STAT5 is abnormally activated in patients with acute lymphoblastic leukemia, and increased STAT5 activation synergizes with PAX5 and EBF1 to induce disease.
Abstract
As STAT5 is critical for the differentiation, proliferation, and survival of progenitor B cells, this transcription factor may play a role in acute lymphoblastic leukemia (ALL). Here, we show increased expression of activated signal transducer and activator of transcription 5 (STAT5), which is correlated with poor prognosis, in ALL patient cells. Mutations in EBF1 and PAX5, genes critical for B cell development have also been identified in human ALL. To determine whether mutations in Ebf1 or Pax5 synergize with STAT5 activation to induce ALL, we crossed mice expressing a constitutively active form of STAT5 (Stat5b-CA) with mice heterozygous for Ebf1 or Pax5. Haploinsufficiency of either Pax5 or Ebf1 synergized with Stat5b-CA to rapidly induce ALL in 100% of the mice. The leukemic cells displayed reduced expression of both Pax5 and Ebf1, but this had little effect on most EBF1 or PAX5 target genes. Only a subset of target genes was deregulated; this subset included a large percentage of potential tumor suppressor genes and oncogenes. Further, most of these genes appear to be jointly regulated by both EBF1 and PAX5. Our findings suggest a model whereby small perturbations in a self-reinforcing network of transcription factors critical for B cell development, specifically PAX5 and EBF1, cooperate with STAT5 activation to initiate ALL.
Transformation of lymphoid progenitor cells leads to the development of acute lymphoblastic leukemia (ALL). The majority of ALL cases involve transformation of B lineage cells, with only a small percentage of cases resulting from T cell transformation. ALL is thought to be caused by genetic lesions in genes critical for proliferation, differentiation, and survival of progenitor cells (Pui et al., 2008). These lesions include chromosomal translocations such as BCR-ABL or E2A-PBX. BCR-ABL represents a fusion gene from a chromosomal translocation called the Philadelphia Chromosome and often results in a poor outcome (Milone and Enrico, 2009; Fielding, 2010). The BCR-ABL translocation results in the activation of signal transducer and activator of transcription (STAT5), a transcription factor that is important for B cell development (Carlesso et al., 1996; Ilaria and Van Etten, 1996). Recent work by Mullighan et al. (2007) has provided new insight into genetic lesions that may be involved in ALL and further illustrated the importance of several additional genes known to be critical in early stages of B cell development and differentiation. In that study, they found that 40% of the genetic alterations identified were in principal regulators of B cell development including early B cell factor 1 (EBF1) and paired box gene 5 (PAX5) with PAX5 modifications accounting for >30% of the cases. This indicates that disruption of genes involved in critical stages of B cell development may lead to B cell leukemia.
STAT5 plays a critical role in both B and T cell development and is encoded by two closely linked genes, Stat5a and Stat5b, which give rise to functionally redundant proteins. Conditional STAT5 deletion strategies found reduced numbers of common lymphoid progenitors and pre/pro–B cells in Stat5−/− mice, whereas pro–B cells and later stages of B cell differentiation were absent (Yao et al., 2006; unpublished data). Thus, STAT5 plays a critical role in early B cell differentiation. We previously generated mice expressing a constitutively active form of STAT5 (Stat5b-CA) throughout B and T cell development (Burchill et al., 2003). These mice exhibit a marked expansion of pro–B and pre–B cells (∼5-fold increase). Despite this increase in B cell progenitors, the number of mature B cells in Stat5b-CA mice is not significantly different from that observed in wild–type littermate controls. However, we found that our Stat5b-CA transgenic mice develop a disease resembling human ALL, although with low penetrance (∼1 to 2%; Burchill et al., 2003; Nakayama et al., 2009). Previous research has suggested that STAT5 may play an important role in cancer, including ALL. For example, work by Weber-Nordt et al. (1996) found constitutive STAT5 activation in the majority of ALL samples they examined, the caveat being that only 3 live and 12 fixed samples were examined in this study. Additional studies have shown that STAT5 is activated by several oncogenic proteins including BCR-ABL (Xie et al., 2001; Buettner et al., 2002). This was accomplished by engineering bone marrow–derived cells to express BCR-ABL or TEL-JAK2 fusion proteins that initiate leukemia upon transfer into histocompatible recipient mice. However, if the engineered bone marrow cells also lack the Stat5a and Stat5b genes, leukemia does not result (Schwaller et al., 2000; Hoelbl et al., 2006). These findings suggest a possible role for STAT5 activation in initiating ALL.
Transcriptional regulation plays a critical role in B cell differentiation with expression of distinct sets of genes at discrete stages resulting in the initiation of lineage differentiation. Two genes crucial for initiating and maintaining B lineage specificity are Ebf1 and Pax5 (Hagman and Lukin, 2006; Nutt and Kee, 2007). Loss of the Ebf1 gene in mice revealed a block in B cell differentiation before the development of prepro–B cells (Lin and Grosschedl, 1995b). In addition, mice heterozygous for Ebf1 show a 50% reduction in mature B cells but normal levels of pro–B cells (Lin and Grosschedl, 1995b). EBF1 is clearly involved in the expression of many B cell–specific genes, including the transcription factor PAX5 (Månsson et al., 2004; Nutt and Kee, 2007). More recent studies have documented that EBF1 also represses several genes that interfere with B cell development (Pongubala et al., 2008; Treiber et al., 2010). As a multifunctional transcriptional regulator, PAX5 represses expression of genes involved in commitment to other lineages while activating B cell–specific genes such as Cd19 and Blnk (Nutt et al., 1999; Cobaleda et al., 2007; Schebesta et al., 2007). In the absence of PAX5 expression, B cell development is arrested at the late pro–B cell stage in the bone marrow. Moreover, Pax5−/− pro–B cells are not committed to the B cell lineage and are able to convert to other hematopoietic cell types such as T cells or myeloid cells (Nutt et al., 1999). EBF1 has been shown to bind to the Pax5 promoter and induce Pax5 expression (Cobaleda et al., 2007). Interestingly, although EBF1 is expressed earlier than PAX5, PAX5 binds the Ebf1 promoter and is required to maintain normal levels of Ebf1 expression (Cobaleda et al., 2007; Roessler et al., 2007). Thus, PAX5 and EBF1 act as part of a self-reinforcing network that induces genes critical for B cell differentiation and represses genes involved in differentiation of other cell lineages.
Though a correlation has been identified between transcription factors critical for early B cell development and ALL, little is known regarding the mechanism by which this promotes transformation. Moreover, data supporting these mutations as drivers in the transformation process rather than just passengers are also absent. In this paper, we demonstrate that elevated levels of STAT5 phosphorylation (phospho-STAT5) are found in certain subgroups of human ALL, and these elevated levels of pSTAT5 correlate with poor disease outcome after treatment. Using Stat5b-CA mice, we demonstrate that STAT5 synergizes with haploinsufficiency of either Ebf1 or Pax5 to rapidly induce progenitor B-ALL. Stat5b-CA x Ebf1+/− and Stat5b-CA x Pax5+/− leukemias show reduced expression of both Ebf1 and Pax5 with effects on a subset of EBF1 and PAX5 responsive genes. The subset of EBF1/PAX5-responsive genes includes potential tumor suppressor genes, oncogenes, and B cell growth factors. Therefore, changes in Ebf1 and Pax5 expression, such as those caused by haploinsufficiency, can result in changes in gene expression leading to induction of oncogenes and repression of tumor suppressor genes involved in ALL. Further, we demonstrate that the development of leukemia requires continued expression of IL-7 or thymic stromal lymphopoietin (TSLP) through use of a mouse model and a newly created ALL mouse cell line. Collectively, our data support a model in which multiple small defects in a network of factors that govern B cell development lead to a loss of tumor suppressor function and aberrant induction of oncogenes that cooperate with STAT5 activation to initiate progenitor B cell leukemia.
RESULTS
Increased STAT5 activation in adult human ALL patients correlates with poor outcome
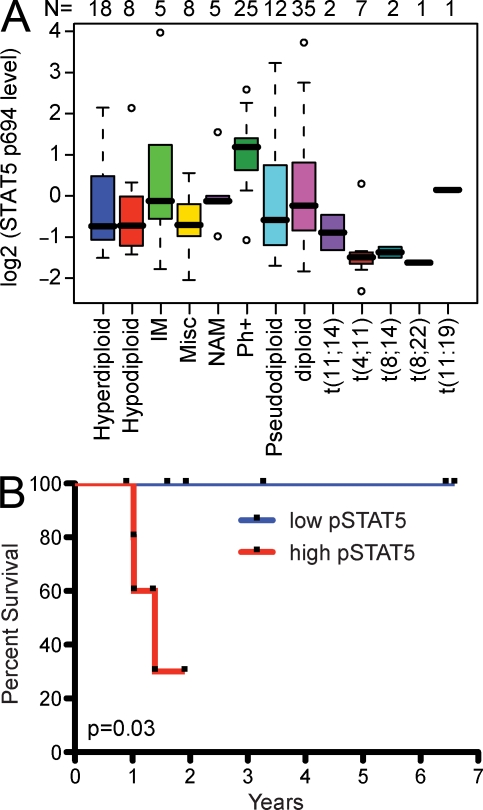
To determine whether STAT5 plays a critical role in human ALL, we examined ALL patient samples using a newly described reverse phase proteomics approach (Kornblau et al., 2009). Consistent with previous microarray studies (Yeoh et al., 2002), we found no increase in total STAT5 protein in human ALL (Table S1). In contrast, some of the ALL patient samples (n = 129; Table S1) exhibited clearly elevated levels of phosphorylated STAT5 (phospho-STAT5; Fig. 1 A). The level of phospho-STAT5 varied significantly between ALL cytogenetic subsets with the highest expression observed in BCR-ABL+ ALL (Fig. 1 A). Historically, the overall survival of BCR-ABL+ patients is quite poor, although the introduction of imatinib as a therapy has improved survival (Thomas et al., 2004). Intriguingly, we found that phospho-STAT5 status before treatment with imatinib predicted the response to subsequent imatinib therapy, with higher levels of phospho-STAT5, correlating with significantly poorer overall survival (Fig. 1 B). These results demonstrate that a significant fraction of patients with ALL exhibit elevated levels of activated STAT5 and that the degree of STAT5 activation correlates with outcome in at least one subset of ALL.
Figure 1.
Increased STAT5 activation in adult human ALL patients correlate with poor prognosis. (A) Shown are the relative levels of phospho-STAT5 (tyrosine 694/699) in cells from indicated human ALL cytogenetic subtypes (hyperdiploid, hypodiploid, immature B cell [IM], miscellaneous [Misc], no analyzable metaphases [NAM], Bcr-Abl [Ph+], pseudodiploid, diploid, t[11;14], t[4;11], t[8,14], t[8;22], and t[11;19]). Solid black bars represent the median and solid boxes represent the 25–75% range; dashed lines give ± 2 SD and open circles represent outliers. The number of samples for each subset is listed above the plot. (B) Bcr-Abl patients were separated into two equal groups representing higher (red line, n = 5) and lower (blue line, n = 6) levels of phospho-STAT5 (pSTAT5). All of these patients were subsequently treated with combination chemotherapy, including a tyrosine kinase inhibitor imatinib, and overall survival was determined. (P = 0.03, Log Rank [Mantle-Cox] test).
Ebf1 and Pax5 heterozygosity cooperate with STAT5 activation to initiate highly penetrant ALL
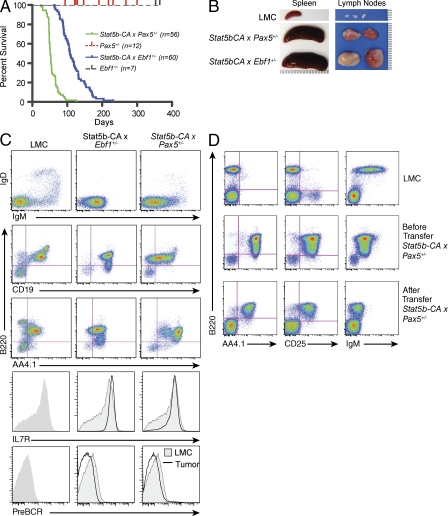
Recent analyses of ALL samples have identified a large number of mutations in transcription factors required for B cell development, including PAX5 and EBF1 (Mullighan et al., 2007). As STAT5 has been implicated in governing EBF1 and PAX5 expression and plays a possible role in ALL, we were interested in studying what affect loss of EBF1 or PAX5 would have on mice expressing an activated form of STAT5. We have previously described transgenic mice that express a constitutively active form of STAT5 (Stat5b-CA) and develop a disease that mimics human ALL, albeit at very low penetrance (∼1 to 2%; Burchill et al., 2003; Nakayama et al., 2009). To examine potential cooperativity between STAT5 activation and defects in Ebf1 or Pax5 in initiating leukemia, we generated Stat5b-CA x Ebf1+/− and Stat5b-CA x Pax5+/− mice. Loss of both alleles of Pax5 in mature B cells has been shown to result in back differentiation and development of progenitor B cell leukemias with a median age of onset of ∼9 mo (Cobaleda et al., 2007). However, Ebf1+/− and Pax5+/− mice never develop leukemia (Fig. 2 A; Urbánek et al., 1994; Lin and Grosschedl, 1995a). In contrast, loss of one allele of either Pax5 or Ebf1 in combination with constitutively active STAT5b resulted in rapid induction of leukemia with complete penetrance (median onset 55 and 109 d, respectively; Fig. 2 A). Leukemic Stat5b-CA x Ebf1+/− or Stat5b-CA x Pax5+/− displayed grossly enlarged lymph nodes and spleen (Fig. 2 B). Further, leukemic cell infiltration was found in the spleen and lymph nodes as well as in bone marrow, blood, thymus, and liver (unpublished data). To determine the phenotype, cells from spleen, lymph node, and bone marrow were removed from wild-type and leukemic mice and stained for markers expressed throughout B cell development. The leukemic cells express AA4.1, IL-7R, B220 (intermediate levels), and CD19 but do not express IgM, pre–BCR, or IgD, indicating that they are progenitor B cells (Fig. 2 C and Fig. S1 A).
Figure 2.
Ebf1 and Pax5 heterozygosity cooperates with STAT5 activation to initiate highly penetrant ALL. (A) Kaplan-Meier survival analysis of mice of the indicated genotype. (B) Pictures of spleens and lymph nodes from leukemic Stat5b-CA x Ebf1+/− and Stat5b-CA x Pax5+/− tumor mice compared with a wild-type C57BL/6 control littermate (LMC) mouse. Tick marks represent millimeter measurement. (C) Flow cytometric analysis of lymph node cells from Stat5b-CA x Ebf1+/− and Stat5b-CA x Pax5+/− tumor mice. Representative flow cytometric analysis of B220, AA4.1, IL-7R, pre–BCR, CD19, IgM, and IgD expression on lymph node cells is shown. Doublets were gated out and a lymphocyte gate was set based on side and forward scatter properties. All gates shown are based on bone marrow isolated from LMC. In histogram plots, LMC is represented as shaded plot with line and the black line referred to as tumor represents cells isolated from the lymph nodes of indicated leukemic mouse. Each mouse designated in A was analyzed. (D) Flow cytometric analysis of Stat5b-CA x Pax5+/− leukemia cells before and after transfer (2–3 wk) into wild-type C57BL/6 recipients. At least three independent experiments were completed with at least one mouse per group per experiment.
To determine if activation of STAT5b coupled with Ebf1 or Pax5 heterozygosity is sufficient to induce ALL, we tested clonality by assessing VDJ rearrangements in leukemic cells. Although the resolution of our assay does not allow us to discriminate between oligoclonal versus monoclonal leukemia, our analysis of VDJ rearrangements indicates that the leukemic cells are clearly not polyclonal (Fig. S1 B). Thus, leukemia initiates from either a single clone or a small number of clones in our model. This finding indicates that although STAT5 activation and loss of one allele of Ebf1 or Pax5 is critical for transformation, additional mutations are also necessary.
Constitutive activation of STAT5 leads to increased numbers of pro–B and pre–B cells as well as CD8+ memory-like T cells and CD4+ regulatory T cells (Burchill et al., 2003). To rule out any cell-extrinsic effect of the environment in our Stat5b-CA transgenic model, leukemic cells were transferred intravenously into recipient mice. The transferred cells induced leukemia in 100% of the recipient mice (unpublished data). These experiments were initially done by injecting a large number of leukemic cells (5 × 106 cells), both by intravenous and intraperitoneal injection, into immunocompromised mice containing no B, T, or NK cells (Rag2−/− x Il2rg−/−). As this procedure induced rapid onset of leukemia (<2 wk; unpublished data), we next transferred the leukemia cells into unmanipulated C57BL/6 syngeneic hosts. Again, 5 × 106 cells were injected, this time by intravenous injection. The mice again succumbed to disease rapidly. As the syngeneic host did not reject the leukemia cells, we continued by titrating down to 1,000 leukemic cells per mouse, again transferring the cells intravenously. In all cases, the animals succumbed to disease in <30 d. In addition, we were able to serially transfer the leukemia cells from one mouse recipient to a new mouse recipient with a similar outcome. Onset of leukemia did not seem to be dependent on cell number, suggesting that even fewer transferred cells would be sufficient to induce death from leukemia. All recipient mice were characterized by enlarged lymph nodes and spleen with leukemic cell infiltration in other tissues including liver, thymus, and bone marrow. Cells recovered from spleen, lymph nodes, and bone marrow were analyzed and compared with wild-type pro–B cells and preinjection leukemic cells. We found that the cells retained their preinjection phenotype including expression of AA4.1, B220, CD19, and IL-7R (Fig. 2 D). This suggests that genetic alterations necessary for leukemic induction are intrinsic to the transformed cell.
A block in B cell development influences, but does not recapitulate, leukemia development seen in Stat5b-CA x Ebf1+/− and Stat5b-CA x Pax5+/− mice
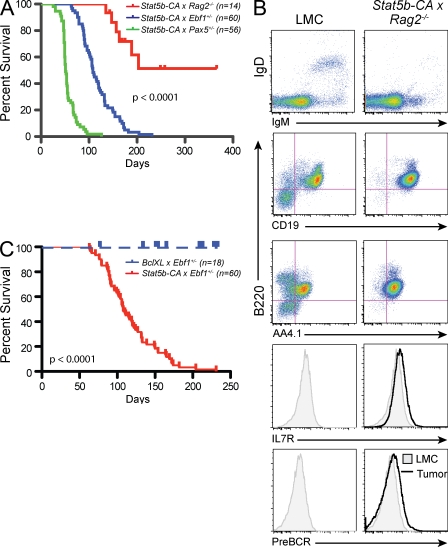
We next wanted to determine if leukemic transformation was caused by a block in early B cell differentiation. As both EBF1 and PAX5 are critical for B cell development and homozygous loss of either results in a complete disruption of B cell development, we generated Stat5b-CA x Rag2−/− mice. The Rag2 mutation blocks development beyond the late pro–B cell stage similar to Pax5 mutation, but later than the Ebf1 mutation. Approximately half of the Stat5b-CA x Rag2−/− mice came down with progenitor B cell leukemia. However, the median age of tumor onset was significantly later in these mice (203 d) as compared with either Stat5b-CA x Ebf1+/− or Stat5b-CA x Pax5+/−, and the overall frequency of leukemia was significantly reduced (P < 0.0001; Fig. 3 A). Both microarray and flow cytometric analysis indicated that most Stat5b-CA x Rag2−/− leukemias resembled Stat5b-CA x Ebf1+/− and Stat5b-CA x Pax5+/− leukemias (Fig. 3 B; Fig. S2; and not depicted). Similar results were seen when crossing Stat5b-CA mice to μMT−/− mice, which also exhibit a complete block in B cell differentiation caused by the absence of the immunoglobulin heavy chain gene (unpublished data). This finding suggests that although inhibiting B cell differentiation is important, other effects also contribute to transformation.
Figure 3.
Role of proliferation and survival in tumor induction. (A) Kaplan-Meier survival analysis of mice of the indicated genotypes. (B) Flow cytometric analysis of lymph node cells from Stat5b-CA x Rag2−/− tumor mice. Representative flow cytometric analysis of B220, AA4.1, IL-7R, pre–BCR, CD19, IgM, and IgD expression on lymph node cells is shown. In histogram plots, LMC is represented as shaded plot with line and the black line referred to as tumor represents cells isolated from the lymph nodes of indicated leukemic mouse. Doublets were gated out and a lymphocyte gate was set based on side and forward scatter properties. All gates shown are based on bone marrow isolated from control C57BL/6 mice (LMC). Each mouse indicated in A was analyzed. (C) Kaplan-Meier survival analysis of mice of the indicated genotypes.
Increased survival or proliferation does not recapitulate STAT5 effects on initiating leukemia
STAT5 is known to induce several genes involved in promoting cell proliferation and survival. Recently, Malin et al. (2010) have suggested that the role of STAT5 in pro–B cells is to provide survival signals through the regulation of the antiapoptotic protein MCL1. Specifically, they demonstrated that overexpression of the prosurvival factor BCL2 largely rescued pro–B cell development in mice in which Stat5 had been deleted throughout B and T cell development. Therefore, we examined whether enhancing cell survival is sufficient to account for the affect of STAT5 on leukemia induction by crossing Ebf1+/− mice to mice expressing a Bcl-xl transgene (Fang et al., 1998). If the role of STAT5b-CA were to promote survival, then simply replacing activated STAT5 with BCL-XL should also induce leukemia in our model. Importantly, our Bcl-xl transgenic mice have an increase in pro–B cell numbers similar to that seen in our Stat5b-CA model. Moreover, it has previously been shown that crossing these Bcl-xl mice to c-Myc transgenic mice greatly accelerates the onset of ALL (Swanson et al., 2004). In contrast, we found that enforced Bcl-xl expression in Ebf1+/− mice did not lead to leukemia (Fig. 3 C). Thus, STAT5 effects on survival may be necessary, but are clearly not sufficient to induce leukemia.
Increased proliferation could also account for the increased cellularity found in our leukemic mice. To determine if the leukemic cells were proliferating more rapidly than wild-type pro–B and pre–B cells, we injected mice displaying visible signs of leukemia with BrdU. In both Stat5b-CA x Ebf1+/− and Stat5b-CA x Pax5+/− mice we found that the leukemic cells proliferate more rapidly than mature B cells but proliferate similarly to wild-type cells at the same stage of B cell development (Fig. S3). Therefore, the development of leukemia does not appear to be caused by an effect of STAT5 on cell proliferation above and beyond that observed in wild-type progenitor B cells.
The IL-7Rα chain is required for induction and maintenance of leukemia
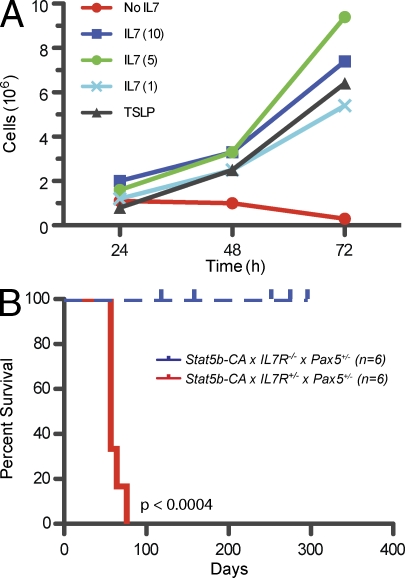
To further study the leukemic cells, we established a cell line from Stat5b-CA x Pax5+/− leukemic lymph node cells. Proliferation of this cell line in vitro was dependent on either IL-7 or TSLP (Fig. 4 A and Fig. S4); none of the other cytokines tested, including SCF and FLT3L, could sustain proliferation of this cell line (Fig. S4). Next, we titrated IL-7 and found that a relatively low dose (1 ng/ml) of IL-7 was all that was necessary for cell survival (Fig. 4 A). To address the issue of TSLP promoting leukemia survival, flow cytometric analysis of surface expression of TSLP receptor on preleukemic progenitor B cells and leukemic lymph node cells was done. In these experiments, we found TSLP receptor expression at higher levels on leukemic cells than on wild-type cells (Fig. S4 B). To determine whether this cell line could cause leukemia in vivo, we transferred 106 cells into wild-type mice and monitored the mice for leukemia. The Stat5b-CA x Pax5+/− cell line was capable of inducing leukemia in wild-type mice. Cells isolated from these new tumors maintained their preinjection phenotype and were found in the lymph nodes, spleen, and bone marrow of the recipient animals (unpublished data).
Figure 4.
Role of IL-7 in tumor induction. (A) Titration of IL-7 on Stat5b-CA x Pax5+/− cell line (SP1). Concentrations of IL-7 were 1, 5, and 10 ng/ml. Cells were counted at each time point and graphed. Graph is representative of two independent experiments. (B) Kaplan-Meier survival analysis of Stat5b-CA x IL-7R+/− x Pax5+/− and Stat5b-CA x IL-7R−/− x Pax5+/− mice.
To further confirm the importance of the IL-7R in progenitor B-ALL, we made use of IL-7R knockout mice (IL-7r−/−), which are defective in both IL-7- and TSLP-dependent signaling. We have previously demonstrated that expression of our Stat5b-CA transgene in IL-7r−/− mice can restore B cell development to near normal levels (Goetz et al., 2005). We crossed Stat5b-CA x Pax5+/− mice to IL-7r −/− mice to generate Stat5b-CA x Pax5+/− x IL-7r +/− or Stat5b-CA x Pax5+/− x IL-7r −/− mice. As seen in Fig. 4 B, mice lacking the IL-7r do not come down with ALL, whereas mice heterozygous for the IL-7r come down with leukemia similarly to Stat5b-CA x Pax5+/− mice. This suggests that signaling through the IL-7R plays a critical role in the induction of ALL.
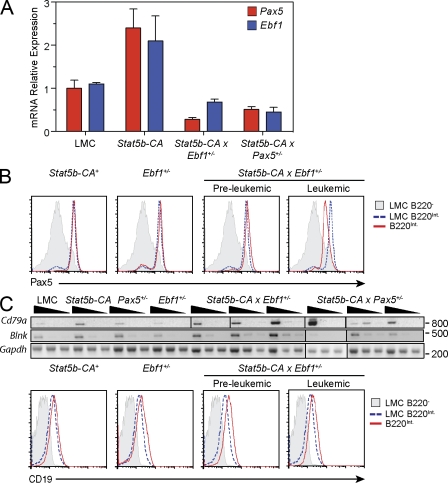
Reduced expression of Ebf1 and Pax5 in both Stat5b-CA x Ebf1+/− and Stat5b-CA x Pax5+/− leukemias
As both EBF1 and PAX5 play a critical role in B cell development, and we are using Ebf1 and Pax5 haploinsufficient mice, we set out to determine the expression levels of each of these transcription factors. We purified B220lowCD19+IgM−IgL− cells from the bone marrow of control C57BL/6 and Stat5b-CA mice and leukemic cells from the lymph nodes of Stat5b-CA x Pax5+/− and Stat5b-CA x Ebf1+/− leukemic mice. Expression was analyzed using quantitative real-time PCR (qRT-PCR). Consistent with our previous observation (Goetz et al., 2005) progenitor B cells from Stat5b-CA mice show increased expression of Pax5 relative to wild-type progenitor B cells (Goetz et al., 2005; Fig. 5 A). Furthermore, we also found increased expression of Ebf1 in Stat5b-CA progenitor B cells (Fig. 5 A). In contrast, both the Stat5b-CA x Pax5+/− and Stat5b-CA x Ebf1+/− leukemic cells expressed reduced levels of Ebf1 and Pax5 when compared with wild-type and Stat5b-CA progenitor B cells.
Figure 5.
Regulation of B cell developmental genes. (A) Ebf1 and Pax5 expression in progenitor B cells from LMC and Stat5b-CA mice and B220+, CD19+ leukemic cells from lymph nodes of Stat5b-CA x Ebf1+/− and Stat5b-CA x Pax5+/− mice were measured by real-time RT-PCR. Blue bars represent expression of Ebf1 and red bars represent expression of Pax5. Each bar is representative of at least three mice from independent experiments. Error bars represent the SEM. (B) Intracellular flow cytometric analysis of PAX5 expression in B220Int bone marrow cells from Stat5b-CA nonleukemic mice, Ebf1+/− nonleukemic mice, Stat5b-CA x Ebf1+/− preleukemic mice, and lymph node cells from Stat5b-CA x Ebf1+/− leukemic mice. LMC B220 negative cells (PAX5−) and LMC B220Int B cells (PAX5+) are controls. All gates shown are based on bone marrow isolated from control C57BL/6 mice. Doublets were gated out and a lymphocyte gate was set based on side and forward scatter properties. Flow plots are representative of three independent experiments for all but the leukemic cells (n = 2). (C) Semiquantitative PCR of CD79a, Blnk, and Gapdh. Serial dilutions were done at 1:5. This figure is representative of three independent experiments representing at least one mouse per experiment. Molecular weights are indicated (base pairs). Flow cytometric analysis of CD19 expression on bone marrow cells from Stat5b-CA nonleukemic mice, Ebf1+/− nonleukemic mice, Stat5b-CA x Ebf1+/− preleukemic mice, and lymph nodes cells from Stat5b-CA x Ebf1+/− leukemic mice. The controls and gates are as in panel B. Flow plots are representative of at least three independent experiments.
EBF1 and PAX5 are known to regulate each other’s expression. Therefore, we set out to determine whether Stat5b-CA x Ebf1+/− mice expressed reduced levels of PAX5 protein before leukemia onset. To address this issue, we made use of PAX5 intracellular staining of cells isolated from the bone marrow of wild-type, preleukemic, and leukemic mice. Using progenitor B cells from wild-type mice as a control for normal levels of PAX5 expression, we found that PAX5 was expressed at normal to slightly higher than normal levels in Stat5b-CA, Stat5b-CA x Ebf1+/− preleukemic, and Ebf1+/− mice (Fig. 5 B). As seen in the PCR experiments assessing transcript level, the Stat5b-CA x Ebf1+/− leukemic cells also expressed reduced levels of PAX5 protein. This indicates that haploinsufficiency for Ebf1 does not result in reduced levels of PAX5 in nonleukemic cells. Thus, independent mutations in Pax5 itself or factors that control its expression are required to reduce Pax5 expression in Stat5b-CA x Ebf1+/− leukemia cells.
A subset of Ebf1- and Pax5-induced genes are deregulated in Stat5b-CA x Ebf1+/− and Stat5b-CA x Pax5+/− leukemias
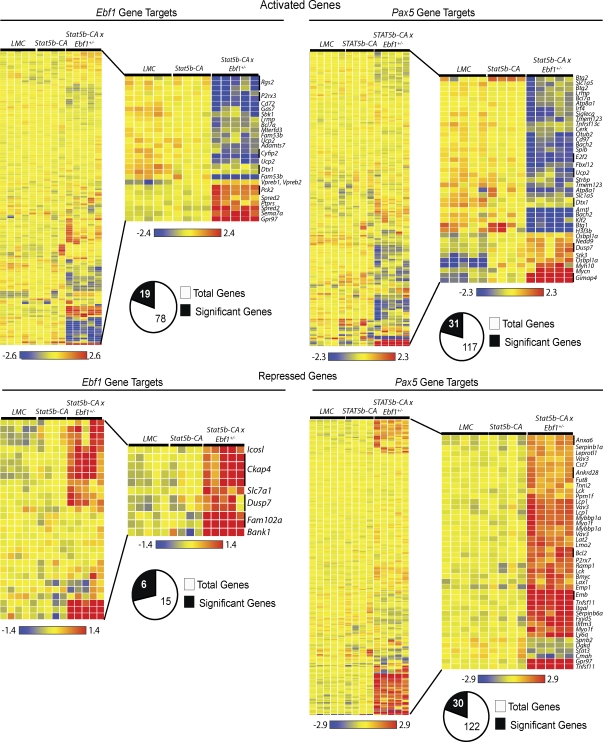
Although both Ebf1 and Pax5 levels are reduced in Stat5b-CA x Ebf1+/− and Stat5b-CA x Pax5+/− leukemias, the reduction was quite modest (approximately twofold). Therefore, we were interested in determining if there were perturbations in known gene targets of either EBF1 or PAX5. To this end, we used RT-PCR and flow cytometry to analyze leukemic cells for expression of several known up-regulated gene targets including Cd79a, Blnk, and Cd19. We found that reduced expression of Pax5 and Ebf1 did not affect expression of Cd79a, Blnk, or Cd19 (Fig. 2 C and Fig. 5 C). To further investigate changes in gene expression, we performed microarray analysis on samples from wild-type and Stat5b-CA progenitor B cells and Stat5b-CA x Ebf1+/− leukemic B220+CD19+ lymph node cells. We then compared our results with previously published datasets of EBF1- and PAX5-activated genes (Schebesta et al., 2007; Pongubala et al., 2008; Pridans et al., 2008; Treiber et al., 2010) and found that the majority of EBF1 and PAX5-induced genes (75.6 and 73.5%, respectively) were unaffected by twofold reductions in Ebf1 and Pax5 expression (Fig. 6 A and Table S2). However, ∼25% of EBF1-induced genes were down-regulated, suggesting that they were sensitive to small perturbations in Ebf1 expression (Table S3). These include several potential tumor suppressor genes including Bcl7a, Cyfip2, and Rgs2 (Zani et al., 1996; Jackson et al., 2007; Schwäble et al., 2005; van Doorn et al., 2005). Likewise, ∼25% of PAX5-induced genes were also down-regulated in Stat5b-CA x Ebf1+/− leukemias, including potential tumor suppressor genes or genes involved in suppressing cell proliferation such as Bach2, Btg1, Btg2, and Ucp2 (Table S3; Rouault et al., 1992, 1996; Sasaki et al., 2000; Lim, 2006; Baffy, 2010). Interestingly, in the case of PAX5-induced genes we could further demonstrate that the majority of these genes (∼64%) are also potential EBF1 targets, as previous studies have shown EBF1 binding to these same genes (Treiber et al., 2010). Thus, these data demonstrate that haploinsufficiency of Ebf1 in the presence of activated STAT5b results in reduced expression of a small subset of genes activated by EBF1 and PAX5.
Figure 6.
EBF1 and PAX5 Gene Targets. Microarray analysis of leukemia samples from progenitor B cells from C57BL/6 (LMC) mice (n = 5), Stat5b-CA nonleukemic mice (n = 4), and Stat5b-CA x Ebf1+/− mice (n = 5). Cluster analysis was performed using gene lists from previously published tables of PAX5 or EBF1 gene targets. The heatmap on the left represents all of the genes, regardless of significance, and the enlarged region contains the genes that were significant by ANOVA analyses (P < 0.01) and displayed a twofold difference in expression from LMC cells (n = 5). The analysis was performed as described in the Materials and methods.
Reduced expression of EBF1 and PAX5 results in derepression of genes that promote proliferation or survival
In addition to their role in activating genes required for B cell development, EBF1 and PAX5 have both been shown to play a key role in suppressing the expression of genes associated with other cell fates such as Notch1 or Tnsf11 (Delogu et al., 2006; Pridans et al., 2008; Treiber et al., 2010). These factors also appear to restrain expression of genes involved in cell survival, such as Bcl2, or cell proliferation, such as c-Myc (Nutt et al., 1998). In examining expression of EBF1- and PAX5-repressed genes, we found that the majority of these repressed genes were unaffected by reductions in EBF1 and PAX5 (60 and 75%, respectively; Fig. 6 B; Table S3). However, a clear subset of EBF1 and PAX5 repressed genes were derepressed. This list of gene targets includes several potential oncogenes such as Bcl2, Lck, Lmo2, and Vav3 (Marth et al., 1985, 1988; Voronova and Sefton, 1986; Boehm et al., 1991; Royer-Pokora et al., 1991; Movilla and Bustelo, 1999; Frenzel et al., 2009; Hirose et al., 2010; Table S3). We confirmed derepression of several target genes including Bcl2, Tnfsf11, and Itgal by RT-PCR and/or flow cytometry (Fig. S5, A and B). c-Myc was not included in this initial analysis, as the datasets we used did not include it because of ambiguity of whether c-Myc is directly repressed by PAX5 or is an indirect PAX5 target. Regardless of the exact mechanism by which PAX5 represses c-Myc expression in wild-type progenitor B cells, it is clear that c-Myc is strongly derepressed (>11-fold increase in expression) in Stat5b-CA x Ebf1+/− leukemias (Table S2). The most strongly derepressed of all genes in our analysis is Tnfsf11 (44-fold increase in expression), which encodes RANKL, a factor known to promote progenitor B cell proliferation (Kong et al., 1999). Both EBF1 and PAX5 repress expression of Tnfsf11 (Delogu et al., 2006; Pongubala et al., 2008; Pridans et al., 2008; Treiber et al., 2010). This prompted us to ask whether the list of significantly derepressed genes in our leukemias was enriched in genes regulated by both EBF1 and PAX5. Recent work by Treiber et al. (2010) have found that 30 out of 120 PAX5-repressed genes (∼25%) also exhibit binding of EBF1, suggesting that EBF1 contributes to their regulation, either via direct effects on transcription or by altering the chromatin configuration of the gene locus. When we examined the list of 30 significantly derepressed PAX5 target genes in Stat5b-CA x Ebf1+/− leukemias we found that 15 of these genes (50%) had been reported to bind to EBF1 as well (Table S3). Thus, there is a significant enrichment in the percentage of PAX5 repressed genes that are derepressed in Sta5b-CA x Ebf1+/− leukemias relative to the entire dataset of known PAX5-repressed genes (P = 0.006; χ2). Collectively, our results demonstrate that reduced expression of Ebf1 and Pax5 correlates with derepression of a significant set of EBF1- and PAX5-repressed genes including several well-characterized oncogenes such as Bcl2 and c-Myc.
DISCUSSION
Previous studies have demonstrated that a substantial percentage of patients with ALL exhibit loss-of-function mutations in a set of genes governing B cell development, including Ebf1 and Pax5 (Mullighan et al., 2007). However, whether these mutations actually play an important role in driving the development of ALL, and if so what factors they cooperate with to do so, has not been determined. Using our newly described mouse model of leukemia we demonstrate that haploinsufficiency of either Ebf1 or Pax5 plays a critical role in initiating ALL. Specifically, we demonstrate that loss of one allele of Ebf1 or Pax5, in combination with STAT5 activation, is sufficient to rapidly initiate B cell leukemia with complete penetrance. Thus, although other genetic changes likely occur in these leukemias, alterations in just the STAT5 and EBF1–PAX5 pathways are sufficient to ensure the subsequent development of leukemia.
The role of STAT5 activation in leukemia has been somewhat controversial. Initial studies by Danial et al. (1995) demonstrated that v-Abl induced JAK/STAT signaling in pre–B cell lines. Subsequent work with a rather limited number of patient samples found elevated STAT5 activation in both CML (Chai et al., 1997) and ALL (Weber-Nordt et al., 1996). However, studies with STAT5 hypomorphic mice argued that this may not be important for transformation as progenitor B cells from such animals were still susceptible to transformation by Bcr-Abl–expressing retroviruses (Sexl et al., 2000). In contrast, we previously demonstrated that mice expressing a constitutively active form of STAT5 develop ALL, albeit with low penetrance (Burchill et al., 2003; Nakayama et al., 2009). More recent data using complete STAT5-null mice has demonstrated that STAT5 is required for transformation by Bcr-Abl–expressing retroviruses (Hoelbl et al., 2006, 2010). Supporting these previous findings in mouse models of leukemia, our data examining 129 patients with ALL confirm that STAT5 activation occurs in a substantial fraction of ALL cases. The BCR-ABL+ subset of ALL exhibited the highest degree of STAT5 activation, although other ALL subsets also exhibited samples with high levels of STAT5 activation. Most importantly, we found that STAT5 activation levels before treatment with imatinib (which blocks BCR-ABL–dependent STAT5 activation) predicted subsequent outcome with high STAT5 activation correlating with poor outcome. This result suggests that in patients with lower levels of phospho-STAT5, STAT5 activation is driven entirely by BCR-ABL. However, in patients with higher levels of phospho-STAT5, other mechanisms must contribute to STAT5 activation. What these mechanisms are remain unclear. One potential mechanism we explored is whether haploinsufficiency for either Ebf1 or Pax5 resulted in increased STAT5 phosphorylation. However, we observed no difference in the level of STAT5 phosphorylation in progenitor B cells isolated directly from WT, Ebf1+/−, and Pax5+/− mice (P = 0.75, analysis of variance [ANOVA]; unpublished data). It remains possible that compound haploinsufficiency for Ebf1 and Pax5 is required to see such an effect; alternatively, other mechanisms may account for the differences in STAT5 activation observed in BCR-ABL patients. Thus, identifying these other mechanisms by which STAT5 can be activated in ALL should prove valuable for developing more effective therapies, especially for refractory forms of ALL such as BCR-ABL+ ALL or the recently described BCR-ABL–like form of ALL (Den Boer et al., 2009), both of which have a poor prognosis.
There are several ways in which STAT5 could be activated in leukemia. First, it is clear that fusion proteins such as BCR-ABL can directly phosphorylate, and hence activate, STAT5 (Carlesso et al., 1996; Ilaria and Van Etten, 1996). Alternatively, as shown in our model, direct mutation of STAT5 can promote constitutive STAT5 activation, although it remains unknown how often that actually occurs in patients with ALL. In contrast, recent studies have shown that activating mutations in the upstream Jak kinases are frequently observed in ALL, particularly refractory subsets of ALL (Mullighan et al., 2009b). Interestingly, cases of ALL with Jak mutations exhibit overexpression of the TSLPRα chain. This raises the possibility that endogenous TSLP or possibly IL-7, which both activate STAT5, may be important for transformation. Consistent with this idea, we demonstrated that Stat5b-CA x Pax5+/− mice fail to develop leukemia if they lack the IL-7Rα chain, which is required to transduce signals from both TSLP and IL-7. In addition, we found increased expression of TSLPR on leukemic cells, but not on wild-type or preleukemic control cells. Finally, both IL-7 and TSLP act as growth factors for Stat5b-CA x Pax5+/− leukemias in vitro. Whether IL-7 or TSLP are needed to increase STAT5 signaling to higher levels than that generated by our relatively weak constitutively active STAT5b transgene or to activate other IL-7R–dependent signaling pathways remains to be determined. The results from our mouse model, combined with data showing a correlation with increased TSLPR expression in some subsets of ALL, suggest that mechanisms to inhibit IL-7 and/or TSLP signaling could be of value in treating patients with high-risk forms of ALL.
EBF1 and PAX5 potentially promote transformation by blocking B cell differentiation. Our finding that Stat5b-CA x Rag2−/− and Stat5b-CA x μMT−/− mice also develop ALL supports this notion. Importantly, the microarray gene signature for Stat5b-CA x Rag2−/− leukemia closely resembles that seen in Stat5b-CA x Ebf1+/− mice (unpublished data), suggesting a common mechanism of transformation. In fact, the entire set of PAX5 and EBF1 target genes described in Fig. 6 are similarly de-regulated in Stat5b-CA x Rag2−/− mice. Consistent with this observation, our microarray analysis of Stat5b-CA x Rag2−/− leukemias also indicated approximately twofold reductions in Ebf1 and Pax5 expression (unpublished data). Interestingly, previous studies have found mutations in the Rag genes in human ALL, whereas others have suggested that the pre–BCR acts as a tumor suppressor (Mullighan et al., 2007; Trageser et al., 2009; Duy et al., 2010). Thus, it is possible that repressing pre–BCR expression contributes to the onset of leukemia. However, the reduced penetrance and delayed onset of the leukemia in Stat5b-CA x Rag2−/− mice suggests that EBF1 and PAX5 play a more fundamental role.
Our data clearly demonstrate that STAT5 activation cooperates very effectively with partial loss-of-function mutations in Ebf1 or Pax5. STAT5 appears to act in multiple ways to drive transformation. First, nontransformed progenitor B cells from Stat5b-CA mice exhibit a substantial up-regulation of Ccnd2, indicating that STAT5 can up-regulate genes involved in the cell cycle independent of effects on Ebf1 and Pax5. In contrast, several STAT5 target genes are only induced in mice with defects in Ebf1 or Pax5. For example, c-Myc and Bcl2 have both been reported to be STAT5 target genes (Lord et al., 2000). However, although c-Myc and Bcl2 levels are significantly increased in Sta5b-CA x Ebf1+/− and Stat5b-CA x Pax5+/− leukemias, and likely play an important role in transformation, neither of these genes is significantly increased in nontransformed Stat5b-CA progenitor B cells (Table S2). A potential explanation for this observation is that both Bcl2 and c-Myc are repressed by PAX5 (as well as EBF1 in the case of Bcl2; Delogu et al., 2006; Pongubala et al., 2008). We find here that reduced expression of Ebf1 and Pax5 enhances the expression Bcl2 and c-Myc, suggesting cooperativity between STAT5 activation and loss of EBF1 and PAX5 expression in turning on genes that may help promote leukemia.
Similarly, Tnfsf11, which is the most highly derepressed gene in our leukemias, is a gene that has previously been reported to be repressed by both EBF1 and Pax5 (Delogu et al., 2006; Pongubala et al., 2008). Although STAT5 has not been reported to affect Tnfsf11 expression in lymphocytes, it clearly does so in other cell types such as in the developing mammary gland (Srivastava et al., 2003). Once again, despite documented binding sites for STAT5 in the Tnfsf11 promoter (Srivastava et al., 2003), we found that STAT5 activation does not induce Tnfsf11 in developing progenitor B cells by itself. These findings suggest that cooperation between the positive effects of STAT5 activation combined with the loss of repression mediated by reduced EBF1 and PAX5 function lead to high levels of Tnfsf11 expression. Tnfsf11, which encodes RANKL, is particularly interesting in the context of transformation, as it has been previously shown to act as a growth factor for developing B cells. Thus, antagonists of RANKL may be beneficial in limiting proliferation of ALL cells.
Using microarray analysis and previously published lists of EBF1 and PAX5 gene targets, we identified modified expression of several known tumor suppressor genes and oncogenes. Interestingly, several of these genes have been identified in human B-ALL. One of these genes, BTG1 is commonly mutated in human ALL, resulting in loss of expression (Kuiper et al., 2007; Mullighan et al., 2007). In one study, loss of BTG1 expression was linked to leukemias with the TEL-AML1 translocation, a common form of pediatric ALL. This study showed that enforced expression of BTG1 could slow the growth of Reh, a TEL-AML1 cell line. This suggested that loss of expression of BTG1 enhances cell growth. (Tsuzuki et al., 2007). Further, recent data from van Galen et al. (2010) demonstrated that loss of BTG1 resulted in resistance of leukemia to glucocorticoid therapy. Another of the genes identified, LMO2, has been implicated in both B-ALL and T-ALL (Royer-Pokora et al., 1991; Cobanoglu et al., 2010; Hirose et al., 2010). By reducing expression of LMO2 in an E2A-HLF ALL cell line, researchers found increased apoptosis, suggesting that increased expression of LMO2 could protect cells from cell death (Hirose et al., 2010). Finally, loss of BACH2 expression, a known tumor suppressor gene, has been linked to BCR-ABL+ ALL (Vieira et al., 2001). In this study, loss of BACH2 expression was seen in BCR-ABL+ ALL samples, suggesting that BCR-ABL can regulate BACH2 expression. Together, these data correlating human ALL with genes regulated by EBF1 and PAX5 further illustrate the role these two proteins play in the induction of ALL and suggest that further examination of genes targeted by EBF1 and PAX5 will be useful in understanding ALL.
A key question is what accounts for the altered expression of certain EBF1 and PAX5 target genes in Stat5b-CA x Ebf1+/− and Stat5b-CA x Pax5+/− leukemias. Our data clearly demonstrate that both Ebf1 and Pax5 continue to be expressed in these leukemias. Moreover, the vast majority of EBF1 and PAX5 target genes continue to be expressed at wild-type levels, suggesting that additional mutations in the remaining Ebf1 or Pax5 allele do not account for this effect. The observation that both Ebf1 and Pax5 levels are reduced in Stat5b-CA x Ebf1+/− and Stat5b-CA x Pax5+/− leukemias suggests that the target genes that are affected may be those that require inputs from both EBF1 and PAX5 for either maximal induction or repression. Consistent with this hypothesis, over two-thirds of Pax5-induced genes were shown by chromatin immunoprecipitation assays to be potential EBF1 targets (Treiber et al., 2010); many of those genes have been independently confirmed to be regulated by EBF1 (Treiber et al., 2010). The case for this is even stronger with regard to PAX5-repressed genes. The overlap with EBF1 binding is much lower for the majority of PAX5-repressed target genes identified by previous studies, as only 25% also have validated EBF1 binding sites (Treiber et al., 2010). In contrast, in Stat5b-CA x Ebf1+/− leukemia cells, 50% of the derepressed PAX5 target genes also have validated EBF1 binding sites. Thus, there is a clear enrichment in the set of derepressed genes in our leukemias that are likely to be regulated by both EBF1 and PAX5.
It is still unclear why both Ebf1 and Pax5 levels are reduced in Stat5b-CA x Ebf1+/− and Stat5b-CA x Pax5+/− leukemias. This is an important question, as considerable effort was spent in the initial publications describing the mutations in PAX5 to determine whether both alleles were affected or whether the PAX5 mutant encoded a dominant-negative form of PAX5 that would inhibit function of the remaining wild-type allele (Mullighan et al., 2007). The conclusion from those studies was that homozygous mutations in PAX5 (or EBF1) were quite rare, although it was possible that in some cases the one mutant PAX5 allele might exhibit a dominant-negative effect. Importantly, our data demonstrate that haploinsufficiency of Pax5 is sufficient to drive ALL without invoking a dominant negative effect. Because this effect is likely caused by reduction of both Pax5 and Ebf1 expression acting on key targets (such as Bcl2 and Tnfsf11), it is important to understand what accounts for reduced expression of both Ebf1 and Pax5. One potential explanation is that in Stat5b-CA x Ebf1+/− mice, the reduction in Ebf1 expression is sufficient to reduce Pax5 expression as well. This would be consistent with the modest defect observed in B cell development in Ebf1+/− mice (O’Riordan and Grosschedl, 1999). However, when we directly monitored PAX5 expression levels in Ebf1+/− and Stat5b-CA x Ebf1+/− preleukemic mice, we did not observe reduced PAX5 levels. Moreover, this explanation fails to account for the reduced expression of Ebf1 and Pax5 in Stat5b-CA x Pax5+/− leukemias. Specifically, Pax5+/− mice have not been reported to exhibit any haploinsufficient effect (Nutt et al., 1997), and therefore cannot exhibit reduced Ebf1 expression, as that would have given rise to a clear defect in pre–B cell differentiation. Therefore, we favor a model in which an additional mutation in either one allele of Ebf1, or one allele of other genes that encode the transcription factor network that governs B cell development (including Ikzf1, Tcfe2a, Runx1, Pbx1, and Sfpi1), cooperate with Pax5 haploinsufficiency to reduce expression of Ebf1 and Pax5 factors and thereby drive ALL.
Consistent with our mouse model, previous studies have also found multiple cases of human ALL with mutations in two or more genes that make up this network (Mullighan et al., 2007, 2009a). For example, 85% of BCR-ABL leukemias exhibit mutations in one allele of IKAROS; 50% of these leukemias also show mutations in PAX5 (Mullighan et al., 2009a). Likewise, TEL-AML leukemias, which essentially have one defective copy of RUNX1 (i.e., AML) frequently exhibit mutations in PAX5 as well (Mullighan et al., 2007). Finally, ALL with E2A-PBX1 fusions essentially have one defective TCFE2A allele and one defective PBX1 allele; they also frequently exhibit mutations in PAX5 (Mullighan et al., 2007). Importantly, in this model, EBF1 and PAX5 do not act as classical tumor suppressor genes in which both alleles must be lost, but rather function as part of a tumor suppressor gene network in which mutations in any two alleles of the network is sufficient to destabilize the tumor suppressor function of the entire network. Such a model could explain the relatively high frequency of mutations in genes that make up this network (EBF1, PAX5, IKZF1, TCFE2A, etc.) as transformation would not require mutations in both alleles of any one of these genes, but rather mutations in any two alleles of any of those genes (for example, in one allele of PAX5 and one allele of EBF1). Thus, strategies aimed at enhancing the function of this gene network might be efficacious in treating B-ALL.
MATERIALS AND METHODS
Mice and cell line.
All mice have been previously described (Shinkai et al., 1992; Urbánek et al., 1994; Lin and Grosschedl, 1995a; Fang et al., 1998; Burchill et al., 2003), and the University of Minnesota IACUC approved all animal experiments. Tumors were identified by visual examination and palpation; mice were euthanized upon tumor identification. Spleen, lymph nodes, bone marrow, and peripheral blood were isolated from tumor-bearing mice and used for further experiments. Cells were either used directly upon isolation or purified using magnetic bead separation for CD19 and B220 (Miltenyi Biotech).
Kaplan-Meier survival curves were created using Prism software (V5; GraphPad Software). Survival of Stat5b-CA x Rag2−/−, Stat5b-CA x Ebf1+/−, and Stat5b-CA x Pax5+/− was compared with Stat5b-CA–negative littermate controls; all mice were monitored for at least 1 yr or until death. Deaths were indicative of tumor development.
For transfer, recipient mice (either C57BL/6 or Rag2−/− x Il2rg−/−) were injected with the indicated number of cells i.p., i.v., or via both routes of injection.
The ALL cell line (SP1) was created by culturing cells isolated from the lymph nodes of a Stat5b-CA x Pax5+/− mouse with leukemia in Opti-MEM medium (Invitrogen) supplemented with 4% FBS, 50 µM 2-mercaptoethanol, 2 mM l-glutamine, 5 I.U. penicillin, 50 µg/ml streptomycin, and 10 ng/ml IL-7 (PeproTech). Other cytokines tested were GM-CSF, SCF, FLT3L, TSLP, G-CSF, and WEHI supernatant (IL-3). All cytokines were obtained from PeproTech and used at 10 ng/ml, except for WEHI supernatant, which was made by collecting from active cultures of WEHI cells. For titration of IL-7, the concentrations used were 1, 5, and 10 ng/ml.
Flow cytometry.
Single-cell suspensions were prepared from the aforementioned tissues and stained with the following antibodies: α-IgM (Jackson ImmunoResearch Laboratories), α-IgD (11–26), α-BP-1 (FG35.4), α-CD4 (L3T4), α-CD4 (GK1.5; BioLegend), α-CD8 (53–6.7), α-CD11a (LFA-1, M17/4), α-CD19 (1D3), α-CD24 (M1/69), α-CD25 (PC61.5), α-CD43 (S7), α-CD45R (RA3-6B2), α-CD93 (AA4.1), α-CD117 (2B8), α-CD127 (A7R34), α-CD135 (A2F10), α-CD254 (RankL; IK22/5), α-pre–BCR (SL165), α-Bcl2 (3F11; BD), α-Pax5 (1H9), α-pStat5 (pY694), α-TSLPR (R&D Systems), and α-GR-1 (RB6-8C5). All antibodies were obtained from eBioscience unless otherwise indicated. SA-PerCP-Cy5.5 (eBioscience) was used to detect biotinylated antibodies. Cells were assayed on an LSRII flow cytometer (BD Biosciences); data were analyzed using FlowJo software (Tree Star). Intracellular staining for PAX5 was performed as described by eBioscience using their Foxp3 Staining kit (eBioscience). BCL2 staining was done using the BD Cytofix/Cytoperm Fixation/Permeabilization kit (BD).
BrdU assays were done as described by the manufacturer (BD). In brief, tumor-bearing mice were injected with 1 mg BrdU per mouse and allowed to rest for 24 h. Tissues were treated as described in the previous paragraph. The cells were stained according the manufacturer. Cells were assayed on a LSRII flow cytometer and data were analyzed using FlowJo software and Prism Graphing Software.
V(D)J recombination.
DNA recombination of the Igh locus was detected by PCR, as previously described (Reynaud et al., 2008). DNA was isolated from B220+CD19+ B cells from tumor lymph node tissue using a High Pure PCR Template Preparation kit (Roche). DNA from tumor cells was amplified by PCR using Ex Taq polymerase (Takara) using primers specific for VHJ558, VH7183, and VHQ52. The resulting PCR products were separated by electrophoresis on a 1.5% agarose gel. Southern blot was performed using a digoxigenin-labeled DHFL16-JH4 probe and detected using chemiluminescent anti-digoxigenin (Roche).
TaqMan and semiquantitative PCR.
Complementary DNA was synthesized from 200 ng of total RNA using Superscript III reverse transcription (Invitrogen) or qScript cDNA synthesis kit (Quanta Biosciences) and a random hexamer primer as described in the instructions provided. 1 µl of cDNA template was used in a 20 µl reaction. Primers for Ebf1, Pax5, and Hprt have been previously described (Goetz et al., 2004). Reactions using Platinum quantitative PCR SuperMix-UDG with ROX were set up according to the instructions provided by the manufacturer (Invitrogen) and performed on a 7000 Sequence Detection PCR System (Applied Biosystems). Amplification conditions were as follows: 50°C for 2 min; 95°C for 10 min; and 40 cycles of 95°C for 15 s and 60°C for 60 s. Normalized values were calculated as previously described (Livak and Schmittgen, 2001).
For semiquantitative PCR, cDNA described in the previous paragraph was used. Serial dilutions (1:10) of cDNA were used in 20 µl reaction using ExTaq (Takara). Primer sequences were taken from the literature, RankL (Totsuka et al., 2009), Blnk, Cd79a, and Gapdh (Kwon et al., 2008). Samples were run on a 1.5% agarose gel and imaged on UVP EC3 imaging system
Microarray.
Microarray analysis was performed on total RNA extracted from either sorted pre–B control cells (C57BL/6 or Stat5b-CA) or B220+CD19+ leukemic cells from lymph nodes of tumor-bearing mice using an RNEasy kit (QIAGEN). cRNA probes were synthesized and hybridized to Mouse 430 2.0 arrays following Affymetrix protocols, and statistical analyses were performed using GeneSpringGX 11.0 (Agilent). Samples were normalized using RMA, filtered on expression (20.0–100.0)th percentile. Significant gene lists were generated using one-way ANOVA with a corrected P value (P < 0.01) with a 2.0 fold change in gene expression (Table S2). Clustering was performed using hierarchical clustering both on entities and conditions, using Euclidean distance metric, and Centroid Linkage rule. Sample Sizes: C57BL/6 pre–B = 5; Stat5b-CA pre–B = 4; Stat5b-CA x Ebf1+/− = 5; and Stat5b-CA x Rag2−/− = 4. Gene array data are accessible through GEO accession no. GSE25645.
Reverse phase protein assays (phospho-STAT5).
This study used 129 samples collected from the blood and/or bone marrow of ALL patients. The sample information is described in Table S1. Samples were collected for the Leukemia Sample Bank at the University of Texas M.D. Anderson Cancer Center between 1992 and 2007. The BCR-ABL+, tyrosine kinase inhibitor–treated samples were from October 2001 through May 2007. These samples were collected on institutional review board (IRB)–approved protocol Lab01-473, and consent was obtained in accordance with the Declaration of Helsinki. Samples were analyzed under an IRB-approved laboratory protocol (Lab05-0654). These samples were analyzed using reverse phase protein arrays, as previously described (Kornblau et al., 2009).
Online supplemental material.
Fig. S1 shows that increased STAT5 activation in adult human ALL patients correlate with poor prognosis. Fig. S2 includes the rest of the phenotypic analysis of Stat5b-CA x Rag2−/− leukemic cells. Fig. S3 shows that the leukemic cells are proliferating at a rate similar to littermate control progenitor B cells. Fig. S4 shows that a leukemic cell line produced from a Stat5b-CA x Pax5+/− mouse requires either IL-7 or TSLP for growth and that leukemic cells express increased levels of TSLPR as compared with nonleukemic mice. Fig. S5 shows increased expression of RANKL, CD11a, and BCL2 on leukemic cells as compared with nonleukemic cells. Table S1 contains demographic data for human patient samples. Table S2 provides a list of significant genes (P < 0.01 and FC ≥ 2) from microarray analysis. Table S3 contains lists of genes generated from our microarray analysis and comparison with EBF1- and PAX5-regulated genes. Both activated and repressed gene targets of EBF1 and PAX5 that were significant (P < 0.01) and displayed a twofold change in expression were included. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20101947/DC1.
Acknowledgments
We are grateful to A. Vegoe, R. Agneberg, J. Bednar, C. Anderson, and P. Schoettler for technical assistance with mouse breeding; P. Champoux and N. Shah for assistance with cell sorting; the University of Minnesota’s Supercomputing Institute for providing computing and bioinformatic resources; Dr. Tim Otter and Crowley Davis Research Inc. for suggesting the initial Stat5b-CA x Ebf1+/− studies; Drs D. Reynaud and H. Singh for DHFL16-JH4 probe; Dr. Meinrad Busslinger and Dr. Rudolf Grosschedl for providing Pax5−/− and Ebf1−/− mice, respectively; and Dr. Michael Burke for critical review of the manuscript.
This work was supported by a Cancer Research Institute Investigator award, a Leukemia and Lymphoma Society Scholar award, and grants from the National Institutes of Health (NIH) to M.A. Farrar. M.J.L. Willette was supported by an NIH training grant (T32-AI07313) and L.B. Ramsey was supported by a University of Minnesota doctoral dissertation fellowship.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- ALL
- acute lymphoblastic leukemia
- ANOVA
- analysis of variance
- Ebf1
- early B cell factor
- Pax5
- paired box protein 5
- STAT5
- signal transducer and activator of transcription 5
- TSLP
- thymic stromal lymphopoietin
References
- Baffy G. 2010. Uncoupling protein-2 and cancer. Mitochondrion. 10:243–252 10.1016/j.mito.2009.12.143 [DOI] [PubMed] [Google Scholar]
- Boehm T., Foroni L., Kaneko Y., Perutz M.F., Rabbitts T.H. 1991. The rhombotin family of cysteine-rich LIM-domain oncogenes: distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13. Proc. Natl. Acad. Sci. USA. 88:4367–4371 10.1073/pnas.88.10.4367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner R., Mora L.B., Jove R. 2002. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin. Cancer Res. 8:945–954 [PubMed] [Google Scholar]
- Burchill M.A., Goetz C.A., Prlic M., O’Neil J.J., Harmon I.R., Bensinger S.J., Turka L.A., Brennan P., Jameson S.C., Farrar M.A. 2003. Distinct effects of STAT5 activation on CD4+ and CD8+ T cell homeostasis: development of CD4+CD25+ regulatory T cells versus CD8+ memory T cells. J. Immunol. 171:5853–5864 [DOI] [PubMed] [Google Scholar]
- Carlesso N., Frank D.A., Griffin J.D. 1996. Tyrosyl phosphorylation and DNA binding activity of signal transducers and activators of transcription (STAT) proteins in hematopoietic cell lines transformed by Bcr/Abl. J. Exp. Med. 183:811–820 10.1084/jem.183.3.811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai S.K., Nichols G.L., Rothman P. 1997. Constitutive activation of JAKs and STATs in BCR-Abl-expressing cell lines and peripheral blood cells derived from leukemic patients. J. Immunol. 159:4720–4728 [PubMed] [Google Scholar]
- Cobaleda C., Jochum W., Busslinger M. 2007. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 449:473–477 10.1038/nature06159 [DOI] [PubMed] [Google Scholar]
- Cobanoglu U., Sonmez M., Ozbas H.M., Erkut N., Can G. 2010. The expression of LMO2 protein in acute B-cell and myeloid leukemia. Hematology. 15:132–134 10.1179/102453309X12583347113618 [DOI] [PubMed] [Google Scholar]
- Danial N.N., Pernis A., Rothman P.B. 1995. Jak-STAT signaling induced by the v-abl oncogene. Science. 269:1875–1877 10.1126/science.7569929 [DOI] [PubMed] [Google Scholar]
- Delogu A., Schebesta A., Sun Q., Aschenbrenner K., Perlot T., Busslinger M. 2006. Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity. 24:269–281 10.1016/j.immuni.2006.01.012 [DOI] [PubMed] [Google Scholar]
- Den Boer M.L., van Slegtenhorst M., De Menezes R.X., Cheok M.H., Buijs-Gladdines J.G., Peters S.T., Van Zutven L.J., Beverloo H.B., Van der Spek P.J., Escherich G., et al. 2009. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 10:125–134 10.1016/S1470-2045(08)70339-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duy C., Yu J.J., Nahar R., Swaminathan S., Kweon S.M., Polo J.M., Valls E., Klemm L., Shojaee S., Cerchietti L., et al. 2010. BCL6 is critical for the development of a diverse primary B cell repertoire. J. Exp. Med. 207:1209–1221 10.1084/jem.20091299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W., Weintraub B.C., Dunlap B., Garside P., Pape K.A., Jenkins M.K., Goodnow C.C., Mueller D.L., Behrens T.W. 1998. Self-reactive B lymphocytes overexpressing Bcl-xL escape negative selection and are tolerized by clonal anergy and receptor editing. Immunity. 9:35–45 10.1016/S1074-7613(00)80586-5 [DOI] [PubMed] [Google Scholar]
- Fielding A.K. 2010. How I treat Philadelphia chromosome positive acute lymphoblastic leukaemia. Blood. 116:3409–3417 10.1182/blood-2010-01-242750 [DOI] [PubMed] [Google Scholar]
- Frenzel A., Grespi F., Chmelewskij W., Villunger A. 2009. Bcl2 family proteins in carcinogenesis and the treatment of cancer. Apoptosis. 14:584–596 10.1007/s10495-008-0300-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz C.A., Harmon I.R., O’Neil J.J., Burchill M.A., Farrar M.A. 2004. STAT5 activation underlies IL7 receptor-dependent B cell development. J. Immunol. 172:4770–4778 [DOI] [PubMed] [Google Scholar]
- Goetz C.A., Harmon I.R., O’Neil J.J., Burchill M.A., Johanns T.M., Farrar M.A. 2005. Restricted STAT5 activation dictates appropriate thymic B versus T cell lineage commitment. J. Immunol. 174:7753–7763 [DOI] [PubMed] [Google Scholar]
- Hagman J., Lukin K. 2006. Transcription factors drive B cell development. Curr. Opin. Immunol. 18:127–134 10.1016/j.coi.2006.01.007 [DOI] [PubMed] [Google Scholar]
- Hirose K., Inukai T., Kikuchi J., Furukawa Y., Ikawa T., Kawamoto H., Oram S.H., Göttgens B., Kiyokawa N., Miyagawa Y., et al. 2010. Aberrant induction of LMO2 by the E2A-HLF chimeric transcription factor and its implication in leukemogenesis of B-precursor ALL with t(17;19). Blood. 116:962–970 10.1182/blood-2009-09-244673 [DOI] [PubMed] [Google Scholar]
- Hoelbl A., Kovacic B., Kerenyi M.A., Simma O., Warsch W., Cui Y., Beug H., Hennighausen L., Moriggl R., Sexl V. 2006. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood. 107:4898–4906 10.1182/blood-2005-09-3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelbl A., Schuster C., Kovacic B., Zhu B., Wickre M., Hoelzl M.A., Fajmann S., Grebien F., Warsch W., Stengl G., et al. 2010. Stat5 is indispensable for the maintenance of bcr/abl-positive leukaemia. EMBO Mol Med. 2:98–110 10.1002/emmm.201000062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilaria R.L., Jr, Van Etten R.A. 1996. P210 and P190(BCR/ABL) induce the tyrosine phosphorylation and DNA binding activity of multiple specific STAT family members. J. Biol. Chem. 271:31704–31710 10.1074/jbc.271.49.31704 [DOI] [PubMed] [Google Scholar]
- Jackson R.S., II, Cho Y.J., Stein S., Liang P. 2007. CYFIP2, a direct p53 target, is leptomycin-B sensitive. Cell Cycle. 6:95–103 10.4161/cc.6.1.3665 [DOI] [PubMed] [Google Scholar]
- Kong Y.Y., Yoshida H., Sarosi I., Tan H.L., Timms E., Capparelli C., Morony S., Oliveira-dos-Santos A.J., Van G., Itie A., et al. 1999. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 397:315–323 10.1038/16852 [DOI] [PubMed] [Google Scholar]
- Kornblau S.M., Tibes R., Qiu Y.H., Chen W., Kantarjian H.M., Andreeff M., Coombes K.R., Mills G.B. 2009. Functional proteomic profiling of AML predicts response and survival. Blood. 113:154–164 10.1182/blood-2007-10-119438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper R.P., Schoenmakers E.F., van Reijmersdal S.V., Hehir-Kwa J.Y., van Kessel A.G., van Leeuwen F.N., Hoogerbrugge P.M. 2007. High-resolution genomic profiling of childhood ALL reveals novel recurrent genetic lesions affecting pathways involved in lymphocyte differentiation and cell cycle progression. Leukemia. 21:1258–1266 10.1038/sj.leu.2404691 [DOI] [PubMed] [Google Scholar]
- Kwon K., Hutter C., Sun Q., Bilic I., Cobaleda C., Malin S., Busslinger M. 2008. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 28:751–762 10.1016/j.immuni.2008.04.014 [DOI] [PubMed] [Google Scholar]
- Lim I.K. 2006. TIS21 (/BTG2/PC3) as a link between ageing and cancer: cell cycle regulator and endogenous cell death molecule. J. Cancer Res. Clin. Oncol. 132:417–426 10.1007/s00432-006-0080-1 [DOI] [PubMed] [Google Scholar]
- Lin H., Grosschedl R. 1995a. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 376:263–267 10.1038/376263a0 [DOI] [PubMed] [Google Scholar]
- Lin H., Grosschedl R. 1995b. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 376:263–267 10.1038/376263a0 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 25:402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lord J.D., McIntosh B.C., Greenberg P.D., Nelson B.H. 2000. The IL-2 receptor promotes lymphocyte proliferation and induction of the c-myc, bcl-2, and bcl-x genes through the trans-activation domain of Stat5. J. Immunol. 164:2533–2541 [DOI] [PubMed] [Google Scholar]
- Malin S., McManus S., Cobaleda C., Novatchkova M., Delogu A., Bouillet P., Strasser A., Busslinger M. 2010. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat. Immunol. 11:171–179 10.1038/ni.1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Månsson R., Tsapogas P., Akerlund M., Lagergren A., Gisler R., Sigvardsson M. 2004. Pearson correlation analysis of microarray data allows for the identification of genetic targets for early B-cell factor. J. Biol. Chem. 279:17905–17913 10.1074/jbc.M400589200 [DOI] [PubMed] [Google Scholar]
- Marth J.D., Peet R., Krebs E.G., Perlmutter R.M. 1985. A lymphocyte-specific protein-tyrosine kinase gene is rearranged and overexpressed in the murine T cell lymphoma LSTRA. Cell. 43:393–404 10.1016/0092-8674(85)90169-2 [DOI] [PubMed] [Google Scholar]
- Marth J.D., Overell R.W., Meier K.E., Krebs E.G., Perlmutter R.M. 1988. Translational activation of the lck proto-oncogene. Nature. 332:171–173 10.1038/332171a0 [DOI] [PubMed] [Google Scholar]
- Milone J.H., Enrico A. 2009. Treatment of Philadelphia chromosome-positive acute lymphoblastic leukemia. Leuk. Lymphoma. 50:9–15 10.3109/10428190903370395 [DOI] [PubMed] [Google Scholar]
- Movilla N., Bustelo X.R. 1999. Biological and regulatory properties of Vav-3, a new member of the Vav family of oncoproteins. Mol. Cell. Biol. 19:7870–7885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan C.G., Goorha S., Radtke I., Miller C.B., Coustan-Smith E., Dalton J.D., Girtman K., Mathew S., Ma J., Pounds S.B., et al. 2007. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 446:758–764 10.1038/nature05690 [DOI] [PubMed] [Google Scholar]
- Mullighan C.G., Su X., Zhang J., Radtke I., Phillips L.A., Miller C.B., Ma J., Liu W., Cheng C., Schulman B.A., et al. ; Children’s Oncology Group 2009a. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N. Engl. J. Med. 360:470–480 10.1056/NEJMoa0808253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan C.G., Zhang J., Harvey R.C., Collins-Underwood J.R., Schulman B.A., Phillips L.A., Tasian S.K., Loh M.L., Su X., Liu W., et al. 2009b. JAK mutations in high-risk childhood acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA. 106:9414–9418 10.1073/pnas.0811761106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J., Yamamoto M., Hayashi K., Satoh H., Bundo K., Kubo M., Goitsuka R., Farrar M.A., Kitamura D. 2009. BLNK suppresses pre-B-cell leukemogenesis through inhibition of JAK3. Blood. 113:1483–1492 10.1182/blood-2008-07-166355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt S.L., Kee B.L. 2007. The transcriptional regulation of B cell lineage commitment. Immunity. 26:715–725 10.1016/j.immuni.2007.05.010 [DOI] [PubMed] [Google Scholar]
- Nutt S.L., Urbánek P., Rolink A., Busslinger M. 1997. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 11:476–491 10.1101/gad.11.4.476 [DOI] [PubMed] [Google Scholar]
- Nutt S.L., Morrison A.M., Dörfler P., Rolink A., Busslinger M. 1998. Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO J. 17:2319–2333 10.1093/emboj/17.8.2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt S.L., Heavey B., Rolink A.G., Busslinger M. 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 401:556–562 10.1038/44076 [DOI] [PubMed] [Google Scholar]
- O’Riordan M., Grosschedl R. 1999. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 11:21–31 10.1016/S1074-7613(00)80078-3 [DOI] [PubMed] [Google Scholar]
- Pongubala J.M., Northrup D.L., Lancki D.W., Medina K.L., Treiber T., Bertolino E., Thomas M., Grosschedl R., Allman D., Singh H. 2008. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat. Immunol. 9:203–215 10.1038/ni1555 [DOI] [PubMed] [Google Scholar]
- Pridans C., Holmes M.L., Polli M., Wettenhall J.M., Dakic A., Corcoran L.M., Smyth G.K., Nutt S.L. 2008. Identification of Pax5 target genes in early B cell differentiation. J. Immunol. 180:1719–1728 [DOI] [PubMed] [Google Scholar]
- Pui C.H., Robison L.L., Look A.T. 2008. Acute lymphoblastic leukaemia. Lancet. 371:1030–1043 10.1016/S0140-6736(08)60457-2 [DOI] [PubMed] [Google Scholar]
- Reynaud D., Demarco I.A., Reddy K.L., Schjerven H., Bertolino E., Chen Z., Smale S.T., Winandy S., Singh H. 2008. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat. Immunol. 9:927–936 10.1038/ni.1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler S., Györy I., Imhof S., Spivakov M., Williams R.R., Busslinger M., Fisher A.G., Grosschedl R. 2007. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol. Cell. Biol. 27:579–594 10.1128/MCB.01192-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault J.P., Rimokh R., Tessa C., Paranhos G., Ffrench M., Duret L., Garoccio M., Germain D., Samarut J., Magaud J.P. 1992. BTG1, a member of a new family of antiproliferative genes. EMBO J. 11:1663–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault J.P., Falette N., Guéhenneux F., Guillot C., Rimokh R., Wang Q., Berthet C., Moyret-Lalle C., Savatier P., Pain B., et al. 1996. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat. Genet. 14:482–486 10.1038/ng1296-482 [DOI] [PubMed] [Google Scholar]
- Royer-Pokora B., Loos U., Ludwig W.D. 1991. TTG-2, a new gene encoding a cysteine-rich protein with the LIM motif, is overexpressed in acute T-cell leukaemia with the t(11;14)(p13;q11). Oncogene. 6:1887–1893 [PubMed] [Google Scholar]
- Sasaki S., Ito E., Toki T., Maekawa T., Kanezaki R., Umenai T., Muto A., Nagai H., Kinoshita T., Yamamoto M., et al. 2000. Cloning and expression of human B cell-specific transcription factor BACH2 mapped to chromosome 6q15. Oncogene. 19:3739–3749 10.1038/sj.onc.1203716 [DOI] [PubMed] [Google Scholar]
- Schebesta A., McManus S., Salvagiotto G., Delogu A., Busslinger G.A., Busslinger M. 2007. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity. 27:49–63 10.1016/j.immuni.2007.05.019 [DOI] [PubMed] [Google Scholar]
- Schwäble J., Choudhary C., Thiede C., Tickenbrock L., Sargin B., Steur C., Rehage M., Rudat A., Brandts C., Berdel W.E., et al. 2005. RGS2 is an important target gene of Flt3-ITD mutations in AML and functions in myeloid differentiation and leukemic transformation. Blood. 105:2107–2114 10.1182/blood-2004-03-0940 [DOI] [PubMed] [Google Scholar]
- Schwaller J., Parganas E., Wang D., Cain D., Aster J.C., Williams I.R., Lee C.K., Gerthner R., Kitamura T., Frantsve J., et al. 2000. Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol. Cell. 6:693–704 10.1016/S1097-2765(00)00067-8 [DOI] [PubMed] [Google Scholar]
- Sexl V., Piekorz R., Moriggl R., Rohrer J., Brown M.P., Bunting K.D., Rothammer K., Roussel M.F., Ihle J.N. 2000. Stat5a/b contribute to interleukin 7-induced B-cell precursor expansion, but abl- and bcr/abl-induced transformation are independent of stat5. Blood. 96:2277–2283 [PubMed] [Google Scholar]
- Shinkai Y., Rathbun G., Lam K.-P., Oltz E.M., Stewart V., Mendelsohn M., Charron J., Datta M., Young F., Stall A.M., Alt F.W. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 68:855–867 10.1016/0092-8674(92)90029-C [DOI] [PubMed] [Google Scholar]
- Srivastava S., Matsuda M., Hou Z., Bailey J.P., Kitazawa R., Herbst M.P., Horseman N.D. 2003. Receptor activator of NF-kappaB ligand induction via Jak2 and Stat5a in mammary epithelial cells. J. Biol. Chem. 278:46171–46178 10.1074/jbc.M308545200 [DOI] [PubMed] [Google Scholar]
- Swanson P.J., Kuslak S.L., Fang W., Tze L., Gaffney P., Selby S., Hippen K.L., Nunez G., Sidman C.L., Behrens T.W. 2004. Fatal acute lymphoblastic leukemia in mice transgenic for B cell-restricted bcl-xL and c-myc. J. Immunol. 172:6684–6691 [DOI] [PubMed] [Google Scholar]
- Thomas D.A., Faderl S., Cortes J., O’Brien S., Giles F.J., Kornblau S.M., Garcia-Manero G., Keating M.J., Andreeff M., Jeha S., et al. 2004. Treatment of Philadelphia chromosome-positive acute lymphocytic leukemia with hyper-CVAD and imatinib mesylate. Blood. 103:4396–4407 10.1182/blood-2003-08-2958 [DOI] [PubMed] [Google Scholar]
- Totsuka T., Kanai T., Nemoto Y., Tomita T., Okamoto R., Tsuchiya K., Nakamura T., Sakamoto N., Akiba H., Okumura K., et al. 2009. RANK-RANKL signaling pathway is critically involved in the function of CD4+CD25+ regulatory T cells in chronic colitis. J. Immunol. 182:6079–6087 10.4049/jimmunol.0711823 [DOI] [PubMed] [Google Scholar]
- Trageser D., Iacobucci I., Nahar R., Duy C., von Levetzow G., Klemm L., Park E., Schuh W., Gruber T., Herzog S., et al. 2009. Pre–B cell receptor–mediated cell cycle arrest in Philadelphia chromosome-positive acute lymphoblastic leukemia requires IKAROS function. J. Exp. Med. 206:1739–1753 10.1084/jem.20090004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber T., Mandel E.M., Pott S., Gyory I., Firner S., Liu E.T., Grosschedl R. 2010. Early B cell factor 1 regulates B Cell gene networks by activation, repression, and transcription-independent poising of chromatin. Immunity. 32:714–725 10.1016/j.immuni.2010.04.013 [DOI] [PubMed] [Google Scholar]
- Tsuzuki S., Karnan S., Horibe K., Matsumoto K., Kato K., Inukai T., Goi K., Sugita K., Nakazawa S., Kasugai Y., et al. 2007. Genetic abnormalities involved in t(12;21) TEL-AML1 acute lymphoblastic leukemia: analysis by means of array-based comparative genomic hybridization. Cancer Sci. 98:698–706 10.1111/j.1349-7006.2007.00443.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbánek P., Wang Z.Q., Fetka I., Wagner E.F., Busslinger M. 1994. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 79:901–912 10.1016/0092-8674(94)90079-5 [DOI] [PubMed] [Google Scholar]
- van Doorn R., Zoutman W.H., Dijkman R., de Menezes R.X., Commandeur S., Mulder A.A., van der Velden P.A., Vermeer M.H., Willemze R., Yan P.S., et al. 2005. Epigenetic profiling of cutaneous T-cell lymphoma: promoter hypermethylation of multiple tumor suppressor genes including BCL7a, PTPRG, and p73. J. Clin. Oncol. 23:3886–3896 10.1200/JCO.2005.11.353 [DOI] [PubMed] [Google Scholar]
- van Galen J.C., Kuiper R.P., van Emst L., Levers M., Tijchon E., Scheijen B., Waanders E., van Reijmersdal S.V., Gilissen C., van Kessel A.G., et al. 2010. BTG1 regulates glucocorticoid receptor autoinduction in acute lymphoblastic leukemia. Blood. 115:4810–4819 10.1182/blood-2009-05-223081 [DOI] [PubMed] [Google Scholar]
- Vieira S.A., Deininger M.W., Sorour A., Sinclair P., Foroni L., Goldman J.M., Melo J.V. 2001. Transcription factor BACH2 is transcriptionally regulated by the BCR/ABL oncogene. Genes Chromosomes Cancer. 32:353–363 10.1002/gcc.1200 [DOI] [PubMed] [Google Scholar]
- Voronova A.F., Sefton B.M. 1986. Expression of a new tyrosine protein kinase is stimulated by retrovirus promoter insertion. Nature. 319:682–685 10.1038/319682a0 [DOI] [PubMed] [Google Scholar]
- Weber-Nordt R.M., Egen C., Wehinger J., Ludwig W., Gouilleux-Gruart V., Mertelsmann R., Finke J. 1996. Constitutive activation of STAT proteins in primary lymphoid and myeloid leukemia cells and in Epstein-Barr virus (EBV)-related lymphoma cell lines. Blood. 88:809–816 [PubMed] [Google Scholar]
- Xie S., Wang Y., Liu J., Sun T., Wilson M.B., Smithgall T.E., Arlinghaus R.B. 2001. Involvement of Jak2 tyrosine phosphorylation in Bcr-Abl transformation. Oncogene. 20:6188–6195 10.1038/sj.onc.1204834 [DOI] [PubMed] [Google Scholar]
- Yao Z., Cui Y., Watford W.T., Bream J.H., Yamaoka K., Hissong B.D., Li D., Durum S.K., Jiang Q., Bhandoola A., et al. 2006. Stat5a/b are essential for normal lymphoid development and differentiation. Proc. Natl. Acad. Sci. USA. 103:1000–1005 10.1073/pnas.0507350103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh E.J., Ross M.E., Shurtleff S.A., Williams W.K., Patel D., Mahfouz R., Behm F.G., Raimondi S.C., Relling M.V., Patel A., et al. 2002. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 1:133–143 10.1016/S1535-6108(02)00032-6 [DOI] [PubMed] [Google Scholar]
- Zani V.J., Asou N., Jadayel D., Heward J.M., Shipley J., Nacheva E., Takasuki K., Catovsky D., Dyer M.J. 1996. Molecular cloning of complex chromosomal translocation t(8;14;12)(q24.1;q32.3;q24.1) in a Burkitt lymphoma cell line defines a new gene (BCL7A) with homology to caldesmon. Blood. 87:3124–3134 [PubMed] [Google Scholar]