Abstract
Delta-like 1 (Dlk1, also known as fetal antigen-1, FA1) is a member of Notch/Delta family that inhibits adipocyte and osteoblast differentiation; however, its role in chondrogenesis is still not clear. Thus, we overexpressed Dlk1/FA1 in mouse embryonic ATDC5 cells and tested its effects on chondrogenic differentiation. Dlk1/FA1 inhibited insulin-induced chondrogenic differentiation as evidenced by reduction of cartilage nodule formation and gene expression of aggrecan, collagen Type II and X. Similar effects were obtained either by using Dlk1/FA1-conditioned medium or by addition of a purified, secreted, form of Dlk1 (FA1) directly to the induction medium. The inhibitory effects of Dlk1/FA1 were dose-dependent and occurred irrespective of the chondrogenic differentiation stage: proliferation, differentiation, maturation, or hypertrophic conversion. Overexpression or addition of the Dlk1/FA1 protein to the medium strongly inhibited the activation of Akt, but not the ERK1/2, or p38 MAPK pathways, and the inhibition of Akt by Dlk1/FA1 was mediated through PI3K activation. Interestingly, inhibition of fibronectin expression by siRNA rescued the Dlk1/FA1-mediated inhibition of Akt, suggesting interaction of Dlk1/FA1 and fibronectin in chondrogenic cells. Our results identify Dlk1/FA1 as a novel regulator of chondrogenesis and suggest Dlk1/FA1 acts as an inhibitor of the PI3K/Akt pathways that leads to its inhibitory effects on chondrogenesis.
Keywords: Akt PKB, Cell Differentiation, Insulin, Signal Transduction, Stem Cells, Dlk1/FA1
Introduction
Chondrogenesis is an essential process in vertebrates. It leads to the formation of cartilage growth plates, which drive linear body growth through the process of endochondral ossification. Disorders of the growth plate result in a large number of skeletal dysplasia. Additionally, chondrogenesis leads to the formation of permanent cartilaginous tissues that provide structural support in the articular joints throughout life. Cartilage degeneration and damage that are due to the common condition of age-related osteoarthritis represents a major health care problem. Thus, understanding the molecular control of chondrogenesis is an important topic in developmental biology as well as in clinical medicine, and there is current interest in identifying novel regulators of chondrogenesis that could be targeted to enhance cartilage regeneration and repair. One of the molecules that has been implicated in the control of regeneration of a number of mesodermal tissues is delta-like factor 1 (Dlk1)2 (1, 2).
Delta-like factor 1 (Dlk1) (also known as preadipocyte factor 1, pref-1) gene was cloned in the preadipocyte cells 3T3-L1 cDNA library. It is synthesized as a membrane-bound precursor with a signal peptide, six EGF-like extracellular repeats, a juxta-membrane region and a short intracellular tail (2). Dlk1 can be proteolytically cleaved within the juxta-membrane region by ADAM17/tumor necrosis factor alpha converting enzyme (TACE) (3), resulting in the release of the extracellular part of Dlk1 as a soluble fragment known as fetal antigen 1 (FA1). FA1 was initially identified in the amniotic fluid of pregnant women, but it is additionally present in all body fluids including serum, urine, and seminal fluids (4, 5). Thus, Dlk1/FA1 can function either as a membrane-bound (Dlk1) or soluble circulating protein (FA1) (6).
Dlk1 is encoded by a paternally imprinted gene located on human chromosome 14 and chromosome 12 in mice (7, 8) and is highly expressed during embryonic development as demonstrated by immune-staining or in situ hybridization (4, 9, 10), suggesting a potential role for cell fate decisions. This hypothesis has been supported by a number of studies demonstrating a regulatory role for Dlk1 in a number of mesoderm differentiation processes including adipogenesis (11), hematopoiesis (12), myogenesis (13), and osteoblastogenesis (14, 15). The importance of Dlk1 in the normal skeletal physiology has been demonstrated by studying human syndromes of unipaternal disomy (overexpression) or unimaternal disomy (deficiency) of the Dlk1 gene. These patients exhibit growth disturbances as well as adipose and skeletal tissues abnormalities (16, 17). Similarly, growth abnormalities and skeletal tissues malformations have been observed in Dlk1-deficient mice (18) and mice with Dlk1 general overexpression (19).
We have recently reported that Dlk1/FA1 is highly expressed in human embryonic stem cells (hESC) committed to the chondrogenic lineage in vitro (20). Similarly, other investigators have reported that Dlk1/FA1 promotes early commitment of skeletal (mesenchymal) stem cells (MSC) into the chondrogenic lineage through enhanced Sox9 transcription (21). These data suggested a possible role for Dlk1/FA1 as a regulator for chondrocyte differentiation.
To examine the role of Dlk1/FA1 in chondrogenesis, as well as the intracellular signaling pathways mediating its effects, we employed a well-established mouse chondrogenic cell line, ATDC5, which in monolayer cultures, undergoes a sequence of cell proliferation, chondrocyte differentiation, maturation, and hypertrophic conversion (22–24). We demonstrate that Dlk1/FA1 acts as a negative regulator for chondrogenic differentiation through suppression of insulin-induced PI3K/Akt activation; and that fibronectin is involved in Dlk1/FA1-mediated inhibition of the Akt pathway in chondrogenic cells.
EXPERIMENTAL PROCEDURES
Collection of Mouse Embryonic Cartilage Samples
Mouse embryonic samples were collected by microdissection and contained whole hind limbs at embryonic days E10.5 and 11.5, knee epiphyseal cartilage at E12.5, 14.5, 16.5, and 18.5 pc, and knee epiphyseal/articular cartilage of newborn (E20.5), 15 day-old, and 2 month-old mice. Knee cartilage samples were dissected free of skin and muscle. Embryonic samples were pooled from 5 to 18 animals to minimize the sampling variation between animals and to obtain enough tissue for RNA isolation. Tissue samples were frozen immediately after collection in liquid nitrogen.
Cell Culture and Differentiation
The mouse chondrogenic ATDC5 cell line was obtained from the RIKEN cell bank (Tsukuba, Japan). Cells were maintained in DMEM/F12 (1:1) medium with 5% FCS, 10 μg/ml human transferrin (Invitrogen A/S, Tastrup, Denmark), and 3 × 10−8 m sodium selenite (Sigma-Aldrich, Copenhagen, Denmark) at 37 °C in a humidified atmosphere containing 5% CO2. Chondrogenic differentiation of ATDC5 cells was performed as previously described (23, 24). Briefly, ATDC5 cells were seeded at a density of 6 × 103 cells/cm2 in 6-well plates or 24-well plates, and grown for 4 days. At the time the cells reached confluence, the medium was replaced by fresh medium supplemented with insulin (10 μg/ml), and the medium was changed every other day for 24 days.
Cell Transfection
The construct encoding the entire mouse Dlk1 gene, cloned into the mammalian expression vector pCD2, was a gift from Dr. J. Battey (NIH, Bethesda, MD). Cells were seeded 1 day before transfection at 70–80% confluence. Transfections were performed using LipofectamineTM 2000 (Invitrogen, Gaithersburg, MD) according to the manufacturer's recommendations. 48 h post-transfection, the cells were passaged and selected using 600 μg/ml G418 (Sigma-Aldrich, Vallensbaek Strand, Denmark) for one month. The selected clones were pooled and used for further experiments. For siRNA transfection, ATDC5 cells at 95% confluence were transfected with 25 nm fibronection small interfering RNA (siRNA), integrin β1 (Itgb1) siRNA or control non-targeting siRNA (Applied Biosystems/Ambion, Denmark) using LipofectamineTM 2000.
Alcian Blue Staining
To evaluate the synthesis of proteoglycans in chondrogenic differentiation, sulfated glycosaminoglycans (GAGs) were stained with Alcian blue. Cells in monolayer cultures were rinsed twice with phosphate buffered saline (PBS), fixed in cold Kahle's fixative for 10 min at room temperature, and stained with Alcian blue overnight and then rinsed twice with distilled water. Results were scanned and recorded using either photomicroscopy or whole wells from the monolayer cultures.
Real-time RT-PCR
Total RNA was isolated from cartilage tissue using TRIzol® reagent (Invitrogen, Tastrup, Denmark). Briefly, samples were pooled prior to snap freezing in liquid nitrogen, frozen pieces of cartilage were ground to a powder before homogenization in TRIzol®. RNA isolation continued according to the manufacturer's instructions (Invitrogen). RNA isolation from cultured cells was performed using the RNAElute kit (Sigma-Aldrich) according to the manufacturer's instructions. All RNA samples were treated with DNase I (Invitrogen). cDNA was prepared from 1 μg of total RNA with the iScriptionTM cDNA synthesis kit (Bio-Rad), in a final volume of 20 μl. The primers for genes were designed using the PrimerSelect program from the LaserGene software package (DNASTAR, Madison, WI) (supplemental Table S1). IQ SYBR Green supermix (Bio-Rad) was used to visualize RT-PCR products. A three-temperature cycling program consisting of a denaturation step at 95 °C for 15 s and annealing/extension step at 55 °C to 68 °C for 20 s, followed by 72 °C for 20 s, was carried out in an iCycler PCR machine (Bio-Rad). The lengths of PCR products were verified by agarose gel electrophoresis, and melting curve analysis was used to assess the specificity of each PCR reaction. For quantitative analysis of individual genes, two independent cDNA samples were prepared and each of the cDNAs was tested in duplicate or triplicate. β-Actin or 18 S rRNA were used as the reference gene. Data were analyzed using the optical system software version 3.0 (Bio-Rad) and Microsoft Excel® to generate relative expression values from the standard curve assay.
Measurements of FA1 in Conditioned Medium
The same number of Dlk1 over-expression and parallel control cells were seeded and cultured to 90–100% confluence. Cells were washed in PBS twice, changed to fresh serum-free medium and incubated at 37 °C overnight. The medium was collected and filtered for Dlk1/FA1 measurement. ELISA was performed as previously described (25) with biotinylated affinity-purified rabbit anti-mFA1 as the capture antibody, and peroxidase-conjugated streptavidin, H2O2, and O-phenylenediamine were used to develop the reaction. 5% serum was added to the collected medium to create conditioned medium (CM) for further cell culture.
Generation and Purification of Recombinant Dlk1/FA1
Plasmid containing the large secreted form of Dlk1 (1–302 amino acids) was fused with the human Fc region encoding the C-terminal 235 amino acids of FA1-Fc (termed here FA1-Fc construct) (provided by Sul H.S.; University of California, Berkeley). Control human Fc-pcDNA3.1 plasmid was constructed by subcloning the Fc fragment into pcDNA3.1 pre-cut using EcoRI and XhoI. The sequence of the construct was verified by sequencing (DNA Technology, Denmark). Free style 293 cells (Invitrogen) were grown in 293 expression medium (Invitrogen) and transfected with the FA1-Fc construct using 293fectin (Invitrogen). Purification of secreted FA1-hFc fusion protein and control hFc fragment from serum-free culture medium were carried out by affinity chromatography using NAbTM Spin kits (Thermo Scientific) according to the manufacturer's recommendations.
Western Blotting and Immunoprecipitation Assays
Antibodies (total or phosphor) specific for Akt (Ser-473, Thr-307), p38, p85, PDK1, p70SK6, SK6, rictor, raptor, mTOR (Ser-2448) were obtained from Cell Signaling Technology. Antibodies for phosphor Erk1/Erk2 (sc-7383), anti ERK2 (C-14, sc-154), PP1, and α-tubulin were purchased from Santa Cruz Biotechnology, Inc. For Western blot analysis, cells were collected at specific time points post-treatment, and washed in cold PBS buffer before being lysed in cell lysis buffer (10 mm Tris-HCl, pH 7.4, 150 mm sodium chloride, 1% Nonidet P-40, 0.1% SDS, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 1 mm NaF, 1 mm Na3VO4), supplemented with protease inhibitor mixture (Roche Diagnostics, Mannheim, Germany). 20–50 μg of protein was separated on 8% to 12% NuPAGE® Novex® Bis-Tris gel systems (Invitrogen), then transferred to PVDF membranes (Millipore). The membrane was blocked and probed with antibodies as indicated and then incubated with peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology). Bound proteins were visualized by ECL chemiluminescence (Thermo Scientific). For the immunoprecipitation assay, 2–5 μg of specific antibody was added to 300–500 μg cleared lysates in RIPA buffer (25 mm Tris-HCl, pH7.6, 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate). Overnight incubation at 4 °C allowed complexes to form, after which 20 μl of 50% slurry protein A/G-agarose (Santa Cruz Biotechnology) was added and incubated at 4 °C for 3 h. Immunoprecipitates were washed four times in RIPA buffer and analyzed by Western blots.
Statistics
Results are reported as mean ± S.E. Comparisons were made using a two-tail Student's t test, where p < 0.05 was considered as significant.
RESULTS
Dlk1/FA1 Is Expressed during Mouse Embryonic Chondrogenesis
To verify the physiological relevance of Dlk1/FA1 in cartilage differentiation, we examined its in vivo expression pattern during mouse embryonic developing limb chondrogenesis. The real time qPCR data demonstrated that expression of Dlk1 mRNA was low during mesenchymal condensation (E10, E11), expression increased at E14.5 to reach the highest expression levels at E16.5; this level then remained high until birth. Thereafter, Dlk1 mRNA expression rapidly decreased and 15 days postnatal, the expression had decreased to the same level as at E10 (Fig. 1).
FIGURE 1.
Dlk1/FA1 expression was detected in cartilage development sites. RNA was isolated from mouse embryonic cartilage tissue at E11.5, 12.5, 14.5, 16.5, and 18.5 pc. and knee epiphyseal cartilage of newborn, 15-day-old, and 2-month-old mice were dissected free of skin and extra muscle. Samples were pooled from 5 to 18 animals to minimize the sampling variation between animals. Expression of Dlk1 was quantified by qPCR analysis. Data were presented after normalization to β-actin. Values are presented as mean (n = 5–18) ± S.D.
Expression of Dlk1/FA1 in ATDC5 Cells Inhibits Chondrocyte Differentiation
To study the role of Dlk1/FA1 during chondrogenesis, we employed ATDC5, a mouse embryonic chondroprogenitor cell model. ATDC5 cells were transfected with mouse plasmid expressing full-length Dlk1/FA1 and cells stably expressing high levels of Dlk1 were selected (termed here as ATDC5-Dlk1). ATDC5-Dlk1 cells exhibited a high Dlk1 mRNA expression level and protein production, as detected by real time PCR, immunocytochemical staining, and Western blot as compared with control ATDC5 cells transfected with vector alone (Fig. 2, A and B and supplemental Fig. S1). During the 24 day in vitro chondrogenesis, FA1 (soluble extracellular domain of Dlk1) was detected in the conditioned media and ranged from 50 to 320 ng/ml (Fig. 2C). There were no significant differences in the rate of cell proliferation between ATDC5-Dlk1 and control cells during differentiation (Fig. 2D). Compared with the control cells, chondrogenic differentiation was impaired in ATDC5-Dlk1 cells as evidenced by formation of less Alcian blue-positive cartilage nodules (Fig. 2E), and decreased mRNA expressions of the chondrogenic marker genes: Col2a1, Agc1, and Col10a1 (Fig. 2F).
FIGURE 2.
Forced expression of mouse Dlk1 inhibits chondrogenesis in mouse ATDC5 cells. Chondrogenic ATDC5 cells were transfected by pCD2-Dlk1 and pCD2 plasmids, selected by G418 for one month to acquire stable clones. A and B, expression of Dlk1 was observed by real time PCR and immunocytochemical staining in selected clones. C, secreted form of the Dlk1 protein (FA1) in culture medium from ATDC5-Dlk1 cells during the 24-day induction was measured by ELISA. D, proliferation rates of ATDC5-Dlk1 cells (black bars) and control cells (ATDC5-vehicle) were compared in medium containing insulin. Cell numbers were counted on 1, 5, and 10 days after seeding. E, cartilage nodule formations were confirmed by Alcian blue staining on the indicated days post induction. F, expression of chondrocyte differentiation markers, such as type II and type X collagen (Col2a1, Col10a1) and aggrecan (Agc1), were detected by real-time PCR. Expression of each target gene was normalized to β-actin and represented as a fold induction over control non-induced cells. Vehicle: ATDC5 cells transfected with empty vector; ATDC5-Dlk1: ATDC5 cells transfected by the Dlk1-expression vector and stably expressing the dlk1/FA1 protein. Representative data from three independent experiments are shown. Values are presented as mean ± S.D. *, p < 0.05; **, p < 0.01, as compared with vehicle control cells.
FA1, the Soluble Form of Dlk1, Inhibits Chondrocyte Differentiation in ATDC5 Cells
In 3T3-L1 cells, the extracellular form of Dlk1, i.e. FA1, has been reported to mediate Dlk1 inhibition of adipocyte differentiation (26). Thus, we studied the effect of FA1 on chondrocyte differentiation. Conditioned media (CM) containing FA1, inhibited the chondrogenic differentiation of ATDC5 cells in a dose-dependent manner (concentration range: 50–250 ng/ml) (Fig. 3, A and B). Similar inhibition of chondrogenesis in ATDC5 cells was observed when using purified FA1-Fc protein (supplemental Fig. S2) verifying that FA1, was the factor responsible for the observed inhibitory effects in the ATDC5-Dlk1 CM. However, the dose-response curve was shifted to the right with the inhibitory effects detectable at a concentration range of 250 ng to 2 μg/ml (Fig. 3, C and D and supplemental Fig. S3A).
FIGURE 3.
The soluble form of Dlk1 (FA1) inhibits insulin-induced chondrogenic differentiation of ATDC5 cells in a dose-dependent manner. Serum-free conditioned medium (CM) was collected from Dlk1 overexpressing cells or control cells, and FA1 concentrations in medium were determined by ELISA. Differentiation of ATDC5 cells was induced in the presence of insulin (10 μg/ml) in CM containing varying amounts of FA1. CM from control cells was used as basal culture medium, subsequently different ratios of FA1 CM from Dlk1 cells was added to achieve FA1 at a range of concentrations (undetectable, 50, 100, or 200 ng/ml). Media was replaced every other day during the 24-day induction of chondrogenesis. A, cartilage nodule formations were detected by Alcian blue staining after 21 days of chondrogenic induction. B, temporal expression levels of type II and type X collagen (Col2a1, Col10a1), and aggrecan (Agc1) mRNAs were quantified by real-time PCR. C and D, effect of adding purified FA1 on chondrogenic differentiation of ATDC5 cells. FA1-Fc proteins were purified (see “Experimental Procedures”), and added at different concentrations to the chondrogenic induction medium. Alcian blue staining was performed at day 24 after induction of chondrogenesis (D), and chondrocyte marker genes were confirmed at day 21 by real time PCR (D). Data shown are representative of at least two independent experiments. Values are expressed as mean ± S.D. *, p < 0.05, compared with control group (containing 2 μg of Fc protein).
Inhibition of Chondrogenic Differentiation of ATDC5 by FA1 Is Differentiation Stage-independent
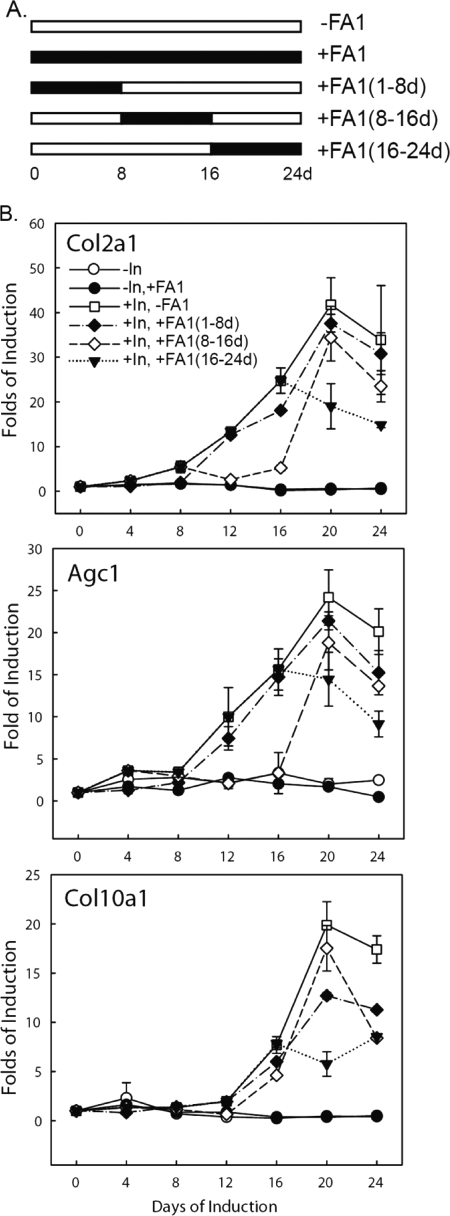
Chondrogenic differentiation in ATDC5 takes place in a sequential manner starting from cell proliferation to chondrogenic commitment and differentiation followed by hypertrophic conversion (27). To examine the effects of Dlk1/FA1 on the stages of chondrogenic differentiation, ATDC5-Dlk CM was added at different time points during the 24-day chondrogenic induction (24). Results showed that ATDC5-Dlk1 CM inhibited chondrogenesis irrespective of the stage of chondrogenic differentiation of ATDC5 (Fig. 4, A and B and supplemental Fig. S3B).
FIGURE 4.
Soluble FA1 inhibits the insulin-induced chondrocyte differentiation of ATDC5 cells at all differentiation stages. Serum-free conditioned medium (CM) was collected from Dlk1 over-expressing cells or parallel control cells. ATDC5 cells were cultured and induced to chondrogenesis in conditioned medium in the presence of insulin (10 μg/ml). CM containing FA1 (250 ng/ml) was replaced at the indicated time intervals (1–24 days, 1–8 days, 8–16 days, 16–24 days) on or after chondrogenic induction of ATDC5 cells. A, schematic representation of the different treatments with FA1-containing conditioned mediums post chondrogenic induction of ATDC5 cells. The periods in which FA1 was added to the differentiation medium are indicated by full bars (200 ng/ml). B, time-course expression levels of type II and type X collagens (Col2a1 and Col10a1) and aggrecan (Agc1) gene markers as measured by real-time PCR. Expression of each target gene was normalized to β-actin and represented as a fold induction over control non-induced cells. Representative data obtained in three independent experiments are shown. Values are expressed as mean ± S.D.
Dlk1/FA1 Inhibits Insulin-mediated Akt Activation during Chondrogenesis
To determine the mechanism of Dlk1/FA1 mediated-inhibition of chondrogenesis, we investigated factors and pathways related to chondrogenesis. We did not detect significant differences in mRNA expression of the Sox9 transcription factor between ATDC5-Dlk1 and control cells (supplemental Fig. S6). Because chondrogenesis in ATDC5 cells is induced by insulin, we examined the effect of Dlk1 on kinases involved in insulin signaling. The addition of insulin to ATDC5 cell cultures stimulated the phosphorylation of ERK1/2 and Akt, but had no effect on p38 activation (Fig. 5A). Insulin-induced Akt phosphorylation was completely abolished in ATDC5-Dlk1 cells and did not affect the activation of ERK1/2 or p38 MAPK (Fig. 5A). Moreover, adding ATDC5-Dlk1 CM (FA1 conc. 250 ng/ml) or the purified recombinant FA1-Fc protein (conc. 2–4 μg/ml) reproduced these effects (Fig. 5, B–D).
FIGURE 5.
Dlk1/FA1 inhibited the insulin-induced Akt activation in ATDC5 cells. A, Western blot analysis of Akt, ERK1/2, and p38 MAP kinase activation in response to insulin (10 μg/ml) in ATDC5 control cells and ATDC5-Dlk1 cells. B, Western blot analysis of Akt activation by insulin in ATDC5 cells pretreated with conditioned medium from ATDC5 control cells (V-CM) and ATDC5-Dlk1 cells (FA1-CM, FA1 ≅ 250 ng/ml). C, Western blot analysis of Akt activation was compared after insulin stimulation in cells pretreated by Fc peptides or purified FA1-Fc protein. ATDC5 cells were cultured to near confluence, and starved in a serum-free medium for 18 h with purified Fc or FA1-Fc protein at different concentrations, cells were then stimulated with insulin for 15 min. Proteins were harvested to perform Western blots analysis. D, Akt activation during a time course in ATDC5 cells pretreated with purified FA1-Fc. ATDC5 cells were cultured to near confluence, then starved in a serum-free medium for 24 h, purified FA1-Fc were added to the starved medium to pretreat cells at different time points. Cells were stimulated with insulin (10 μg/ml) for 15 min; proteins were harvested and subjected to Western blot analysis. Results are representative of at least three independent experiments. α-Tubulin was utilized as the loading control.
To verify the significance of the Akt signaling pathway in chondrogenesis, a number of kinase inhibitors were examined for their effects on insulin-induced chondrocyte differentiation in ATDC5 cells. Specific inhibitors of Akt e.g. triciribine (28) and BML-257 (29), inhibited the chondrogenic differentiation of ATDC5. PI3K inhibitors e.g. LY294002 and Wortmannin demonstrated some inhibition of chondrogenic differentiation but the effects were less potent. In contrast, two MEK inhibitors: PD98059 and U0126, had no affect on the chondrogenic differentiation (supplemental Fig. S4). These results indicate that the Akt-dependent pathway plays an important role in chondrogenic differentiation of ATDC5 cells and corroborates that Dlk1/FA1 inhibited chondrogenesis through the inhibition of Akt activation.
Dlk1 Inhibits PI3K Activation
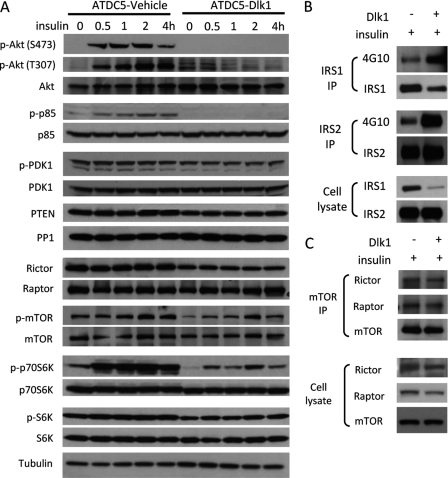
Potential mechanisms of Dlk1/FA1 mediated Akt inhibition are through the deactivation of upstream proteins or the activation of an Akt-specific phosphatase. We checked the expression levels of a series of signaling proteins related to the regulation of Akt signaling. As shown in Fig. 6A, both Ser-473 and Thr-307 sites of Akt phosphorylation were inhibited by Dlk1/FA1. Activation of the regulatory subunit of PI3K (p85) was clearly inhibited, but not phosphoinositide-dependent protein kinase 1 (PDK-1). The expression of PTEN, a negative regulator of PI3K/Akt signaling, was decreased in ATDC5-Dlk1 cells; while the total level of protein phosphatase 1 (PP1), a protein that regulates the dephosphorylation of Akt, was not increased in ATDC5-Dlk1 cells (Fig. 6A). Instead, Dlk1/FA1 inhibited some of the downstream targets of Akt activation as evidenced by reduction in the activation of mTOR, p70S6K, and, to a less degree, S6K (Fig. 6A). Immunoprecipitation experiments revealed that inhibition of insulin-induced p85 activation in ATDC5-Dlk1 cells was not associated with changes in tyrosine phosphorylation of insulin receptor substrate proteins (IRS1 or IRS2) after insulin treatment (Fig. 6B), even though the IRS1 expression was much less in Dlk1 cells. Since we detected clear inhibition at the Ser-473 phosphorylation site of Akt, and to determine whether formation of mTOR complexes (mTORC2 and mTORC1, especially for mTORC2) contributes to inhibition of Akt phosphorylation, we compared the bindings of Rictor and Raptor with mTOR in ATDC5-Dlk1 cells and control cells. Western blot and immunoprecipitation showed slightly decreased expression of Rictor and Raptor in ATDC5-Dlk1 cells, additionally there were no significant alternation of binding of Rictor and Raptor to mTOR in the presence of Dlk1 (Fig. 6C).
FIGURE 6.
Dlk1/FA1 inhibited PI3K activation by insulin, but not IRS tyrosine phosphorlyation or mTOR complex formation. A, Western analysis for different kinases involved in Akt pathway. Confluent ATDC5 control and ATDC5-Dlk1 cells were starved 18 h in serum free medium, then treated with 10 μg/ml insulin from 30 min to 4 h. Protein samples were collected post-treatment and subjected to Western blot analysis for different proteins involved in the insulin pathways. B, immunoprecipitation of IRS1 and IRS2 in ATDC5 control cells and ATDC5-Dlk1 cells, then tyrosine phosphorylations were verified by the 4G10 antibody. Upper group: IRS1 immunoprecipitation; middle group: IRS2 immunoprecipitation; lower group: IRS1 and IRS2 expression in loading samples. C, immunoprecipitation of mTOR in ATDC5 control cells and ATDC5-Dlk1 cells, binding of rictor and raptor to mTOR was checked by the individual antibodies (upper group). Expressions of mTOR, rictor, and raptor in the loading samples are shown in the lower group.
Down-regulation of Fibronectin Rescues Dlk1/FA1-mediated Inhibition of Akt
Dlk1/FA1 inhibited PI3K/PDK/Akt activation, but not through inhibition of insulin-mediated activation of IRS1 or IRS2. This suggests that Dlk1/FA1 interacts with other signaling pathways that converge on the PI3K/Akt pathway. One of these signaling pathways is integrin signaling which is mediated by fibronectin (30). Fibronectin (Fn) and its receptor, integrin β (Itgb), are highly expressed in ATDC5 cells and their expression is up-regulated during chondrogenesis (24). We carried out siRNA knockdown experiments of Fn and Itgb expression in ATDC5-Dlk1 and control cells. RT-qPCR demonstrated that baseline levels of Fn and Itgb mRNA expression were similar in ATDC-Dlk1 and control cells, and siRNA transfection inhibited 80–95% expressions of Fn and Itgb1 in both cells (Fig. 7, A and B). In control ATDC5 cells, inhibition of Fn expression did not affect the insulin-induced Akt activation; but in ATDC5-Dlk1 cells, it rescued the inhibition of Akt activation (Fig. 7A). Inhibition of Itgb did not affect Akt activation in either cell line (Fig. 7B). Additionally, we examined the activation of focal adhesion kinase (FAK), a kinase mediating integrin and PI3K activation in some cell models (31) but we did not detect clear insulin-mediated activation of FAK (Fig. 7C); and siRNA inhibition of expression of Fn or Itgb1 did not change FAK activation in the presence of insulin (Fig. 7, A and B).
FIGURE 7.
Fn, but not integrin or FAK kinase, interfered with the Dlk1/FA1 inhibition of insulin-induced Akt activation. A and B, ATDC5 control cells and ATDC-Dlk1 cells were transfected with either non-targeting (NT), fibronectin (Fn), or integrin β (Itgb) siRNAs. 48 h after transfection, cells were starved for 18 h, and treated with 10 μg/ml insulin for 20 min. Western blots were performed using phosphorylation Akt and FAK, total Akt and FAK antibodies, α-Tubulin was detected as loading control (upper lane). Real time PCRs were performed to verify the inhibition efficiency of siRNA on target genes (lower lane). Results are representative of at least three independent experiments. C, FAK activation is not involved in insulin-induced Akt activation. ATDC5 control cells and ATDC-Dlk1 cells were cultured to near confluence, starved in serum-free medium for 18 h, then stimulated with insulin for different times. Cell lysates were harvested to perform Western blots using phosphorylated Akt and FAK, total Akt and FAK antibodies, α-tubulin was utilized as the loading control. Results are representative of at least three independent experiments.
DISCUSSION
Delta-like factor 1 (Dlk1/FA1) was initially cloned as a negative regulator of adipocyte cell differentiation (32) and we have previously demonstrated that Dlk1/FA1 also inhibits differentiation of human skeletal MSC into osteoblasts (14). In the present study, we demonstrated clear inhibitory effects of Dlk1/FA1 on chondrogenic differentiation as evidenced by strong inhibition of mature chondrocyte markers (ColII, aggrecan, ColX) during chondrogenic differentiation. Therefore, Dlk1/FA1 functions as a universal factor inhibiting committed mesoderm progenitor cell differentiation into adipocytes, osteoblasts, and chondrocytes.
We detected Dlk1/FA1 expression at different levels during embryonic cartilage development, and its mRNA expression reached the highest levels at E16.5 followed by decreased expression during the postnatal period. These results corroborate our previous immunohistochemical stainings (20), where Dlk1/FA1 was co-expressed with type IIA procollagen in proliferating chondrocytes in the developing limb epiphyses and was absent from the hypertrophic zones. The high levels of Dlk1/FA1 gene expression and its presence in the proliferating chondrocytes suggest a biological role during in vivo chondrogenesis and endochondral ossification.
During in vitro chondrogenic differentiation, we detected that Dlk1/FA1 expression level is down-regulated and it did not show reciprocal changes with another imprinted gene Gtl2 (supplemental Fig. S5). In addition, we demonstrated that Dlk1/FA1 exhibits clear inhibitory effects on chondrogenic differentiation of ATDC5 cells, which represent a model of chondroprogenitor cell populations (27), and that these inhibitory effects were differentiation-stage independent. Interestingly, Dlk1/FA1 increased mesenchymal condensations of the cells in culture but their chondrogenic conversion was inhibited. Additionally, Dlk1/FA1 inhibited mature chondrogenic markers (ColII, aggrecan, ColX) but not the chondrogenic transcription factor Sox9. Thus, our data suggest that Dlk1 inhibits the transition of committed progenitor cells to differentiated cell types. It is plausible that this is a general cellular mechanism for Dlk1/FA1 action since similar observations have been reported in skeletal (mesenchymal) stem cells (14), human embryonic stem cells (20) and adipocyte progenitor cells (33).
Dlk1/FA1 inhibition of chondrocytic differentiation of ATDC5 was not associated with significant changes in the expression of the chondrogenic transcription factor Sox9 (supplemental Fig. S6). This seems at variance with the recent study of Wang et al. (21) where the authors reported that Dlk1/FA1 in transgenic mice inhibited chondrocytic differentiation of murine multipotent stromal cells by its induction of Sox9 expression. This difference may be related to biological differences in the cell models employed because ATDC5 cells represent chondroprogenitor cells (27). It is plausible that Sox9 induction by Dlk1/FA1 plays a role in the transition from multipotent stem cells to chondroprogenitor cells, but not in the following differentiation progress. Similarly, our previous studies in human MSC demonstrated that Dlk1/FA1 did not affect osteoblastic transcription factor: Runx2 or adipocytic transcription factor: PPARγ (14).
Skeletal progenitor cell differentiation is usually accompanied by reduced cell proliferation (34). Dlk1 was originally cloned as a tumor protein (35) and Dlk1/FA1 has been reported to either promote cell proliferation under certain conditions e.g. in hepatocellular carcinoma cells (36) and neuroendocrine tumors (35), or inhibit cell proliferation e.g. in renal carcinoma cells (37). Additionally, Dlk1/FA1 has been reported to have interactions with cell growth-regulating proteins (38). Although we observed higher proliferation rates in ATDC-Dlk1 cells, as compared with the control cells (data not shown), we did not detect significant effects of Dlk1/FA1 on cell proliferation during chondrogenic differentiation of ATDC5 cells. Similarly, in our previous study, human MSC overexpressing Dlk1/FA1 exhibited normal growth rates as compared with control hMSC cells (14). Thus, the regulatory role of Dlk1/FA1 in cell proliferation requires further investigation.
Dlk1 is cleaved by the tumor necrosis factor alpha converting enzyme (TACE/ADAM17) (3), resulting in the release of a 50-KDa heterogeneous protein. This protein is identical to a protein that was first isolated and characterized from the amniotic fluid of pregnant women and named FA1 (fetal antigen 1) (4, 5, 25). In preadipocytic cells, the 50-kDa FA1 soluble fragment is responsible for inhibition of adipocyte differentiation (26), osteoblast and adipocyte differentiation of human MSC in vitro (14) as well as decreased bone and fat mass in vivo (39). Similarly, in the current study, the inhibitory effects on chondrogenic differentiation were detected either by using only the FA1 conditioned medium or the secreted part of the Dlk1 protein (e.g. FA1). We observed that the conditioned medium from Dlk1/FA1 cells worked more efficiently in regulating Akt activation and chondrogenic differentiation than the purified FA1. There are several possible explanations for this: 1) Dlk1/FA1 activity may be more stable in the conditioned medium compared with that of the purified FA1 protein; 2) Dlk1/FA1 might interact with other factors in the medium which cooperate in the regulation of cell signaling (15).
Dlk1 has six EGF repeats and belongs to a family of epidermal growth factor (EGF)-like repeat-containing proteins that include Notch/Delta/Serrate. However, unlike other delta-family molecules, Dlk1 lacks the DSL motif in the extracellular domain that appears to be required for the interaction with Notch family receptors thus suggesting possible involvement of other signaling pathways. Several signaling pathways are known to be involved in insulin-induced chondrogenic differentiation of ATDC5 cells including p38, ERK1/2 and PI3K (40, 41) and that PI3K/Akt pathway is indicated as a central regulatory pathway (42, 43). We observed that Dlk1/FA1 presence impaired insulin-dependent PI3K/Akt activation, but did not affect ERK1/2 or p38 MAPK. Interestingly, the similar inhibitions of activation of insulin-induced PI3K/Akt were also detected in human muscle satellite cells, some mouse stromal cell clones, primary mouse embryonic fibroblast (MEF) cells, and mouse cranial cells (data not shown), these suggested that Dlk1/FA1 might be a novel regulator for insulin/Akt pathway in some tissue cells. In addition, we found that Dlk1/FA1 inhibited the activation of PI3K (p85), and this effect was not due to enhanced expressions of PTEN and PP1, nor reduction of IRS tyrosine phosphorylation after insulin induction. Moreover, Dlk1/FA1 inhibited the mTOR/P70S6K/S6K pathway and thus contributed to the downstream regulation of chondrogenesis by mTOR (44) and the release of the feedback inhibition of the IRS phosphorylation by mTOR and p70S6K in cells (45–47). These data might explain why we detected increased levels of IRS phosphorylation in Dlk1 cells after insulin stimulation. Additionally, mTORC2 or mTORC1 formations have no significant change in Dlk1 cells compared with control cells. It is plausible that Dlk1/FA1 induced inhibition of PI3K/Akt activation might not act directly through inhibition of the insulin receptor (IR) and IRS activation, or through reduction of mTORC2 formation, but through alternative pathways targeting PI3K/Akt activation.
Recently, fibronectin (Fn) was reported to interact with Dlk1/FA1 resulting in the activation of the insulin-induced ERK/MAPK pathway (48). We observed that suppression of fibronectin expression in ATDC5-Dlk1 cells, rescued the inhibition of insulin-induced Akt activation. Interestingly, these effects were not related to the fibronectin receptor integrin beta 1 (Itgb) or the canonical target of integrin signaling, FAK (focal adhesion kinase) which is one of the upstream regulators of Akt kinases (49). These results suggest that Dlk1/FA1 may exert its effects through interaction with fibronectin, but that the signaling is not mediated though Fn/Itgb/FAK1, but possibly through its unknown cognate receptor. Further investigations to identify the receptors of Dlk1/FA1 should give us a better understanding of the roles of the Dlk1/FA1 in cell differentiation.
Supplementary Material
Acknowledgments
We thank Tina Kamilla Nielsen, Lone Christiansen, and Bianca Jorgensen for excellent technical assistance and Linda Harkness for English editing.
This work was supported by grants from the Karen Elise Jensen's Foundation, The Novo Nordisk foundation, and a local grant from the region of Southern Denmark, Denmark, King Abdulaziz City for Science and Technology (09-BIO740-20), KSA.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- Dlk1
- delta-like factor 1
- FA1
- fetal antigen-1
- hESC
- human embryonic stem cell
- MSC
- mesenchymal stem cells
- Fn
- fibronectin.
REFERENCES
- 1. Sul H. S. (2009) Mol. Endocrinol. 23, 1717–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Laborda J. (2000) Histol. Histopathol. 15, 119–129 [DOI] [PubMed] [Google Scholar]
- 3. Wang Y., Sul H. S. (2006) Mol. Cell Biol. 26, 5421–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fay T. N., Jacobs I., Teisner B., Poulsen O., Chapman M. G., Stabile I., Bohn H., Westergaard J. G., Grudzinskas J. G. (1988) Eur. J. Obstet. Gynecol. Reprod. Biol. 29, 73–85 [DOI] [PubMed] [Google Scholar]
- 5. Jensen C. H., Krogh T. N., Højrup P., Clausen P. P., Skjødt K., Larsson L. I., Enghild J. J., Teisner B. (1994) Eur. J. Biochem. 225, 83–92 [DOI] [PubMed] [Google Scholar]
- 6. Garcés C., Ruiz-Hidalgo M. J., Bonvini E., Goldstein J., Laborda J. (1999) Differentiation 64, 103–114 [DOI] [PubMed] [Google Scholar]
- 7. Wylie A. A., Murphy S. K., Orton T. C., Jirtle R. L. (2000) Genome. Res. 10, 1711–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takada S., Paulsen M., Tevendale M., Tsai C. E., Kelsey G., Cattanach B. M., Ferguson-Smith A. C. (2002) Hum. Mol. Genet. 11, 77–86 [DOI] [PubMed] [Google Scholar]
- 9. Floridon C., Jensen C. H., Thorsen P., Nielsen O., Sunde L., Westergaard J. G., Thomsen S. G., Teisner B. (2000) Differentiation 66, 49–59 [DOI] [PubMed] [Google Scholar]
- 10. Yevtodiyenko A., Schmidt J. V. (2006) Dev. Dyn. 235, 1115–1123 [DOI] [PubMed] [Google Scholar]
- 11. Smas C. M., Sul H. S. (1997) Crit. Rev. Eukaryot. Gene Expr. 7, 281–298 [DOI] [PubMed] [Google Scholar]
- 12. Sakajiri S., O'Kelly J., Yin D., Miller C. W., Hofmann W. K., Oshimi K., Shih L. Y., Kim K. H., Sul H. S., Jensen C. H., Teisner B., Kawamata N., Koeffler H. P. (2005) Leukemia 19, 1404–1410 [DOI] [PubMed] [Google Scholar]
- 13. Crameri R. M., Langberg H., Magnusson P., Jensen C. H., Schrøder H. D., Olesen J. L., Suetta C., Teisner B., Kjaer M. (2004) J. Physiol. 558, 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abdallah B. M., Jensen C. H., Gutierrez G., Leslie R. G., Jensen T. G., Kassem M. (2004) J. Bone Miner. Res. 19, 841–852 [DOI] [PubMed] [Google Scholar]
- 15. Abdallah B. M., Boissy P., Tan Q., Dahlgaard J., Traustadottir G. A., Kupisiewicz K., Laborda J., Delaisse J. M., Kassem M. (2007) J. Biol. Chem. 282, 7339–7351 [DOI] [PubMed] [Google Scholar]
- 16. Berends M. J., Hordijk R., Scheffer H., Oosterwijk J. C., Halley D. J., Sorgedrager N. (1999) Am. J. Med. Genet. 84, 76–79 [DOI] [PubMed] [Google Scholar]
- 17. Hordijk R., Wierenga H., Scheffer H., Leegte B., Hofstra R. M., Stolte-Dijkstra I. (1999) J. Med. Genet. 36, 782–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moon Y. S., Smas C. M., Lee K., Villena J. A., Kim K. H., Yun E. J., Sul H. S. (2002) Mol. Cell. Biol. 22, 5585–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee K., Villena J. A., Moon Y. S., Kim K. H., Lee S., Kang C., Sul H. S. (2003) J. Clin. Invest. 111, 453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harkness L., Taipaleenmaki H., Mahmood A., Frandsen U., Saamanen A. M., Kassem M., Abdallah B. M. (2009) Stem Cell Rev. 5, 353–368 [DOI] [PubMed] [Google Scholar]
- 21. Wang Y., Sul H. S. (2009) Cell Metab. 9, 287–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fujita T., Fukuyama R., Enomoto H., Komori T. (2004) J. Cell Biochem. 93, 374–383 [DOI] [PubMed] [Google Scholar]
- 23. Shukunami C., Shigeno C., Atsumi T., Ishizeki K., Suzuki F., Hiraki Y. (1996) J. Cell Biol. 133, 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen L., Fink T., Zhang X. Y., Ebbesen P., Zachar V. (2005) Differentiation 73, 350–363 [DOI] [PubMed] [Google Scholar]
- 25. Jensen C. H., Krogh T. N., Støving R. K., Holmskov U., Teisner B. (1997) Clin. Chim. Acta. 268, 1–20 [DOI] [PubMed] [Google Scholar]
- 26. Mei B., Zhao L., Chen L., Sul H. S. (2002) Biochem. J. 364, 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Atsumi T., Miwa Y., Kimata K., Ikawa Y. (1990) Cell Differ. Dev. 30, 109–116 [DOI] [PubMed] [Google Scholar]
- 28. Yang L., Dan H. C., Sun M., Liu Q., Sun X. M., Feldman R. I., Hamilton A. D., Polokoff M., Nicosia S. V., Herlyn M., Sebti S. M., Cheng J. Q. (2004) Cancer Res. 64, 4394–4399 [DOI] [PubMed] [Google Scholar]
- 29. Lundholt B. K., Linde V., Loechel F., Pedersen H. C., Møller S., Praestegaard M., Mikkelsen I., Scudder K., Bjørn S. P., Heide M., Arkhammar P. O., Terry R., Nielsen S. J. (2005) J. Biomol. Screen 10, 20–29 [DOI] [PubMed] [Google Scholar]
- 30. Clark E. A., Brugge J. S. (1995) Science 268, 233–239 [DOI] [PubMed] [Google Scholar]
- 31. Xia H., Nho R. S., Kahm J., Kleidon J., Henke C. A. (2004) J. Biol. Chem. 279, 33024–33034 [DOI] [PubMed] [Google Scholar]
- 32. Smas C. M., Sul H. S. (1993) Cell 73, 725–734 [DOI] [PubMed] [Google Scholar]
- 33. Smas C. M., Sul H. S. (1996) Int. J. Obes. Relat. Metab. Disord. 20, Suppl. 3, S65–S72 [PubMed] [Google Scholar]
- 34. Stein G. S., Lian J. B., Gerstenfeld L. G., Shalhoub V., Aronow M., Owen T., Markose E. (1989) Connect Tissue Res. 20, 3–13 [DOI] [PubMed] [Google Scholar]
- 35. Laborda J., Sausville E. A., Hoffman T., Notario V. (1993) J. Biol. Chem. 268, 3817–3820 [PubMed] [Google Scholar]
- 36. Huang J., Zhang X., Zhang M., Zhu J. D., Zhang Y. L., Lin Y., Wang K. S., Qi X. F., Zhang Q., Liu G. Z., Yu J., Cui Y., Yang P. Y., Wang Z. Q., Han Z. G. (2007) Carcinogenesis 28, 1094–1103 [DOI] [PubMed] [Google Scholar]
- 37. Kawakami T., Chano T., Minami K., Okabe H., Okada Y., Okamoto K. (2006) Hum. Mol. Genet. 15, 821–830 [DOI] [PubMed] [Google Scholar]
- 38. Baladrón V., Ruiz-Hidalgo M. J., Bonvini E., Gubina E., Notario V., Laborda J. (2002) Biochem. Biophys. Res. Commun. 291, 193–204 [DOI] [PubMed] [Google Scholar]
- 39. Abdallah B. M., Ding M., Jensen C. H., Ditzel N., Flyvbjerg A., Jensen T. G., Dagnaes Hansen F., Gasser J. A., Kassem M. (2007) Endocrinology 148, 3111–3121 [DOI] [PubMed] [Google Scholar]
- 40. Oh C. D., Chun J. S. (2003) J. Biol. Chem. 278, 36563–36571 [DOI] [PubMed] [Google Scholar]
- 41. Nakamura K., Shirai T., Morishita S., Uchida S., Saeki-Miura K., Makishima F. (1999) Exp. Cell Res. 250, 351–363 [DOI] [PubMed] [Google Scholar]
- 42. Hidaka K., Kanematsu T., Takeuchi H., Nakata M., Kikkawa U., Hirata M. (2001) Int. J. Biochem. Cell Biol. 33, 1094–1103 [DOI] [PubMed] [Google Scholar]
- 43. Starkman B. G., Cravero J. D., Delcarlo M., Loeser R. F. (2005) Biochem. J. 389, 723–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Phornphutkul C., Wu K. Y., Auyeung V., Chen Q., Gruppuso P. A. (2008) Dev. Dyn. 237, 702–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ozes O. N., Akca H., Mayo L. D., Gustin J. A., Maehama T., Dixon J. E., Donner D. B. (2001) Proc. Natl. Acad. Sci. U S A 98, 4640–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harrington L. S., Findlay G. M., Gray A., Tolkacheva T., Wigfield S., Rebholz H., Barnett J., Leslie N. R., Cheng S., Shepherd P. R., Gout I., Downes C. P., Lamb R. F. (2004) J. Cell Biol. 166, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li Y., Soos T. J., Li X., Wu J., Degennaro M., Sun X., Littman D. R., Birnbaum M. J., Polakiewicz R. D. (2004) J. Biol. Chem. 279, 45304–45307 [DOI] [PubMed] [Google Scholar]
- 48. Wang Y., Zhao L., Smas C., Sul H. S. (2010) Mol. Cell. Biol. 30, 3480–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guan J. L. (1997) Int. J. Biochem. Cell Biol. 29, 1085–1096 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.