Abstract
Background. Invasive nontyphoid Salmonella (iNTS) disease is common and severe in adults with human immunodeficiency virus (HIV) infection in Africa. We previously observed that ex vivo macrophages from HIV-infected subjects challenged with Salmonella Typhimurium exhibit dysregulated proinflammatory cytokine responses.
Methods. We studied the transcriptional response in whole blood from HIV-positive patients during acute and convalescent iNTS disease compared to other invasive bacterial diseases, and to HIV-positive and -negative controls.
Results. During iNTS disease, there was a remarkable lack of a coordinated inflammatory or innate immune signaling response. Few interferon γ (IFNγ)--induced genes or Toll-like receptor/transcription factor nuclear factor κB (TLR/NFκB) gene pathways were upregulated in expression. Ex vivo lipopolysacharide (LPS) or flagellin stimulation of whole blood, however, showed that convalescent iNTS subjects and controls were competent to mount prominent TLR/NFκB-associated patterns of mRNA expression. In contrast, HIV-positive patients with other invasive bacterial infections (Escherichia coli and Streptococcus pneumoniae) displayed a pronounced proinflammatory innate immune transcriptional response. There was also upregulated mRNA expression in cell cycle, DNA replication, translation and repair, and viral replication pathways during iNTS. These patterns persisted for up to 2 months into convalescence.
Conclusions. Attenuation of NFκB-mediated inflammation and dysregulation of cell cycle and DNA-function gene pathway expression are key features of the interplay between iNTS and HIV.
Invasive nontyphoid Salmonella (iNTS) disease is one of the commonest causes of bloodstream infection in adults and children across sub–Saharan Africa where human immunodeficiency virus (HIV) infection is prevalent [1]. Case series have demonstrated a 22%–47% acute mortality among adults, in whom coinfection with HIV is present in >95% of cases, and a 20%–40% rate of relapse among survivors despite antibiotic treatment [2–4]. A varied clinical presentation makes iNTS diagnostically challenging [3], and a rapid diagnostic biomarker test would be valuable. There is a paucity of human data to explain the pathogenesis and severity of this infection [5].
In the context of HIV in adults, there is a significant intracellular stage during the pathogenesis of iNTS [6]. We have previously observed dysregulated proinflammatory cytokine responses from ex vivo macrophages from HIV-infected adults challenged with Salmonella Typhimurium, with responses declining in late HIV disease [7]. We hypothesized that the peripheral blood mononuclear cell (PBMC) response during acute disease would provide further useful insights into pathogenesis by comparison with other infections. This study reports the first attempt to profile the global host responses to iNTS in vivo in a large HIV patient group, with the key aim of providing novel insights clarifying the nature of iNTS disease. We utilized microarray technology and advanced systems biology analyses [8] to dissect the transcriptional host responses during acute and convalescent iNTS in the context of HIV, and compared this to other acute invasive bacterial infections in HIV-positive patients and to baseline asymptomatic HIV-positive controls.
In addition, we used an ex vivo whole-blood stimulation assay based on lipopolysacharide (LPS) and flagellin to provide further insight into host responsiveness. We discuss the results of these novel in vivo and ex vivo studies of iNTS infection in comparison with previous transcriptional studies in tissue models, animal models, and human Salmonella disease [9–13].
SUBJECTS, MATERIALS, AND METHODS
Blood Collection and Sample Processing
Venous blood was taken from consecutive consenting febrile (>37.5°C axilla) adults (>14 years of age) who were admitted to Queen Elizabeth Central Hospital (QECH) in Malawi for routine aerobic blood culture (5 mL, BacT/Alert, BioMerieux) [4] and whole blood RNA stabilization. Patients were recruited before antibiotic treatment. Subjects whose blood culture was positive for growth of nontyphoid Salmonella (NTS) were treated with ciprofloxacin (500 mg twice a day [bd]) for 10–14 days, and invited to return for convalescent venesection 4–6 weeks later. Consenting asymptomatic, afebrile HIV-positive antiretroviral-naive controls, without other chronic or active disease and who were matched for CD4 cell count, were recruited at the QECH antiretroviral clinic. Consenting healthy HIV-negative adult controls were recruited among hospital staff and unrelated hospital visitors. Blood (2.5 mL) was taken into PAXgene RNA tubes (PreAnalytiX, Qiagen/BD) and left at room temperature for 2 hours before being stored at −80°C. Full blood count (FBC; Beckman Coulter), thick-film microscopy for malaria parasites, HIV testing (Unigold, Trinity Biotech; and Determine, Inverness Medical), and CD4+ cell counts (Trucount, Becton Dickinson) were performed.
This study was approved by the Research Ethics Committee of the Liverpool School of Tropical Medicine, United Kingdom (ref 07.14) and by the Malawi College of Medicine Research Ethics Committee (ref P.03/07/501). All participating subjects gave written informed consent.
For ex vivo stimulation assays, 3 mL fresh blood from afebrile convalescent NTS cases and from controls was collected into sodium heparin (Vacutainer, Becton Dickinson). Blood was stimulated with either S. Typhimurium LPS (1 μg/mL, Sigma) or S. Typhimurium flagellin (1 μg/mL, Autogen Bioclear), or mock-stimulated with PBS for 4 hours at 37°C on a roller, then put in PAXgene RNA tubes, left at room temperature for 2 hours and stored at −80°C.
Microarrays and Determination of Differentially Expressed Genes
RNA was extracted (PaxGene Blood RNA Extraction kit, PreAnalytiX, BD/Qiagen) according to the manufacturer’s instructions. After quality checks, RNA was hybridized on the Illumina Human WG-6_V3 array (48,803 probes). Data were normalized (quantile algorithm for between-array normalization, and median of all samples baseline within-array correction), and analyzed using GeneSpring software (Agilent Technologies). Adjusted P values were calculated using the Benjamini and Hochberg (BH) method [14]. For each comparison, differentially expressed (DE) genes were defined as having a fold change in gene expression >±2 and a false discovery rate (FDR)–corrected P value of <0.05. Microarray data were deposited at ArrayExpress, EMBL-EBI (accession number E-TABM-856).
Pathway and Gene Ontology Analysis
Illumina probe sequence ID numbers were mapped to NCBI RefSeq IDs, and uploaded to InnateDB (www.innatedb.com) [15] with their associated expression data. InnateDB is a publicly available, manually curated molecular interaction and pathway database and computational analysis platform for all known human and mouse genes, designed to enable systems biology approaches to investigate all cellular signaling responses, including innate immunity. For each comparison, InnateDB pathway and gene ontology (GO) analyses were undertaken to determine which pathways or GO terms were statistically significantly associated with DE genes using the hypergeometric test. The BH FDR correction was applied to correct for multiple testing.
Molecular Interaction Network Analysis
InnateDB contains a uniquely comprehensive, manually annotated database of experimentally supported molecular interactions, allowing complex interaction networks to be constructed [8, 15]. Using the list of DE genes uploaded to InnateDB, different molecular interaction networks were constructed, consisting of only the interactions between DE genes first, followed by inclusion of all non-DE interacting partners of the DE genes. Networks were then submitted to Hub Objects Analyser (http://hub.iis.sinica.edu.tw/Hubba/) [16] to identify hub proteins (network nodes that are the most highly connected to other DE genes), bottleneck proteins (network nodes that are key connector proteins in a network, thus being the “shortest path”), and differentially expressed subnetworks.
RESULTS
Subjects and Controls
From 112 febrile acute adult admissions, we diagnosed 25 HIV-infected Malawian adults with acute iNTS disease (median age 30 years, 15 males), and 6 HIV-infected adults with “other” acute bacterial bloodstream infections (3 E. coli, 3 S. pneumoniae, median age 41 years, 2 males). Fourteen matched HIV-infected controls (median age 39 years, 5 males) and 18 healthy HIV-negative controls (median age 40 years, 9 males) were recruited. Five patients with iNTS died during hospital admission. Another 13 were followed up and studied in convalescence (median 42 days, range 7–154 days). None of the follow-up cases had a recrudescence of iNTS. No subjects or controls had started antiretroviral treatment. Clinical data for subjects are summarized in Table 1.
Table 1.
Summary of Subject and Control Blood Count Results
| Subject group | Hemoglobin g/dL median (95% CI) | Platelets x 106/L median (95% CI) | White cell counts x 106/L median (95% CI) | Neutrophils x 106/L median (95% CI) | Lymphocytes x 106/L median (95% CI) | Monocytes x 106/L median (95% CI) | CD4 cell count cells/μL median (range) |
| iNTS acute | 8.6 (6.1–12.2) P = .0027a | 106 (59–269) P = .086 | 3.7 (2.7–9.2) P = .16 | 2.8 (1.7–6.1) P = .27 | 1.0 (.7–2.1) P = .029 | 0.3 (.2–1.2) P = .18 | 147 (1–400) P = .061 |
| HIV+ controls | 13 (11.7–13.6) | 225 (152–282) | 5.4 (4.1–7.6) | 2.0 (1.2–3.2) | 2.3 (1.5–3.8) | 0.6 (.4–.8) | 204 (24–534) |
| HIV–controls | 14.1 (12.6–14.5) P = .0.02a | 188 (165–248) P = .59 | 5.3 (4.6–6.2) P = .95 | 2.6 (1.8–4.8) P = .27 | 2.1 (1.9–2.5) P = .91 | 0.4 (.4–.5) P = .31 | 814 (564–1148) P = .000a |
Abbreviations: CI, confidence interval.
P values are for comparison to the HIV-positive control group, by Mann--Whitney 2-sample rank sum analysis.
The mRNA Transcriptional Response Attributable to Acute iNTS Disease
Unsupervised Hierarchical Clustering of All Acute and Convalescent Events and Controls.
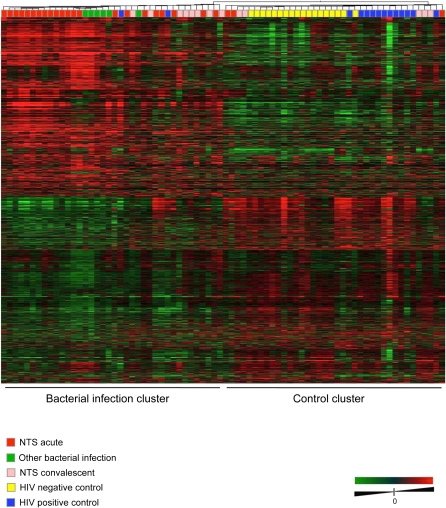
Unsupervised hierarchical clustering of all acute and convalescent iNTS and other infection samples, HIV-positive controls, and HIV-negative controls were conducted to produce a dendrogram based on similarities in gene expression profiles across all groups, and is shown in Figure 1. This analysis revealed strong groupings of iNTS-infected patients, other bacterially infected patients, HIV-positive controls, and HIV-negative controls. The 13 iNTS convalescent cases were distributed among the bacterial infection and control clusters according to length of follow up; the 8 convalescent cases in the bacterial infection cluster were sampled 1–2 months after acute infection (designated “FUearly”), while 4/5 convalescent cases in the control cluster (designated “FUlate”) were sampled after 3–5 months. The FUearly and FUlate groups were therefore analyzed separately. There was no clustering relationship to clinical parameters, such as inpatient death, hemoglobin, cell counts, age, multidrug antibiotic resistance of isolates, or reported use of antibiotics prior to admission.
Figure 1.
Heat map showing unsupervised hierarchical clustering according to the similarity of overall gene expression patterns for all acute and convalescent cases of iNTS, acute “other” bacterial bloodstream infections (E. coli and S. pneumoniae), and HIV-positive and HIV-negative controls.
Acute iNTS mRNA Profiles Lacked Evidence of a Coordinated Inflammatory or Immune Profile.
InnateDB analysis was used to examine the functional attributes of DE genes among patient groups [8, 15]. This included analysis of GO terms, signaling and metabolic pathways, and network attributes that were statistically significantly overrepresented among DE genes associated with the different patient conditions.
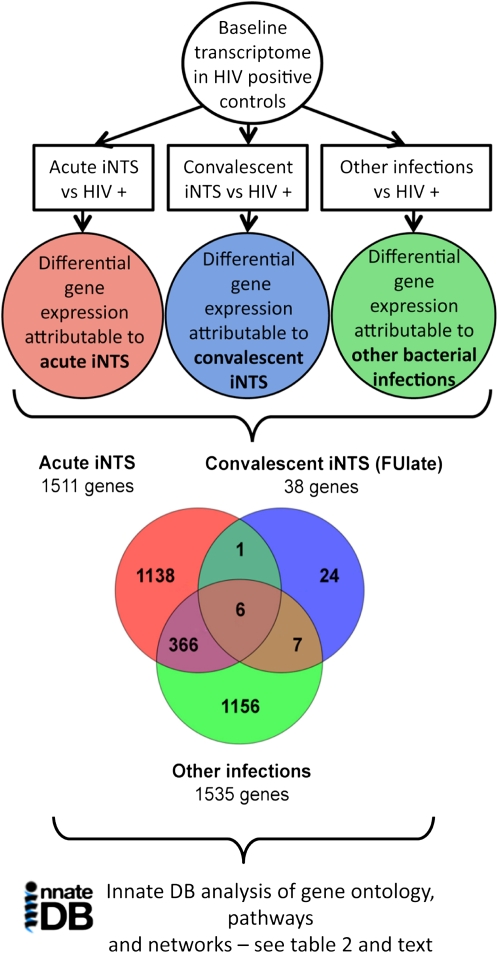
Figure 2 summarizes the analysis pipeline used to describe the differential gene expression that was attributable to invasive bacterial disease states on a preexisting background of HIV disease. There were 1511 DE genes attributable to acute iNTS and 1535 DE genes attributable to other acute bacterial invasive disease, but only 24% overlap (372 genes) between these groups. Thus, although the responses were of similar magnitude in terms of the number of DE genes, they differed markedly in the type of gene that was differentially regulated.
Figure 2.
The analysis pipeline used to describe the pathways, biological processes, and molecular interaction networks associated with differentially expressed (DE) genes in acute invasive nontyphoidal salmonella (iNTS) infected patients, convalescent iNTS patients, and other (E. coli or S. pneumoniae) infected patients. Comparison to matched HIV-positive control patients was used to dissect the differential gene expression attributable to different bacterial infections on a background of HIV infection. This is displayed as a Venn diagram in the lower panel. Only the ”late” convalescent group is shown here (see Table 2 and text for further description). The analysis of the different groups of DE genes, using InnateDB analysis, is shown in Table 2 and described in the text.
The DE genes attributable to iNTS disease, examined using InnateDB, exhibited surprisingly little evidence of a common substantive or coordinated innate immune or inflammatory response (Table 2). Although some individual genes relating to inflammatory processes were upregulated in their expression, classical GO and pathway analysis revealed few annotated biological processes or pathways related to immunity or inflammation as being significantly overrepresented. Although IFNγ mRNA was modestly upregulated, few downstream IFNγ-inducible genes showed upregulation. Thus, we were unable to identify during iNTS any coordinated host immune response beyond what is already expressed during underlying HIV infection. The only statistically significantly downregulated GO term in this comparison was “chemotaxis” (involving genes BDKRB1, CCR3, CMTM3, CXCL5, GPR44, GPR68, HRAS, IL8RB, and TTC7B).
Table 2.
GO and Pathway Analysis of Transcriptional Responses Attributable to Acute and Convalescent iNTS Disease and “Other” Infections
| Datasets compared | DE genes up/down regulated | Gene ontology terms in DE gene datasets (number of genes) | Pathways in DE gene datasets |
| Acute iNTS vs HIV+ (1511 DE genes) | 1075 upregulated | Cell cycle (55); DNA replication (25); DNA translation (18); DNA repair (26); Mitosis (39); Cell division (36) | Viral mRNA translation; Viral replication; Cell cycle; Nucleotide excision repair; Aurora B signalling |
| 436 downregulated | Chemotaxis (10) | None | |
| Acute “other” infections vs HIV+ (1535 DE genes) | 1017 upregulated | Innate immune response (51); Inflammatory response (24) | IL-1 mediated signalling; IL-4 mediated signalling; Complement and coagulation; Atypical NFκB |
| 518 downregulated | Transmembrane receptor activity (13); Immune response (26) | NK cell mediated cytotoxicity; TCR signalling in naive CD4 T-cells; 2nd messenger molecules in TCR signalling; Translocation of ZAP to immonological synapse; Antigen processing and presentation; Cytokine-cytokine receptor interaction | |
| FU early (1–2 months) vs HIV+ (418 DE genes) | 257 upregulated | Cell cycle; Cell division; Translation | Cell cycle; Viral mRNA translation; Influenza life cycle; Influenza infection |
| 161 downregulated | None | None | |
| FU late (3-5 months) vs HIV+ (38 DE genes) | 19 upregulated | None | None |
| 19 downregulated | None | None |
All cases are compared with stage-matched HIV-infected controls in order to demonstrate the transcriptional responses attributable to bacterial infection, against a background of HIV-infection (see Figure 2).
Abbreviations: DE, differentially expressed; FUearly, the 8 convalescent cases in the bacterial infection cluster that were sampled 1–2 months after acute infection; FUlate, the 4/5 convalescent cases in the control cluster that were sampled after 3–5 months; GO, gene ontology.
Acute iNTS Cases Showed Transcriptional Upregulation of Cell Cycle and DNA Function Genes.
The term “cell cycle” was significantly overrepresented in the dataset of genes upregulated in acute iNTS in both GO and pathway analyses (Table 2). Other statistically overrepresented GO terms included “DNA replication,” “DNA translation,” and “DNA repair.” Pathway analysis similarly identified viral mRNA translation (14 ribosomal proteins), and DNA replication as significantly upregulated pathways. This profile of gene expression may be indicative of cell cycle dysregulation and possibly increased HIV viral replication in iNTS patients. Indeed, there are known direct interactions between HIV viral proteins (Vpr, Tat, and Vif) and 4 of the cell cycle proteins that were upregulated at the gene expression level (CCNB1, CDC25C, YWHAG, and MDM2) [17–19].
Other Genes With Altered Transcription During Acute iNTS Disease.
Although the GO and pathway analyses did not identify specific immune-related pathways as being statistically significantly overrepresented in iNTS, there were several individual immune-relevant genes of interest that were significantly upregulated during iNTS cases. Notably, while REL (c-Rel), an alternative component of the NFκB complex, and TIRAP, an adaptor protein involved in TLR4-NFκB signaling [20] were upregulated, so were several important regulators or inhibitors of NFκB and inflammatory pathways, including NFKBIB (IκBβ, 3-fold), STAP2 (2.7-fold) [21, 22], TRAIP (2.4-fold) [23], inhibitory regulators of cytokine signaling SOCS4 and SOCS7, and PI3K (phosphatidyl-3–phosphate kinase), suggesting mechanisms for the downregulation of inflammatory responses.
A small number of individual immune-relevant genes were also significantly downregulated in their expression, including SIGIRR (a regulator of the TLR and IL-1R pathways), CSF1 (MCSF, a key IFNγ-inducible cytokine for macrophage activation and differentiation), IFNA13 (IFNα, a key cytokine for the control of HIV viral replication during acute infection [24, 25]), and TNFRSF10B and TNFRSF6B (TNF-superfamily receptors).
Overall however, although some individual immune-related genes were differentially expressed in iNTS, there was a notable lack of a coordinate immune/inflammatory response that would be commonly observed in other infections [26]
Molecular Interaction Network Analysis in Acute iNTS Cases.
Pathway and GO analyses rely on known biological pathways and processes to identify coordinated transcriptional profiles. Network analysis, however, uses known direct molecular interactions to model how differentially expressed genes and their encoded products may interact with each other; for example, through protein–protein interactions. Network analysis identifies key nodes (genes/proteins/RNAs) that are highly interconnected with other nodes and thus may be substantially responsible for driving the observed patterns of gene expression. Molecular interaction network analysis of DE genes during iNTS infection, using InnateDB [15] and Hubba [16], identified NFKBIB (IκBβ), a key NFκB inhibitor and negative regulator of inflammatory signaling [27], as a highly ranked hub protein in the network (3-fold upregulated), further supporting the suggestion that suppression of NFκB-mediated transcriptional responses may explain the lack of a coordinated inflammatory/immune response observed during iNTS disease. YWHAG, a 14-3-3γ protein that has potential interactions with HIV Vpr protein [18], was also a highly ranked hub (3-fold upregulated). Other hubs related to the cell cycle, in agreement with the pathway and GO analyses.
A Signature mRNA Profile Can Persist for up to 2 Months After iNTS.
The distribution and pattern of DE genes during early and late convalescence suggested that near-complete resolution eventually occurred, but took up to 3 months to achieve (Figure 2; Table 2). Late convalescent patients, at 3–5 months, had gene expression patterns that were nearly identical to those seen in matched HIV-infected controls. This provides a reassuring internal control to confirm that iNTS cases and HIV-infected controls were well matched for the biological effects of underlying HIV, and that any changes observed during acute iNTS were truly attributable to iNTS and not to poor matching of controls. Patients in the first 2 months of convalescence, however (Table 2), still had 418 DE genes differentially regulated compared with HIV-infected controls, and the pattern of expression revealed by GO and pathway analyses was very similar to that seen in acute disease, suggesting a slow return to baseline despite antibiotic treatment and clinical resolution of their presenting febrile illness.
Transcriptional Response Attributable to “Other” Invasive Bacterial Infections.
In this study, we also profiled the host response in 6 cases of “other” bacterial infections in febrile HIV-infected patients (3 E. coli, 3 S. pneumoniae) who presented consecutively alongside iNTS cases. The global expression profiles of these patients, based on similarities across all groups, showed that iNTS and other bacterial infections clustered together, to the exclusion of controls, and that “other” infections formed a subcluster (Figure 1); InnateDB GO and pathway analyses of DE genes in these other bacterial infections, compared with HIV-positive controls, revealed a transcriptional profile that was indicative of a pronounced inflammatory/immune response, in sharp contrast to observations described above for iNTS patients.
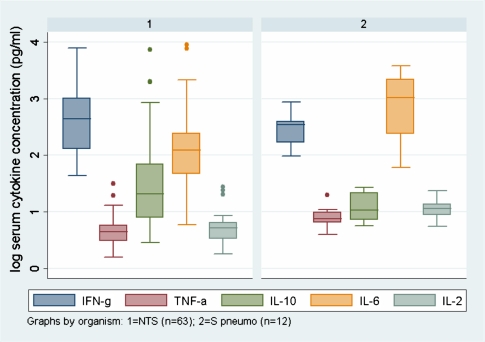
GO terms that were statistically significantly overrepresented among upregulated DE genes in “other” bacterial infections included “immune response” (51 genes) and “inflammatory response” (24 genes). Pathway analysis demonstrated that the IL-1 and IL-4 pathways were overrepresented, with IL1R1, IL1R2, and IL1RN upregulated 5-, 10- and 3-fold, respectively. Complement and coagulation, and the atypical NFκB pathway were also statistically overrepresented (Table 2). Network analysis identified that the top hub nodes were all different from those seen during iNTS disease, and included SRC (a tyrosine kinase), AKT1 (a protein kinase), and HSP90AA1 (a heat shock protein), all of which are involved in regulation of inflammatory responses. These “other” acute bacterial infections during HIV therefore caused a marked stereotypical and well-coordinated immune and inflammatory transcriptional pattern that was lacking during iNTS infection. The contrast of the host response in iNTS compared with “other” infections is supported by proinflammatory cytokine protein levels measured from consecutive patients with iNTS and invasive S pneumoniae disease (Figure 3), recruited using the same protocols as for this study as described [6].
Figure 3.
Box-and-whisker plots showing median and interquartile ranges of serum cytokine levels during acute first-event invasive bacterial disease caused by NTS (graph 1; n = 63) and S. pneumoniae (graph 2; n = 12), in consecutive, febrile HIV-positive adults admitted to QECH. Cytokines were measured using the Becton Dickinson Cytometric Bead Array according to manufacturer’s instructions. Serum levels of the proinflammatory cytokines TNFα (P < .0001), IL-6 (P = .0005), and IL-2 (P < .0001) were significantly lower among patients with iNTS compared with those with invasive S. pneumoniae disease, in keeping with the findings at transcriptional level. There was no significant difference in IFNγ (P = .45) or IL-10 (P = .16) levels (Mann-Whitney signed rank test, corrected).
Transcriptional Response Attributable to Underlying HIV Infection.
HIV-positive controls were also compared with HIV-negative controls to confirm that the expression profiles in HIV-positive controls were representative of known gene expression profiles [28]. There were 374 DE genes attributable to underlying HIV disease, and the GO terms “response to virus”, “immune response,” “innate immune response,” and “inflammatory response” were significant among upregulated DE genes. Forty-three percent of upregulated genes were IFN-inducible. The “RIG-I–like receptor signaling pathway” and “cytosolic DNA sensing pathway” were upregulated, both of which are involved in the detection of cytosolic viral nucleic acids. Network analysis of DE genes and their interacting partners identified a significant subnetwork involving the STAT1 and IRF7 transcription factors and many of the IFN-inducible genes, confirming the known central importance of IFN responses during HIV infection. These same underlying HIV-related transcriptional patterns were also clearly identified when iNTS cases themselves were compared with HIV-negative controls, again confirming adequate biological matching of iNTS cases with HIV-positive controls.
The Transcriptional mRNA Response During Ex Vivo Stimulation of Whole Blood
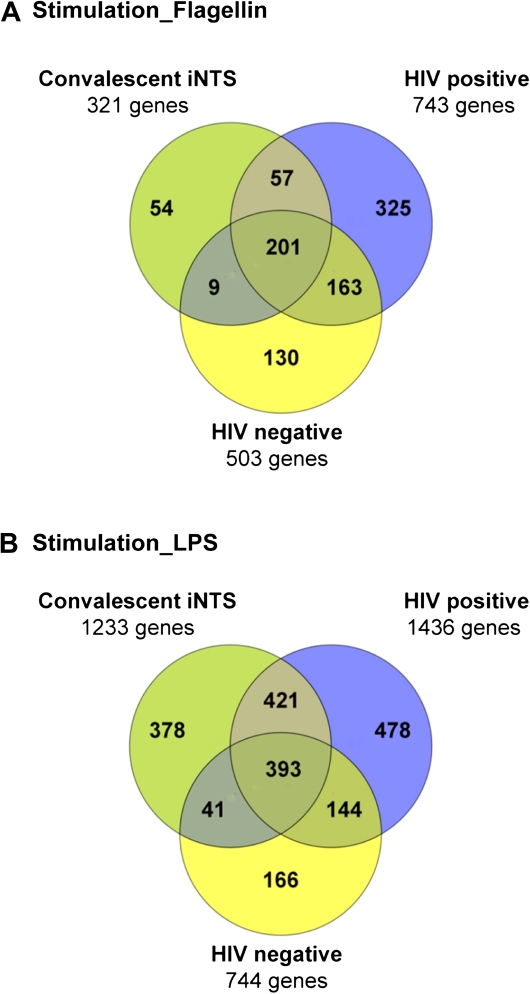
Whole blood from 3 subject groups, HIV-infected convalescent iNTS cases (n = 6), HIV-positive controls (n = 9), and HIV-negative controls (n = 11), were stimulated with either LPS or flagellin, or mock stimulated with PBS, to assess whether iNTS convalescent cases were capable of mounting a TLR-driven transcriptional response. On hierarchical clustering analysis, samples grouped strongly into stimulated and mock-stimulated clusters (Supplementary Figure 1). For both stimuli, the majority of DE genes were in common between at least 2 subject groups, suggesting a common response for all subject groups (Figure 4). There was a greater magnitude of overall response, as measured by numbers of DE genes, for LPS compared with flagellin stimulation of blood cells. GO and pathway analysis (Supplementary Table 1) confirmed that ex vivo stimulation of whole blood with either LPS or flagellin elicited a strong common inflammatory response pattern. Immune-related pathways such as the IKK-NFκB cascade, IL-1 pathway and the inflammasome, TNF pathway, apoptosis and TLR signaling were statistically overrepresented in all 3 subject groups for both stimuli compared with mock stimulation with PBS.
Figure 4.
Venn diagram showing the distribution of differentially expressed (DE) genes following ex vivo whole blood stimulation with (A) S. typhimurium flagellin or (B) S. Typhimurium LPS. All groups were compared with HIV-negative subjects. The description of the different groups of DE genes, analyzed using InnateDB, is shown in Supplementary Table 1 and described in the text.
Molecular interaction network analysis showed that for both stimulants, and in all subject groups, the 10 top hub (highly connected) nodes were the same—IKBKE, NFKB1, NFKB2, RELA, SRC, TRAF1, TRAF2, NFKBAI, MAPK1, and SMAD3—all of which are involved in signal transduction during immune responses, particularly through the NFκB pathway. This confirms that PBMC from convalescent iNTS cases were capable of mounting a highly stereotypical NFκB-mediated response, in keeping with the transcriptional responses reported in published studies [9–11]. This is in sharp contrast to our observation that the same patients did not mount such responses during iNTS disease, indicating that other factors, perhaps endotoxin tolerance-like responses [29], caused suppression or attenuation of typical immune responses.
DISCUSSION
In the present study, we used microarray analysis to describe the differential transcription of mRNA expression associated with iNTS in adults with HIV infection. It is remarkable that in the context of this life-threatening bloodstream infection, there was no evidence of a coordinated immune and proinflammatory response over and above the background transcriptional perturbations associated with underlying HIV. Specifically, there was an underrepresentation of the TLR-signaling and NFκB pathways that are prominent in previously reported studies of transcription during Salmonella infection, including the interactions of Salmonella with epithelial cells and macrophages in vitro [9, 10], in an infection model exploiting zebrafish [11], and in human invasive typhoid fever caused by S. Typhi in HIV-uninfected patients [12]. In addition, while network hub proteins following ex vivo stimulation were dominated by multiple components of the NFκB pathway, during iNTS disease itself, the NFκB inhibitor IκBβ was an upregulated hub protein, and several other negative regulators of innate immunity were also upregulated.
Although IFNγ mRNA was modestly upregulated during iNTS, there was a notable lack of measurable downstream effects on IFNγ-inducible genes. This is consistent with our previous observations during intracellular NTS infection of ex vivo primary macrophages from HIV-infected subjects, in which we found that the marked decline of cytokine responses in late HIV was not corrected by ex vivo priming with IFNγ [7]. An intrinsic host defect associated with HIV infection is, however, unlikely to fully explain these differences. In contrast to patients with iNTS, other bacterial bloodstream infections in HIV-infected adults caused a prototypical proinflammatory transcriptional response, which showed only 24% overlap with iNTS among differentially expressed genes. In addition, convalescent iNTS cases were able to mount NFκB responses comparable in magnitude and pattern to control groups following ex vivo stimulation with LPS or flagellin, suggesting that their capacity to respond to inflammatory stimuli was not simply “exhausted” by underlying HIV disease.
HIV infection is thought to dysregulate the NFκB pathway, which may be “hijacked” to activate HIV transcription [30]. Noursadeghi et al. [31] have demonstrated that HIV infection of macrophages may selectively attenuate elements of the NFκB cascade without compromising overall immune or inflammatory responses. Endotoxin tolerance responses have also been associated with many clinical syndromes, including sepsis [29]. We hypothesize that the effects of late HIV and iNTS coinfection may specifically interact to result in a more profound attenuation and unique lack of coordinated gene expression in NFκB-related proinflammatory and immune pathways.
We also found that iNTS infection in HIV-infected individuals enhanced the transcription of genes related to the cell cycle, including some known to specifically interact with HIV proteins [17–19]. Other upregulated pathways were associated with viral replication and cellular repair. This contrasts with our previous observations that cell cycle regulator genes are downregulated during in vitro Salmonella infection of macrophages in the absence of HIV [10]. Cell cycle dysregulation, with unbalanced proliferation and apoptosis of T cells during HIV infection, is a recognized contributor to HIV immunopathology [17], and antagonism of MDM2 has been explored as a therapeutic tool in HIV [27, 32]. Intracellular S. Typhimurium have been shown to reactivate latent HIV infection in chronically infected cells [33, 34]. We therefore postulate that the interaction between iNTS and HIV contributes to worsened HIV-related cell cycle dysfunction and possibly to heightened HIV viral replication. The persistence of this signature into convalescence may therefore compromise effective immune reconstitution once antiretroviral drugs are commenced after an episode of iNTS.
The use of whole blood is important because of its accessibility and potential for future diagnostics, but it is not possible to comment on the relative contribution of different cell types or tissues to the observed transcriptional profile. The characteristic transcriptional patterns we observed may provide useful biomarkers for diagnosis. The apparent unique interaction between HIV and NTS, both intracellular pathogens, therefore merits further clinical study to determine the impact of iNTS on the course of HIV disease, and further ex vivo study to understand the cellular basis and molecular mechanisms of these observations and to identify potential therapeutic targets for this neglected human disease.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://www.oxfordjournals.org/our_journals/jid/).
Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Acknowledgments
The authors thank the staff and patients of Queen Elizabeth Central Hospital and the University of Malawi College of Medicine for their cooperation and participation in this study. They also thank Prof Neil French and Prof Stephen Gordon for making shared study samples available for analysis (Figure 3).
Financial support. This work was supported by a Grand Challenges in Global Health Grant from the Foundation for the National Institutes of Health and Canadian Institutes for Health Research (ID:419 to B. B. F.). M. A. G. was supported by a research fellowship from the Wellcome Trust, UK. R. E. W. H. holds a Canada Research Chair, and F. S. L. B. is a Michael Smith Foundation for Health Research Senior Scholar. The Wellcome Trust Sanger Institute and the Malawi-Liverpool-Wellcome Trust (MLW) Major Overseas Programme are supported by the Wellcome Trust, UK.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Reddy EA, Shaw AV, Crump JA. Community-acquired bloodstream infections in Africa: a systematic review and meta-analysis. Lancet Infect Dis. 2010;10:417–32. doi: 10.1016/S1473-3099(10)70072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon MA, Walsh AL, Chaponda M, et al. Bacteraemia and mortality among adult medical admissions in Malawi—predominance of non-typhi salmonellae and Streptococcus pneumoniae. J Infect. 2001;42:44–9. doi: 10.1053/jinf.2000.0779. [DOI] [PubMed] [Google Scholar]
- 3.Gordon MA, Banda HT, Gondwe M, et al. Non-typhoidal salmonella bacteraemia among HIV-infected Malawian adults: high mortality and frequent recrudescence. AIDS. 2002;16:1641. doi: 10.1097/00002030-200208160-00009. [DOI] [PubMed] [Google Scholar]
- 4.Gordon MA, Graham SM, Walsh AL, et al. Epidemics of invasive Salmonella enterica serovar enteritidis and Salmonella enterica serovar typhimurium infection associated with multidrug resistance among adults and children in Malawi. Clin Infect Dis. 2008;46:963–9. doi: 10.1086/529146. [DOI] [PubMed] [Google Scholar]
- 5.Gordon MA. Salmonella infections in immunocompromised adults. J Infect. 2008;56:413–22. doi: 10.1016/j.jinf.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Gordon MA, Kankwatira AM, Mwafulirwa G, et al. Invasive non-typhoid salmonellae establish systemic intracellular infection in HIV-infected adults: an emerging disease pathogenesis. Clin Infect Dis. 2010;50:953–62. doi: 10.1086/651080. [DOI] [PubMed] [Google Scholar]
- 7.Gordon MA, Gordon SB, Musaya L, Zijlstra EE, Molyneux ME, Read RC. Primary macrophages from HIV-infected adults show dysregulated cytokine responses to Salmonella, but normal internalization and killing. AIDS. 2007;21:2399–408. doi: 10.1097/QAD.0b013e3282f25107. [DOI] [PubMed] [Google Scholar]
- 8.Gardy JL, Lynn DJ, Brinkman FS, Hancock RE. Enabling a systems biology approach to immunology: focus on innate immunity. Trends Immunol. 2009;30:249–62. doi: 10.1016/j.it.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Eckmann L, Smith JR, Housley MP, Dwinell MB, Kagnoff MF. Analysis by high density cDNA arrays of altered gene expression in human intestinal epithelial cells in response to infection with the invasive enteric bacteria Salmonella. J Biol Chem. 2000;275:14084–94. doi: 10.1074/jbc.275.19.14084. [DOI] [PubMed] [Google Scholar]
- 10.Rosenberger CM, Scott MG, Gold MR, Hancock RE, Finlay BB. Salmonella typhimurium infection and lipopolysaccharide stimulation induce similar changes in macrophage gene expression. J Immunol. 2000;164:5894–904. doi: 10.4049/jimmunol.164.11.5894. [DOI] [PubMed] [Google Scholar]
- 11.Stockhammer OW, Zakrzewska A, Hegedus Z, Spaink HP, Meijer AH. Transcriptome profiling and functional analyses of the zebrafish embryonic innate immune response to Salmonella infection. J Immunol. 2009;182:5641–53. doi: 10.4049/jimmunol.0900082. [DOI] [PubMed] [Google Scholar]
- 12.Thompson LJ, Dunstan SJ, Dolecek C, et al. Transcriptional response in the peripheral blood of patients infected with Salmonella enterica serovar typhi. Proc Natl Acad Sci USA. 2009;106:22433–8. doi: 10.1073/pnas.0912386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Detweiler CS, Cunanan DB, Falkow S. Host microarray analysis reveals a role for the Salmonella response regulator phoP in human macrophage cell death. Proc Natl Acad Sci USA. 2001;98:5850–5. doi: 10.1073/pnas.091110098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–8. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 15.Lynn DJ, Winsor GL, Chan C, et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol Syst Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin CY, Chin CH, Wu HH, Chen SH, Ho CW, Ko MT. Hubba: hub objects analyzer—a framework of interactome hubs identification for network biology. Nucleic Acids Res. 2008;36:W438–43. doi: 10.1093/nar/gkn257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galati D, Bocchino M. New insights on the perturbations of T cell cycle during HIV infection. Curr Med Chem. 2007;14:1920–4. doi: 10.2174/092986707781368559. [DOI] [PubMed] [Google Scholar]
- 18.Kino T, Gragerov A, Valentin A, et al. Vpr protein of human immunodeficiency virus type 1 binds to 14-3-3 proteins and facilitates complex formation with Cdc25C: implications for cell cycle arrest. J Virol. 2005;79:2780–7. doi: 10.1128/JVI.79.5.2780-2787.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izumi T, Takaori-Kondo A, Shirakawa K, et al. MDM2 is a novel E3 ligase for HIV-1 Vif. Retrovirology. 2009;6:1. doi: 10.1186/1742-4690-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verstak B, Nagpal K, Bottomley SP, Golenbock DT, Hertzog PJ, Mansell A. MyD88 adapter-like (Mal)/TIRAP interaction with TRAF6 is critical for TLR2- and TLR4-mediated NF-κB proinflammatory responses. J Biol Chem. 2009;284:24192–203. doi: 10.1074/jbc.M109.023044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeda O, Sekine Y, Yasui T, et al. STAP-2 negatively regulates both canonical and noncanonical NF-kappaB activation induced by Epstein-Barr virus-derived latent membrane protein 1. Mol Cell Biol. 2008;28:5027–42. doi: 10.1128/MCB.00194-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sekine Y, Yumioka T, Yamamoto T, et al. Modulation of TLR4 signaling by a novel adaptor protein signal-transducing adaptor protein-2 in macrophages. J Immunol. 2006;176:380–9. doi: 10.4049/jimmunol.176.1.380. [DOI] [PubMed] [Google Scholar]
- 23.Lee SY, Lee SY, Choi Y. TRAF-interacting protein (TRIP): a novel component of the tumor necrosis factor receptor (TNFR)- and CD30-TRAF signaling complexes that inhibits TRAF2-mediated NF-kappaB activation. J Exp Med. 1997;185:1275–85. doi: 10.1084/jem.185.7.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hosmalin A, Lebon P. Type I interferon production in HIV-infected patients. J Leukoc Biol. 2006;80:984–93. doi: 10.1189/jlb.0306154. [DOI] [PubMed] [Google Scholar]
- 25.Jacquelin B, Mayau V, Targat B, et al. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest. 2009;119:3544–55. doi: 10.1172/JCI40093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tegner J, Nilsson R, Bajic VB, Bjorkegren J, Ravasi T. Systems biology of innate immunity. Cell Immunol. 2006;244:105–9. doi: 10.1016/j.cellimm.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coley W, Kehn-Hall K, Van DR, Kashanchi F. Novel HIV-1 therapeutics through targeting altered host cell pathways. Expert Opin Biol Ther. 2009;9:1369–82. doi: 10.1517/14712590903257781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van den BR, Florence E, Vlieghe E, et al. Transcriptome analysis of monocyte-HIV interactions. Retrovirology. 2010;7:53. doi: 10.1186/1742-4690-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biswas SK, Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 2009;30:475–87. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 30.Hiscott J, Kwon H, Genin P. Hostile takeovers: viral appropriation of the NF-kappa B pathway. J Clin Invest. 2001;107:143–51. doi: 10.1172/JCI11918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noursadeghi M, Tsang J, Miller RF, et al. Genome-wide innate immune responses in HIV-1-infected macrophages are preserved despite attenuation of the NF-kappa B activation pathway. J Immunol. 2009;182:319–28. doi: 10.4049/jimmunol.182.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vassilev LT, Vu BT, Graves B, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 33.Andreana A, Gollapudi S, Gupta S. Salmonella typhimurium induces expression of P glycoprotein (multidrug resistance 1 gene product) in a promonocytic cell line chronically infected with human immunodeficiency virus type 1. J Infect Dis. 1994;169:760–5. doi: 10.1093/infdis/169.4.760. [DOI] [PubMed] [Google Scholar]
- 34.Gollapudi S, Gupta S, Thadepalli H. Salmonella typhimurium–induced reactivation of latent HIV-1 in promonocytic U1 cells is inhibited by trovafloxacin. Int J Mol Med. 2000;5:615–8. doi: 10.3892/ijmm.5.6.615. [DOI] [PubMed] [Google Scholar]