bladed β-propeller fold with a central, solvent-accessible channel having openings on opposite faces of the molecule. Each blade consists of a four-stranded β-sheet with the possible exception of the first blade, in which the fourth strand forms only two backbone hydrogen bonds with strand 3 from the same blade (Fig. 4A). The fourth β-strand of each blade connects with the first strand of the subsequent one, forming part of a series of large loops on the “top” side of the molecule that surround the entry to the central channel (Fig. 4A). The remaining loops on this side of the protein are made up of the connections between strands 2 and 3 of each β-sheet. The C-terminal blade of the propeller structure contains the three most C-terminal β-strands with strand 4 being contributed by the N terminus of the protein, producing what has been referred to as a Velcro closure to the propeller (Fig. 4, A and B) (34). The wall of the central channel consists of a parallel arrangement of the first strands from each blade of the propeller (Fig. 4A). Potential ligand binding sites occur at each end of the channel, with one being located on the “top” of the molecule and surrounded by the series of loops described above. The second potential entry point is at the “bottom” side of the protein. The loops surrounding the bottom entry are made up of the connections between β-strands 1 and 2 along with those of strands 3 and 4 from each β-sheet. Approximately midway through the central channel, the side chains of Gln-91 and Asp-328 are positioned by hydrogen bonds to partially occlude the channel, dividing it into two distinct pockets. Two disulfide bonds are present in LJM11, which link Cys-97 with Cys-168 and Cys-301 with Cys-377 (Fig. 4B).
FIGURE 4.
The crystal structure of LJM11. A, side (left) and top (right) views of LJM11. The top side view represents an approximate 90° rotation around the horizontal axis. The color scheme represents a gradient from blue at the N terminus to red at the C terminus. B, stereoview of Cα trace of the LJM11-serotonin complex. The positions of the two disulfide bonds are labeled DS, followed by the residue numbers of the two cysteine residues participating in the bond. The cysteine side chains are colored in cyan and yellow. The serotonin ligand (SRO) is colored red and shown as a stick diagram. The N and C termini of the model are indicated.
The Ligand-binding Site of LJM11
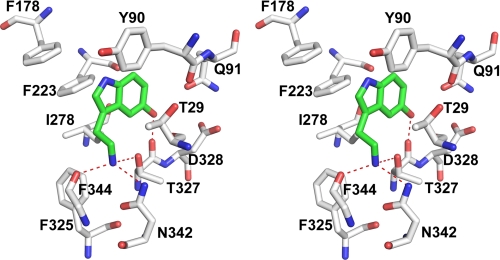
Data were collected from LJM11 crystals to which serotonin had been added via soaking in ligand-containing cryoprotectant solution or by adding to the crystallization drop the day before freezing (Table 2). With both conditions, a single large peak of electron density was observable in 2Fo − Fc maps in the top side binding pocket consistent with the 1:1 binding stoichiometry observed in the ITC experiments (supplemental Fig. S2). The addition of ligand to the crystallization drop produced more interpretable electron density for the ligand, whereas the addition of ligand to the cryoprotectant produced higher diffraction resolution and superior overall structure quality. Serotonin was modeled into the pocket, and after refinement, its binding mode could be determined (Fig. 5). In the refined structure, the ligand is oriented with its amino group positioned to form hydrogen bonds with the side chains of Asn-342 and Thr-327 as well as with the carbonyl oxygen of Phe-344. The indole portion of the ligand is surrounded by a number of hydrophobic and aromatic side chains, including Phe-178, Phe-223, Ile-278, Phe-325, Phe-344, and Tyr-90 (Fig. 5). The phenolic hydroxyl group of serotonin forms a hydrogen bond with the carbonyl oxygen of Thr-327 and possibly the side chain of Gln-91 (Fig. 5).
FIGURE 5.
Stereoview of the biogenic amine-binding pocket of LJM11. Carbon atoms of the ligand are shown in green, and those of the protein are shown in white. Oxygen and nitrogen are colored in red and blue, respectively. Hydrogen bonds are shown as red dashed lines.
The validity of the binding pocket identification was tested by substitution of Thr-327 and Asn-342 for alanine using site-directed mutagenesis. Mutation of Asn-342 resulted in a nearly complete loss of detectable serotonin, epinephrine, and norepinephrine binding as measured by ITC (supplemental Fig. S1). This is consistent with the participation of this residue in a hydrogen bond with the amino group of the ligand. Substitution of Thr-327 by alanine had little effect on the binding of serotonin (Kd = 3.9 nm; supplemental Fig. S1) but caused a large reduction in the affinity of LJM11 for norepinephrine, epinephrine, and dopamine (supplemental Fig. S1). This result suggests that Thr-327 is not essential for binding of the amino group of serotonin but may interact with the secondary hydroxyl group or amino group of norepinephrine and epinephrine.
Modeling of LJM11 and LJM111
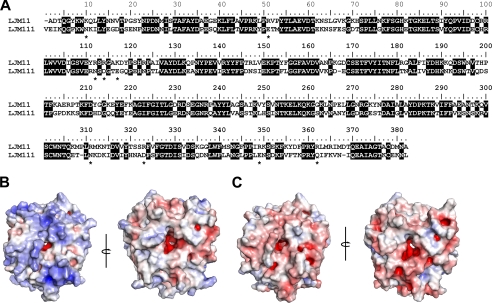
LJM11 and LJM111 are ∼60% identical at the amino acid level, and alignment of the two reveals no major sequence gaps (Fig. 6A). Importantly, LJM11 produces a DTH response to L. longipalpis saliva in immunized animals and confers protection against L. major infection as opposed to LJM111, which did not induce a DTH response or antibodies (Fig. 1). Because of their sequence similarities, the structure of LJM111 could be simply modeled using the crystal structure of LJM11 as a homology template. Electrostatic calculations (35) showed LJM11 and LJM111 to have differing surface charge characteristics, a feature that may be related to the different immunological properties of the two proteins. The surface of the top face of LJM11, including the area surrounding the entrance to the ligand binding pocket, carries a strong positive charge (Fig. 6B). In comparison, the corresponding face of LJM111 is not positively charged (Fig. 6C). The bottom surfaces of both proteins are neutral to weakly negative in charge, whereas the interior of their ligand binding pockets has a negative surface potential, consistent with the binding of positively charged (at physiological pH) biogenic amines. These features are reflected in differences in the calculated isoelectric points for LJM11 and LJM111, which are 9.3 and 4.7, respectively. The basic residues found in these charged surface regions of LJM11 but not LJM111 are highlighted in Fig. 6A.
FIGURE 6.
LJM11 and LJM111 display different electrostatic surface potentials. A, amino acid sequence alignment showing basic residues found in the positively charged surface regions of LJM11 (asterisk below alignment). These are Lys-9, Arg-50, Arg-111, Arg-113, Arg-116, Arg-180, Arg-310, Arg-322, Arg-348, and Arg-361. Identical sequence positions are shaded black. Shown are electrostatic potential along surfaces of LJM11 (B) and a hypothetical model of LJM111 (C) as determined by the Adaptive Poisson-Bolzmann Solver software, with blue being positive and red being negative. Left panels represent a view looking into the top surface of the proteins, and the panels to the right represent a view looking into the bottom surface following a 180° rotation around the vertical axis shown.
Sequence Relationships in the Yellow Protein Family
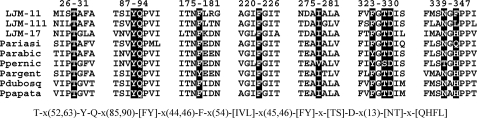
Sequence comparisons of salivary yellow proteins with other family members show that they are most closely related to yellow proteins from other species of Diptera and more distantly related to MRJPs from Hymenoptera (36). Within the sand flies, yellow proteins are known to occur in the salivary secretions of the genera Lutzomyia and Phlebotomus. We constructed a sequence motif from the LJM11 structure using only residues having side chains lining the binding pocket and lying within 5 Å of the ligand in the serotonin complex (Fig. 5). This motif was used to search the non-redundant GenBankTM protein data base. Only salivary yellow proteins from Lutzomyia and Phlebotomus sp. were retrieved in this search, suggesting that biogenic amine binding is restricted to this group and that salivary yellow proteins in both genera share this function (Fig. 7 and supplemental Fig. S3). Five of 11 residues in this motif are absolutely conserved (45%), whereas the overall sequence identity for this group of proteins is 17.9% (supplemental Fig. S3).
FIGURE 7.
Sequence alignment showing conserved amino acids contained in the ligand binding pocket of yellow protein family members. Only the segments surrounding these residues in selected yellow family members are shown, whereas the complete alignments are shown in the supplemental materials. The residues contained in the search motif are highlighted by shading, and the corresponding LJM11 amino acid positions for each sequence block are shown above the block. Below the alignment is the motif used for searching the non-redundant data base using Seedtop. The motif is shown in PROSITE notation (30). Pariasi, Phlebotomus ariasi gi 61373243; Parabic, Phlebotomus arabicus gi 242564631; Ppernic, Phlebotomus perniciosus gi 76446591; Pargent, Phlebotomus argentipes gi 74486543; Pdubosq, Phlebotomus dubosqi gi 112361963; Ppapata, Phlebotomus papatasi gi 15963519.
DISCUSSION
In vaccine development, understanding the biological role and structure of a candidate antigen facilitates prediction and resolution of any unwanted effects on host physiology. Despite the fact that several sand fly salivary molecules have been identified as protective against leishmaniasis (10, 11), the significance of their physiological activity and the structural basis for their ability to induce a DTH response have not been elucidated. Here, we have identified LJM11 as a DTH-inducing protein that confers protection against cutaneous leishmaniasis in a mouse model of infection and determined that this protein binds biogenic amines, a group of prohemostatic and proinflammatory mediators potentially facilitating feeding.
Robust IFN-γ production by splenocytes of sand fly-exposed mice stimulated with BMDCs pulsed with LJM11HEK demonstrates that immunity to LJM11 following exposure to bites is systemic and as such potentially inducible at any site of bite. This is critical when considering LJM11 as a potential vaccine for inhabitants of endemic regions. Importantly, LJM11 is immunogenic in humans and dogs, as demonstrated by its strong induction of a specific humoral response in both species (12) as well as a cellular skin reaction in dogs (37). LJM111, a related yellow protein from L. longipalpis, binds biogenic amines in a manner similar to LJM11 but does not produce a significant DTH response. Additionally, it is non-immunogenic in humans and dogs (12, 37). Sharing a similar global structure and function, the most conspicuous difference between LJM11 and LJM111 is surface charge distribution. The surface of the LJM11 protein carries a positive electrostatic potential on the face containing entry to the ligand binding pocket, whereas a hypothetical model of LJM111 is negative to neutral over its entire surface. Positively charged nanoparticles have been shown to be much more effectively phagocytosed by macrophages and dendritic cells than negatively charged or neutral particles (38, 39). This suggests that the attribute responsible for the DTH-inducing ability of LJM11, and therefore its ability to protect animals against infection by Leishmania, is independent of its function as a biogenic amine-binding protein and could be due more to its surface structural features. However, salivary proteins are known to be multifunctional in some cases, and it is possible that the protective ability of the protein could be influenced by a function that has yet to be discovered.
Yellow protein family representatives are found in a taxonomically diverse group of insects as well as a variety of bacteria, fungi, and two non-arthropod metazoans, but little is known about the function of most family members (36, 40). The limited distribution of the family in animals suggests that in insects, it originated by lateral transfer from bacteria (36) and has subsequently taken on essential roles in melanization of the cuticle. Clearly, additional functions will be uncovered for these proteins because D. melanogaster possesses 13 different forms, the silk moth Bombyx mori contains 7, the flour beetle Tribolium castaneum contains 14, and three species of the parasitic wasp Nasonia sp. contain 25 (36). In the honey bee, nine MRJP members have been identified along with a number of other yellow family members (36). A core group of yellow proteins appears to be conserved in the four largest orders of insects. The function of the yellow family member has only been firmly established for the dopachrome conversion enzymes (DCEs) of Drosophila and mosquitoes that are involved in melanization (41, 42) and here for the Lutzomyia salivary yellow proteins. The molecular mechanism of each protein involves binding of a small molecule. This raises the possibility that ligand binding is a common functional theme within the family as a whole.
The identification and structural characterization of the LJM11 binding pocket located on the top surface of the six-bladed β-propeller structure has allowed us to examine other members of the yellow family for conservation of amino acid sequence in this region and take the first step toward identifying possible functional groupings within the family. A motif based on the binding pocket of LJM11 retrieved only 14 salivary yellow protein family members from the genera Lutzomyia and Phlebotomus, strongly suggesting a common function of biogenic amine binding for all of these proteins. It is likely that the active site of the DCEs is coincident with that of the biogenic amine-binding pocket of LJM11 because ligand binding sites of β-propeller proteins are normally found on the top side of the protein structure (43, 44) in a location similar to that of LJM11. The structures of serotonin and dopachrome are similar, and it is plausible that they would bind in a similar manner. It may be possible to identify likely DCE proteins in additional insect species based on conservation of hypothetical binding site residues.
Like many other salivary proteins, the yellow proteins of L. longipalpis occur as a multigene family arising as a result of apparent gene duplication events (1). In other salivary protein families, the acquisition of new functions by individual gene cluster members, beyond the common function of the group, has been demonstrated (1). These functional differences can be relatively small, resulting in variations in affinity for a particular ligand, or large, in which completely new binding partners are acquired. The data presented here suggest that L. longipalpis yellow proteins have distinctly different binding selectivity for biogenic amines, indicating that functional divergence has occurred within the family. LJM11 shows very high affinity for serotonin and moderate to low affinity for catecholamines, whereas LJM111 binds catecholamines with very high affinity and shows moderate affinity for serotonin (Table 1). LJM17 binds serotonin very tightly and also binds histamine, suggesting that the protein helps to dampen the proinflammatory effects of this compound in the skin during feeding. This system provides another example of gene duplication followed by divergence and modification of function in the evolution of salivary protein families. In this case, a possible progenitor of the salivary yellow proteins is apparent. As discussed above, yellow family members play an important role in the melanization of cuticle (41, 42). The structural similarity of the DCE substrate and ligands bound by LJM11 suggests that a DCE protein served as a progenitor to the salivary yellow proteins.
It is also possible that the multiple yellow protein forms are maintained through selection pressure imposed by the host immune system. LJM11 and LJM111 belong to the same family of proteins but have distinct immunogenicity. One may speculate that antibodies generated against LJM11 may inhibit the ability of this protein to bind biogenic amines. The maintenance of LJM111, which is very silent immunologically but retains the ability to bind biogenic amines, suggests that LJM11 may act as a decoy system to deviate the immune system to one molecule while preserving the needed activity with a similar molecule.
LJM11 has critical attributes that make it a good candidate for further development as a component of a Leishmania vaccine: 1) it is immunogenic, driving a Th1 type immune response in mice that protects against Leishmania infection; 2) it is recognized by the human and dog immune systems, suggesting that it is immunogenic across species; 3) it can be produced in recombinant form in mammalian cells and bacteria; 4) it has no mammalian homologues; 5) following elucidation of its function and structure, it is now amenable to manipulation without compromising the ability to elicit a DTH response. Overall, we conclude that the rich mixture of salivary proteins found in blood feeding insects is an important aspect in the development of vaccines against vector-borne diseases. Here, we identify LJM11 as an immunogenic sand fly salivary protein that confers protection against Leishmania parasites; we also describe the crystal structure and function of LJM11 and show for the first time that the DTH-inducing ability of a sand fly salivary protein is independent of a known biological function.
Supplementary Material
Acknowledgments
We thank D. Garboczi and A. Gittis for discussions. We also thank the staffs of the Structural Biology Center and the Southeast Regional Collaborative Access Team, Advanced Photon Source, Argonne National Laboratory for assistance with x-ray data collection.
This work was supported, in whole or in part, by the National Institutes of Health, NIAID, Intramural Research Program. Use of the Advanced Photon Source beamlines was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38.
The atomic coordinates and structure factors (codes 3Q6T, 3Q6P, and 3Q6K) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
- DTH
- delayed type hypersensitivity
- MRJP
- major royal jelly protein
- SGH
- salivary gland homogenate
- BMDC
- bone marrow-derived dendritic cell
- ITC
- isothermal titration calorimetry
- DCE
- dopachrome conversion enzyme.
REFERENCES
- 1. Ribeiro J. M., Francischetti I. M. (2003) Annu. Rev. Entomol. 48, 73–88 [DOI] [PubMed] [Google Scholar]
- 2. Ribeiro J. M., Mans B. J., Arcà B. (2010) Insect Biochem. Mol. Biol. 40, 767–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belkaid Y., Valenzuela J. G., Kamhawi S., Rowton E., Sacks D. L., Ribeiro J. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6704–6709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gomes R., Teixeira C., Teixeira M. J., Oliveira F., Menezes M. J., Silva C., de Oliveira C. I., Miranda J. C., Elnaiem D. E., Kamhawi S., Valenzuela J. G., Brodskyn C. I. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 7845–7850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kamhawi S., Belkaid Y., Modi G., Rowton E., Sacks D. (2000) Science 290, 1351–1354 [DOI] [PubMed] [Google Scholar]
- 6. Morris R. V., Shoemaker C. B., David J. R., Lanzaro G. C., Titus R. G. (2001) J. Immunol. 167, 5226–5230 [DOI] [PubMed] [Google Scholar]
- 7. Oliveira F., Lawyer P. G., Kamhawi S., Valenzuela J. G. (2008) PLoS Negl. Trop. Dis. 2, e226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thiakaki M., Rohousova I., Volfova V., Volf P., Chang K. P., Soteriadou K. (2005) Microbes Infect. 7, 760–766 [DOI] [PubMed] [Google Scholar]
- 9. Valenzuela J. G., Belkaid Y., Garfield M. K., Mendez S., Kamhawi S., Rowton E. D., Sacks D. L., Ribeiro J. M. (2001) J. Exp. Med. 194, 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bethony J. M., Cole R. N., Guo X., Kamhawi S., Lightowlers M. W., Loukas A., Petri W., Reed S., Valenzuela J. G., Hotez P. J. (2011) Immunol. Rev. 239, 237–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oliveira F., Jochim R. C., Valenzuela J. G., Kamhawi S. (2009) Parasitol. Int. 58, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Teixeira C., Gomes R., Collin N., Reynoso D., Jochim R., Oliveira F., Seitz A., Elnaiem D. E., Caldas A., de Souza A. P., Brodskyn C. I., de Oliveira C. I., Mendonca I., Costa C. H., Volf P., Barral A., Kamhawi S., Valenzuela J. G. (2010) PLoS Negl. Trop. Dis. 4, e638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Andersen J. F., Francischetti I. M., Valenzuela J. G., Schuck P., Ribeiro J. M. (2003) J. Biol. Chem. 278, 4611–4617 [DOI] [PubMed] [Google Scholar]
- 14. Calvo E., Mans B. J., Ribeiro J. M., Andersen J. F. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3728–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mans B. J., Ribeiro J. M. (2008) Insect Biochem. Mol. Biol. 38, 862–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mans B. J., Ribeiro J. M., Andersen J. F. (2008) J. Biol. Chem. 283, 18721–18733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ribeiro J. M., Walker F. A. (1994) J. Exp. Med. 180, 2251–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oliveira M. C., Pelegrini-da-Silva A., Parada C. A., Tambeli C. H. (2007) Neuroscience 145, 708–714 [DOI] [PubMed] [Google Scholar]
- 19. Xanthos D. N., Bennett G. J., Coderre T. J. (2008) Pain 137, 640–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Calvo E., Mans B. J., Andersen J. F., Ribeiro J. M. (2006) J. Biol. Chem. 281, 1935–1942 [DOI] [PubMed] [Google Scholar]
- 21. Gudderra N. P., Ribeiro J. M., Andersen J. F. (2005) J. Biol. Chem. 280, 25022–25028 [DOI] [PubMed] [Google Scholar]
- 22. Oliveira F., Kamhawi S., Seitz A. E., Pham V. M., Guigal P. M., Fischer L., Ward J., Valenzuela J. G. (2006) Vaccine 24, 374–390 [DOI] [PubMed] [Google Scholar]
- 23. Lima H. C., Bleyenberg J. A., Titus R. G. (1997) Parasitol. Today 13, 80–82 [DOI] [PubMed] [Google Scholar]
- 24. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 25. Schneider T. R., Sheldrick G. M. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 26. Sheldrick G. M. (2002) Z. Kristallogr. 217, 644–650 [Google Scholar]
- 27. CCP4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 28. Cowtan K. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 29. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30. Sigrist C. J., Cerutti L., Hulo N., Gattiker A., Falquet L., Pagni M., Bairoch A., Bucher P. (2002) Brief. Bioinform. 3, 265–274 [DOI] [PubMed] [Google Scholar]
- 31. de Moura T. R., Oliveira F., Novais F. O., Miranda J. C., Clarêncio J., Follador I., Carvalho E. M., Valenzuela J. G., Barral-Netto M., Barral A., Brodskyn C., de Oliveira C. I. (2007) PLoS Negl. Trop. Dis. 1, e84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kamhawi S. (2000) Microbes Infect. 2, 1765–1773 [DOI] [PubMed] [Google Scholar]
- 33. Valenzuela J. G., Garfield M., Rowton E. D., Pham V. M. (2004) J. Exp. Biol. 207, 3717–3729 [DOI] [PubMed] [Google Scholar]
- 34. Fülöp V., Jones D. T. (1999) Curr. Opin. Struct. Biol. 9, 715–721 [DOI] [PubMed] [Google Scholar]
- 35. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ferguson L. C., Green J., Surridge A., Jiggins C. D. (2011) Mol. Biol. Evol. 28, 257–272 [DOI] [PubMed] [Google Scholar]
- 37. Collin N., Gomes R., Teixeira C., Cheng L., Laughinghouse A., Ward J. M., Elnaiem D. E., Fischer L., Valenzuela J. G., Kamhawi S. (2009) PLoS Pathog. 5, e1000441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roser M., Fischer D., Kissel T. (1998) Eur. J. Pharm. Biopharm. 46, 255–263 [DOI] [PubMed] [Google Scholar]
- 39. Thiele L., Rothen-Rutishauser B., Jilek S., Wunderli-Allenspach H., Merkle H. P., Walter E. (2001) J. Control Release 76, 59–71 [DOI] [PubMed] [Google Scholar]
- 40. Drapeau M. D., Albert S., Kucharski R., Prusko C., Maleszka R. (2006) Genome Res. 16, 1385–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fang J., Han Q., Johnson J. K., Christensen B. M., Li J. (2002) Biochem. Biophys. Res. Commun. 290, 287–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Han Q., Fang J., Ding H., Johnson J. K., Christensen B. M., Li J. (2002) Biochem. J. 368, 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Blum M. M., Löhr F., Richardt A., Rüterjans H., Chen J. C. (2006) J. Am. Chem. Soc. 128, 12750–12757 [DOI] [PubMed] [Google Scholar]
- 44. Dai J., Liu J., Deng Y., Smith T. M., Lu M. (2004) Cell 116, 649–659 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.