Abstract
Background
Streptococcus gallolyticus subsp. gallolyticus is an important causative agent of infectious endocarditis, while the pathogenicity of this species is widely unclear. To gain insight into the pathomechanisms and the underlying genetic elements for lateral gene transfer, we sequenced the entire genome of this pathogen.
Results
We sequenced the whole genome of S. gallolyticus subsp. gallolyticus strain ATCC BAA-2069, consisting of a 2,356,444 bp circular DNA molecule with a G+C-content of 37.65% and a novel 20,765 bp plasmid designated as pSGG1. Bioinformatic analysis predicted 2,309 ORFs and the presence of 80 tRNAs and 21 rRNAs in the chromosome. Furthermore, 21 ORFs were detected on the plasmid pSGG1, including tetracycline resistance genes telL and tet(O/W/32/O). Screening of 41 S. gallolyticus subsp. gallolyticus isolates revealed one plasmid (pSGG2) homologous to pSGG1. We further predicted 21 surface proteins containing the cell wall-sorting motif LPxTG, which were shown to play a functional role in the adhesion of bacteria to host cells. In addition, we performed a whole genome comparison to the recently sequenced S. gallolyticus subsp. gallolyticus strain UCN34, revealing significant differences.
Conclusions
The analysis of the whole genome sequence of S. gallolyticus subsp. gallolyticus promotes understanding of genetic factors concerning the pathogenesis and adhesion to ECM of this pathogen. For the first time we detected the presence of the mobilizable pSGG1 plasmid, which may play a functional role in lateral gene transfer and promote a selective advantage due to a tetracycline resistance.
Background
Streptococcus gallolyticus subsp. gallolyticus (formerly known as S. bovis biotype I) is a gram-positive bacterium belonging to the Lancefield Group D streptococci. Over the last ten years, the classification of S. gallolyticus subsp. gallolyticus has been revised several times [1-4]. S. bovis was previously divided into three biotypes, designated as biotype I, biotype II/1, and biotype II/2. The majority of isolates associated with human endocarditis have been assigned to biotype I, which was recently reclassified as Streptococcus gallolyticus subsp. gallolyticus [5]. Furthermore, S. gallolyticus subsp. gallolyticus is a common member of the microflora and appears in approximately 2.5 to 15% of the gastrointestinal tract of healthy human [6,7]. It is an opportunistic human pathogen which can cause several bacterial infections, including septicemia and endocarditis. Over the last few years, the percentage of cases of endocarditis caused by group D streptococci has significantly increased [8-10]. Recently, Russel et al. estimated that S. gallolyticus subsp. gallolyticus is the causative agent in 24% of streptococcal endocarditis cases [11]. In addition, several studies present strong correlations between appearance of colon neoplasms and S. gallolyticus subsp. gallolyticus infection [7,12], while the underlying pathomechanisms are still unknown. Sillanpää et al. suggest that premalignant and malignant lesions in the intestinal tract could facilitate translocation of S. gallolyticus subsp. gallolyticus through the disrupted mucosal barrier and provide access to blood circulation [13]. Furthermore, studies have suggested a linkage between inflammation by S. bovis and colon carcinogenesis [14]. In addition, a variety of animal infections, such as mastitis, septicemia in poultry, lactic acidosis and infections of various ruminant animals are caused by S. gallolyticus subsp. gallolyticus [15-17]. However, the exact pathomechanisms of S. gallolyticus subsp. gallolyticus or S. bovis infections remain unclear.
S. gallolyticus subsp. gallolyticus shares its environment with numerous other potentially pathogenic bacteria, such as S. agalactiae, Enterococcus faecalis or others. This implies the possibility of horizontal gene transfer of antimicrobial resistance genes or genomic islands, e.g. phage-related clusters, by transposons, plasmids or phages, within the human gut or the animal rumen [18]. Several studies have reported the occurrence of competence-stimulating peptides in S. bovis [19]. These factors facilitate the acquisition of novel genes, resistance islands or virulence-associated regions [20], in particular when several species coexist within biofilms [21]. Recently we were able to show the capability of biofilm formation on polystyrene surfaces for S. gallolyticus subsp. gallolyticus [22]. Nevertheless, most of the mechanisms of transfer and insertion are poorly understood [23,24].
In vitro studies have demonstrated the adhesion and invasion of S. gallolyticus subsp. gallolyticus to extracellular matrix proteins [22,25], virulence associated proteins [13,26,27], as well as EA.hy926 or HUVEC cells [22]. Furthermore, studies have addressed biosynthesis of capsular polysaccharides [28] and fimbriae-like structures on the bacterial surface in S. gallolyticus subsp. gallolyticus [29]. It has been demonstrated that S. gallolyticus subsp. gallolyticus has 11 cell wall-anchored proteins with "microbial surface component recognizing matrix molecules" (MSCRAMM) characteristics, including a collagen-binding adhesin and proteins with similarities to pilus subunits [13].
Recently, Rusinok et al. published the first whole genome sequence of S. gallolyticus subsp. gallolyticus strain UCN34 and analyzed the main metabolic and cell surface features, particularly with regard to adaptation to the rumen and the virulence association of polysaccharide capsule, glucan mucopolysaccharides, different types of pili and collagen binding proteins [30].
Here we present the whole genome sequence of a not described, considerably divergent S. gallolyticus subsp. gallolyticus strain. The strain under study was the tetracycline resistant strain ATCC BAA-2069, isolated from a patient with infectious endocarditis. We demonstrate the occurrence of a previously undescribed plasmid (pSGG1) which carries genes for tetracycline resistance (tetL, tet(O/W/32/O)) and reveals strong sequence similarities to plasmids and chromosomes from several ruminal and gastrointestinal bacteria, indicating that pSGG1 may act as a native carrier for horizontal gene transfer.
Results
General genome properties
The whole genome sequence of S. gallolyticus subsp. gallolyticus was determined by pyrosequencing using the 454 GS FLX Titanium technique (Roche, Mannheim, Germany) and, after assembly of the 454 reads, remaining gaps were closed by PCR and conventional Sanger sequencing. The genome contains a 2,356,444 bp circular DNA molecule with a G+C-content of 37.65% and a previously undescribed 20,765 bp plasmid designated as pSGG1. Mapping of gene set was performed against S. gallolyticus subsp. gallolyticus genome UCN34 (GenBank Acc. No.: FN597254) [30]. Bioinformatic analysis predicted 2,309 open reading frames (ORFs), the presence of 80 tRNAs and 21 rRNAs in the chromosome, as well as 21 ORFs on the plasmid pSGG1.
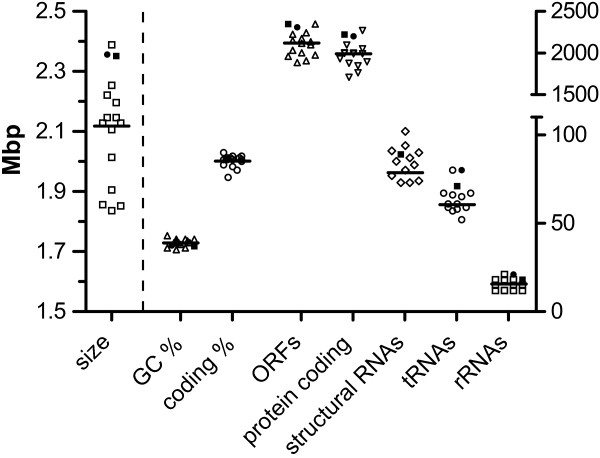
The size of the BAA-2069 circular chromosome (2,356,444 bp) exceeds the average of other previously published streptococcal genomes by 12% (mean: 2.1 mb; n = 15) (Table 1, Figure 1). Direct comparison shows that only the S. sanguinis SK36 genome is larger (2,388,435 bp), and the G+C-content is 1.7% lower than average (range from 35.3 to 43.4%; n = 15). Altogether 2,309 ORFs were automatically annotated, which is 10% higher than the average of all complete sequenced Streptococcus genomes (2,107 ORFs). In direct comparison to the S. gallolyticus subsp. gallolyticus genome UCN34, the BAA-2069 genome is 5.5 kb larger (2,356,444 to 2,350,911 bp), has 70 fewer CDS (2,309 to 2,239) and contains the 20,765 bp plasmid pSGG1.
Table 1.
Comparison of Streptococcus/Enterococcus species with S. gallolyticus subsp. gallolyticus BAA-2069
| Strain | GenBank Acc No: | GC % | Coding % | Size bp | ORFs | tRNAs | rRNAs |
|---|---|---|---|---|---|---|---|
| S. gallolyticus subsp. gallolyticus BAA-2069 | FR824043 | 38 | 87 | 2,356,444 | 2,309 | 80 | 21 |
| S. gallolyticus subsp. gallolyticus UCN34 | FN597254 | 37 | 86 | 2,350,911 | 2,349 | 71 | 18 |
| S. agalactiae A909 | CP000114 | 35 | 86 | 2,127,839 | 2,136 | 80 | 21 |
| S. dysgalactiae subsp. equisimilis GGS_124 | AP010935 | 39 | 86 | 2,106,340 | 2,174 | 57 | 15 |
| S. equi subsp. equi 4047 | FM204883 | 41 | 80 | 2,253,793 | 2,243 | 66 | 18 |
| S. sanguinis SK36 | CP000387 | 43 | 88 | 2,388,435 | 2,348 | 61 | 12 |
| S. suis BM407 | FM252032 | 41 | 83 | 2,146,229 | 2,118 | 52 | 12 |
| S. uberis 0140J | AM946015 | 36 | 87 | 1,852,352 | 1,908 | 59 | 15 |
| S. pyogenes MGAS9429 | CP000259 | 38 | 87 | 1,836,467 | 1,962 | 67 | 18 |
| S. pneumoniae ATCC 700669 | FM211187 | 39 | 82 | 2,221,315 | 2,224 | 58 | 12 |
| S. mutans NN2025 | AP010655 | 36 | 85 | 2,013,587 | 1,976 | 65 | 15 |
| S. mitis B6 | FN568063 | 39 | 86 | 2,146,611 | 2,098 | 61 | 12 |
| S. thermophilus LMD-9 | CP000419 | 39 | 76 | 1,856,368 | 2,002 | 67 | 18 |
| S. gordonii str. challis substr. CH1 | CP000725 | 40 | 87 | 2,196,662 | 2,150 | 59 | 12 |
| S. oralis ATCC 35037 | AEDW00000000 | 41 | 90 | 1,905,531 | 1,886 | n.d. | n.d. |
| S. salivarius SK126 | ACLO00000000 | 40 | 88 | 2,128,332 | 2,034 | n.d. | n.d. |
| Enterococcus faecalis V583 | AE016830 | 37 | 85 | 3,359,974 | 3,417 | 68 | 12 |
Figure 1.
Distribution of whole genome characteristics. Black dot represents S. gallolyticus subsp. gallolyticus strain Isolate BAA-2069. Black square represents S. gallolyticus subsp. gallolyticus strain UCN-34. Symbols represent genomes of S. agalactiae A909, S. dysgalactiae subsp. equisimilis GGS_124, S. equi subsp. equi 4047, S. sanguinis SK36, S. suis BM407, S. uberis 0140J, S. pyogenes MGAS9429, S. pneumoniae ATCC 700669, S. mutans NN2025, S. mitis B6, S. thermophilus LMD-9, S. gordonii str. challis substr. CH1, S. oralis ATCC 35037, S. salivarius SK126.
The sequences and annotations of chromosome and plasmid have been deposited at the NCBI GenBank (Acc. No. FR824043, FR824044).
Comparative genomics
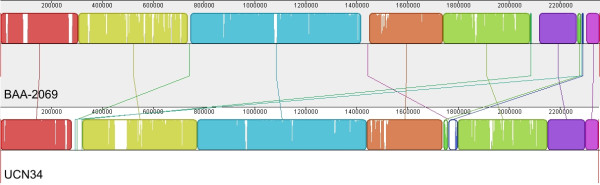
In a direct comparison of genome BAA-2069 to UCN34, we noted various ORFs and regions inserted or deleted scattered along the genomes; nonetheless the majority of genetic information is shared by both strains. The BAA-2069 genome contains 2040 (87%) ORFs which are predicted to be common in BAA-2069 and UCN34. The arrangement of genetic information is very similar overall, based on alignment of the genomes and the synteny plot (Figure 2, Additional file 1: Figure S1). The comparison of the BAA-2069 genome with UCN34 showed about 224 kb (9.5%) of unmatched genetic information. In the UCN34 genome, 199 (9%) unique genes are present, the BAA-2069 genome contains 269 (12%) unique or weak similar genes. There are numerous strain-specific regions with functional genes originated by genetic evolution or lateral gene transfer (LGT). Due to the high number of genomic differences, we focused on genes and regions relating to putative virulence-associated functions or genes affected by habitant adaptation. All unique genes and corresponding islands calculated by EDGAR analysis are summarized in Additional file 2: Table S1 (BAA-2069) and Additional file 3: Table S2 (UCN34)
Figure 2.
Whole genome alignment of the two strains of S. gallolyticus subsp. gallolyticus. Representation of 13 local collinear blocks (LCB) between chromosomal sequences of the S. gallolyticus subsp. gallolyticus BAA-2069 and UCN34 was generated by MAUVE 2.3.1 software [31] with a minimum weight of 355. Sequence of BAA-2069 (top) is the reference against UCN34. The connecting lines between blocks indicate the location of each block in the two genomes. Each colored block represents a homologous region without rearrangements, although white areas within blocks are strain-specific.
Comparison of whole chromosome sequences by MAUVE software [31] reveals an alignment consisting of 13 local collinear blocs (LCB) (Figure 2). No significant inversions or displacements of large regions between the S. gallolyticus subsp. gallolyticus genomes of BAA-2069 and UCN34 were obvious. Regions with low similarity to the corresponding genome occur frequently and their distribution is almost random, although the region from base 2,117,000 bp to the end of the genome seems to be more conserved.
Virulence factors
The BAA-2069 genome contains a 34 kb unique insertion comprising 35 ORFs (SGGBAA2069_c20310-c20660), including the putative major cell surface adhesin pac. This gene is a major colonization factor in S. mutants [32] and may play a similar role in BAA-2069, in addition, it has a 84% similarity to a gene in UCN34 (Gallo_1675). Almost identical to this region is another 30 kb large section in the BAA-2069 genome (SGGBAA2069_c13640-c13980). Both described genetic islands could be functionally virulence-associated, comprising several proteins for cell adhesion and other virulence-determining factors.
In addition, we found a unique 23 kb genetic island in the BAA-2069 genome, coding for bacteriocin-associated genes (SGGBAA2069_c00810-c00960). This region contains genes for lanthionine biosynthesis and for a bacteriocin/lanthionine exporter orthologous to genes described in S. mutants and S. ratti. Lanthionine is a lantibiotic (bacteriocin), a unique class of peptide antibiotic substances [33]. Conducting an agar overlay experiment, we revealed an inhibited growth of Lactococcus lactis, resulting in a zone of clearing around BAA-2069 (data not shown).
Three genes (SGGBAA2069_ c05730, c12530, c17410) are partly homologue to hemolysin A, hemolysin III and an undefined hemolysin-like protein, although group D streptococci are usually non-hemolytic or eventually display weak alpha hemolysis. Moreover, BAA-2069 does show alpha-hemolysis on Schaedler Agar with 5% sheep blood.
The polysaccharide capsule coding region, contains 12 genes (cpsA - cpsM/SGGBAA2069_c09190 - c09300). The genes are located in a 13.5 kb region and are identical to the UCN34 genome.
Comparison of surface proteins
We predicted 21 proteins with C-terminal LPxTG motif by in silico analysis. Additionally, we found orthologous or similar genes to all the proteins with MSCRAMMS characteristics described by Sillanpää et al. regarding the S. gallolyticus subsp. gallolyticus TX20005 genome ("Sbs" genes) and to genes mentioned by Rusinok et al. regarding the UCN34 genome ("Gallo_"-genes) [13,30]. All genes with the LPxTG motif and their best hits in related genomes are listed in Table 2.
Table 2.
Overview and comparison of S. gallolyticus subsp. gallolyticus genes containing the LPxTG DNA motif
| BAA-2069 | UCN34 (Acc. No: FN597254.1) | TX20005 (Acc. No: AEEM00000000.1) | Annotation |
|---|---|---|---|
| SGGBAA2069_c01280 | Gallo_0112 | Sbs10 | (fruA) fructan beta-fructosidase |
| SGGBAA2069_c05110 | Gallo_0577 | Sbs16 (64/98%) | (cna) collagen adhesin |
| SGGBAA2069_c07210 | Gallo_0748 | Sbs6 | (prtS) lactocepin |
| SGGBAA2069_c10430 | Gallo_1058 | Sbs2 | (spaP) glucan binding protein C |
| SGGBAA2069_c13880 | Gallo_1675 (88/89%) | unknown function | |
| SGGBAA2069_c13900 | unknown function | ||
| SGGBAA2069_c14850 | Gallo_1462 | (pulA) pulluanase/glycosidase | |
| SGGBAA2069_c15950 | Gallo_1569 | Sbs11 (100/66%) | (fszB) fimbrial subunit type 2 |
| SGGBAA2069_c15960 | Gallo_1570 | Sbs12 (69/76%) | (cna) collagen adhesin |
| SGGBAA2069_c16150 | Gallo_1578 | (nanA) peptidoglycan linked protein | |
| SGGBAA2069_c16640 | Gallo_1636 | Sbs1 | (pmrB) major facilitator superfamily permease |
| SGGBAA2069_c19780 | Gallo_2018 | (blpT) putative immunity/modification protein | |
| SGGBAA2069_c19910 | Gallo_2032 | Sbs13 | (cna) collagen adhesin |
| SGGBAA2069_c19970 | Gallo_2039 | Sbs14 | major pilus subunit |
| SGGBAA2069_c19980 | Gallo_2040 | Sbs15 | (FN1) peptidoglycan linked protein |
| SGGBAA2069_c20560 | Gallo_1675 (91/84%) | (pac) major cell-surface adhesin | |
| SGGBAA2069_c20580 | unknown function | ||
| SGGBAA2069_c21750 | Gallo_2178 | Sbs7 | backbone pilus subunit |
| SGGBAA2069_c21760 | Gallo_2179 | Acb | (cna) collagen adhesin |
| SGGBAA2069_c22120 | unknown function | ||
| SGGBAA2069_c22310 | Gallo _0272 (98/95%) | (sspA) putative agglutinin receptor |
The indicated percentage in brackets represents the query coverage to the orthologous gene and the identities revealed by blastn (two sequence alignment)
Within the analysis, we found three proteins containing the LPxTG motif carried by genomic islands specific to strain BAA-2069. The gene SGGBAA2069_c13880 and its paralog SGGBAA2069_c20560 have only very weak similarities to Gallo_1675 and code for a putative major cell surface adhesin (pac). The gene SGGBAA2069_c13900 and its paralog SGGBAA2069_c20580 have cell anchor characteristics but no similarities to functional genes. Furthermore, the unique protein SGGBAA2069_c22120 comprising the LPxTG motif is another gene with putative function in virulence.
Protective elements
In comparison to S. gallolyticus subsp. gallolyticus UCN34, the BAA-2069 holds two more restriction enzyme genes. The type III enzyme SthIR (SGGBAA2069_c10290) is located on a 9.9 kb unique island (SGGBAA2069_c10280 - c10350), together with the corresponding restriction-methylation subunit and an integrase gene. Another type II restriction endonuclease Eco47II and its modification methylase is encoded on a 9.7 kb region (SGGBAA2069-c22460 - c22570).
Mentionable regions missing in BAA-2069, but present in the UCN-34 genome, are a 46 kb phage-associated region containing a putative phage-associated cell wall hydrolase. A "cluster regulatory interspaced short palindromic repeats" (CRISPR) element is sited between 1,507,890 - 1,508,913 bp and containing 16 repetitions of a 36 bp consensus sequence. Another 5.6 kb CRISPR associated region is sited at 1,515,490 - 1,516,317 bp but mostly conserved between the two strains (BAA2069 1,517,213 - 1,518,237 bp). A unique CRISPR locus for BAA-2069 is between 1,515,726 - 1,516,570 bp. Corresponding cas genes are for BAA-2069 SGGBAA2069_c14660 and c14670 (cas2), c14670 (cas1), respectively Gallo_1437, Gallo_1444 (cas2) and Gallo_1438, Gallo_1439 (cas1) for UCN34. CRISPR data of both genomes are also accessible by CRISPRs web server http://crispr.u-psud.fr.
Genome comparison to related species
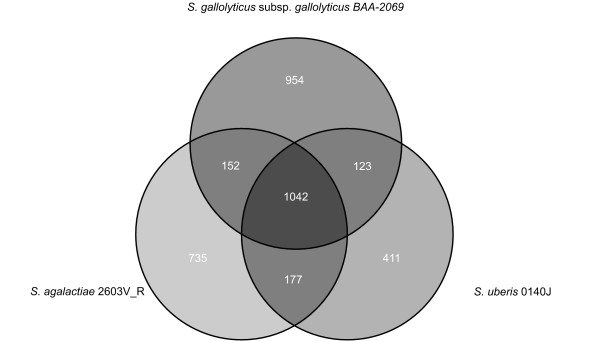
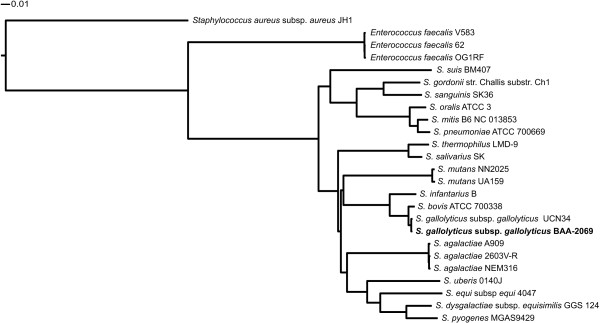
To evaluate the genetic distance to related species, a direct comparison to the taxonomically most closely related species with available whole genome sequences, in particular S. uberis 0140J and S. agalactiae 2603V_R was conducted. The analysis revealed a core genome consisting of 1118 genes common to all three species, whereas S. gallolyticus subsp. gallolyticus BAA-2069 has 804 unique genes (Figure 3). Furthermore, we included three Enterococcus faecalis genomes (V583, OG1RF and 62 [34-36]). Comparison analysis revealed a set of 825 common genes, including a putative hemolysin A gene (SGGBAA2069_c05730), a fibronectin/fibrinogen binding protein (SGGBAA2069_c08170) and a sortase A gene (SGGBAA2069_c11150) which could have a possible conserved role in virulence (Additional file 4: Table S3). A complete list of common or unique ORFs in comparison to BAA-2069, considering all known Streptococcus genomes, is shown in Additional file 5: Table S4. Furthermore, a taxonomic analysis based on alignment of core genomes was performed (Figure 4). The calculation includes the total number of coding sequences common to all analyzed species [37]. The revealing phylogenetic tree indicates a huge genomic diversity between S. gallolyticus subsp. gallolyticus and related whole genome sequenced species.
Figure 3.
Venn diagram of different streptococcal genomes. Venn diagram revealed by EDGAR analysis [37]. Numbers in intersections represent the number common to two or three species. Venn diagram representing common and strain-specific genes of S. gallolyticus subsp. gallolyticus BAA-2069, S. uberis 0140J and S. agalactiae 2603V_R.
Figure 4.
Phylogenetic tree of different streptococcal genomes. Tree was calculated by alignment of core genomes. Non-matching parts of the alignment were masked and subsequently removed. For further calculation details see material and methods.
Plasmid
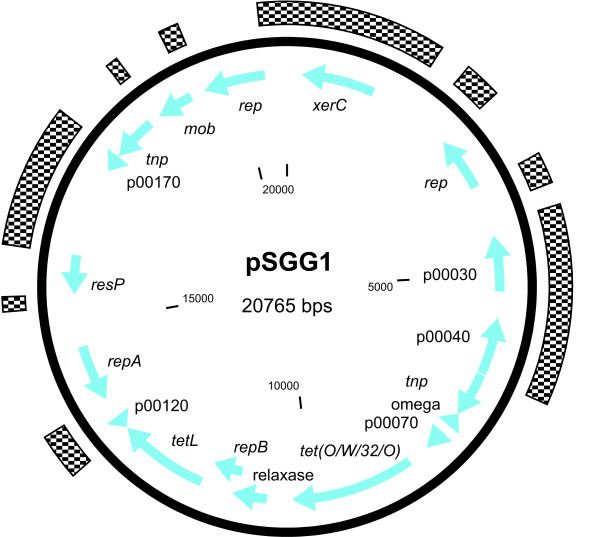
A plasmid designated as pSGG1 was identified by sequence analysis and later isolated from S. gallolyticus subsp. gallolyticus BAA-2069 (Figure 5). The plasmid pSGG1 consists of 20,765 bp and contains 21 ORFs, of which 14 genes code for proteins with similarities to sequence databases including the tetracycline resistance gene tetL (SGGBAA2069_p00110) and the mosaic tetracycline resistance gene tet(O/W/32/O), which are common in plasmids of gram-positive pathogens. Two insertion sequence IS1216 elements and a putative resolvase and a relaxase gene were identified. The relaxase gene has similarities to plasmid pTet35 from Campylobacter jejuni subsp. jejuni 81-176, which suggests its classification of the conjugative transfer system in clade MOBP4. Although, it is more likely that it belongs to the MOBV cluster, which is still ancestrally related to MOBP [38]. The replication is probably regulated by one of four putative rep elements, belonging to rep_1 superfamily (SGGBAA-2069_p00100) and rep_3 superfamily (SGGBAA-2069_p00020, p00140, p00200). The repA (SGGBAA-2069_p00140) element has 78% sequence identity to that of the cryptic plasmid pSBO1 isolated from S. equinus [39]. However, we were not able to determine the functional rep gene by in silico analysis. The plasmid pSGG1 seems to be incapable of conjugal self-transfer since it encodes no tra protein and only a putative resolvase, although it was not tested experimentally. Moreover, a mobilization region orthologous to a mob gene in Streptococcus ferrus was found (SGGBAA2069_p00200), which is a necessary feature for transmissible plasmids and therefore promotes the ability for LGT transfer in presence of a helper conjugative plasmid. Five ORFs were assigned to encode proteins with unknown functions and no significant sequence similarities to previously described genes exist in these cases (Figure 5). The analysis of sequence identity to other plasmids or genomes reveals a mosaic-like structure representing a high number of similarities with common habitants of the rumen or the gastrointestinal tract, including different streptococcal species as well as Enterococcus and others. In particular, the tetracycline resistance genes, which are very common among streptococci, are partly identical among many different plasmids and species, although no similarities in arrangement of resistance genes were observed. To evaluate the distribution of pSGG1 among strains of S. gallolyticus subsp. gallolyticus with different origin (animal, strain collections and human samples), we screened 41 strains by Southern blot hybridization analysis with a digoxygenin nick-labeled probe of pSGG1 (Figure 6). We identified and isolated a plasmid (pSGG2) mainly homologous to pSGG1 in another strain (isolate 010672), originally isolated from the blood culture of a patient with infectious endocarditis. The restriction fragment analysis of pSGG2 revealed a partially different pattern in comparison to pSGG1, indicating sequence variation between both plasmids (Additional file 6: Figure. S2). In further experiments we sequenced the pSGG2 plasmid and revealed only differences in non-coding regions (data not shown).
Figure 5.
Plasmid map of pSGG1. Plasmid pSGG1 isolated from S. gallolyticus subsp. gallolyticus BAA-2069. Unique regions are marked by squared boxes.
Figure 6.
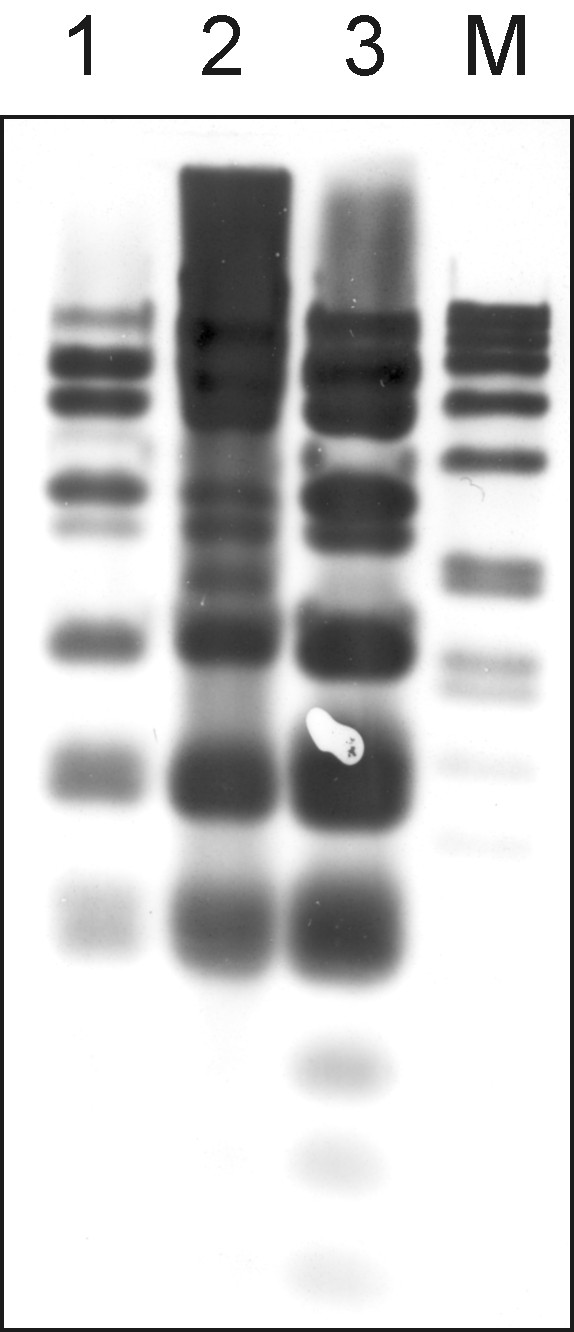
Southern blot analysis of BamHI-digested plasmids from two S. gallolyticus subsp. gallolyticus strains. Total DNA was digested with BamHI and hybridized against a probe consisting of DIG-11-UTP-labeled pSGG1 plasmid DNA. Lane 1: S. gallolyticus subsp. gallolyticus strain 010672 genomic DNA. Lane 2: S. gallolyticus subsp. gallolyticus strain BAA-2069 genomic DNA (positive control). Lane 3: Plasmid DNA of pSGG1. M: DIG DNA Molecular Weight Marker VII, DIG-labeled (Roche, Mannheim, Germany).
In order to analyze whether the frequency and phenotype of tetracycline resistance of strain BAA-2069 is coincident with the presence of pSGG1, we screened 41 S. gallolyticus subsp. gallolyticus strains for presence of tetL and mob genes by PCR. Additionally, we performed a tetracycline susceptibility test. The epidemiological cut-off for the WT of related streptococci is ≤ 1 μg/mL http://eucast.org. About 42% of strains were growth-inhibited by a tetracycline concentration between 0.5-1 μg/mL, and 95% of strains tested were unable to grow at concentrations higher than 256 μg/mL. The two strains which showed a tetracycline MIC value of 512 μg/mL carrying the pSGG plasmid, and only these were screened positively for tetL and mob genes (Additional file 7: Figure. S3).
Discussion
The present study describes the full genome sequence of S. gallolyticus subsp. gallolyticus BAA-2069 and the comparison to related genomes in order to evaluate possible virulence-associated characteristics of this species. Previous publications have shown a significant diversity in adhesion and invasion potential for binding to endothelial host cells, as well as binding to ECM proteins in vitro [22]. Other studies have shown that virulence gene profiles are associated with disease [40]. Therefore genomic comparison analysis provides the basis for understanding pathogenicity.
Within whole genome comparison analysis the "pan-genome" includes a core genome containing genes present in all strains of one species. This is complemented by an individual set of genes unique to a strain or not shared by all strains. With the growing number of sequenced strains, the increasing size of the pan-genome is evidence of the genomic diversity between different isolates of a distinct species. Tettelin et al. have shown that in the case of Streptococcus agalactiae the core genome of eight strains comprises about 80% of genes of any single genome, and exploration of data reveals that the gene reservoir is immense [41], whereas in the case of Bacillus anthracis the number of strain-specific genes after addition of the fourth strain was zero [42]. The number of strain-specific regions in the two analyzed S. gallolyticus subsp. gallolyticus strains is, in contrast to S. agalactiae strains (average 7.27%, maximum ~10%), about 3.5% higher. This could be taken as a hint that, with the increasing number of sequenced strains, the pan-genome of S. gallolyticus subsp. gallolyticus is far larger by proportion. However, these data are preliminary, pending the sequencing of further S. gallolyticus subsp. gallolyticus strains.
In a direct comparison to the recently sequenced strain UCN34 [30], surprisingly many unique genes with putative virulence associated characteristics are present in each strain, which could be an indication that the pathogenicity of S. gallolyticus subsp. gallolyticus is very diverse. The majority of exclusive sequences found in the UCN34 genome are located in three large regions representing 111 kb of sequence information (53%), whereas the three largest unique regions in BAA-2069 constitute only 87 kb (39%) of strain-specific sequence and mostly consist of smaller regions. However, the tendency of virulence factors to be located within genomic islands may lead to a higher ratio of exchangeability of such genes in comparison to other regions [43]. Furthermore, additional restriction enzymes in BAA-2069 may have a function in protection against viral DNA and heritable CRISPR elements are able to mediate immunity against phages and be transmitted to other organisms by genetic transformation events [44].
Surface proteins and in particular proteins belonging to "microbial surface component recognizing matrix molecules" (MSCRAMM) were shown to play a functional role in the pathogenesis of all bacteria. Of specific interest is a group of proteins containing the C-terminal cell wall-sorting motif LPxTG, which serves as a recognition site for the membrane-associated sortase. After sortase-mediated cleavage of the protein, the polypeptide is covalently bound to the peptidoglycan of bacterial cell surface and can therefore promote the first step in bacterial adherence [45,46]. Three of the 21 predicted LPxTG motif genes are unique for BAA-2069 and further studies are required to evaluate their contribution to pathogenicity.
In silico analysis of genome data strongly indicated the presence of a multi-copy plasmid. The purification of plasmid DNA and further analysis of sequence data confirmed these hints and showed a localization of tetracycline resistance genes. Analysis of plasmid distribution shows only two mainly homologous plasmids in 41 strains overall. Therefore, the incidence of the pSGG plasmids among S. gallolyticus isolates does not seem to be widespread. The mosaic tetracycline resistance gene tet(O/W/32/O) is usually chromosomally located and mediates resistance by ribosome protection. It has been shown that the mosaic tet(O/W) genes have a higher level of resistance than the original genes [47]. This could be verified by our experimental data, showing the strains carrying the pSGG plasmid have the highest resistance levels. The tetL gene is generally found on plasmids and coding for a tetracycline efflux protein [48]. In contrast to the BAA-2069 strain, the tetracycline resistance of strain UCN34, mediated by tetL and tetM, was located on the chromosome and adjacent to putative plasmid and transposon Tn916-associated genes [30]. This indicates a strong dependence between high tetracycline resistance mediated by tetL and the occurrence of plasmids of the pSGG family.
Because of antibiotic treatment, gastrointestinal tract and rumen are well-known reservoirs of mobilizable antibiotic resistance genes [49]. Furthermore, the transfer of antibiotic resistance across several species and genera between commensal bacteria is well known, and habitants with a dense population and, in particular, the ability to form biofilms, are optimal for genetic transfer [50]. Especially because, S. gallolyticus subsp. gallolyticus is a commensal and facultative pathogen of animals, the intensive tetracycline treatments in animal husbandry, causes a general advantage regarding evolutionary fitness for pathogenic and natural habitants of the intestinal tract to accommodate resistance genes by LGT [51,52]. Although the plasmid pSGG1 is incapable of conjugal self-transfer, it is mobilizable by a helper conjugative plasmid. These findings suggest that it may play a functional role in LGT between different streptococcal groups and further related species. However, the detection of only two plasmids out of 41 strains is so far not evidence of LGT, but further screening of a huge variety of strains in combination with epidemiological studies should help to evaluate the role of pSGG plasmids.
Conclusion
This study presented the analysis and comparison of the whole genome sequence of S. gallolyticus subsp. gallolyticus strain BAA-2069, a causative agent of infective endocarditis. The results promote identification of genetic factors concerning the pathogenesis and adhesion to ECM. Novel candidate genes were detected probably contributing to the pathogenicity. The comparison to S. gallolyticus subsp. gallolyticus strain UCN34 revealed significant differences concerning virulence factors, surface proteins and protective elements.
Furthermore, we detected for the first time the presence of the pSGG1 plasmid, containing 21 ORFs including mosaic tetracycline resistance genes and may play a functional role in lateral gene transfer.
Methods
Bacterial strains, growth conditions, nucleic acid extraction
The S. gallolyticus subsp. gallolyticus strain was isolated in 2003 at the Herz- und Diabeteszentrum Nordrhein-Westfalen from a blood culture from a 68-year-old woman with aortic heart valve endocarditis and deposited at the American Type Culture Collection (ATCC, Manassas, USA) (BAA-2069). Strain BAA-2069 was confirmed by isolation of the same strain by lesion smear test of aortic heart valve and detection in valve excision material by culture and PCR. The strain was selected because it had been defined as virulent during earlier tests [22] and shows phenotypic resistance against oxacillin, tobramycin, co-trimoxazole, colistin, metronidazole and tetracycline and intermediate resistance against gentamycin (minimal inhibitory concentration (MIC) 8 μg/mL). Isolate 010672 with plasmid pSGG2 was isolated in 2001 at the Herz- und Diabeteszentrum Nordrhein-Westfalen from a blood culture from a 62-year-old man with infectious endocarditis with no obvious connection to the origin of strain BAA-2069. For genomic DNA isolation, cells were grown for 12 h in Brain Heart Infusion Broth (BHI) (Oxoid, Hampshire, United Kingdom) at 37°C, 200 rpm. DNA extraction was performed by the Hopwood alkaline lysis method [53].
Genome sequencing, assembly and gap closure
DNA sequencing was performed using 454 Life Science pyrosequencing technology [54], GS-FLX Titanium produced 455,842 reads of average 329 bp. The reads were assembled using Newbler V2.3, resulting in 38 contigs with 31 contigs larger than 500 bp. The large contigs obtained with 64.9× coverage served as the basis for the gap closure. Gap closure was performed by custom primer walking with long range PCR (using Phusion polymerase, New England Biolabs, Frankfurt (Main), Germany) and subsequent Sanger sequencing, resulting in 62 reads in total (IIT Biotech, Bielefeld, Germany). Long repeat structures (copies of the rrn operon and two repeats of 17.4 and 5 kbp respectively) were resolved by introducing fake reads based on the consensus sequence.
Genome annotation
Curation and annotation of the genome were performed using the genome annotation system GenDB 2.4 [55]. Prediction of coding sequences (CDS) was accomplished using Critica [56], Glimmer [57] and Reganor [58]. All predicted ORFs were automatically submitted to similarity searches against nr, Swissprot, KEGG, InterPro, Pfam and TIGRfam databases. Putative signal peptides, transmembrane helices and nucleic acid binding domains were predicted using SignalP [59], TMHMM [60] and Helix-Turn-Helix [61], respectively. The automatic annotation of each CDS was manually checked and corrected according to the most congruent tool results.
Genome analysis
S. gallolyticus subsp. gallolyticus BAA-2069 gene content was compared to S. gallolyticus subsp. gallolyticus UCN34, S. agalactiae A909, S. dysgalactiae subsp. equisimilis GGS_124, S. equi subsp. equi 4047, S. sanguinis SK36, S. suis BM407, S. uberis 0140J, S. pyogenes MGAS9429, S. pneumoniae ATCC 700669, S. mutans NN2025, S. mitis B6, S. thermophilus LMD-9, S. gordonii str. challis substr. CH1, S. oralis ATCC 35037, S. salivarius SK126 with EDGAR [37], which defines orthologous proteins based on bidirectional best blast hit and then calculates BLASTP score ratio values (SRV). Paralogous genes might be discarded during the analysis. For each comparison, SRV distribution was fitted with binominal or bibeta distribution with a self written R script, and a cutoff was determined at the point where the probability of belonging to one or other peak is equal. Accordingly, a general cutoff of 0.21 was used to retrieve the core genes and singletons. LPxTG-related proteins were searched by screening for [LYF]P[TSA][GANS] motif and using of an LPxTG Hidden Marcov Model for sortase substrates created by Boekhorst et al. [46].
Comparison of whole chromosome sequences
Comparison of whole chromosome sequences was performed by MAUVE software using local collinear blocs (LCB). An LCB is defined as a collinear (consistent) set of multi-MUMs (exact match subsequences shared by all the considered chromosomes that appear once in each chromosome and are bordered on both sides by mismatched nucleotides). The weight (the sum of the lengths of the included multi-MUMs) of an LCB serves as a measure of confidence that it is a true orthologous region instead of a random match and is set to 355. Therefore, the ORFs or sequences between the LCBs and any regions with low similarity (shown as white in LCB) are classified as strain-specific regions.
Calculation of phylogenetic tree
For calculation of phylogenetic tree, EDGAR was used [49]. In detail this means that, for this project comprising 25 genomes 300 core genes (orthology-cutoff 35% Score Ratio Value) of these genomes are computed. In a next step alignments of the core genes are generated using MUSCLE, non-matching parts of the alignment are masked by GBLOCKS and subsequently removed. The remaining parts of all alignments are concatenated to one huge alignment. Based on this alignment, a distance matrix is calculated using the Kimura algorithm, which is used as input for the neighbor joining method (both algorithms realized in the PHYLIP package). This leads to a phylogenetic tree, represented in newick format.
GC skew analysis
The GC skew measures the excess of Gs by calculating the difference between the number of Gs and Cs (G-C) in a sliding window of 1000 nucleotides. The skews were cumulated to obtain the cumulative GC skew that represents the sum of the GC skews from the first to the ith window.
Plasmid screening
Screening of 41 different S. gallolyticus subsp. gallolyticus strains for presence of pSGG1 plasmid or homologs was performed by Southern-hybridization analysis in accordance with standard protocols. The probe was prepared by nick translation DIG labeling of pSGG1 referring to DIG DNA Labeling Kit (Roche Diagnostics, Mannheim, Germany) [62]. Furthermore all strains were screened for the presence of tetL gene by PCR using the whole genome sequence derived primer tet_f (5'-GCTATGGGAGAAGGTATCG-3') and tet_r (5'-GAGACAAACCCTGCTACTG-3'), or mob_f (5'-AAGCTGTACTTGGCTCTC-3') and mob_r (5'-CAGTGGCAGAACTATCTC-3') respectively, by standard methods.
Nucleotide sequence accession number
Whole genome sequence of S. gallolyticus subsp. gallolyticus was deposited at GenBank (Acc. no. FR824043). Sequence of the novel designated plasmid pSGG1 was deposited with accession no. FR824044.
Tetracycline susceptibility testing
For each strain, 200 μL BHI broth (Oxoid, Cambridge, UK) supplemented with indicated tetracycline concentration were inoculated with 1 μL of overnight culture of S. gallolyticus subsp. gallolyticus strains and cultivated in 96 well plates at 37°C. After 16 h incubation, OD 600 was measured and growth was determined as OD 600 > 0.2. The assay was performed in triplicate.
Authors' contributions
DH prepared the DNA and plasmid extraction, carried out the sequence analyses, participated in the gap closure and bioinformatics analysis and wrote the manuscript. TV participated in the design of figures and helped to draft the manuscript. CR performed sequencing and carried out the sequence alignment. JB worked on bioinformatics analysis. JK participated in the design and drafted the manuscript. CK and JD conceived, designed and coordinated the study and helped to draft the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Pairwise synteny plot of the S. gallolyticus subsp. gallolyticus BAA-2069 and UCN34 genome. Every CDS of the first contig is checked for a reziprocal best blast hit. If one is found, the stopposition of both CDS are read from the database and used as coordinates for a dot.
Unique genes of S. gallolyticus subsp. gallolyticus BAA-2069 in relation to S. gallolyticus subsp. gallolyticus UCN34. Unique genes calculated by EDGAR analysis.
Unique genes of S. gallolyticus subsp. gallolyticus UCN34 in relation to S. gallolyticus subsp. gallolyticus BAA-2069. Unique genes calculated by EDGAR analysis.
Core genome set of S. gallolyticus subsp. gallolyticus BAA-2069 and three Enterococcus feacalis strains. Following strains were used for calculation by EDGAR: E. faecalis 62 (Acc. No CP002491), E. faecalis OG1RF (Acc. no. CP002621) and E. faecalis V583 (Acc. no. NC_004668).
Number of unique or common ORFs. Numbers represent the common or unique ORFs in comparison to BAA-2069 and indicated species.
Agarose gel electrophoresis of restriction fragment pattern. Pattern were obtained with seven different enzymes, regarding plasmid pSGG2 (left lane) and pSGG1 (right lane). Ladder marker: 1 kb Ladder plus (Fermentas, St. Leon-Rot, Germany).
Tetracycline susceptibility test. Minimum inhibitory concentration (MIC) was determined growth in liquid cultures with indicated tetracycline concentration.
Contributor Information
Dennis Hinse, Email: dhinse@hdz-nrw.de.
Tanja Vollmer, Email: tvollmer@hdz-nrw.de.
Christian Rückert, Email: Christian.Rueckert@CeBiTec.Uni-Bielefeld.DE.
Jochen Blom, Email: jblom@cebitec.uni-bielefeld.de.
Jörn Kalinowski, Email: joern@cebitec.uni-bielefeld.de.
Cornelius Knabbe, Email: cknabbe@hdz-nrw.de.
Jens Dreier, Email: jdreier@hdz-nrw.de.
Acknowledgements
We thank Sarah L. Kirkby for her linguistic advice and Melanie Weinstock for help with manuscript preparation.
References
- Farrow JAE, Kruze J, Phillips BA, Bramley AJ, Collins MD. Taxonomic studies on Streptococcus bovis and Streptococcus equinus: description of Streptococcus alactolyticus sp. nov. and Streptococcus saccharolyticus sp. nov. Syst Appl Microbiol. 1984;12:467–482. [Google Scholar]
- Osawa R. Formation of a clear zone on tannin-treated brain heart infusion agar by a Streptococcus sp. isolated from feces of koalas. Appl Environ Microbiol. 1990;12:829–831. doi: 10.1128/aem.56.3.829-831.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa R, Fujisawa T, Sly LI. Streptococcus gallolyticus sp. nov.; gallate degrading organisms formerly assigned to Streptococcus bovis. Syst Appl Microbiol. 1995;12:74–78. [Google Scholar]
- Schlegel L, Grimont F, Ageron E, Grimont PA, Bouvet A. Reappraisal of the taxonomy of the Streptococcus bovis/Streptococcus equinus complex and related species: description of Streptococcus gallolyticus subsp. gallolyticus subsp. nov., S. gallolyticus subsp. macedonicus subsp. nov. and S. gallolyticus subsp. pasteurianus subsp. nov. Int J Syst Evol Microbiol. 2003;12:631–645. doi: 10.1099/ijs.0.02361-0. [DOI] [PubMed] [Google Scholar]
- Beck M, Frodl R, Funke G. Comprehensive study of strains previously designated Streptococcus bovis consecutively isolated from human blood cultures and emended description of Streptococcus gallolyticus and Streptococcus infantarius subsp. coli. J Clin Microbiol. 2008;12:2966–2972. doi: 10.1128/JCM.00078-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CA, McCaughey R, Lauter CB. The association of Streptococcus bovis fecal carriage and colon neoplasia: possible relationship with polyps and their premalignant potential. Am J Gastroenterol. 1985;12:42–46. [PubMed] [Google Scholar]
- Klein RS, Recco RA, Catalano MT, Edberg SC, Casey JI, Steigbigel NH. Association of Streptococcus bovis with carcinoma of the colon. N Engl J Med. 1977;12:800–802. doi: 10.1056/NEJM197710132971503. [DOI] [PubMed] [Google Scholar]
- Tripodi MF, Fortunato R, Utili R, Triassi M, Zarrilli R. Molecular epidemiology of Streptococcus bovis causing endocarditis and bacteraemia in Italian patients. Clin Microbiol Infect. 2005;12:814–819. doi: 10.1111/j.1469-0691.2005.01248.x. [DOI] [PubMed] [Google Scholar]
- Hoen B, Chirouze C, Cabell CH, Selton-Suty C, Duchene F, Olaison L, Miro JM, Habib G, Abrutyn E, Eykyn S. et al. Emergence of endocarditis due to group D streptococci: findings derived from the merged database of the International Collaboration on Endocarditis. Eur J Clin Microbiol Infect Dis. 2005;12:12–16. doi: 10.1007/s10096-004-1266-6. [DOI] [PubMed] [Google Scholar]
- Vollmer T, Piper C, Horstkotte D, Körfer R, Kleesiek K, Dreier J. 23S rDNA real-time polymerase chain reaction of heart valves: a decisive tool in the diagnosis of infective endocarditis. Eur Heart J. 2010;12:1105–1113. doi: 10.1093/eurheartj/ehp600. [DOI] [PubMed] [Google Scholar]
- Sillanpää J, Nallapareddy SR, Singh KV, Ferraro MJ, Murray BE. Adherence characteristics of endocarditis-derived Streptococcus gallolyticus ssp. gallolyticus (Streptococcus bovis biotype I) isolates to host extracellular matrix proteins. FEMS Microbiol Lett. 2008;12:104–109. doi: 10.1111/j.1574-6968.2008.01378.x. [DOI] [PubMed] [Google Scholar]
- Arzanauskiene R, Zabiela P, Sakalnikiene M. [Streptococcus bovis endocarditis - predictor of colonic carcinoma?] Medicina (Kaunas) 2003;12:174–176. [PubMed] [Google Scholar]
- Sillanpää J, Nallapareddy SR, Qin X, Singh KV, Muzny DM, Kovar CL, Nazareth LV, Gibbs RA, Ferraro MJ, Steckelberg JM. et al. A collagen-binding adhesin, Acb, and ten other putative MSCRAMM and pilus family proteins of Streptococcus gallolyticus subsp. gallolyticus (Streptococcus bovis Group, biotype I) J Bacteriol. 2009;12:6643–6653. doi: 10.1128/JB.00909-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjalsma H, Scholler-Guinard M, Lasonder E, Ruers TJ, Willems HL, Swinkels DW. Profiling the humoral immune response in colon cancer patients: diagnostic antigens from Streptococcus bovis. Int J Cancer. 2006;12:2127–2135. doi: 10.1002/ijc.22116. [DOI] [PubMed] [Google Scholar]
- Sasaki E, Osawa R, Nishitani Y, Whiley RA. ARDRA and RAPD analyses of human and animal isolates of Streptococcus gallolyticus. J Vet Med Sci. 2004;12:1467–1470. doi: 10.1292/jvms.66.1467. [DOI] [PubMed] [Google Scholar]
- Baele M, Vanrobaeys M, Vaneechoutte M, De Herdt P, Devriese LA, Haesebrouck F. Genomic fingerprinting of pigeon Streptococcus gallolyticus strains of different virulence by randomly amplified polymorphic DNA (RAPD) analysis. Vet Microbiol. 2000;12:103–111. doi: 10.1016/S0378-1135(99)00169-8. [DOI] [PubMed] [Google Scholar]
- Russell JB, Hino T. Regulation of Lactate Production in Streptococcus bovis: A Spiraling Effect That Contributes to Rumen Acidosis. J Dairy Sci. 1985;12:1712–1721. doi: 10.3168/jds.S0022-0302(85)81017-1. [DOI] [PubMed] [Google Scholar]
- Kelly BG, Vespermann A, Bolton DJ. Gene transfer events and their occurrence in selected environments. Food Chem Toxicol. 2009;12:978–983. doi: 10.1016/j.fct.2008.06.012. [DOI] [PubMed] [Google Scholar]
- Ichihara H, Kuma K, Toh H. Positive selection in the ComC-ComD system of Streptococcal Species. J Bacteriol. 2006;12:6429–6434. doi: 10.1128/JB.00484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison J. Genetic exchange between bacteria in the environment. Plasmid. 1999;12:73–91. doi: 10.1006/plas.1999.1421. [DOI] [PubMed] [Google Scholar]
- Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol. 2001;12:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer T, Hinse D, Kleesiek K, Dreier J. Interactions between endocarditis-derived Streptococcus gallolyticus subsp. gallolyticus isolates and human endothelial cells. BMC Microbiol. 2010;12:78. doi: 10.1186/1471-2180-10-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker J, Carniel E. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2001;12:376–381. doi: 10.1093/embo-reports/kve097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger X, Roberts AP, Morel C, Choulet F, Pavlovic G, Mullany P, Decaris B, Guedon G. Conjugative transfer of the integrative conjugative elements ICESt1 and ICESt3 from Streptococcus thermophilus. J Bacteriol. 2009;12:2764–2775. doi: 10.1128/JB.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrobaeys M, Haesebrouck F, Ducatelle R, De Herdt P. Adhesion of Streptococcus gallolyticus strains to extracellular matrix proteins. Vet Microbiol. 2000;12:273–280. doi: 10.1016/S0378-1135(00)00180-2. [DOI] [PubMed] [Google Scholar]
- Boleij A, Schaeps RM, de Kleijn S, Hermans PW, Glaser P, Pancholi V, Swinkels DW, Tjalsma H. Surface-exposed histone-like protein a modulates adherence of Streptococcus gallolyticus to colon adenocarcinoma cells. Infect Immun. 2009;12:5519–5527. doi: 10.1128/IAI.00384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrobaeys M, De Herdt P, Haesebrouck F, Ducatelle R, Devriese LA. Secreted antigens as virulence associated markers in Streptococcus bovis strains from pigeons. Vet Microbiol. 1996;12:339–348. doi: 10.1016/S0378-1135(96)01254-0. [DOI] [PubMed] [Google Scholar]
- De Herdt P, Haesebrouck F, Devriese LA, Ducatelle R. Biochemical and antigenic properties of Streptococcus bovis isolated from pigeons. J Clin Microbiol. 1992;12:2432–2434. doi: 10.1128/jcm.30.9.2432-2434.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanrobaeys M, De Herdt P, Charlier G, Ducatelle R, Haesebrouck F. Ultrastructure of surface components of Streptococcus gallolytics (S. bovis) strains of differing virulence isolated from pigeons. Microbiology. 1999;12(Pt 2):335–342. doi: 10.1099/13500872-145-2-335. [DOI] [PubMed] [Google Scholar]
- Rusniok C, Couve E, Da Cunha V, El Gana R, Zidane N, Bouchier C, Poyart C, Leclercq R, Trieu-Cuot P, Glaser P. Genome sequence of Streptococcus gallolyticus: insights into its adaptation to the bovine rumen and its ability to cause endocarditis. J Bacteriol. 2010;12:2266–2276. doi: 10.1128/JB.01659-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;12:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Nakano Y, Yamashita Y, Oho T, Koga T. Effects of antibodies against cell surface protein antigen PAc-glucosyltransferase fusion proteins on glucan synthesis and cell adhesion of Streptococcus mutans. Infect Immun. 1997;12:2292–2298. doi: 10.1128/iai.65.6.2292-2298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyink O, Balakrishnan M, Tagg JR. Streptococcus rattus strain BHT produces both a class I two-component lantibiotic and a class II bacteriocin. FEMS Microbiol Lett. 2005;12:235–241. doi: 10.1016/j.femsle.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF. et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;12:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- Bourgogne A, Garsin DA, Qin X, Singh KV, Sillanpaa J, Yerrapragada S, Ding Y, Dugan-Rocha S, Buhay C, Shen H. et al. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 2008;12:R110. doi: 10.1186/gb-2008-9-7-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brede DA, Snipen LG, Ussery DW, Nederbragt AJ, Nes IF. Complete Genome Sequence of the Commensal Enterococcus faecalis 62, Isolated from a Healthy Norwegian Infant. J Bacteriol. 2011;12:2377–2378. doi: 10.1128/JB.00183-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom J, Albaum SP, Doppmeier D, Puhler A, Vorholter FJ, Zakrzewski M, Goesmann A. EDGAR: a software framework for the comparative analysis of prokaryotic genomes. BMC Bioinformatics. 2009;12:154. doi: 10.1186/1471-2105-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcillan-Barcia MP, Francia MV, de la Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev. 2009;12:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- Nakamura M, Ogata K, Nagamine T, Tajima K, Matsui H, Benno Y. Characterization of the cryptic plasmid pSBO2 isolated from Streptococcus bovis JB1 and construction of a new shuttle vector. Curr Microbiol. 2000;12:27–32. doi: 10.1007/s002840010086. [DOI] [PubMed] [Google Scholar]
- Silva LM, Baums CG, Rehm T, Wisselink HJ, Goethe R, Valentin-Weigand P. Virulence-associated gene profiling of Streptococcus suis isolates by PCR. Vet Microbiol. 2006;12:117–127. doi: 10.1016/j.vetmic.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS. et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome". Proc Natl Acad Sci USA. 2005;12:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim P, Price LB, Klevytska AM, Smith KL, Schupp JM, Okinaka R, Jackson PJ, Hugh-Jones ME. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J Bacteriol. 2000;12:2928–2936. doi: 10.1128/JB.182.10.2928-2936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Sui SJ, Fedynak A, Hsiao WW, Langille MG, Brinkman FS. The association of virulence factors with genomic islands. PLoS One. 2009;12:e8094. doi: 10.1371/journal.pone.0008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;12:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Navarre WW, Schneewind O. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria. Mol Microbiol. 1994;12:115–121. doi: 10.1111/j.1365-2958.1994.tb01271.x. [DOI] [PubMed] [Google Scholar]
- Boekhorst J, de Been MW, Kleerebezem M, Siezen RJ. Genome-wide detection and analysis of cell wall-bound proteins with LPxTG-like sorting motifs. J Bacteriol. 2005;12:4928–4934. doi: 10.1128/JB.187.14.4928-4934.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson AJ, Rincon MT, Flint HJ, Scott KP. Mosaic tetracycline resistance genes are widespread in human and animal fecal samples. Antimicrob Agents Chemother. 2007;12:1115–1118. doi: 10.1128/AAC.00725-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer BS, Shoemaker NB, Salyers AA. Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin Microbiol Rev. 1992;12:387–399. doi: 10.1128/cmr.5.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004;12:412–416. doi: 10.1016/j.tim.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Mathur S, Singh R. Antibiotic resistance in food lactic acid bacteria--a review. Int J Food Microbiol. 2005;12:281–295. doi: 10.1016/j.ijfoodmicro.2005.03.008. [DOI] [PubMed] [Google Scholar]
- van den Bogaard AE, Stobberingh EE. Epidemiology of resistance to antibiotics. Links between animals and humans. Int J Antimicrob Agents. 2000;12:327–335. doi: 10.1016/S0924-8579(00)00145-X. [DOI] [PubMed] [Google Scholar]
- Chopra I, Roberts M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev. 2001;12:232–260. doi: 10.1128/MMBR.65.2.232-260.2001. second page, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser H, Lydiate DJ, Smith CP, Ward JM, Schrempf H. Genetic Manipulation of Streptomyces: A Laboratory Manual. Cold Spring Harbor Laboratory Press; 1985. [Google Scholar]
- Droege M, Hill B. The Genome Sequencer FLX System--longer reads, more applications, straight forward bioinformatics and more complete data sets. J Biotechnol. 2008;12:3–10. doi: 10.1016/j.jbiotec.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J, Kalinowski J, Linke B, Rupp O, Giegerich R, Puhler A. GenDB--an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003;12:2187–2195. doi: 10.1093/nar/gkg312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger JH, Olsen GJ. CRITICA: coding region identification tool invoking comparative analysis. Mol Biol Evol. 1999;12:512–524. doi: 10.1093/oxfordjournals.molbev.a026133. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;12:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke B, McHardy AC, Neuweger H, Krause L, Meyer F. REGANOR: a gene prediction server for prokaryotic genomes and a database of high quality gene predictions for prokaryotes. Appl Bioinformatics. 2006;12:193–198. doi: 10.2165/00822942-200605030-00008. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;12:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;12:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Dodd IB, Egan JB. Systematic method for the detection of potential lambda Cro-like DNA-binding regions in proteins. J Mol Biol. 1987;12:557–564. doi: 10.1016/0022-2836(87)90681-4. [DOI] [PubMed] [Google Scholar]
- Anderson DG, McKay LL. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;12:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pairwise synteny plot of the S. gallolyticus subsp. gallolyticus BAA-2069 and UCN34 genome. Every CDS of the first contig is checked for a reziprocal best blast hit. If one is found, the stopposition of both CDS are read from the database and used as coordinates for a dot.
Unique genes of S. gallolyticus subsp. gallolyticus BAA-2069 in relation to S. gallolyticus subsp. gallolyticus UCN34. Unique genes calculated by EDGAR analysis.
Unique genes of S. gallolyticus subsp. gallolyticus UCN34 in relation to S. gallolyticus subsp. gallolyticus BAA-2069. Unique genes calculated by EDGAR analysis.
Core genome set of S. gallolyticus subsp. gallolyticus BAA-2069 and three Enterococcus feacalis strains. Following strains were used for calculation by EDGAR: E. faecalis 62 (Acc. No CP002491), E. faecalis OG1RF (Acc. no. CP002621) and E. faecalis V583 (Acc. no. NC_004668).
Number of unique or common ORFs. Numbers represent the common or unique ORFs in comparison to BAA-2069 and indicated species.
Agarose gel electrophoresis of restriction fragment pattern. Pattern were obtained with seven different enzymes, regarding plasmid pSGG2 (left lane) and pSGG1 (right lane). Ladder marker: 1 kb Ladder plus (Fermentas, St. Leon-Rot, Germany).
Tetracycline susceptibility test. Minimum inhibitory concentration (MIC) was determined growth in liquid cultures with indicated tetracycline concentration.