Mice lacking miR-146a exhibit exaggerated inflammatory responses, autoimmunity, and increased rate of tumorigenesis.
Abstract
Excessive or inappropriate activation of the immune system can be deleterious to the organism, warranting multiple molecular mechanisms to control and properly terminate immune responses. MicroRNAs (miRNAs), ∼22-nt-long noncoding RNAs, have recently emerged as key posttranscriptional regulators, controlling diverse biological processes, including responses to non-self. In this study, we examine the biological role of miR-146a using genetically engineered mice and show that targeted deletion of this gene, whose expression is strongly up-regulated after immune cell maturation and/or activation, results in several immune defects. Collectively, our findings suggest that miR-146a plays a key role as a molecular brake on inflammation, myeloid cell proliferation, and oncogenic transformation.
Robust activation of the innate immune response is essential for immediate defense against infection as well as for the generation of a long-lasting adaptive immunity against pathogens. However, if it is not terminated properly, the response can be harmful to the host, leading to the pathological manifestations of acute and chronic inflammatory disorders. Thus, it is not surprising that nature utilizes numerous and sometimes redundant molecular mechanisms to keep inflammation in check. Decades of intense study have uncovered multiple layers of innate immune regulation, from soluble receptors to inducible intracellular protein regulators (Liew et al., 2005). However, the full picture of how innate immune responses are modulated to avoid overreaction has yet to be painted.
MicroRNAs (miRNAs), a family of small noncoding RNAs, have recently emerged as powerful posttranscriptional regulators of various biological processes from cell fate determination to signaling events (Ambros, 2004; Bartel, 2004). A growing body of evidence suggests that the development and function of cells in the immune system is particularly subject to regulation by miRNAs (Baltimore et al., 2008; Tsitsiou and Lindsay, 2009). A few years ago, we postulated that miRNAs might comprise a novel layer of regulation of the innate immune response and performed a systematic effort to identify miRNAs that might be involved in the mammalian response to microbial infection (Taganov et al., 2006). We identified three miRNA genes (miR-146a, miR-132, and miR-155) whose expression is sharply up-regulated in response to LPS, and this list was later expanded further to include miR-9, miR-21, and miR-147 through the work of others (Bazzoni et al., 2009; Liu et al., 2009; Sheedy et al., 2010).
miR-146a is a member of the miR-146 miRNA family, consisting of two evolutionary conserved miRNA genes: miR-146a and miR-146b. In people, these loci are located on separate chromosomes, in quite unrelated sequence contexts, but differ in their mature sequence only by 2 nt at the 3′ end. Previously, we have shown that both genes respond to LPS in human monocytes, but only miR-146a is processed to a mature form, and determined that induction of expression of miR-146a is NF-κB dependent (Taganov et al., 2006). Based on our in vitro results, we proposed a model suggesting that miR-146a acts as a negative feedback regulator of the innate immune response by targeting two adapter proteins, TRAF6 (TNF receptor–associated factor 6) and IRAK1 (IL-1 receptor-associated kinase 1), that are crucial for proinflammatory signaling.
To test this notion experimentally and to further understand the biological role of miR-146a, we have created mutant mice with a targeted deletion of this gene. We have found that in mice, miR-146a is expressed predominantly in immune tissues, and its expression can be induced in immune cells upon cell maturation and/or activation. Ablation of miR-146a expression in mice results in several immune-related phenotypes, correlating well with its localization of expression. Lack of miR-146a expression results in hyperresponsiveness of macrophages to bacterial LPSs and leads to an exaggerated inflammatory response in endotoxin-challenged mice. In contrast, overexpression of miR-146a in monocytes has the opposite effect. Later in life, miR-146a–null mice develop a spontaneous autoimmune disorder, characterized by splenomegaly, lymphadenopathy, and multiorgan inflammation; as a result, many die prematurely. Mechanistically, autoimmunity in the KO mice correlated with the loss of peripheral T cell tolerance. Using a combination of gain and loss of function approaches, we have confirmed TRAF6 and IRAK1 genes as bona fide miR-146a targets, whose derepression in miR-146a KO mice might account for some of the observed immune phenotypes. In addition, we found that miR-146a seems to play a role in the control of immune cell proliferation; aging miR-146a–null mice display an excessive production of myeloid cells and develop frank tumors in their secondary lymphoid organs, suggesting that miR-146a can function as a tumor suppressor in the context of the immune system. Collectively, our results clearly establish miR-146a as an important negative regulator of inflammation, myeloid cell proliferation, and cancer.
RESULTS
miR-146a is up-regulated in immune cells in response to activation and/or upon maturation
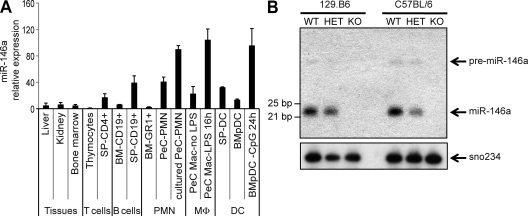
To establish the functional role of miR-146a in the immune system, we first surveyed its expression in mouse hematopoietic tissues. Expression of mature miR-146a was found to be relatively high in dendritic cells, thioglycollate-elicited macrophages and granulocytes, and peripheral B and T cells but low in the corresponding precursor populations and nonimmune tissues (Fig. 1 A). This suggests that miR-146a might be involved in the development and homeostasis of both myeloid and lymphoid cell lineages. Moreover, consistent with our and others’ previous findings (Taganov et al., 2006; Brown et al., 2007; Landgraf et al., 2007), we have also observed an increase in miR-146a expression in mouse primary macrophages, dendritic cells, and neutrophils in response to Toll-like receptor (TLR) agonists (Fig. 1 A and not depicted), suggesting that this miRNA may play an important role in the response of myeloid cells to infection.
Figure 1.
Profile of miR-146a gene expression in normal mouse tissues and abrogation of its expression in miR-146a−/− mice. (A) qRT-PCR analysis of mature miR-146a expression in various mouse tissues (BALB/c strain) and purified hematopoietic cells. Data are the mean of three mouse samples, and miR-146a expression from different tissues was normalized by U6 snRNA level. PMN, polymorphonuclear leukocyte; MΦ, macrophages; BM-Gr1+, purified Gr1+ cells from BM; BM-CD19+, purified CD19+ BM cells; SP-CD4+, purified CD4+ splenocytes; SP-CD19+, purified CD19+ splenocytes; PeC PMN, thioglycollate-elicited neutrophils lysed immediately after isolation or after being cultured in vitro for 3 h; PeC Mac, thioglycollate-elicited macrophages cultured in vitro for 16 h with or without 20 ng/ml LPS; SP DC, CD11c+MHCII+ splenic dendritic cells; BMpDC, BM-derived Flt3L-differentiated B220+Ly6C+PDCA1+CD11b− plasmacytoid dendritic cells; BMpDC 24 h CpG, BMpDCs stimulated with 5 µg/ml CpG for 24 h. Expression level of miR-146a in thymocytes was set arbitrarily to 1. Data are represented as mean ± SD. (B) Northern Blot analysis of miR-146a expression in splenocytes isolated from WT, miR-146a+/− (HET), and miR-146a−/− (KO) mice on two different genetic backgrounds, as indicated. Membrane was probed with DNA oligonucleotide complementary to mature miR-146a and then reprobed with sno234 probe to control for loading (bottom).
Generation of mice with targeted deletion of miR-146a gene
To test experimentally the biological role of miR-146a in the immune system, we have created mutant mice bearing a targeted deletion of the miR-146a gene. The miR-146a locus is situated on mouse chromosome 11, and its precursor is transcribed from an independent transcriptional unit with its own promoter. Therefore, our KO strategy simply called for the removal of an ∼300-nt-long genomic fragment, containing the miR-146a precursor sequence, and replacement of it with a single loxP sequence (Fig. S1, A and B). The absence of miR-146a expression in miR-146a−/− mice was confirmed in several hematopoietic tissues by Northern blotting and quantitative RT-PCR (qRT-PCR; Fig. 1 B and Fig. S1 C). The high homology of miR-146a and miR-146b has made it difficult to determine their individual levels. With miR-146a gone, we examined miR-146b levels, and interestingly, we were able to detect a low level of expression of mature miR-146b in some of the miR-146a KO tissues tested (unpublished data). Future studies will be required to determine the exact profile of miR-146b expression in mouse tissues, but it seems likely that expression of miR-146b is not masking phenotypes caused by the miR-146a KO, at least in most immune cells.
We have independently created miR-146a–null alleles in the context of two mouse genetic backgrounds, pure C57BL/6 (hereafter called B6/miR-146a−/−) and mixed C57BL/6 × 129/sv (hereafter called 129.B6/miR-146a−/−). Both KO lines were born at the expected Mendelian ratios to their littermates, were fertile, displayed no gross morphological abnormalities after birth, and produced very similar phenotypes later in life. However, certain nuances caused by the differences in genetic background were noted and will be discussed in Aging miR-146a–null mice develop autoimmune disorder.
miR-146a–null mice are hypersensitive to LPS
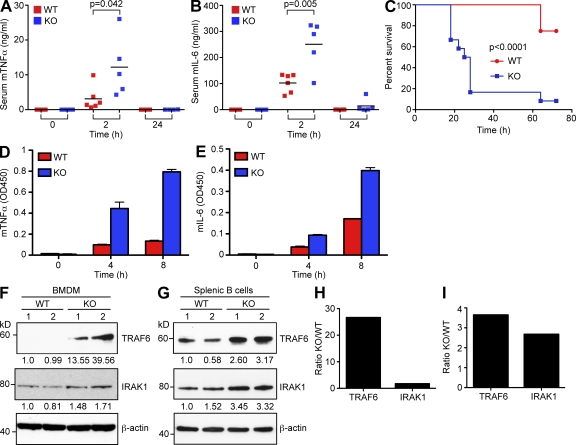
Because our previous findings suggested a negative role for miR-146a in control of TLR signaling (Taganov et al., 2006), we examined the possibility that miR-146a deletion might affect a mouse’s response to bacterial challenge. We found that young miR-146a–null mice systemically overproduced proinflammatory cytokines like TNF, IL-6, and IL-1β in response to injection with a sublethal LPS dose (Fig. 2, A and B; and Fig. S2 A). In addition, we observed elevated serum levels of IL-10 in miR-146a KO animals after LPS challenge, whereas the levels of two other cytokines, IL-12 and IFN-γ, did not change significantly (Fig. S2, B–D). In agreement with these observations, miR-146a–null mice succumbed much faster to the effects of septic shock than their WT littermates when injected with a lethal LPS does (Fig. 2 C) and in the process exhibited signs of a hypercytokinemia (Fig. S2, F and G).
Figure 2.
Exaggerated inflammatory response in miR-146a–null mice. (A and B) Serum levels of TNF (A) and IL-6 (B) in miR-146a−/− (n = 5; KO) and WT (n = 6) animals challenged intraperitoneally with sublethal dose of LPS (1 mg/kg). Peripheral blood was drawn at the times indicated in the graphs, and cytokine concentrations were assessed by ELISA. Results are shown as means; data are representative of two independent experiments. P-value calculations were performed using Student’s t test. (C) Kaplan-Meier survival curves of miR-146a−/− (KO) and WT mice (n = 12 for each group) challenged with a lethal dose of LPS (35 mg/kg). Results are representative of two independent experiments. P-value calculation was performed using log-rank (Mantel-Cox) test. (D and E) Production of TNF (D) and IL-6 (E) by miR-146a−/− (KO) and WT BMDMs in response to 10 ng/ml LPS challenge. Cells were stimulated for the indicated time, and cytokine concentrations were assessed by ELISA. Data are shown as mean ± SD. Results are representative of three independent experiments. (F and G) Western blot analysis of TRAF6 and IRAK1 protein expression in WT and miR-146a−/− (KO) BMDMs (F) and CD19+ splenic B cells (G). Numbers under the blots denote relative expression normalized to β-actin expression for each sample. Two mice per genotype were analyzed. Results are representative of two independent experiments. (H and I) Quantification of Western blots in F and G. Relative TRAF6 and IRAK1 expression in WT and KO BMDMs (H) and CD19+ splenic B cells (I) is plotted. Data from two mice for each genotype were averaged and are expressed as a ratio of protein expression in miR-146a KO to WT cells. All experiments were carried in the B6/miR-146a−/− strain of mice.
miR-146a–null macrophages are hyperresponsive to LPS
Tissue macrophages are considered a primary source of proinflammatory cytokine production. Therefore, to explain the LPS hyperresponsiveness observed in miR-146a mutant mice, we examined the effect of miR-146a deletion on primary macrophage function. We determined the levels of secreted proinflammatory cytokines by miR-146a−/− BM-derived macrophages (BMDMs) after LPS stimulation by ELISA and found a significant increase in the level of production of TNF, IL-6, and IL-1β proteins in comparison with WT cells (Fig. 2, D and E; and Fig. S2 E). miR-146a–null BMDMs also produced more nitric oxide in response to endotoxin than WT cells (unpublished data).
Monocytes overexpressing miR-146a show a dampened inflammatory response
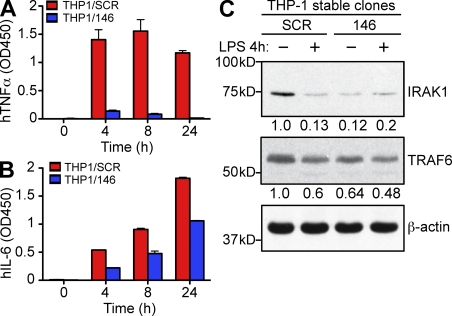
A role for miR-146a in macrophage activation and control of proinflammatory cytokine production was further supported by miR-146a overexpression experiments in human monocytic THP-1 cells. To create cells with ectopic expression of miR-146a (THP1/146), we transduced THP-1 cells with a lentivirus carrying a miR-146a expression cassette under control of the ubiquitin promoter (Fig. S3 A). As a control, we also generated a THP-1 line expressing a scrambled miR-146a sequence (THP1/SCR) using the same lentiviral vector. Analysis of miR-146a expression in THP1/146 cells by qRT-PCR revealed a 10-fold increase in mature miR-146a levels in comparison with THP1/SCR control cells (Fig. S3 B), which is about the level of miR-146a up-regulation seen in THP-1 cells after LPS challenge (Taganov et al., 2006). In striking contrast to primary miR-146a KO macrophages, stimulation of THP1/146 cells with LPS resulted in significantly attenuated production of proinflammatory cytokines, including TNF, IL-6, and IL-12, in comparison with THP1/SCR cells (Fig. 3, A and B; and Fig. S3 C). These results are in agreement with previous studies showing that overexpression of miR-146a in cell lines results in the dampening of proinflammatory response (Perry et al., 2008; Nahid et al., 2009). Collectively, our findings support the proposition (Taganov et al., 2006) that miR-146a plays a negative role in the control of inflammation in primary macrophages.
Figure 3.
Ectopic expression of miR-146a in THP-1 cells results in attenuation of inflammatory response. (A and B) Production of TNF (A) and IL-6 (B) by THP-1 stable cell lines ectopically expressing either miR-146a (THP1/146) or scramble control sequence (THP1/SCR) in response to 1 µg/ml LPS stimulation. Cells were stimulated for the indicated time, and cytokines concentrations were assessed by ELISA. Results are shown as mean ± SD. Data are representative of two independent experiments. (C) Western blot analysis of TRAF6 and IRAK1 protein expression in THP1/146 and THP1/SCR clones stimulated or not with LPS, as indicated. Numbers under the blots denote relative expression normalized to β-actin expression for each sample. Data are representative of two independent experiments.
TRAF6 and IRAK1 are bona fide targets of miR-146a
To understand the molecular mechanism by which miR-146a can regulate macrophage function, we performed expression profiling of primary macrophages derived from WT and KO animals using Affymetrix microarrays. miRNAs are believed to regulate the expression of hundreds of gene targets by binding to sequences within the 3′ untranslated regions (UTRs) of target messenger RNAs (mRNAs) that are complementary to nucleotides 2–8 (seed sequence) of the miRNA. The degree of the target regulation is usually very modest and correlates with the level of destabilization of the mRNA (Baek et al., 2008). Analysis of the expression data from miR-146a–null BMDMs by the Sylamer algorithm (van Dongen et al., 2008) did not reveal global derepression of miR-146a seed-matched transcripts in either unstimulated or LPS-stimulated cells (Fig. S2 J), despite significant changes in the expression of multiple genes in the KO cells (Tables S1 and S2).
To examine whether key miR-146a targets were derepressed primarily at the protein level, we have analyzed expression levels of several predicted molecular targets of miR-146a in primary miR-146a−/− macrophages and in the THP1/146 cell line by Western blot. Consistent with our proposed model (Taganov et al., 2006), analysis of miR-146a–null BMDMs revealed a marked increase in TRAF6 and IRAK1 protein levels in comparison with WT cells (Fig. 2, F and H). The derepression of TRAF6 and IRAK1 genes was not limited to the KO macrophages, as other hematopoietic tissues (e.g., peripheral B cells) also showed qualitatively similar changes (Fig. 2, G and I). Of note, expression of both TRAF6 and IRAK1 genes was not affected at the mRNA level (Fig. S2, H and I).
In contrast, THP1/146 cells expressed decreased amounts of IRAK1 and TRAF6 when compared with THP1/SCR control cells (Fig. 3 C). miR-146a had quantitatively different effects on target regulation in BMDMs and THP-1 cells: TRAF6 was derepressed more strongly than IRAK1 in miR-146a–null BMDMs; in contrast, miR-146a overexpression had a stronger impact on down-regulation of IRAK1 but not TRAF6 levels in THP-1 cells. Despite these differences, both of these molecular outcomes are consistent with our proposed model (Taganov et al., 2006), and the discrepancy in the extent of miR-146a target regulation requires further study.
In addition, LPS treatment of THP1/SCR control cells also resulted in repression of IRAK1 and TRAF6 protein expression (Fig. 3 C). The extent of this repression in THP1/SCR was quantitatively very similar to the level of silencing of the target proteins in THP1/146 cells, suggesting that up-regulation of miR-146a expression in THP1/SCR cells is mediating this effect. Degradation of IRAK1 protein in response to LPS was shown to contribute to the establishment of endotoxin tolerance (Li et al., 2000), in turn suggesting a potential role for miR-146a in this process, as supported by a recent study by Nahid et al. (2009).
Aging miR-146a–null mice develop autoimmune disorder
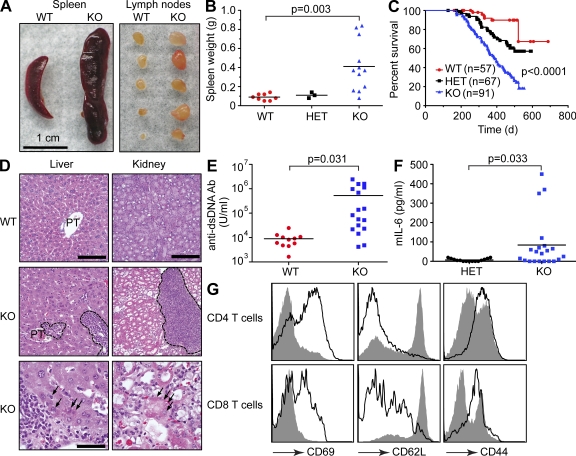
Although gross pathology in young miR-146a–null mice was unremarkable, aged mutant mice began to display several abnormal phenotypes. Starting at 6–8 mo, many 129.B6/miR-146a−/− mice exhibited signs of an immunoproliferative disease, characterized by splenomegaly, lymphadenopathy, and premature death (Fig. 4, A–C). Histological examination of tissues from miR-146a–null mice revealed multiorgan inflammation manifested by lymphocytic and monocytic infiltrates in various organs, including liver, kidneys, and lungs, with some evidence of tissue damage (Fig. 4 D and not depicted). These features were suggestive of autoimmune disease, and testing the serum of aged miR-146a–null mice for the presence of autoantibodies against double-stranded DNA revealed, on average, a 60-fold higher titer in KO animals compared with WT controls (Fig. 4 E). Moreover, aging 129.B6/miR-146a−/− mice displayed moderately elevated serum levels of IL-6, thus supporting the notion of ongoing systemic inflammation in the KO mice (Fig. 4 F). As often observed for autoimmune disorders in people (Dale et al., 2006), miR-146a−/− females were more affected by the disease than male littermates: the death rate in the former was significantly accelerated (Fig. S4 A). The onset of the immunoproliferative and autoimmune phenotypes was somewhat delayed in B6/miR-146a−/− line, and the rate of premature death was lower in this background (Fig. S4 B). In addition, the penetrance of the autoimmune phenotype in both strains of miR-146a–null mice was incomplete, and splenomegaly in KO animals was not always accompanied by lymphadenopathy.
Figure 4.
Deletion of miR-146a in mice results in autoimmune disorder. (A) Splenomegaly and lymphadenopathy observed in miR-146a−/− mice. Photographs of spleens and peripheral lymph nodes removed from a single representative WT and KO mouse. (B) Spleen weights observed in WT (n = 8), miR-146a+/− (HET; n = 3), and miR-146a−/− (KO; n = 12) animals. Results are shown as means and are representative of two independent experiments. P-value calculation was performed using Student’s t test. (C) Kaplan-Meier survival curves of WT, miR-146a+/− (HET), and miR-146a−/− (KO) mouse populations. P-value calculations were performed using log-rank (Mantel-Cox) test. P-value for WT and KO curve comparison is <0.0001. P-value for WT and HET curve comparison is 0.0041. (D) Multiorgan inflammation in miR-146a−/− mice. Hematoxylin- and eosin-stained sections of liver and kidney from miR-146a+/+ (top) and miR-146a−/− (middle, low magnification; bottom, high magnification) mice. Note the leukocytic infiltrates in the KO liver (circled with dashed lines), compared with a relatively spared portal triad (PT) in the WT liver. At higher magnification, degenerated hepatocytes (marked by arrows) displaying fragmented nuclei and eosinophilic cytoplasm indicate tissue damage caused by the inflammatory infiltrates. Note the large infiltrate of inflammatory cells (circled) in the miR-146a−/− kidney. In the low-power image, the KO tubules appear more eosinophilic, and their lumina are dilated. At higher magnification, a degenerated tubule (marked by arrows) indicates tissue damage caused by the inflammatory infiltrates. Bars: (top and middle) 200 µm; (bottom) 40 µm. (E) ELISA-based analysis of autoantibodies against double-stranded DNA (dsDNA) in the serum of WT (n = 11) and miR-146a−/− (KO; n = 17) animals. Ab, antibody. (F) Analysis of IL-6 levels in serum of miR-146a+/− (HET; n = 15) and miR-146a−/− (KO; n = 21) animals by ELISA. (E and F) Results are shown as means. P-value calculation was performed using Student’s t test. (G) FACS analysis of CD4+ and CD8+ splenocytes from miR-146a+/− and miR-146a−/− animals using anti-CD62, -CD44, and -CD69 antibodies. Gray plot indicates miR-146a+/− cells, and the open plot indicates miR-146a−/− cells. Data are representative of three independent experiments. All experiments were performed using the 129.B6/miR-146a−/− strain of mice.
Autoimmunity in miR-146a KO mice correlates mechanistically with a loss of peripheral T cell tolerance
Activation of peripheral T cells in naive mice is often a sign of a break in immune tolerance and is commonly found in mouse models of autoimmune diseases, like that observed in miR-146a−/− mice. Examination of the T cell lineage in miR-146a–null mice revealed that a large fraction of peripheral T cells (both CD8 and CD4 positive) displayed an activated, effector status: they stained positively for classical T cell activation markers CD69 and CD44 and negatively for CD62L (Fig. 4 G). Thus, it appears that lack of miR-146a expression over time can lead to a loss of peripheral T cell tolerance that correlates with the development of an autoimmune disease in miR-146a–null mice.
Massive myeloproliferation in miR-146a KO mice
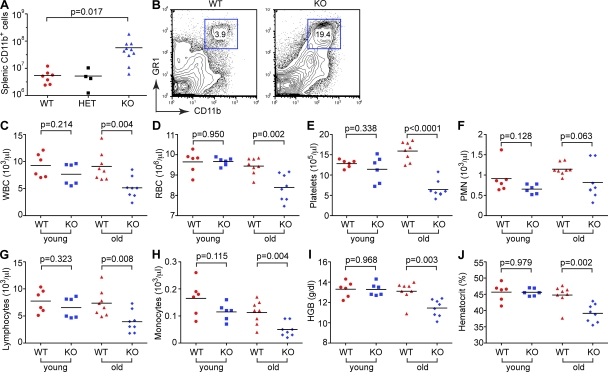
Detailed analysis of the miR-146a–null secondary lymphoid organs demonstrated a range of pathological phenotypes. The splenomegaly noted grossly was found to result mainly from myeloid proliferation. Analysis of hematopoietic cell lineages in the secondary lymphoid organs of miR-146a–null mice by flow cytometry revealed massive myeloproliferation (∼10-fold increase in the number of CD11b+ cells), whereas no significant change in the absolute number of B and T cells was found (Fig. 5 A and Fig. S5, A and B). CD11b+GR1+ blasts were the major cell population of the expanding myeloid compartment in KO spleens (Fig. 5 B). Aging miR-146a KO mice displayed anemia, thrombocytopenia, and lymphopenia (Fig. 5, C–J), as well as extramedullary erythropoiesis (Fig. S5 C), which are abnormalities frequently associated with BM failure, myelodysplastic syndrome (MDS), and myeloproliferative conditions. We also observed a mild and variable expansion of CD11b+ precursor cells in the KO BM (unpublished data).
Figure 5.
Myeloproliferation and BM failure in aging miR-146a−/− mice. (A) Absolute cell counts of myeloid cells (CD11b+) in WT (n = 7), miR-146a+/− (HET; n = 4), and miR-146a−/− (KO; n = 10) spleens. Results are shown as means and are representative of two independent experiments. (B) FACS analysis of myeloid cells in WT and KO spleens. Gated are Gr1 and CD11b double-positive cells. Numbers indicate percentage of cells in the gate. Data are representative of three independent experiments. (C–J) Complete blood count test with differential examination of peripheral blood from young (∼4 mo old; n = 6 for both genotypes) and old (∼14 mo old; n = 8 for both genotypes) WT and miR-146a−/− (KO) mice. (C) Absolute white blood cell counts. (D) Absolute red blood cell counts. (E) Absolute thrombocyte counts. (F) Absolute neutrophil counts. (G) Absolute lymphocyte counts. (H) Absolute monocyte counts. (I) Hemoglobin concentration. (J) Blood hematocrit. (A and C–J) P-value calculations were performed using Student’s t test. Experiments in B–J were performed using the B6/miR-146a−/− strain of mice. The experiment A was performed in 129.B6/miR-146a−/− mice.
miR-146a regulates myeloproliferation by controlling expression of M-CSF receptor
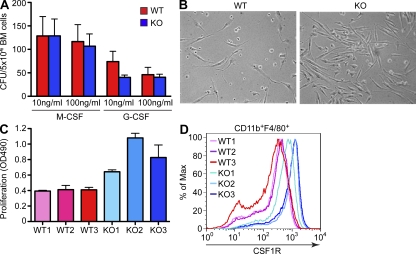
To test for a possibility that miR-146a deletion affects the number of myeloid precursor cells, we have performed colony-forming assays using BM cells and M-CSF or G-CSF as differentiating agents. We found that miR-146a–deficient BM gives rise to a comparable number of granulocyte and macrophage CFUs as observed for the WT marrow (Fig. 6 A), suggesting that miR-146a deletion has no effect on the rate of production of myeloid progenitors. Because M-CSF is known to act as a growth factor for myeloid cells, we have also measured the rate of proliferation of BMDMs differentiated in the presence of M-CSF and observed a markedly accelerated growth of miR-146a–null BMDMs (Fig. 6, B and C). To explain this finding, we have examined the level of cell surface expression of M-CSF receptor (CSF1R) on proliferating miR-146a–null BMDMs by FACS analysis and found it to be significantly elevated in comparison with WT cells (Fig. 6 D). Of note, the level of CSF1R expression on BMDM cells obtained from several KO BMs correlated well with the rate of proliferation of these cell cultures (Fig. 6, C and D), suggesting that CSF1R expression might indeed underlie the differences in proliferation of these cells. Moreover, pharmacological inhibition of CSF1R signaling in this BMDM proliferation assay significantly attenuated the growth of KO macrophages (Fig. S6, A and B). Interestingly, CSF1R receptor is predicted by several computational algorithms to function as a putative miR-146a target because the 3′ UTR of this mouse gene contains one evolutionary conserved and one nonconserved binding site for miR-146a. To determine whether miR-146a can indeed control CSF1R expression posttranscriptionally through binding to its 3′ UTR, we have fused the 3′ UTR of mouse CSF1R to a luciferase gene and tested this construct in a 3′ UTR reporter test. No significant change in Luciferase-CSF1R 3′ UTR reporter expression was observed in the presence of an excess of miR-146a (unpublished data), suggesting that miR-146a may control CSF1R expression in myeloid cells indirectly.
Figure 6.
miR-146a–null BMDMs proliferate more robustly in response to M-CSF and express more CSF1R on their cell surface. (A) Analysis of myeloid precursor cells by colony-forming cell assays. BM cells from WT and miR-146a–null mice (KO) were grown for 10 d in methylcellulose in the presence of the indicated concentrations of recombinant M-CSF or G-CSF, and the number of macrophage and granulocyte CFUs was scored under a microscope. (B) Photographs of proliferating in response to 50 ng/ml M-CSF BMDM cultures, derived from WT and miR-146a−/− (KO) mice. Pictures were taken on day 3 after cell plating and are representative of several biological replicates. Data are representative of two independent experiments. (C) Quantification of cellular proliferation of differentiating BMDM cultures from WT and miR-146a−/− (KO) mice in response to 50 ng/ml M-CSF. Three age- and gender-matched mice were used per genotype. Cell proliferation was assessed by MTS assay on day 3 after cell plating. Data are representative of two independent experiments. (A and C) Data are represented as mean ± SD. (D) FACS analysis of cell surface CSF1R expression on BMDM cells, derived from three WT and three miR-146a−/− (KO) mice as described in C. Cells were pregated for CD11b and F4/80 markers first. Data are representative of two independent experiments. All experiments were performed using the B6/miR-146a−/− strain of mice.
Aging miR-146a–null mice develop tumors
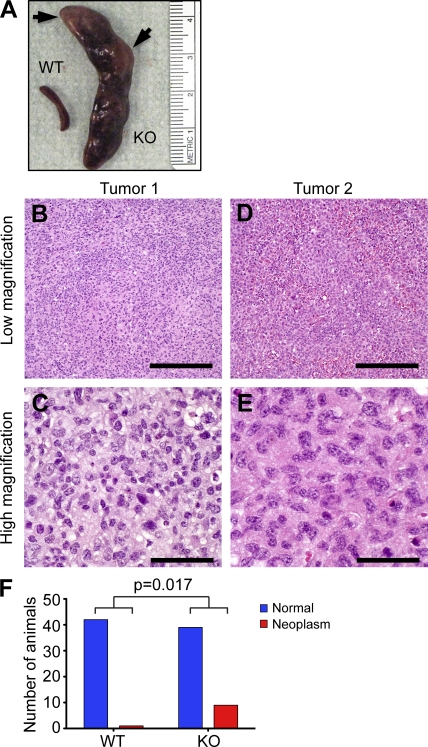
The premise that miR-146a can control cell proliferation/differentiation was further supported by an intriguing observation in some aging (older than 1 yr) 129.B6/miR146a-null mice. These animals developed frank tumors in their secondary lymphoid organs, most frequently in their spleens (Fig. 7 A). Histologically, these neoplasms were characterized as diffuse large cell neoplasms (Fig. 7, B–E) that stained negatively with pan-B, -T, and -myeloid cell markers (CD19, CD3, and CD11b, respectively) while displaying positive immunostaining only with anti-CD45 antibody, indicating a hematolymphoid origin for these tumors. More detailed histological sampling of lymphoid organs in aging 129.B6/miR-146a−/− mice has revealed four distinct temporal stages in the tumorigenesis process. The first manifestation of the disease in fairly young miR-146a−/− mice is displayed as follicular hyperplasia in the spleen (Fig. S7, B and G). Then, at 6–9 mo, when KO mice started to develop splenomegaly, the histological picture became more heterogeneous: follicular hyperplasia remained a frequent abnormality, but many of the animals showed features suggestive of BM failure such as markedly expanded splenic hematopoiesis (Fig. S7, C and H). A significant proportion of disease-afflicted miR-146a−/− mice aged 9–12 mo showed signs of low-grade follicular lymphoma and/or myeloproliferative disease with heterogeneous infiltrates in the spleen composed of lymphocytes and myeloid cells (Fig. S7, D, I, and K). Some cases of high-grade follicular lymphoma, with a preponderance of large cells, were also seen in mice aged around 1 yr (Fig. S7, E, J, and L). The transformation process accelerated after 12 mo, when several instances (n = 9) of high-grade large cell hematolymphoid neoplasms were detected (Fig. 7, B–E). The rate of neoplastic lesions in aging miR-146a–null mice (20%) was significantly higher than in WT animals (2.4%; Fig. 7 F).
Figure 7.
Increased rate of tumorigenesis in mice as a result of miR-146a ablation. (A) Photograph of spleens isolated form representative WT and miR-146a−/− (KO) mice; arrows are pointing at frank tumors. (B–E) Hematoxylin- and eosin-stained sections of two independent tumors isolated from miR-146a−/− mice showing features of diffuse large cell hematolymphoid neoplasms. The lower-power images show diffuse infiltrate without any architecture, whereas at higher magnification, the cells are very large with nuclei approaching 20 µm in size. Bars: (B and D) 400 µm; (C and E) 40 µm. (F) Rates of neoplasm occurrence in cohorts of aging miR-146a+/+ (n = 42; mean age = 537 d) and miR-146a−/− (n = 45; mean age = 571 d) mice. P-value calculation was performed using two-tailed Fisher’s exact test. All experiments were carried with 129.B6/miR-146a−/− mice.
DISCUSSION
Negative regulation of the immune response plays an important role in controlling homeostasis of the immune system and preventing development of autoimmune diseases. Multiple regulatory mechanisms including a complicated network of transcriptional factors and receptors as well as adapter and decoy molecules have evolved to keep activation of the immune system in check. Previously, we have proposed that miRNAs might constitute an integral component of a negative feedback mechanism in the innate immune system (Taganov et al., 2006), and in this study, we have provided functional evidence that miR-146a can serve as a key negative regulator of inflammation/autoimmunity.
Mice deficient in miR-146a expression are over-responsive to bacterial challenge and produce excessive amounts of proinflammatory cytokines (TNF and IL-6) and second messengers (NO) in response to LPS. As a result, miR-146a KO mice are very susceptible to endotoxic shock. In addition, aging miR-146a–null mice die prematurely because they develop a pathological condition that has the classical signs of an autoimmune disease: splenomegaly, lymphadenopathy, multiorgan inflammation with tissue damage, a high titer of autoantibodies, and elevated levels of proinflammatory cytokines in the blood. Collectively, these findings strongly suggest that miR-146a is required for proper termination of the immune response, and its elimination is seriously deleterious to the organism. The late onset of the autoimmune phenotype in miR-146a KO and its incomplete penetrance suggest that the break of immune tolerance in these mice occurs gradually over time, perhaps as a result of progressive exposure to pathogens and/or commensal bacteria in the gut. The autoimmune phenotype of miR-146a–null mice raises a possibility that misregulation of miR-146a expression might contribute to the pathology of human autoimmune diseases, making it relevant to note that the distal region of chromosome 5q, which contains the miR-146a gene (5q33) in man, has been previously suggested to harbor susceptibility loci for autoimmune diseases such as rheumatoid arthritis (Tokuhiro et al., 2003), Crohn’s disease (Rioux et al., 2001), asthma (Kauppi et al., 2001), and psoriasis (Friberg et al., 2006).
We have shown that miR-146a is expressed rather widely in the mouse immune system, and its expression can be up-regulated as a result of activation or maturation of myeloid and lymphoid cells. Therefore, mechanistically, the autoimmune phenotype in miR-146a KO mice can be driven by miR-146a deficiency in several cell lineages, and we have provided evidence of the contribution of at least two different cell types to this phenotype. First, we believe that excessive production of proinflammatory cytokines, a well-established driver of or contributor to autoimmune disease, in miR-146a KO mice can be explained by a functional defect in tissue macrophages as they are often considered to be a major source for proinflammatory cytokines in the body. We have shown that stimulation of miR-146a–deficient macrophages with LPS results in an overproduction of TNF and IL-6 proteins; in contrast, monocytes with ectopic expression of miR-146a display a dampened inflammatory response (Perry et al., 2008; Nahid et al., 2009; this study). Second, we found that a majority of peripheral effector T cells in miR-146a KO display an activated status, suggesting a breakdown in the peripheral T cell tolerance. Further studies will be required to test whether this T effector defect is cell autonomous or is actually a result of a defective function of regulatory T cells observed in miR-146a–null mice (Lu et al., 2010).
To gain further insight into the mechanism of the autoimmune defect in miR-146a–deficient mice, we have compared gene expression profiles of WT and miR-146a–deficient BMDMs as well as CD4+ splenic T cells (unpublished data) but did not detect a significant global derepression of miR-146a targets using computer algorithms, as was reported for some other miRNA KO models (Rodriguez et al., 2007; Baek et al., 2008). Our failure to observe a global change in expression of putative miR-146a targets in the tissues of KO mice suggests that either miR-146a regulates a rather restricted set of targets, which would preclude computer algorithms from detecting statistically significant enrichment, or that the majority of miR-146a targets are regulated by repression of protein synthesis without significant destabilization of the transcripts.
Lack of global derepression of miR-146a targets prompted us to examine expression levels of a few selected miR-146a target proteins, and we were able to observe a significant increase in expression of TRAF6 and IRAK1 at the protein but not at the mRNA level in several KO cell lineages. These two adapter molecules were previously identified as putative miR-146a targets (Taganov et al., 2006). In contrast, overexpression of miR-146a in monocytes resulted in a significant decrease in the levels of these two signaling molecules in our and others’ hands (Nahid et al., 2009). Combined, these findings suggest that TRAF6 and IRAK1 are indeed bona fide miR-146a targets, whose misregulation might very well be behind the autoimmunity and inflammation observed in miR-146a−/− mice because these two genes are required for signal transduction by a very diverse group of immune receptors, including antigen receptors, TLRs, and members of the IL-1 and TNF receptor families. Because these two adapter molecules often work together in a linear cascade, even small changes in their expression levels could have profound effects on downstream NF-κB and MAPK (mitogen-activated protein kinase) signaling. Overexpression of either of these genes in cells was shown to lead to the activation of IKK (IκBα kinase) and MAPK pathways, resulting in overproduction of proinflammatory cytokines (Cao et al., 1996; Ishida et al., 1996; Li et al., 2001). In addition, TRAF6 has been strongly implicated in the development and function of multiple hematopoietic lineages (Choi, 2005), whereas IRAK1 deficiency makes mice more resistant to the onset of autoimmune diseases (Deng et al., 2003; Berglund et al., 2008). Further studies of mice carrying heterozygous alleles of either TRAF6 or IRAK1 genes on an miR-146a–null background might be able to address the exact contribution of each of these miR-146a targets to the autoimmune phenotype of miR-146a KO mice.
Our results suggest that in addition to its anti-inflammatory role, miR-146a plays a negative regulatory role in the development of myeloid cells because its deletion in mice results in a myeloproliferative disorder with a massive accumulation of Gr1+Mac1+ blast cells in the secondary lymphoid organs. This defect seems to result from an excessive proliferative capacity of myeloid cells in the absence of miR-146a caused by increased CSF1R expression, producing a more robust proliferation of myeloid cells in response to M-CSF without a change in the number of myeloid progenitors.
Increased CSF1R expression is strongly associated in the literature with myeloproliferative diseases. Up-regulation of CSF1R expression was noted both in human and mouse acute myeloid leukemia (AML; Gisselbrecht et al., 1987; Rambaldi et al., 1988; Wang et al., 1988). A chromosomal translocation resulting in expression of a fusion protein between RBM6 (RNA-binding motif protein 6) and CSF1R was recently reported to associate with AML and to cause myeloproliferative disorder in mice with an accumulation of Gr1+Mac1+ blasts in spleen and liver (Gu et al., 2007). Moreover, in the AML model resulting from aberrant expression of the leukemia-associated MOZ (monocytic leukemia zinc finger)-TIF2 fusion protein, a high level of CSF1R expression was shown to confer leukemia-initiating activity to myeloid cells (Aikawa et al., 2010). Despite the fact that miR-146a was predicted to target the CSF1R gene by multiple miRNA target prediction algorithms, we failed to detect direct interaction between miR-146a and the 3′ UTR of the CSF1R gene, suggesting that up-regulation of CSF1R expression on macrophage precursors in miR-146a–null mice may be mediated by an intermediate factor. Several putative miR-146a targets with an established role in myeloproliferation disease (e.g., RUNX1T1, CBFA2T2, SPI1, etc.) could potentially serve such a role.
Misregulation of TRAF6 gene expression also can contribute to the myeloproliferative phenotype of miR-146a–null mice because this gene was shown to play an important role in the development of myeloid cells, e.g., osteoclasts and dendritic cells (Lomaga et al., 1999; Naito et al., 1999; Kobayashi et al., 2003). Moreover, chimeric mice overexpressing TRAF6 in the hematopoietic compartment were shown to develop either BM failure or AML starting at 5 mo after BM transplantation (Starczynowski et al., 2010).
It is worth noting that the loss of human chromosome 5 or deletion of the 5q region, which harbors the miR-146a gene, is associated with MDS and AML (Thirman and Larson, 1996). Recently, abrogation of miR-146a expression with consequent up-regulation of TRAF6 expression was strongly implicated in the pathogenesis of MDS (Starczynowski et al., 2010). Although miR-146a–null mice in our examination lack some of the classical features of 5q− syndrome (like neutropenia, thrombocytosis, and megakaryocytic dysplasia), future analysis of the heterozygous mice will be required to establish the exact contribution of miR-146a to this syndrome. In addition, two other miRNAs, miR-155 and miR-223, have also been implicated in the development of myeloid cells in mice (Johnnidis et al., 2008; O’Connell et al., 2008). Interestingly, miR-155, which, like miR-146a, can also be induced by LPS, was shown to cause myeloproliferation, albeit it was overexpression and not deletion that led to the hyperplasia of myeloid cells.
Our data also imply that miR-146a can function as a tumor suppressor gene, because aging miR-146a–null mice develop frank tumors in lymphoid organs. This conclusion is in agreement with previous studies, suggesting that inhibition of miR-146a activity could promote tumor growth and metastasis (Bhaumik et al., 2008; Lin et al., 2008; Hurst et al., 2009; Xia et al., 2009). In addition, down-regulation of miR-146a expression by the c-Myc oncogenic transcription factor was previously observed in human and mouse B cell lymphomas (Chang et al., 2008). However, the observed latency to development of these tumors clearly suggests that loss of miR-146a alone is insufficient for definitive tumorigenesis and that there are other factors that likely collaborate in the development of frankly malignant tumors. Excessive inflammation/autoimmunity seen in miR-146a KO mice can potentially serve as a confounding factor in the development of neoplasms in these animals. A growing body of evidence suggests that inflammation and cancer are intimately linked (Balkwill and Mantovani, 2001; Coussens and Werb, 2002; Karin, 2006; Mantovani et al., 2008). Further research into the molecular mechanism of miR-146a action will be required to understand whether this miRNA truly serves as a bridge between the two phenomena. These efforts will not only provide us with a deeper understanding of inflammatory and cancer induction processes but also might lead to new means for therapeutic intervention in chronic diseases.
MATERIALS AND METHODS
Generation of miR-146a−/− mice.
To generate miR-146a−/− mice, we created a targeting construct in which 295 bp of genomic sequence containing mouse pre–miR-146a were replaced with a floxed neomycin expression cassette. The assembly of the targeting construct was performed by recombineering in bacteria according to the protocol established by Copeland et al. (2001). The linearized targeting construct was then electroporated into CJ7 or Bruce4 embryonic stem cell lines to generate 129.B6/miR-146a−/− or B6/miR-146a−/− mouse strains, respectively. Neomycin-resistant clones that had undergone homologous recombination were identified by Southern blotting and further analyzed for a normal karyotype using DAPI staining. The resulting targeted embryonic stem clones were injected into C57BL/6 or C57BL/6-Tyr blastocysts and transferred into pseudopregnant foster females to produce chimeric mice. Germline-competent chimeras were then used to produce heterozygous F1 litters that were crossed with C57BL/6Tg-EIIa-Cre mice to remove the Neo selection cassette. WT C57BL/6, C57BL/6-Tyr, and C57BL/6Tg-EIIa-Cre mice were obtained from the Jackson Laboratory. Animal experiments were approved by the Institutional Animal Care and Use Committees of the California Institute of Technology and Explora Biolabs.
miRNA and mRNA qRT-PCR.
Total RNA was isolated using TRIZOL reagent (Invitrogen), the mirVana miRNA Isolation kit (Invitrogen), or the miRNeasy kit (QIAGEN). miRNA expression was measured with the mirVana qRT-PCR miRNA Detection kit (Invitrogen) or TaqMan miRNA assays (Applied Biosystems) according to the vendor protocol and normalized by 5S rRNA, U6 small nuclear RNA (snRNA), or sno234 snRNA levels. Expression of TRAF6 and IRAK1 genes was analyzed by TaqMan Gene Expression assays (Applied Biosystems) and normalized by mouse β-actin expression.
Cell culture and reagents.
THP-1 cells were obtained from the American Type Culture Collection and cultured in RPMI medium 1640 supplemented with 10% FBS, 1× nonessential amino acids, 100 U/ml penicillin, 100 U/ml streptomycin, and 2 mM glutamine in a humidified incubator containing 5% CO2 at 37°C. BMDMs were differentiated from BM precursors in the presence of 50 ng/ml recombinant mouse M-CSF (PeproTech) according to protocol (Karincaoglu et al., 2003). BM-derived plasmacytoid dendritic cells were differentiated from BM cells by culturing in the presence of 100 ng/ml recombinant Flt3 protein for 8–10 d. B220+ dendritic cells were then sorted out using Miltenyi Biotech MicroBeads and stimulated with 5 μg/ml CpG for 24 h. LPS (Escherichia coli 055:B5) was obtained from Sigma-Aldrich.
Western blot.
Total protein extracts were resolved on 10% Bis-Tris NuPAGE gel, transferred on nitrocellulose membrane, and blotted for the protein of interest. Anti–mouse IRAK1 antibodies were obtained from Cell Signaling Technology, and anti-mouse TRAF6 were obtained from MBL International; anti–human IRAK1 and TRAF6 were purchased from Santa Cruz Biotechnology, Inc. Quantification of relative expression for each target protein was performed in Photoshop CS4 (Adobe), and β-actin levels were used for normalization.
Northern blot.
Northern blot analysis was performed as described previously (Taganov et al., 2006). Membranes were hybridized with γ-32P–labeled DNA oligonucleotide probes that were perfectly complementary to mature miR-146a or sno234.
Flow cytometry.
Single cell suspensions prepared from various mouse hematolymphoid tissues were treated with red blood cell lysis buffer (BioLegend) and then stained with fluorophore-conjugated monoclonal antibodies against specific cell surface markers (eBioscience). Stained cells were assayed using a FACSCalibur flow cytometer (BD) and analyzed with FlowJo software (Tree Star).
ELISA.
For detection of cytokine levels in tissue culture media, ELISAs using specific combinations of capture and detection antibodies (eBioscience) were performed. Cytokine levels in the serum of LPS-challenged mice were assayed by a mouse proinflammatory 7-plex-panel kit (Meso Scale Discovery). Titers of autoantibodies against double-stranded DNA were calculated using a commercial ELISA test (BioVendor) according to manufacturer’s protocol.
Plasmid construction and generation of THP1/SCR and THP1/146 stable cell lines.
To create miR-146a expression vector, we modified FUW vector (Lois et al., 2002) by inserting the EGFP gene downstream of the ubiquitin promoter as well as puromycin resistance cassette; the cassette for miRNA expression was then cloned immediately downstream of the EGFP gene. The miRNA expression cassette was based on mouse miR-155 precursor sequence, in which mature miR-155 sequence was replaced with miR-146a or scrambled sequences. To produce viral particles, HEK293T cells were transfected with lentiviral expression vectors along with helper plasmids. Supernatant containing virus was used to infect THP-1 cells. Stable clones of transduced THP-1 cells were selected in the presence of puromycin for 14 d. Cells from resulting puromycin-resistant colonies were pooled and designated as THP1/SCR and THP1/146 lines.
Colony-forming cell assays.
Colony-forming cell assays were setup in MBM (methylcellulose base medium; R&D Systems) according to the manufacturer’s protocol. In brief, 5 × 104 BM cells from three age- and gender-matched WT and KO mice were plated in MBM in the presence of various concentrations of recombinant M-CSF and G-CSF proteins. Macrophage and granulocyte CFUs were scored after 10 d under a microscope (Diaphot 200; Nikon). Assays were performed in duplicate.
BMDM proliferation assays.
BM cells from three age- and gender-matched WT and KO mice were plated at a concentration of 2 × 104 per well in a 96-well plate in the presence of 50 ng/ml M-CSF. 3 d later, proliferation was assayed using MTS reagent (Promega) according to the manufacturer’s protocol. Assays were performed in triplicates. For pharmacological inhibition of BMDM proliferation with CSF1R inhibitor, the experiment was repeated in the presence of 500 nM cFMS receptor tyrosine kinase inhibitor (EMD). The rate of cellular proliferation was assessed on day 4 using MTS reagent.
3′ UTR reporter assays.
Genomic DNA fragment (608 bp) corresponding to the mouse 3′ UTR of CSF1R mRNA was obtained by PCR and cloned into pMIR-Report vector (Applied Biosystems). Site-directed mutagenesis was used to mutate both miR-146a binding sequences in the CSF1R 3′ UTR. To perform an assay, 6 × 104 293FT (Invitrogen) cells were plated in a 24-well culture dish and transfected using Attractene reagent (QIAGEN) with 200 ng miR-146a expression plasmid or empty vector, 20 ng 3′ UTR luciferase reporter plasmid, and 10 ng Renilla luciferase reporter plasmid (pRL-SV40; Promega). 48 h later, cell lysates were prepared, and the luciferase activities were measured using a Dual Luciferase Reporter assay system (Promega).
Morphological assessment of hematolymphoid tissues.
For histological sectioning, organs were placed into 10% neutral-buffered formalin immediately after necropsy, fixed for 12–18 h, washed, and transferred to 70% ethanol before standard paraffin embedding, sectioning, and staining with hematoxylin and eosin. All cytological preparations were air-dried and stained with Wright’s stain. Both histological and cytological preparations were examined on a microscope (BX-51; Olympus) and photographed using a color charge-coupled device digital camera (Insight 4 MP; SPOT) and software (SPOT).
Complete blood cell count tests.
Complete blood cell count tests with differential examination were performed by MDS Pharma Services.
In vivo LPS challenges.
For sublethal LPS challenge, mice were injected intraperitoneally with 1 mg/kg LPS. Peripheral blood was collected before the LPS injection as well as at 2 and 24 h after injection by retro-orbital bleeding. For lethal LPS challenge, mice were injected intraperitoneally with 35 mg/kg LPS and then monitored for 72 h. Moribund animals were euthanized immediately.
Tumor experiments.
Cohorts of aging (65–106 wk old) 129.B6/miR-146a−/− (n = 45) and WT (n = 42) animals were sacrificed and examined for the presence of gross abnormalities. Organs with apparent tumor growth as well as enlarged spleens/lymph nodes were collected and submitted for histological analysis.
Statistical tests.
All statistical analyses were performed using Excel (Microsoft) or Prism 5 (GraphPad Software) software. For animal survival curves, p-value was calculated using the Mantel-Cox log-rank test; for all other data, the Student’s two-tailed t test was used. Statistical analysis of neoplasm occurrence was performed using the two-tailed Fisher’s exact test.
Online supplemental material.
Fig. S1 describes the generation of miR-146a–null mice. Fig. S2 shows ELISA analyses of cytokine expression in the serum of LPS-stimulated miR-146a–null mice and qRT-PCR and Sylamer analysis of gene expression in miR-146a–null BMDMs. Fig. S3 describes the generation of THP1/146 and THP1/SCR cells and analysis of IL-12 secretion in these cells in response to LPS. Fig. S4 shows Kaplan-Meier survival curves for males and females from the 129.B6/miR-146a−/− line, as well as lifespan for the B6/miR-146a−/− line. Fig. S5 shows absolute T and B cells counts, as well as extramedullary hematopoiesis in miR-146a–null spleens. Fig. S6 shows that pharmacological inhibition of CSF1R activity blocks BMDM proliferation. Fig. S7 shows the characterization of tumorigenesis stages in miR-146a–null mice. Table S1, included as an Excel file, lists genes that are differentially expressed in miR-146a–deficient BMDMs. Table S2, included as an Excel file, lists genes that are differentially expressed in LPS-treated miR-146a–deficient BMDMs. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20101823/DC1.
Acknowledgments
We thank Shirley Pease and other members of the Caltech Transgenic Core Facility for their technical support. We thank Sergei Grivennikov for critical reading of the manuscript and Matthew Liabson for the generation of 3′ UTR reporter constructs.
This work was supported by National Institutes of Health grant 5R01AI079243-02 and career development award 5K08CA133521 (to D.S. Rao).
M.P. Boldin, K.D. Taganov, J. Xu, G. Sun, J. Tay, and P.S. Linsley are employees and shareholders of Regulus Therapeutics, a biotech company developing miRNA-based drugs. D. Baltimore is a director and Chairman of the Scientific Advisory board of the same company.
Author contributions: M.P. Boldin, K.D. Taganov, D.S. Rao, L. Yang, J.L. Zhao, and D. Baltimore designed the study, analyzed data, and wrote the paper. M.P. Boldin, K.D. Taganov, D.S. Rao, L. Yang, M. Kalwani, Y. Garcia-Flores, M. Luong, A. Devrekanli, J. Xu, G. Sun, and J. Tay collected data. P.S. Linsley performed bioinformatical analyses and wrote the paper.
Footnotes
Abbreviations used:
- AML
- acute myeloid leukemia
- BMDM
- BM-derived macrophage
- MDS
- myelodysplastic syndrome
- miRNA
- microRNA
- mRNA
- messenger RNA
- qRT-PCR
- quantitative RT-PCR
- snRNA
- small nuclear RNA
- TLR
- Toll-like receptor
- UTR
- untranslated region
References
- Aikawa Y., Katsumoto T., Zhang P., Shima H., Shino M., Terui K., Ito E., Ohno H., Stanley E.R., Singh H., et al. 2010. PU.1-mediated upregulation of CSF1R is crucial for leukemia stem cell potential induced by MOZ-TIF2. Nat. Med. 16:580–585: 1p: 585 10.1038/nm.2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. 2004. The functions of animal microRNAs. Nature. 431:350–355 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- Baek D., Villén J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. 2008. The impact of microRNAs on protein output. Nature. 455:64–71 10.1038/nature07242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill F., Mantovani A. 2001. Inflammation and cancer: back to Virchow? Lancet. 357:539–545 10.1016/S0140-6736(00)04046-0 [DOI] [PubMed] [Google Scholar]
- Baltimore D., Boldin M.P., O’Connell R.M., Rao D.S., Taganov K.D. 2008. MicroRNAs: new regulators of immune cell development and function. Nat. Immunol. 9:839–845 10.1038/ni.f.209 [DOI] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bazzoni F., Rossato M., Fabbri M., Gaudiosi D., Mirolo M., Mori L., Tamassia N., Mantovani A., Cassatella M.A., Locati M. 2009. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc. Natl. Acad. Sci. USA. 106:5282–5287 10.1073/pnas.0810909106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund M., Thomas J.A., Hörnquist E.H., Hultgren O.H. 2008. Toll-like receptor cross-hyporesponsiveness is functional in interleukin-1-receptor-associated kinase-1 (IRAK-1)-deficient macrophages: differential role played by IRAK-1 in regulation of tumour necrosis factor and interleukin-10 production. Scand. J. Immunol. 67:473–479 10.1111/j.1365-3083.2008.02096.x [DOI] [PubMed] [Google Scholar]
- Bhaumik D., Scott G.K., Schokrpur S., Patil C.K., Campisi J., Benz C.C. 2008. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 27:5643–5647 10.1038/onc.2008.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B.D., Gentner B., Cantore A., Colleoni S., Amendola M., Zingale A., Baccarini A., Lazzari G., Galli C., Naldini L. 2007. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 25:1457–1467 10.1038/nbt1372 [DOI] [PubMed] [Google Scholar]
- Cao Z., Xiong J., Takeuchi M., Kurama T., Goeddel D.V. 1996. TRAF6 is a signal transducer for interleukin-1. Nature. 383:443–446 10.1038/383443a0 [DOI] [PubMed] [Google Scholar]
- Chang T.C., Yu D., Lee Y.S., Wentzel E.A., Arking D.E., West K.M., Dang C.V., Thomas-Tikhonenko A., Mendell J.T. 2008. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 40:43–50 10.1038/ng.2007.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. 2005. Role of TRAF6 in the immune system. Adv. Exp. Med. Biol. 560:77–82 10.1007/0-387-24180-9_11 [DOI] [PubMed] [Google Scholar]
- Copeland N.G., Jenkins N.A., Court D.L. 2001. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2:769–779 10.1038/35093556 [DOI] [PubMed] [Google Scholar]
- Coussens L.M., Werb Z. 2002. Inflammation and cancer. Nature. 420:860–867 10.1038/nature01322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale E., Davis M., Faustman D.L. 2006. A role for transcription factor NF-kappaB in autoimmunity: possible interactions of genes, sex, and the immune response. Adv. Physiol. Educ. 30:152–158 10.1152/advan.00065.2006 [DOI] [PubMed] [Google Scholar]
- Deng C., Radu C., Diab A., Tsen M.F., Hussain R., Cowdery J.S., Racke M.K., Thomas J.A. 2003. IL-1 receptor-associated kinase 1 regulates susceptibility to organ-specific autoimmunity. J. Immunol. 170:2833–2842 [DOI] [PubMed] [Google Scholar]
- Friberg C., Björck K., Nilsson S., Inerot A., Wahlström J., Samuelsson L. 2006. Analysis of chromosome 5q31-32 and psoriasis: confirmation of a susceptibility locus but no association with SNPs within SLC22A4 and SLC22A5. J. Invest. Dermatol. 126:998–1002 10.1038/sj.jid.5700194 [DOI] [PubMed] [Google Scholar]
- Gisselbrecht S., Fichelson S., Sola B., Bordereaux D., Hampe A., André C., Galibert F., Tambourin P. 1987. Frequent c-fms activation by proviral insertion in mouse myeloblastic leukaemias. Nature. 329:259–261 10.1038/329259a0 [DOI] [PubMed] [Google Scholar]
- Gu T.L., Mercher T., Tyner J.W., Goss V.L., Walters D.K., Cornejo M.G., Reeves C., Popova L., Lee K., Heinrich M.C., et al. 2007. A novel fusion of RBM6 to CSF1R in acute megakaryoblastic leukemia. Blood. 110:323–333 10.1182/blood-2006-10-052282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst D.R., Edmonds M.D., Scott G.K., Benz C.C., Vaidya K.S., Welch D.R. 2009. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 69:1279–1283 10.1158/0008-5472.CAN-08-3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Mizushima S., Azuma S., Kobayashi N., Tojo T., Suzuki K., Aizawa S., Watanabe T., Mosialos G., Kieff E., et al. 1996. Identification of TRAF6, a novel tumor necrosis factor receptor-associated factor protein that mediates signaling from an amino-terminal domain of the CD40 cytoplasmic region. J. Biol. Chem. 271:28745–28748 10.1074/jbc.271.46.28745 [DOI] [PubMed] [Google Scholar]
- Johnnidis J.B., Harris M.H., Wheeler R.T., Stehling-Sun S., Lam M.H., Kirak O., Brummelkamp T.R., Fleming M.D., Camargo F.D. 2008. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 451:1125–1129 10.1038/nature06607 [DOI] [PubMed] [Google Scholar]
- Karin M. 2006. Nuclear factor-kappaB in cancer development and progression. Nature. 441:431–436 10.1038/nature04870 [DOI] [PubMed] [Google Scholar]
- Karincaoglu Y., Kaya E., Esrefoglu M., Aydogdu I. 2003. Development of large genital ulcer due to hydroxyurea treatment in a patient with chronic myeloid leukemia and Behcet’s disease. Leuk. Lymphoma. 44:1063–1065 10.1080/1042819031000067864 [DOI] [PubMed] [Google Scholar]
- Kauppi P., Lindblad-Toh K., Sevon P., Toivonen H.T., Rioux J.D., Villapakkam A., Laitinen L.A., Hudson T.J., Kere J., Laitinen T. 2001. A second-generation association study of the 5q31 cytokine gene cluster and the interleukin-4 receptor in asthma. Genomics. 77:35–42 10.1006/geno.2001.6613 [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Walsh P.T., Walsh M.C., Speirs K.M., Chiffoleau E., King C.G., Hancock W.W., Caamano J.H., Hunter C.A., Scott P., et al. 2003. TRAF6 is a critical factor for dendritic cell maturation and development. Immunity. 19:353–363 10.1016/S1074-7613(03)00230-9 [DOI] [PubMed] [Google Scholar]
- Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 129:1401–1414 10.1016/j.cell.2007.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Cousart S., Hu J., McCall C.E. 2000. Characterization of interleukin-1 receptor-associated kinase in normal and endotoxin-tolerant cells. J. Biol. Chem. 275:23340–23345 10.1074/jbc.M001950200 [DOI] [PubMed] [Google Scholar]
- Li X., Commane M., Jiang Z., Stark G.R. 2001. IL-1-induced NFkappa B and c-Jun N-terminal kinase (JNK) activation diverge at IL-1 receptor-associated kinase (IRAK). Proc. Natl. Acad. Sci. USA. 98:4461–4465 10.1073/pnas.071054198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew F.Y., Xu D., Brint E.K., O’Neill L.A. 2005. Negative regulation of toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 5:446–458 10.1038/nri1630 [DOI] [PubMed] [Google Scholar]
- Lin S.L., Chiang A., Chang D., Ying S.Y. 2008. Loss of mir-146a function in hormone-refractory prostate cancer. RNA. 14:417–424 10.1261/rna.874808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Friggeri A., Yang Y., Park Y.J., Tsuruta Y., Abraham E. 2009. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc. Natl. Acad. Sci. USA. 106:15819–15824 10.1073/pnas.0901216106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C., Hong E.J., Pease S., Brown E.J., Baltimore D. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 295:868–872 10.1126/science.1067081 [DOI] [PubMed] [Google Scholar]
- Lomaga M.A., Yeh W.C., Sarosi I., Duncan G.S., Furlonger C., Ho A., Morony S., Capparelli C., Van G., Kaufman S., et al. 1999. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 13:1015–1024 10.1101/gad.13.8.1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.-F., Boldin M.P., Chaudhry A., Lin L.-L., Taganov K.D., Hanada T., Yoshimura A., Baltimore D., Rudensky A.Y. 2010. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 142:914–929 10.1016/j.cell.2010.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A., Allavena P., Sica A., Balkwill F. 2008. Cancer-related inflammation. Nature. 454:436–444 10.1038/nature07205 [DOI] [PubMed] [Google Scholar]
- Nahid M.A., Pauley K.M., Satoh M., Chan E.K. 2009. miR-146a is critical for endotoxin-induced tolerance: implication in innate immunity. J. Biol. Chem. 284:34590–34599 10.1074/jbc.M109.056317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito A., Azuma S., Tanaka S., Miyazaki T., Takaki S., Takatsu K., Nakao K., Nakamura K., Katsuki M., Yamamoto T., Inoue J. 1999. Severe osteopetrosis, defective interleukin-1 signalling and lymph node organogenesis in TRAF6-deficient mice. Genes Cells. 4:353–362 10.1046/j.1365-2443.1999.00265.x [DOI] [PubMed] [Google Scholar]
- O’Connell R.M., Rao D.S., Chaudhuri A.A., Boldin M.P., Taganov K.D., Nicoll J., Paquette R.L., Baltimore D. 2008. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 205:585–594 10.1084/jem.20072108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry M.M., Moschos S.A., Williams A.E., Shepherd N.J., Larner-Svensson H.M., Lindsay M.A. 2008. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J. Immunol. 180:5689–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaldi A., Wakamiya N., Vellenga E., Horiguchi J., Warren M.K., Kufe D., Griffin J.D. 1988. Expression of the macrophage colony-stimulating factor and c-fms genes in human acute myeloblastic leukemia cells. J. Clin. Invest. 81:1030–1035 10.1172/JCI113413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux J.D., Daly M.J., Silverberg M.S., Lindblad K., Steinhart H., Cohen Z., Delmonte T., Kocher K., Miller K., Guschwan S., et al. 2001. Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat. Genet. 29:223–228 10.1038/ng1001-223 [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Vigorito E., Clare S., Warren M.V., Couttet P., Soond D.R., van Dongen S., Grocock R.J., Das P.P., Miska E.A., et al. 2007. Requirement of bic/microRNA-155 for normal immune function. Science. 316:608–611 10.1126/science.1139253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy F.J., Palsson-McDermott E., Hennessy E.J., Martin C., O’Leary J.J., Ruan Q., Johnson D.S., Chen Y., O’Neill L.A. 2010. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 11:141–147 10.1038/ni.1828 [DOI] [PubMed] [Google Scholar]
- Starczynowski D.T., Kuchenbauer F., Argiropoulos B., Sung S., Morin R., Muranyi A., Hirst M., Hogge D., Marra M., Wells R.A., et al. 2010. Identification of miR-145 and miR-146a as mediators of the 5q-syndrome phenotype. Nat. Med. 16:49–58 10.1038/nm.2054 [DOI] [PubMed] [Google Scholar]
- Taganov K.D., Boldin M.P., Chang K.J., Baltimore D. 2006. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA. 103:12481–12486 10.1073/pnas.0605298103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirman M.J., Larson R.A. 1996. Therapy-related myeloid leukemia. Hematol. Oncol. Clin. North Am. 10:293–320 10.1016/S0889-8588(05)70340-3 [DOI] [PubMed] [Google Scholar]
- Tokuhiro S., Yamada R., Chang X., Suzuki A., Kochi Y., Sawada T., Suzuki M., Nagasaki M., Ohtsuki M., Ono M., et al. 2003. An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat. Genet. 35:341–348 10.1038/ng1267 [DOI] [PubMed] [Google Scholar]
- Tsitsiou E., Lindsay M.A. 2009. microRNAs and the immune response. Curr. Opin. Pharmacol. 9:514–520 10.1016/j.coph.2009.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen S., Abreu-Goodger C., Enright A.J. 2008. Detecting microRNA binding and siRNA off-target effects from expression data. Nat. Methods. 5:1023–1025 10.1038/nmeth.1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Kelleher C.A., Cheng G.Y., Miyauchi J., Wong G.G., Clark S.C., Minden M.D., McCulloch E.A. 1988. Expression of the CSF-1 gene in the blast cells of acute myeloblastic leukemia: association with reduced growth capacity. J. Cell. Physiol. 135:133–138 10.1002/jcp.1041350119 [DOI] [PubMed] [Google Scholar]
- Xia H., Qi Y., Ng S.S., Chen X., Li D., Chen S., Ge R., Jiang S., Li G., Chen Y., et al. 2009. microRNA-146b inhibits glioma cell migration and invasion by targeting MMPs. Brain Res. 1269:158–165 10.1016/j.brainres.2009.02.037 [DOI] [PubMed] [Google Scholar]