Abstract
The XRCC4-like factor (XLF)-XRCC4 complex is essential for nonhomologous end joining, the major repair pathway for DNA double strand breaks in human cells. Yet, how XLF binds XRCC4 and impacts nonhomologous end joining functions has been enigmatic. Here, we report the XLF-XRCC4 complex crystal structure in combination with biophysical and mutational analyses to define the XLF-XRCC4 interactions. Crystal and solution structures plus mutations characterize alternating XRCC4 and XLF head domain interfaces forming parallel super-helical filaments. XLF Leu-115 (“Leu-lock”) inserts into a hydrophobic pocket formed by XRCC4 Met-59, Met-61, Lys-65, Lys-99, Phe-106, and Leu-108 in synergy with pseudo-symmetric β-zipper hydrogen bonds to drive specificity. XLF C terminus and DNA enhance parallel filament formation. Super-helical XLF-XRCC4 filaments form a positively charged channel to bind DNA and align ends for efficient ligation. Collective results reveal how human XLF and XRCC4 interact to bind DNA, suggest consequences of patient mutations, and support a unified molecular mechanism for XLF-XRCC4 stimulation of DNA ligation.
Keywords: DNA Repair, Protein DNA-Interaction, Protein Structure, X-ray Crystallography, X-ray Scattering, DNA Ligase IV, Ku, XLF-XRCC4 Complex, Nonhomologous End Joining (NHEJ), Small Angle X-ray Scattering
Introduction
DNA double strand breaks (DSBs),4 produced by DNA-damaging agents such as ionizing radiation as well as during cellular processes such as V(D)J recombination and during responses to replication fork collapse, are considered the most cytotoxic form of DNA damage (1). The major pathway for DSB repair in human cells is nonhomologous end joining (NHEJ), defects in which lead to radiation sensitivity and immune deficiency as well as chromosomal translocations and genomic instability (2).
NHEJ proceeds in three steps as follows: detection of the lesion, DNA end processing, and finally DNA ligation (3). First, DSBs are detected by the Ku70/80 heterodimer. Ku tethers the DNA ends together and recruits other NHEJ factors, including DNA-PKcs (DNA-dependent protein kinase catalytic subunit), which regulates access of additional factors to the DNA ends through autophosphorylation-dependent mechanisms (4). Subsequently, DNA ends are processed by Artemis, polynucleotide kinase/phosphatase, and/or DNA polymerases and finally ligated by the XRCC4-DNA ligase IV complex in a reaction that is regulated by XRCC4-like factor (XLF, also called Cernunnos) (2, 3).
XLF interacts with XRCC4 (5), and XLF-deficient cell lines display sensitivity to ionizing radiation and defects in DSB repair (5, 6). Moreover, XLF deficiency in patients is characterized by immune deficiency as well as growth retardation and microcephaly (6); however, detailed functions of XLF in NHEJ remain unclear. Like XRCC4, XLF has no known enzymatic activity. Yet, XLF stimulates the rate of DNA ligase IV (LigIV)-mediated ligation in vitro 20–200-fold (7–9), particularly at mis-matched DNA ends (9). One way in which XLF may enhance ligation is by promoting re-adenylation of LigIV (10). In addition, it has been suggested that XLF may play a structural role, either by aligning the DNA (8) or by bridging DNA ends via interaction with Ku (11).
Both XRCC4 and XLF are composed of a globular head domain followed by a coiled-coil or stalk terminating in an unstructured C-terminal region (CTR), but in XLF the stalk region folds back upon itself (12, 13). Like XRCC4, XLF exists in solution as a homodimer and forms higher order oligomers in solution (12). Small angle x-ray scattering (SAXS) analyses suggest that XRCC4 and XLF interact via head-to-head interactions in solution (14). Mutations in XRCC4 and XLF head domains (Lys-63, Lys-65, and Lys-99 of XRCC4 and Leu-115 of XLF) disrupt the XRCC4-XLF interaction in vitro (12), and Arg-64, Leu-65, and Leu-115 of XLF are required for DSB repair in vivo (11) indicating that the interaction between XRCC4 and XLF is critical for NHEJ; however, the nature of and basis for functional interactions between XRCC4 and XLF are unknown.
Like XRCC4, XLF binds dsDNA in a DNA length- and protein concentration-dependent manner in vitro (7, 8, 12). The C-terminal region (CTR) of XLF (XLF(249–299)) has in vitro DNA binding activity (12, 15) and is required for XLF to go to sites of DNA damage in vivo (16), but it is not required for cellular survival after DNA damage, for DSB repair, or for V(D)J recombination (17). Recruitment of XLF to DNA in vitro and to DSBs in vivo requires Ku, whereas XRCC4 helps retain XLF at sites of damage (18). Thus, understanding how XLF interacts with XRCC4 and how this complex interacts with Ku and DNA and activates ligase IV is critical to understanding the role of XLF in NHEJ.
Here, we characterize the molecular basis of the stabilization of DSBs by XLF-XRCC4 complexes and the importance of this interface for NHEJ ligation. To elucidate XLF and XRCC4 interactions and conformational changes that affect DSB stabilization, we solved the crystal structure of XRCC4(1–140) in complex with XLF(1–224). Using a combination of crystallography, SAXS, and hydrogen-deuterium exchange mass spectrometry (HDX), we characterize the XRCC4-XLF interface and reveal conformational changes in both XRCC4 and XLF upon complex formation that permits multiple protein interactions and the formation of larger macromolecular assemblies. Furthermore, we identify new DNA-binding sites at the XLF-XRCC4 interface, and we propose a model to explain how a channel formed through XRCC4 and XLF interactions promotes alignment of long DNA substrates to facilitate ligation. Together with our SAXS solution structure of the Ku-XLF-XRCC4-DNA complex, our combined results explain synergetic effects of Ku-XLF-XRCC4-DNA complexation leading to DSB stabilization and enhancement of LigIV targeting and efficiency for NHEJ DSB repair.
EXPERIMENTAL PROCEDURES
Crystallization and Structural Analysis of XLF(1–224)- XRCC4(1–140)
Crystals were grown by hanging drop vapor diffusion. Equal volumes of XLF(1–224) and XRCC4(1–140) (∼10 mg/ml), in 50 mm Tris-HCl, pH 7.0, 300 mm NaCl, 1 mm DTT, 0.2 mm PMSF, 0.2 μg/ml pepstatin, 0.2 μg/ml leupeptin, and crystallization solution (100 mm HEPES, pH 7.8, 13% (w/v) PEG 3350, 300 mm NaCl with additives containing a mixture of 2 mm ADP, 7 mm NaF, and 3 mm BeCl), were dehydrated over 1000 μl of 19% PEG 3350, 300 mm NaCl, 100 mm HEPES, pH 7.8, under argon. Crystals (500 × 50 × 50 μm) grew after 2–3 days at 30 °C in a space group of P6522, with cell dimensions a = b = 110.0 Å and c = 763.7 Å and two complexes in the asymmetric unit, which corresponded to a solvent content of 69%. The structure of XLF(1–224)-XRCC4(1–140) was solved by molecular replacement with x-ray diffraction data to a limiting resolution of 3.97 Å collected from frozen crystals at the SIBYLS Beamline BL 12.3.1 and BL 8.3.1 of the Advanced Light Source (Berkeley, CA). After data processing with the HKL2000 (19), molecular replacement solution was accomplished with the refined structure of XLF(1–224) as a search model (PDB code 2r9a (12)) with the PHASER program (20). A resolution limit of 4.50 Å was used in the molecular replacement. The PHASER score of the solution was 70.6. The initial phase allowed visualization of electron density for XRCC4(1–140) (supplemental Fig. S1). The truncated structure of XRCC4(1–140) (PDB code 1fu1) was manually placed in the electron density map (supplemental Fig. S1) to construct the initial XLF(1–224)-XRCC4(1–140) structure. Initial phases were refined and extended from 4.50 to 3.97 Å with solvent flattening and rigid body refinement by the PHENIX (21) program. Before the first run of the refinement, all atomic B factors of the model were set to 80 Å2. A typical refinement protocol consisted of rigid body refinement. The rigid body groups included individual helices and sheets using secondary structure restraints. The model was then analyzed visually in COOT (22), and the ideal conformations of the side chains were fit to the resulting SigmaA-weighted 2Fo − Fc, Fo − Fc maps. Such refinement/rebuilding stages were carried out, giving final R, Rfree values of 35.7, 36.9%, respectively, for the resolution range 67–3.97 Å. Atomic coordinates and structure factors are deposited in the Protein Data Bank (PDB code 3SR2).
SAXS Data Collection and Evaluation
SAXS data were collected at the ALS beamline 12.3.1 (SIBYLS) Lawrence Berkeley National Laboratory, Berkeley, CA (23). The wavelength λ = 1.0 Å and sample-to-detector distances were set to 1.5 m, resulting in scattering vectors, q, ranging from 0.01 to 0.32 Å−1. The scattering vector is defined as q = 4π sinθ/λ, where 2θ is the scattering angle. All experiments were performed at 20 °C, and data were processed as described (23, 24). The buffer used in SAXS experiments was 50 mm Tris-HCl, pH 7.8, 150 mm NaCl, 1 mm DTT, 1 mm PMSF. All proteins and protein complexes were re-purified using size exclusion column chromatography (SEC) (25, 26) prior to SAXS experiments. Pair distribution functions (P(r)) for the experimental SAXS data for different protein concentrations, different molar ratios of protein/DNA, and different time points in time-resolved experiments were calculated by the program GNOM (27). The maximal dimension for the P(r) calculation for the filaments was set to the limit of our experimental setup, ∼400 Å. Aggregation-free states for nonfilament samples (XLFFL, XLF(1–224), XRCC4(1–140), and XLFFL-XRCC4(1–140)-Ku-DNA) were investigated using Guinier plots (27). In our rigid body modeling strategy BILBOMD (29), molecular dynamics simulations were used to explore conformational space adopted by the C terminus in XLFFL. For each registered conformation, the theoretical SAXS profile and the corresponding fit to the experimental data were calculated using the program FoXS (30). Because of the dynamic character of the XRCC4-XLF and XRCC4-XLF-DNA complexes, samples represented equilibria of different oligomerization states, and a minimal ensemble search (MES) was used to identify the fraction ratios of the components required to best fit the experimental data (29). Comparison of the lengths of the selected filaments in the ensemble subset allowed us to determine filament elongation without determining the maximal dimension from the P(r).
Other
Methods for protein purification, hydrogen-deuterium exchange mass spectrometry, fluorescence quenching, circular dichroism spectroscopy, DNA docking, and DNA oligonucleotide sequences are provided in the supplemental material.
RESULTS
Structure of the XLF-XRCC4 Complex
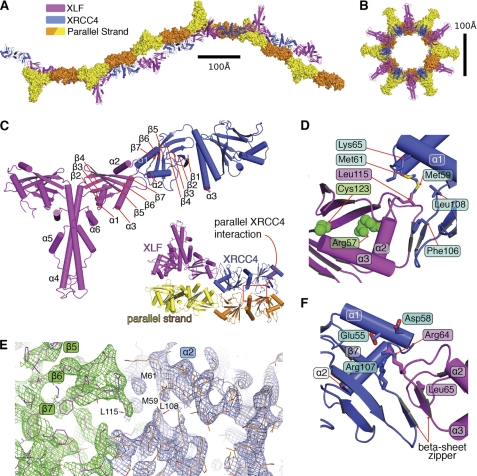
To characterize the molecular interaction of XLF with XRCC4 and the structural basis for their involvement in NHEJ, we purified, crystallized, and determined the crystal structure of the human XLF(1–224)-XRCC4(1–140) complex. We solved and refined the complex structure at 3.97 Å resolution (Fig. 1, A–F, and supplemental Table S1). The crystals belong to the hexagonal space group P6522: a = 110, b = 110, c = 764 Å. The extraordinarily long C axis results from filaments in the crystal lattice. Furthermore, two molecules of the XLF-XRCC4 complex in the asymmetric unit resulted in a high crystal solvent content (∼70%). However, screening of many crystals ultimately provided sufficient data for phasing by molecular replacement and a refined model with an R value of 35.7% and an Rfree of 36.9%.
FIGURE 1.
Crystal structural of XLF(1–224)-XRCC4(1–140). A, two orthogonal views of the biological unit of the XLF(1–224)-XRCC4(1–140) complex (magenta and blue, respectively), as seen in the crystal lattice. Parallel strands are in surface representation. B, view of the biological unit rotated by 90°. C, overall architecture of the XLF(1–224)-XRCC4(1–140) complex, colored magenta and blue. Critical secondary structure elements are indicated. Inset, close-up view of the parallel interaction of XLF(1–224)-XRCC4(1–140) in the biological unit. D, XLF(1–224)-XRCC4(1–140) interface highlighting the interaction of the XLF β6-β7 loop (Leu-115) with an XRCC4 cavity formed by β6-β7 (Leu-108 and Phe-106) and α1-α2 (Met-59, Met-61, and Lys-65). E, electron density of the 2Fo − Fc map for the XLF(1–224)-XRCC4(1–140) interaction region (green and blue) is shown contoured at 1.5σ. Crystallographic statistics are in supplemental Table S1. F, XLF(1–224)-XRCC4(1–140) interface highlighting the interaction of the XRCC4 β6-β7 loop (Arg-107) and α2 helix (Glu-55 and Asp-58) (blue) with XLF α2-α3 loop (Arg-64 and Leu-65, magenta).
In the structure of the complex, XLF and XRCC4 are structurally similar, and both are composed of two domains. The N-terminal globular head domain consists of a seven-stranded anti-parallel β-sandwich with a helix-turn-helix motif (α2, α3 for XLF and α1, α2 for XRCC4) inserted between β-strands 4 and 5 (Fig. 1C). The second domain is formed by the elongated α-helical stalks (α4 in XLF and α3 in XRCC4) and brings two head domains together. Unlike XRCC4, the XLF stalk region doubles back on itself, positioning the C-terminal helices α5, α6 toward the head domains. These XLF and XRCC4 structures resemble their unbound structures (12, 13, 31), except for a localized disorder-to-order transition and other changes noted below.
The XRCC4-XLF interface is mediated by a head-to-head interaction that buries ∼600 Å2 of interface. The XLF β6, β7 patch (Fig. 1 and supplemental Fig. S2), containing residue Leu-115 that is critical for binding to XRCC4 (12), functions as a “Leu-lock” by inserting Leu-115 into a complementary keyhole on XRCC4 formed by conserved helices α1, α2, and sheets β7 (Fig. 1, C and D). Leu-115 interacts directly with XRCC4 residues Met-59, Met-61, Lys-65, Lys-99, Leu-108, and Phe-106 (Fig. 1D). Comparison of XLF and XRCC4 in their free versus bound (XLF-XRCC4) states showed structural changes consistent with cooperative interactions. In particular, the XLF patch between β6, β7 and the XRCC4 patch between β6, β7 were displaced by ∼3 Å (Fig. 2A). This conformational change promotes insertion of the XLF Leu-lock into an XRCC4 hydrophobic pocket. Conservation of these residues among species supports their biological importance and the specificity of this XLF-XRCC4 interaction (supplemental Fig. S2A).
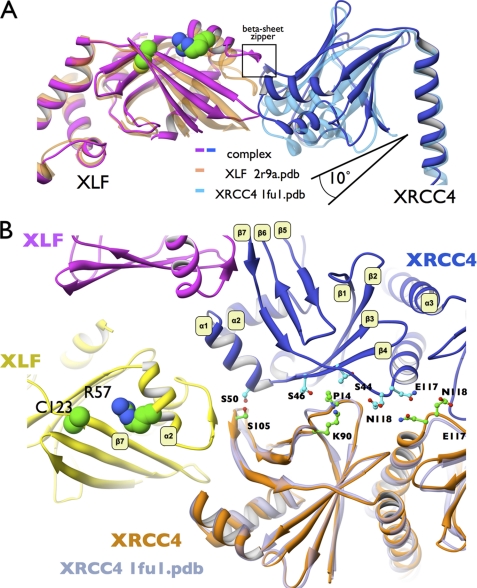
FIGURE 2.
Conformational changes upon XLF-XRCC4 complexation initiate XRCC4 site-to-site interaction. A, superposition of XLF(1–224) and XRCC4 structures with the XLF(1–224)-XRCC4(1–140) complex. The stalk regions of XLF (orange) or XRCC4 (light blue) superimposed with the stalks in the XLF-XRCC4 complex (magenta and blue). The angle between the head domains and stalks in the complex is larger than in free XLF or free XRCC4. B, close up view of the XRCC4(1–140)-XRCC4(1–140) parallel binding site (blue, orange) highlighting conformational changes and interface symmetry upon complexation. XRCC4 (PDB code 1fu1, transparent gray) have been superimposed on the complexed XRCC4 structure to show conformational changes at the parallel interface β1, β2-β4, and β6,β7-α1 sheets. Interaction residues are highlighted. XLF mutations residues R57G and C123R found in patients are highlighted.
XLF residues at the loop between α2 and α3 (Thr-66 and Ala-67) form backbone hydrogen bonds with XRCC4(1–140) β7 (Ser-105 and Phe-106). This anti-parallel β-zipper serves an important function in stabilization of the XLF-XRCC4 complex through the close interaction of the polypeptide backbone (Fig. 1F). Formation of the β-zipper propagates conformational changes along the β7-sheet (Fig. 2B). Structure-based sequence alignment of XLF and XRCC4 (supplemental Fig. S2A) reveals pseudo-symmetry of this interaction, where the XLF β6-β7 loop interacts with XRCC4 α1-α2, and the XRCC4-related β6-β7 loop interacts with XLF α2-α3, which is related to α1-α2 in XRCC4 (supplemental Fig. S2A).
To test the structural interface in solution, we assessed the XRCC4-binding properties of two site-directed mutants of XLF, XLF(1–224)(L115D) and XLF(1–224)(R64E, L65D), by monitoring time-dependent changes in the P(r) function calculated from SAXS (supplemental Fig. S2, B and C, see also Fig. 4E). The P(r) functions of the XLFmutant-XRCC4(1–140) mixture shows no filament formation (supplemental Fig. S2B). The SAXS results support the XLF-XRCC4 interactions determined from the crystal structure, as neither mutant had detectable binding to XRCC4, despite each protein being properly folded (supplemental Fig. S2C). The finding that XLF(1–224)(L115D) demonstrated no binding to XRCC4(1–140) emphasizes the importance of the specific Leu-lock interaction formed (Fig. 1D) and suggests that contacts formed between other XLF-XRCC4 interface residues are insufficient for complex formation. Yet, the absence of binding of the XLF(1–224)(R64E,L65D) mutant to XRCC4(1–140) supports the complementary importance of the anti-parallel β-zipper. These results thus reveal the synergy of both interaction regions, where specificity is driven by the Leu-lock hydrophobic interaction, as supported by specific intramolecular hydrogen bonds.
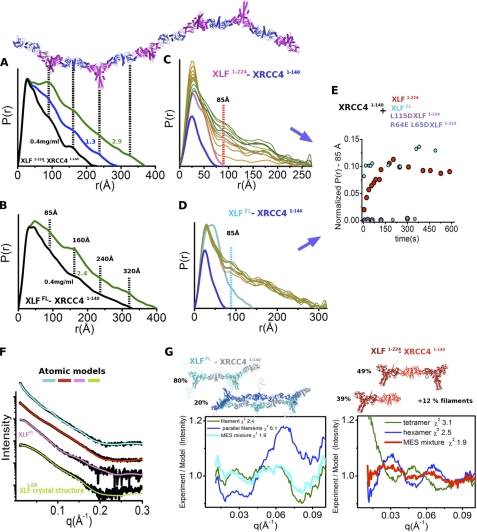
FIGURE 4.
Overall arrangement of XRCC4-XLF filaments in solution. P(r) functions of XLF(1–224)-XRCC4(1–140) (A) and XLFFL-XRCC4(1–140) (B) for dilution series (0.4, 2.3, 2.4, and 2.9 mg/ml as indicated). The P(r) functions are normalized to unity at their maxima. Four distinct distances at 85, 160, 240, and 320 Å are indicated, together with the XLF(1–224)-XRCC4(1–140) filament as shown in the crystal structure (Fig. 1A). C, P(r) of XLF(1–224) and XRCC4(1–140) (pink and blue) in comparison with a mixed sample observed 10–600 s after extensive mixing (from orange to green). The P(r) functions are not normalized. D, P(r) of XLFFL and XRCC4(1–140) (cyan and blue) in comparison with a P(r) observed 10–600 s after extensive mixing (from orange to green). E, normalized intensity of the P(r) at r = 85 Å observed in time-dependent SAXS measurements: C, XLF(1–224)-XRCC4(1–140) (red); D, XLFFL-XRCC4(1–140) (cyan); XLF(1–224) (L115D)-XRCC4(1–140) (magenta) and XLF(1–224)(R64E,L65D) (blue). F, experimental scattering profiles of the collected SEC peak fractions at ∼0.4 mg/ml or XLFFL-XRCC4(1–140) (cyan), XLF(1–224)-XRCC4(1–140) (red), and XLFFL (pink) and XLF(1–224) (green). The theoretical scattering from the final MES model matching the experimental data (χ2 = 1.9, XLFFL-XRCC4(1–140) (cyan); χ2 = 1.9, XLF(1–224)-XRCC4(1–140) (red); χ2 = 1.3, XLFFL; χ2 = 1.3 XLF(1–224)). G, MES atomic model of XLFFL-XRCC4(1–140) and XLF(1–224)-XRCC4(1–140) along with respective percentages. Bottom, plots show residual for the fit of the MES-atomistic models with corresponding χ2 values.
The long crystallographic C axis results from formation of a filament containing alternating dimers of XLF and XRCC4 in the biological unit of the crystal structure (Fig. 1, A and B). A tilt angle of ∼30° between XRCC4 and XLF supports formation of an L-shaped super-helical filament of ∼760 Å in length and ∼100-Å inner diameter. In the crystal, two super-helical filaments are aligned through interaction of two XRCC4 molecules, side by side, to form parallel filaments. Contact between parallel XRCC4 molecules is maintained by the loop located between β7 and the central stalk helix of XRCC4 and interaction of the β1-β2 loop with the α1 region of XRCC4 (Figs. 1C and 2B).
Formation of XLF-XRCC4 Filaments in Solution
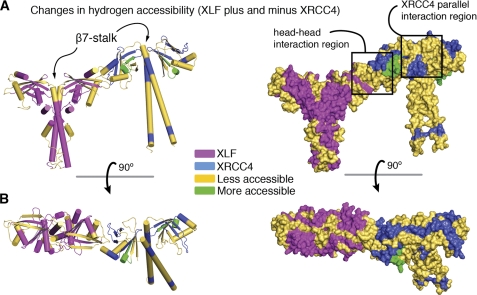
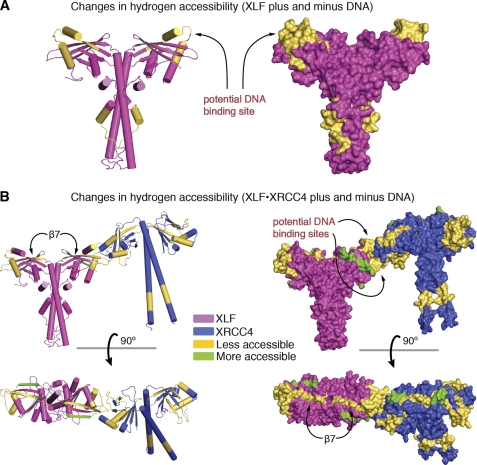
To test further whether the XLF-XRCC4 interactions and conformational changes in solution resemble those observed in the crystal structure, we initiated HDX with XLF(1–248) and XRCC4(1–200) in the complexed state. We identified 13 peptides in XLF and 11 in XRCC4 that showed a significant shift, representing regions with more or less exposure to the solvent upon complexation (supplemental Fig. S3). The observed shifts suggest that these regions are located at the XLF-XRCC4 interface or undergo conformational changes upon complexation. Our crystal structure of XLF(1–224)-XRCC4(1–140) allowed us to map the regions that showed a shift following complexation (Fig. 3). The identified regions match the interface observed in the crystal structure and the structural differences between the free and complexed structures (Figs. 2 and 3). We observed less exposure in the XRCC4 region responsible for the formation of the parallel filaments (XRCC4 loop β7-α3 and XRCC4 loop β1-β2). The entire XRCC4-XLF interaction region also showed also less exposure to the solvent upon complexation. Less accessibility along the entire XLF, XRCC4 β7 sheet and the beginning of the central stalk region confirm propagation of conformational changes from the interface to the stalks, as seen in the crystal structure. Furthermore, the parallel XRCC4 interaction may bring neighboring XRCC4 stalks in close proximity, in agreement with the observed reduced accessibility along the XRCC4 stalk. The XLF α4-stalk region also shows decreased accessibility that may also be explained by the close proximity of the neighboring XLF stalks (Fig. 3).
FIGURE 3.
HDX analysis of XRCC4-XLF complexation. A, superposition of the complexation-induced mass shifts on the XLF-XRCC4 model. Significant reductions (yellow) and increases (green) in deuteration upon complexation are indicated. B, the structures are rotated by 90°. See supplemental Fig. S3 for details.
Interestingly, R57 and C123, which are mutated in patients with microcephaly (6) maintain the integrity of the XLF-β7 sheet (Fig. 1D and supplemental Fig. S2C). In our structure, the identified patient mutations, R57G and C123R, are predicted to not only destabilize the XLF head (11, 13) but also disrupt conformational changes along XLF- β7, which are required for XRCC4 interaction. Indeed, XLF-R57G does not interact with XRCC4 (11).
XLF-β7 also shows an increase in random coil, allowing the head domains to swing out further from the stalks (Fig. 2A). This transformation of β-sheet to random coil upon XLF-XRCC4 complexation was also supported by circular dichroism (CD) experiments that show a slight increase in random coil structure at the expense of β-sheet (supplemental Fig. S4). HDX revealed less exposure to the solvent over the entire XRCC4 C-terminal stalk region (Fig. 3), indicating that the XRCC4 stalk may interact with another nearby stalk in a parallel XRCC4 arrangement. Thus, propagation of conformational changes from the XLF-XRCC4 interaction interface to the stalks of XRCC4 influence both elongation and parallel interaction of the XLF-XRCC4 assemblies.
As a further independent test of the implications from the crystal structure in solution, we investigated the dynamic structure of XLF(1–224)-XRCC4(1–140) in solution by SAXS (32) in combination with sample re-purification by SEC (24). Pair distribution functions (P(r)) calculated from the SAXS profiles (supplemental Fig. S5, A–D) of XLF(1–224)-XRCC4(1–140) over a concentration range of 0.4–3 mg/ml indicate filaments of different lengths that exist in a concentration-dependent equilibrium in solution (Fig. 4, A and B). Distinct scattering intensity peaks at 85, ∼160, ∼240, and ∼320 Å in the P(r) functions indicate that repeating units of the XLF(1–224)-XRCC4(1–140) complex adopt a persistent length of 85 Å consistent with measurements described for similar complexes (Fig. 4, A and B) (14). Higher peak intensities in samples at higher protein concentration indicate increasingly frequent repeat units for longer filaments (Fig. 4, A and B). Given the co-existence of filaments of different lengths as well as parallel filaments, we applied an MES to identify the ratio of oligomers required to best fit the experimental data (29). Assembly of three selected filament-lengths closely fit the data from SAXS at low protein concentration (χ2 = 1.9), indicating co-existence of filaments with differing lengths with a predominant ratio of shorter filaments (Fig. 4, F and G, red). The longest filament (760 Å) used in the MES matches the experimental SAXS for the highest protein concentration. However, the experimental intensities at very small q (q <0.05) are higher than the theoretical intensities, indicating the existence of even longer filaments in solution than the selected filament (supplemental Fig. S5E).
Unfolded XLF CTR Supports Formation of Parallel XLF-XRCC4 Filaments
To test the predicted conformational disorder of the XLFFL C terminus, we visualized the unfolded C terminus by SAXS. In this experiment, all selected XLFFL MES conformers from the pool of models with different compactness displayed the unfolded character of the C terminus, with an excellent match to the SAXS profile (χ2 = 1.3) (Fig. 4F and supplemental Fig. S5F). Next, we examined full-length XLF (XLFFL) in complex with XRCC4(1–140). Similar to the complex with truncated XLF(1–224), the P(r) function shows that the XLFFL-XRCC4(1–140) complex forms filaments with alternating repeating units of XLF and XRCC4. The shape of the P(r) with peaks at ∼85, ∼160, ∼240, and ∼320 Å resembles that of the truncated complex. However, at low concentration, the solution is dominated by the presence of long filaments, which were not present in the truncated XLF(1–224)-XRCC4(1–140) sample at the same concentration (Fig. 4B). Thus, the unfolded C-terminal region of XLFFL stabilizes the XLF-XRCC4 interaction and shifts the equilibrium toward longer filaments.
To identify the ratio of the filaments of different lengths and parallel filaments, we applied MES. Assembly of two selected filaments fit the SAXS data closely (χ2 = 1.9), indicating co-existence of parallel filaments as well as single-stranded filaments (Fig. 4, F and G). To characterize the kinetics of filament formation, we employed time-resolved SAXS (for details see supplemental material). Time-dependent changes in the P(r) peak at r = 85 Å demonstrate formation of protein filaments in solution. Equilibrium of XLF(1–224)-XRCC4(1–140) complexation was reached in ∼200 s, whereas XLFFL-XRCC4(1–140) filament formation reached equilibrium in less than 10 s under the same conditions (Fig. 4, C–E). Slower filament formation for truncated XLF suggests more transient complexation. Thus, stabilization of the XLF-XRCC4 interaction through the XLF C terminus (residues 249–299) leads to stabilization of longer filaments (Fig. 4, A and B) and faster equilibration of filament formation (Fig. 4, C–E). The association of XRCC4 with C-terminally truncated XLF is markedly weakened despite the presence of an intact XRCC4 interaction site on the N-terminal globular domain. This explains and extends the finding that the XLF CTR is required for recruitment of XLF to DSBs in vivo (16).
XLF-XRCC4 Interaction with DNA
To assess the DNA interaction properties of the XLF-XRCC4 complex, we first determined the binding constants for interaction of a 40-bp duplex DNA molecule (40-bp DNA) with XLFFL, XLF(1–248), XRCC4FL, and the XRCC4FL-XLF(1–248) complex. Addition of 40-bp DNA to these proteins resulted in quenching of tryptophan fluorescence at 335 nm, when excited at 295 nm. Fluorescence titrations determined binding constants (Kd) for DNA as 1.6 ± 0.1 μm for XLFFL and 2.9 ± 0.2 μm for XLF(1–248). Kd values were 0.95 ± 0.05 μm for XRCC4FL and 0.70 ± 0.05 μm for XLF(1–248)-XRCC4FL. These measurements show comparative binding and that XLFFL binds DNA better than XLF(1–224), but these values are low on an absolute scale. Yet in the cell, more than two species come together and likely form a more stable complex. Furthermore, given the cooperative nature of XLF and XRCC4 DNA binding (7, 8, 12), the effective protein concentration at a DSB may be quite high so that the measured differences in DNA binding affinity could have important consequences in the cell.
Fluorescence polarization measurements also revealed an interaction between XRCC4-XLF and DNA. Polarization values for XRCC4 increased from 0.180 ± 0.002 to 0.205 ± 0.002 upon the addition of XLF(1–248) and the subsequent addition of 40-bp DNA resulted in a further increase in polarization to 0.226 ± 0.002, providing evidence for DNA binding to XRCC4 in the presence of XLF(1–248), and formation of a ternary complex. This is consistent with elongation and parallel interaction of the filaments through DNA interaction.
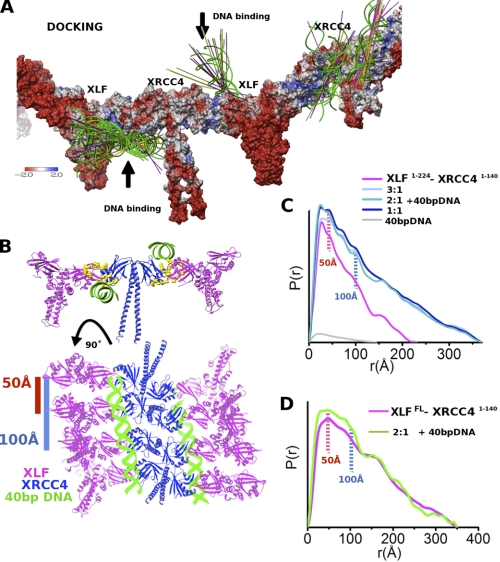
We identified potential DNA binding sites by performing HDX mass spectrometry on XLF and the XRCC4-XLF complex alone and with 40-bp DNA. Monitoring changes in the hydrogen exchange between the DNA-free and DNA-bound states allowed us to identify protein regions protected by DNA and conformational changes induced by DNA complexation. Identified DNA-binding sites in XLF alone are located at the top of the XLF head covering the β4-α2-α3 region (Fig. 5A). Interestingly, a small region in the stalk (α5) was also identified as a potential DNA-binding site. Complexation with XRCC4 transposed the DNA binding regions from the top of the XLF molecule to the bottom (α3-β6-β7), whereas the stalk region was no longer protected (Fig. 5B). Together, these data indicate different DNA-binding sites for XLF compared with XLF in complex with XRCC4. Protected regions in XRCC4 (α2-α1 and -β6-β7 loop) are structurally related to regions in XLF (Fig. 5B and supplemental Fig. S2). HDX also identified structurally related regions in both proteins with less DNA protection at the beginning of β5 (Fig. 5B) and indicates conformational change upon DNA interaction. Interestingly, the XRCC4 stalk was less protected compared with the DNA-free complex, indicating release of the XRCC4 stalks upon DNA complexation (Figs. 3 and 5B). To assess further the DNA-binding sites, we computationally docked atomic models for 20-bp DNA (Fig. 6A), 11-bp DNA, and 40-bp DNA (supplemental Fig. S6, E and F) to the structure of the XLF(1–224)-XRCC4(1–178) filament. All top-ranked placements predict DNA binding in the XLF-XRCC4 interface, where the DNA interaction site involves both proteins. We identify two favorable binding sites at the bottom (XLF-α3 and XRCC4-α2) and top (XLF-α2 and XRCC4-α1) of the XLF-XRCC4 interface (Fig. 6B). Both predicted DNA-binding sites are characterized by positively charged surfaces formed by conserved lysine/arginine clusters (XRCC4: Arg-71, Lys-72, Lys-99, Lys-102, and Arg-107; XLF: Lys-31, Arg-57, Lys-59, Lys-63, Arg-64, Arg-81, Arg-107, and Arg-109) (12, 31), which co-localize at the XLF-XRCC4 interface (Fig. 6B).
FIGURE 5.
HDX analysis of XRCC4-XLF-DNA complexation. Superposition of the complexation-induced mass shifts on the XLF(1–224) structure arising from DNA binding (A) and the XLF(1–224)-XRCC4(1–178) structure arising from DNA binding (B). Significant reductions (yellow) and increases (green) in deuteration upon complexation are indicated. For additional details see supplemental Fig. S3.
FIGURE 6.
Stabilization of XLF-XRCC4 filaments through DNA bridging. A, 20 top-ranked 20-bp DNA docking placements are shown as green helices and the 99 top-ranked placements as axis lines colored by helix direction. The protein surface is colored by electrostatic potential. B, atomistic model of XLFFL-XRCC4(1–140)-40-bp DNA built based on the crystal structure of XLF(1–224)-XRCC4(1–140), the HDX, and DNA docking. C, P(r) functions of XLF(1–224)-XRCC4(1–140)-40-bp DNA assemblies calculated for samples with molar ratios of XLF(1–224)-XRCC4(1–140): 40-bp DNA of 1:0, 3:1, 2:1, 1:1, and 0:0.3 (magenta, light blue, blue, dark-blue, and gray). In the presence of 40-bp DNA, the P(r) function shows distinct distances at r = 50 and 100 Å, indicating new order in the filaments. Elongation in the P(r) indicates stabilization of the filaments. D, P(r) functions of XLFFL-XRCC4(1–140)-40-bp DNA assemblies for samples with high protein concentration (∼3.0 mg/ml) and molar ratio of 40-bp DNA 2:1 (magenta and green) show changes and indicate new order in the filaments after DNA binding.
Thus, both HDX and computational docking identified symmetrical DNA-binding sites at the surface of the XLF-XRCC4 interaction interface. Placement of the 40-bp DNA on these sites is predicted to bridge two parallel filaments. Indeed, stabilization of parallel filaments in the DNA-XLF-XRCC4 complex was also predicted from the solution scattering experiments (Fig. 6C). We also examined scattering results in the DNA titration experiment. DNA (40 bp) was added to XLF(1–224)-XRCC4(1–140) (protein concentration 0.4 mg/ml) at different molar ratios. Elongation of the P(r) reveals stabilization of longer filaments upon DNA binding (Fig. 6C). Besides the elongation of the P(r) function, P(r) peaks at r ∼50 and r ∼100 Å were observed. These distances were less significant in the absence of DNA. The titration of DNA to XLFFL-XRCC4(1–140) also shows changes in the P(r) function, showing more short order interactions (50–100 Å) than before (Fig. 6D). We interpret the 50-Å peak as the distance between parallel filaments; although 100 Å represents the distance between a third parallel filament formed in solution (Fig. 6B).
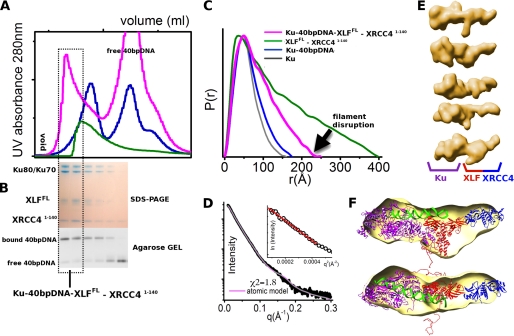
XLF-XRCC4 assembly with Ku recognizes DSBs, and its interaction with XLF stimulates the ligation step of NHEJ (16). Within the Ku-XLF complex, interactions map to 10 terminal residues in XLF(289–299) and the central DNA binding domain of the Ku heterodimer (16). To determine the overall arrangement of the XLF-XRCC4-Ku-DNA assembly in solution, we combined XLFFL-XRCC4(1–140) with an excess of DNA-loaded Ku and examined scattering results of the SEC re-purified XLFFL-XRCC4(1–140)-Ku-40-bp DNA assembly (Fig. 7). The SEC peak, in which all three proteins and 40-bp DNA co-eluted (Fig. 7, A and B), shows a shift in comparison to the SEC peak observed for Ku-DNA or XLFFL-XRCC4(1–140). The experimental P(r) function from SAXS collected for the peak fraction of Ku-DNA-XLFFL-XRCC4(1–140) shows dramatic shape changes in comparison to Ku-DNA or XLFFL-XRCC4(1–140), indicating compactness of the assembly. Notably, the complex was re-purified from the mixture containing excess of DNA-loaded Ku, which caused disruption of XLF-XRCC4 filaments and allowed formation of a XLFFL-XRCC4(1–140)-Ku-DNA (40 bp) assembly with a well defined molar ratio (Fig. 7, A and B). The maximal dimension of the assembly determined from the P(r) and the radius of gyration decreased relative to values for the XLFFL-XRCC4(1–140) filaments at a similar protein concentration (Dmax 225 versus ∼400 Å; Rg 64.5 versus ∼100 Å) (Fig. 7, C and D). The SAXS envelope revealed an elongated shape with a bulky density belonging to the Ku heterodimer and a side protrusion consistent with the XLF-XRCC4(1–140) complex (Fig. 7, E and F). Based on the Ku-DNA complex solution structure (33) and the XLF-XRCC4(1–140) crystal structure described here, we built a model for the XLFFL-XRCC4(1–140)-Ku-DNA (40 bp) assembly, which matches the experimental data well (χ2 = 1.8) (Fig. 7D) and confirms the existence of a well defined assembly with XLF located between Ku and XRCC4 (Fig. 7F). Our results suggest a Ku-DNA-XLF-XRCC4 complex with DNA positioned on one side of the XLF-XRCC4 interface providing a mechanistic basis for Ku recruitment of the XLF-XRCC4 complex to dsDNA in vivo (34).
FIGURE 7.
Overall arrangement of the Ku-XLF-XRCC4-DNA assembly. A, SEC of Ku-XLFFL-XRCC4(1–140)-40-bp DNA (magenta) in comparison with Ku-40-bp DNA (blue) and XLFFL-XRCC4(1–140) (green). The fraction collected for SAXS measurements is highlighted. B, SDS-PAGE analysis and agarose gel for DNA analysis of the peak fractions. C, P(r) of Ku-XLFFL-XRCC4(1–140)-40-bp DNA (magenta) in comparison with Ku-40-bp DNA (blue), Ku (gray), and XLFFL-XRCC4(1–140) (green). The P(r) functions are normalized to unity at their maxima. D, experimental scattering profiles of the collected Ku-XLFFL-XRCC4(1–140)-40-bp DNA fraction (black) with its atomic model matching the experimental data (χ2 = 1.8, magenta). The atomic model is shown in F. A Guinier plot with linear fit in the limit qRG >1.6 is shown in the inset. E, five representative SAXS envelopes of Ku-XLFFL-XRCC4(1–140)-40-bp DNA were calculated by DAMMIF (51). F, average SAXS envelope is superimposed on the atomic model of Ku-XLFFL-XRCC4(1–140)-40-bp DNA.
LigIV BRCT Domain Aligns the Catalytic Domain with DNA
DNA LigIV is composed of an N-terminal catalytic domain, an unstructured linker, and a C-terminal BRCT domain that interacts with the stalk of XRCC4 (35, 36). Examination of the amino acid sequence of LigIV reveals a series of basic amino acids (residues 626–629 and 633–636) located N-terminal to the tandem BRCT domain (residues 654–911). To determine whether these basic patches might have a role in DNA binding, we expressed BRCT LigIV fragments 618–911, 631–911, and 653–911, and we measured their ability to bind 45-bp duplex DNA by monitoring intrinsic tryptophan fluorescence quenching. BRCT LigIV(618–911) and BRCT LigIV(631–911) bound DNA with Kd values of 0.18 ± 0.01 and 1.60 ± 0.20 μm, whereas no DNA binding was detected for BRCT LigIV(653–911) (supplemental Fig. S7), indicating that the basic patches at residues 626–629, and to a lesser extent 633–636, may align LigIV with DNA in the XRCC4/XLF channel, positioning the catalytic domain for end ligation.
To further assess the implied relationships of the ligase DNA-binding site to XLF-XRCC4-bound DNA, we examined the top solutions for computationally docked atomic models for 20-bp DNA (Fig. 4E), 11-bp DNA, and 40-bp DNA (supplemental Fig. S6, E and F) to the structure of the XLF(1–224)-XRCC4(1–178) filament. In the top ranked solutions, DNA was oriented with the major groove facing XLF-XRCC4, resembling the dsDNA binding orientation seen for human FEN1 (37). This dsDNA binding orientation implies that the minor groove that is bound by the ligase DNA binding domain in ligase I and III complexes with DNA (38, 39) would be exposed on the XLF-XRCC4 grooved filament. If the DNA binding domain of LigIV acts similarly to that of ligase I and ligase III, then our DNA docking results suggest that XLF-XRCC4 and LigIV may simultaneously bind to opposite sides of dsDNA, which would allow a handoff of DNA from XLF-XRCC4 to ligase without the need to release DNA ends.
DISCUSSION
Although structural biology has revealed much about the role of Ku and DNA-PKcs in the control of the initial stages of NHEJ (4, 33, 40, 41), the function of XLF and XRCC4 in NHEJ and in the critical ligation step of NHEJ has remained obscure. The interaction interface between XLF and XRCC4 investigated and defined here shows the importance of XRCC4 and XLF head domains. Our crystal structure reveals the basis for the critical role of XLF Arg-64, Leu-65, and Leu-115. The fact that the R64E, L65D, and L115D mutants are structurally intact (supplemental Fig. S2C), their expression levels and cellular localization are unperturbed (11), but they are unable to complex with XRCC4 (supplemental Fig. S2B) or support DNA repair (11) reveals the importance of the XLF-XRCC4 interaction in DSB repair. Interestingly, R57 and C123, which are mutated in patients (6) support the arrangement of the XLF Leu-lock region (α2 and β7) that binds into the XRCC4 hydrophobic pocket (Fig. 1D) and further underscores the importance of these interactions in the function of XLF in vivo.
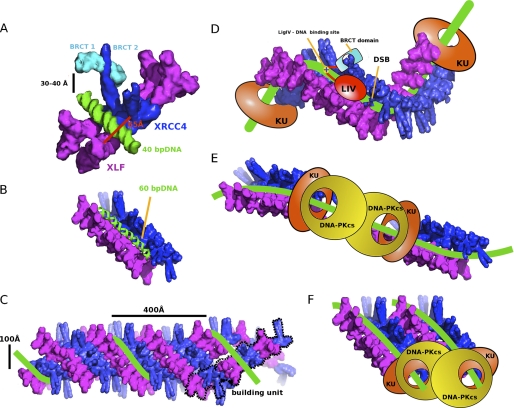
Our identification of long XRCC4-XLF super-helical filaments explains and extends biochemical data showing that XLF and XRCC4 interact with DNA in a DNA length and protein concentration-dependent manner (8, 15, 42). Based on our collective results from crystallography, HDX, SAXS, and computer modeling, we propose that XRCC4 and XLF interact to form long super-helical filaments that interact in parallel to form a grooved U-shaped channel ∼65 Å wide (Figs. 6C and 8, A and B). The base of the channel is formed by XRCC4/XLF head domains; the sides are formed by the stalks. Sixteen parallel XLF-XRCC4 complexes form one superhelical turn, allowing for alignment of ∼160-bp dsDNA (Fig. 8C) located at the base of the grooved DNA-binding surface (Fig. 8B). The proposed DNA alignment channel reveals new architectural roles for XLF, XRCC4, and LigIV in positioning DNA ends for ligation. Our results support a comprehensive model for NHEJ in which DSBs are first bound by the Ku heterodimer, which contains a preformed channel that encircles the DNA (41). Inward translocation of Ku from each DSB leads to recruitment of DNA-PKcs, which binds to the Ku-DNA complex. DNA-PKcs thus occupies the extreme termini of each side of the DSB, and tethers DNA ends together through a synaptic complex (reviewed in Ref. 3). Autophosphorylation results in release of DNA-PKcs from DNA ends, which may then be susceptible to nucleolytic attack or processing (3, 4). We propose that the movement of Ku away from the DNA ends creates a binding platform for XLF-XRCC4 to interact simultaneously with DNA and Ku. Ku translocation could proceed until it reaches flanking nucleosomes, as Ku binds nucleosomes (43, 44). Ku also binds to telomeres. Thus, the ability of Ku to move away from ends and provide a binding platform for XLF-XRCC4 that regulates end ligation may allow ligation to occur for NHEJ without allowing for illegitimate fusion of telomere ends where Ku migration is likely blocked by T-loop formation. For NHEJ, the Ku nucleated XLF-XRCC4 filament is suitable to maintain DNA end alignment via its grooved DNA-binding surface. DNA wrapped around these filaments is thus protected from nuclease attack and aligned for ligation by LigIV, which is recruited to DSBs through its interaction with the stalk of XRCC4 (35, 36). Yet this surface evidently provides exposed minor groove dsDNA faces for a handoff to LigIV, analogously to the FEN1 handoff to ligase where the minor grooves of the dsDNA, which are bound by the ligase I DNA binding domain, are exposed for ligase interaction in the FEN1-DNA complex (37). As the XLF-XRCC4 channel can bridge two DNA strands separated by a DSB, the position of LigIV would permit joining of one or both strands, depending on the structure of the DNA ends (9). Our identification of a basic patch at LigIV residues 626–629 that interacts with DNA reveals an additional mechanism for tethering LigIV to dsDNA to facilitate positioning of the N-terminal catalytic domain for catalysis. This newly discovered architectural channel created by the interface defined here explains the synergy of Ku-XLF-XRCC4 interaction in stabilizing DSBs and provides a possible molecular mechanism for the activation of LigIV by the XLF-XRCC4-DNA assembly (Fig. 8, D–F).
FIGURE 8.
Combined crystallographic and SAXS structures explain the synergy of Ku-XLF-XRCC4 interactions in ligating DSBs. A, molecular surface of the XLF-XRCC4-BRCT-DNA assembly (magenta-blue-cyan-green). The XRCC4-BRCT structure (PDB code 3ii6, cyan) (35, 36) was superimposed with XRCC4. B, half-turn of the superhelical channel (8 units of XLF-XRCC4 dimers) with 60-bp DNA (green). C, superhelical channel molecular surface. The parallel XLF-XRCC4 filaments are shown as seen in the crystal structure (building unit, dotted line). D, Ku-nucleated XLF-XRCC4 filament appears suitable to maintain DNA end alignment via its grooved DNA-binding surface. This model, which utilizes the filament groove, is distinguished by having Ku moved distal to the DSB, as shown with BRCT-bound LigIV (LIV, cyan) with its identified DNA-binding site (+) at the break. Two alternative testable models would keep Ku (orange) and DNA-PKcs (yellow) proximal to the DSB, allowing possible DNA-PK activation of partners and providing steric access to processing enzymes and LigIV with the DNA either anti-parallel (E) or parallel (F).
The established function of XLF-XRCC4 as an important NHEJ ligation factor raises the possibility of targeting this interaction for new anti-cancer compounds. Because of the high XRCC4 concentration in nucleus (45), therapeutic intervention targeting this molecule would need to be potent; however, the structures presented here show that the XLF-XRCC4 interaction occurs in a relatively discrete region that may facilitate inhibitor design. The existence of a well defined binding site with a hydrophobic Leu-lock makes this interface a promising starting point for high throughput screening or for the design of new bioactive peptides, peptidomimetics, or other small molecules as therapeutic candidates. Currently, the specific interface and resulting extended grooved scaffold for DNA ligation and DSB repair provides a testable structural mechanism for the biologically important roles of XLF-XRCC4 in the response to DSBs in humans. Such scaffolding functions are often biologically key but challenging to characterize compared with enzymes because of their more extended structures, lack of localized active sites, and typically dynamic assembly features. Consequently, there are many compelling structural and mechanistic questions that have remained unanswered for scaffolding complexes. The XLF-XRCC4 complex structure provided here may guide the design of chemical inhibitors to this scaffolding complex suitable to probe its roles in cell biology for diverse cell types and stress conditions.
In general, the spatial organization of DNA and proteins within the nucleus is critical for cell biology, and their dynamic assemblies impact the timing and coordination of replication, transcription, and repair events. During DNA base repair, intermediates are protected by tight product binding and by handoffs from one repair step to the next by the enzymes involved (46). Similarly specific binding to damaged DNA can create a platform to direct pathway selection (47). The critical roles of targeting DNA repair interactions is underscored by defects in processes, such as small ubiquitin-like modifier-targeted ubiquitin ligases, that display genomic instability and hypersensitivity to endogenous genotoxic stress (48) as well as by the use of structural mimicry to control biological outcomes (49). Such spatial control is especially essential for efficient managing of DNA ends and their error-free ligation in the context of many cellular proteins, including nucleases that would otherwise bind and degrade DNA ends and helicases that might open duplex allowing mutations. Completion of NHEJ requires LigIV to join DNA ends, but paradoxically initial binding of Ku to the DSB and assembly of the DNA-PK complex would not only protect DNA ends from nucleases but also block access to LigIV, which also encircles DNA ends for ligation based upon the human ligase I and ligase III structures (38, 39, 50). The structure of the XLF-XRCC4-DNA channel and the unified NHEJ model proposed here reveals a specific molecular mechanism whereby cells may provide dynamic scaffolding assembly to maintain and control DNA end architecture while providing sufficient accessibility for ligation initiation.
Supplementary Material
Acknowledgments
We thank the Berkeley Laboratory Advanced Light Source, SIBYLS beamline (BL 12.3.1) staff; Dr. Robert Rambo for helpful advice; Dr. Aaron Goodarzi for comments on the manuscript; James Holton and George Meigs at beamline 8.3.1 for aiding SAXS and PX data collection; and Wayne Moffat (Department of Chemistry, University of Alberta) for CD analysis. SIBYLS beamline efforts to combine SAXS and crystallography at the Advanced Light Source of Lawrence Berkeley National Laboratory are supported in part by United States Department of Energy Program Integrated Diffraction Analysis Technologies under Contract DE-AC02-05CH11231.
This work was supported, in whole or in part, by National Institutes of Health Grant P01 CA92584 (Structural Cell Biology of DNA Repair Machines) (to J. A. T. and S. P. L.-M.). This work was also supported by Grant 69139 from the Canadian Institutes of Health Research and Grant 23817 from Alberta Cancer Foundation (to S. P. L.-M. and M. W.).
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” “Results,” Figs. S1–S8, Tables S1, and additional references.
The atomic coordinates and structure factors (code 3SR2) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- DSB
- DNA double strand break
- CTR
- C-terminal region
- DNA-PKcs
- DNA-dependent protein kinase catalytic subunit
- HDX
- hydrogen-deuterium exchange mass spectrometry
- MES
- minimal ensemble search
- NHEJ
- nonhomologous end joining
- SAXS
- small angle x-ray scattering
- SEC
- size exclusion chromatography
- XLF
- XRCC4-like factor
- PDB
- Protein Data Bank.
REFERENCES
- 1. Ciccia A., Elledge S. J. (2010) Mol. Cell 40, 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lieber M. R. (2010) Nat. Struct. Mol. Biol. 17, 393–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mahaney B. L., Meek K., Lees-Miller S. P. (2009) Biochem. J. 417, 639–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dobbs T. A., Tainer J. A., Lees-Miller S. P. (2010) DNA Repair 9, 1307–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ahnesorg P., Smith P., Jackson S. P. (2006) Cell 124, 301–313 [DOI] [PubMed] [Google Scholar]
- 6. Buck D., Malivert L., de Chasseval R., Barraud A., Fondanèche M. C., Sanal O., Plebani A., Stéphan J. L., Hufnagel M., le Deist F., Fischer A., Durandy A., de Villartay J. P., Revy P. (2006) Cell 124, 287–299 [DOI] [PubMed] [Google Scholar]
- 7. Hentges P., Ahnesorg P., Pitcher R. S., Bruce C. K., Kysela B., Green A. J., Bianchi J., Wilson T. E., Jackson S. P., Doherty A. J. (2006) J. Biol. Chem. 281, 37517–37526 [DOI] [PubMed] [Google Scholar]
- 8. Lu H., Pannicke U., Schwarz K., Lieber M. R. (2007) J. Biol. Chem. 282, 11155–11162 [DOI] [PubMed] [Google Scholar]
- 9. Tsai C. J., Kim S. A., Chu G. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7851–7856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Riballo E., Woodbine L., Stiff T., Walker S. A., Goodarzi A. A., Jeggo P. A. (2009) Nucleic Acids Res. 37, 482–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Malivert L., Ropars V., Nunez M., Drevet P., Miron S., Faure G., Guerois R., Mornon J. P., Revy P., Charbonnier J. B., Callebaut I., de Villartay J. P. (2010) J. Biol. Chem. 285, 26475–26483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andres S. N., Modesti M., Tsai C. J., Chu G., Junop M. S. (2007) Mol. Cell 28, 1093–1101 [DOI] [PubMed] [Google Scholar]
- 13. Li Y., Chirgadze D. Y., Bolanos-Garcia V. M., Sibanda B. L., Davies O. R., Ahnesorg P., Jackson S. P., Blundell T. L. (2008) EMBO J. 27, 290–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hammel M., Yu Y., Fang S., Lees-Miller S. P., Tainer J. A. (2010) Structure 18, 1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu Y., Mahaney B. L., Yano K., Ye R., Fang S., Douglas P., Chen D. J., Lees-Miller S. P. (2008) DNA Repair 7, 1680–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yano K., Morotomi-Yano K., Lee K. J., Chen D. J. (2011) FEBS Lett. 585, 841–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Malivert L., Callebaut I., Rivera-Munoz P., Fischer A., Mornon J. P., Revy P., de Villartay J. P. (2009) Mol. Cell. Biol. 29, 1116–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yano K., Chen D. J. (2008) Cell Cycle 7, 1321–1325 [DOI] [PubMed] [Google Scholar]
- 19. Otwinowski Z., Minor W. (1997) in Methods in Enzymology (Carter C. W., Jr., Sweet R. M. eds) pp. 307–326, Academic Press, New York: [DOI] [PubMed] [Google Scholar]
- 20. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 23. Hura G. L., Menon A. L., Hammel M., Rambo R. P., Poole F. L., 2nd, Tsutakawa S. E., Jenney F. E., Jr., Classen S., Frankel K. A., Hopkins R. C., Yang S. J., Scott J. W., Dillard B. D., Adams M. W., Tainer J. A. (2009) Nat. Methods 6, 606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Classen S., Rodic I., Holton J., Hura G. L., Hammel M., Tainer J. A. (2010) J. Synchrotron Radiat. 17, 774–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rambo R. P., Tainer J. A. (2010) Curr. Opin. Struct. Biol. 20, 128–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rambo R. P., Tainer J. A. (2010) RNA 16, 638–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Svergun D. (1992) J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 28. Guinier A., Fournet F. (1955) Small Angle Scattering of X-rays, Wiley Interscience, New York [Google Scholar]
- 29. Pelikan M., Hura G. L., Hammel M. (2009) Gen. Physiol. Biophys. 28, 174–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schneidman-Duhovny D., Hammel M., Sali A. (2010) Nucleic Acids Res. 38, (suppl.) W540–W544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Junop M. S., Modesti M., Guarné A., Ghirlando R., Gellert M., Yang W. (2000) EMBO J. 19, 5962–5970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Putnam C. D., Hammel M., Hura G. L., Tainer J. A. (2007) Q. Rev. Biophys. 40, 191–285 [DOI] [PubMed] [Google Scholar]
- 33. Hammel M., Yu Y., Mahaney B. L., Cai B., Ye R., Phipps B. M., Rambo R. P., Hura G. L., Pelikan M., So S., Abolfath R. M., Chen D. J., Lees-Miller S. P., Tainer J. A. (2010) J. Biol. Chem. 285, 1414–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yano K., Morotomi-Yano K., Wang S. Y., Uematsu N., Lee K. J., Asaithamby A., Weterings E., Chen D. J. (2008) EMBO Rep. 9, 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sibanda B. L., Critchlow S. E., Begun J., Pei X. Y., Jackson S. P., Blundell T. L., Pellegrini L. (2001) Nat. Struct. Biol. 8, 1015–1019 [DOI] [PubMed] [Google Scholar]
- 36. Wu P. Y., Frit P., Meesala S., Dauvillier S., Modesti M., Andres S. N., Huang Y., Sekiguchi J., Calsou P., Salles B., Junop M. S. (2009) Mol. Biol. Cell 29, 3163–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tsutakawa S. E., Classen S., Chapados B. R., Arvai A. S., Finger L. D., Guenther G., Tomlinson C. G., Thompson P., Sarker A. H., Shen B., Cooper P. K., Grasby J. A., Tainer J. A. (2011) Cell 145, 198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cotner-Gohara E., Kim I. K., Hammel M., Tainer J. A., Tomkinson A. E., Ellenberger T. (2010) Biochemistry 49, 6165–6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pascal J. M., O'Brien P. J., Tomkinson A. E., Ellenberger T. (2004) Nature 432, 473–478 [DOI] [PubMed] [Google Scholar]
- 40. Sibanda B. L., Chirgadze D. Y., Blundell T. L. (2010) Nature 463, 118–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Walker J. R., Corpina R. A., Goldberg J. (2001) Nature 412, 607–614 [DOI] [PubMed] [Google Scholar]
- 42. Tsai A. G., Lieber M. R. (2010) BMC Genomics Suppl. 11 1, S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Park E. J., Chan D. W., Park J. H., Oettinger M. A., Kwon J. (2003) Nucleic Acids Res. 31, 6819–6827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Roberts S. A., Ramsden D. A. (2007) J. Biol. Chem. 282, 10605–10613 [DOI] [PubMed] [Google Scholar]
- 45. Mani R. S., Yu Y., Fang S., Lu M., Fanta M., Zolner A. E., Tahbaz N., Ramsden D. A., Litchfield D. W., Lees-Miller S. P., Weinfeld M. (2010) J. Biol. Chem. 285, 37619–37629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huffman J. L., Sundheim O., Tainer J. A. (2005) Mutat. Res. 577, 55–76 [DOI] [PubMed] [Google Scholar]
- 47. Tubbs J. L., Latypov V., Kanugula S., Butt A., Melikishvili M., Kraehenbuehl R., Fleck O., Marriott A., Watson A. J., Verbeek B., McGown G., Thorncroft M., Santibanez-Koref M. F., Millington C., Arvai A. S., Kroeger M. D., Peterson L. A., Williams D. M., Fried M. G., Margison G. P., Pegg A. E., Tainer J. A. (2009) Nature 459, 808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Prudden J., Pebernard S., Raffa G., Slavin D. A., Perry J. J., Tainer J. A., McGowan C. H., Boddy M. N. (2007) EMBO J. 26, 4089–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Prudden J., Perry J. J., Arvai A. S., Tainer J. A., Boddy M. N. (2009) Nat. Struct. Mol. Biol. 16, 509–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ellenberger T., Tomkinson A. E. (2008) Annu. Rev. Biochem. 77, 313–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Franke D., Svergun D. I. (2009) J. Appl. Crystallogr. 42, 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.