1. Overview
Over the last five decades, the overuse of antibiotics has led to widespread resistance in pathogenic bacteria. This resistance, combined with slow progress in designing new therapeutics has prompted a crisis in the treatment of many once easily curable diseases. Great effort has been put into developing new compounds that effectively target essential features of microbial life, but for the majority, only their growth inhibitory effects have been extensively studied. How low concentrations of antimicrobials affect bacterial survival and most importantly, whether or not the molecules might have other functions at sub-inhibitory concentrations (SIC) are outstanding questions. Pioneering studies on the global transcriptome response of many important human pathogens to SICs of antibiotics have demonstrated that these molecules can affect the expression of genes related to virulence, colonization, motility, stress response and/or biofilm formation. Further research has shown that some of these effects result from interference with bacterial cell-cell communication. This observation has prompted the idea that antibiotics might actually act as signal molecules in natural environments, facilitating intra- or interspecies interactions within microbial communities. In this review, we highlight examples of the effect of diverse clinically relevant antimicrobials and other natural products on bacterial developmental programs, with an emphasis on biofilm formation. How this knowledge can be used to improve the current use of antibiotics and how new therapies that target alternative microbial processes might be developed will also be discussed.
2. Introduction
Antibiotics have been extensively used in the treatment of infectious diseases. The lethality of these compounds has been exploited in clinical and laboratory approaches and their specific targets in bacterial physiology elucidated. However, along with this development of drugs has come the problem of increasing resistance in microbes, resulting in dramatically reduced therapeutic effectiveness 1. Pathogenic microbes have rapidly evolved efficient mechanisms of resistance, including increased efflux, enzymatic inactivation, target modification, or biofilm formation 2.
The concentrations of these molecules required to achieve an antimicrobial effect are likely extremely high compared to the concentrations in which these molecules can be found in natural environments. While we know their effect at lethal concentrations, the activities of these molecules at concentrations below the inhibitory limit needs deeper investigation 3. The findings of the nineteenth-century pharmacologist Hugo Schulz, who noted that certain disinfectants could have stimulatory effect on yeast growth at low concentrations, could be considered the first evidence that the action of an antimicrobial can cause a differential response depending on the concentration. His observation was the first example of what it would be later called hormesis. This term was coined by Chester Southam and John Ehrlich in the mid-1920s and it is used to refer to the ability of certain molecules to induce diverse responses depending on the concentration used 3. The vast amount of information related to the lethal concentration of antibiotics, targets, or side effects, contrasts dramatically with the relatively few studies focused on their effect at concentrations below the MIC (minimal inhibitory concentration). As pointed by Davies, only a small fraction of natural products that have antimicrobial activity have been extensively studied, and their role in natural settings is poorly understood. Thus it is possible that many of these molecules formerly considered antibiotics might have a different function in nature. It is now believed that many of these compounds might act as signaling molecules that modulate gene expression in microbial populations, or physiological functions such as motility, pigmentation, and production of metabolites, and thus facilitate inter- and intra-species communication 4.
This fundamental lack of understanding may be rooted in our conception of the microbial world as single and separated species, as they are usually studied under laboratory conditions. However, in nature, each niche is complex, and to different extents, variable in microbial community composition 5,6–8. Therefore it is conceivable that molecules fluctuate in concentration and diversity, thus facilitating communication among species 9. Many interesting lines of research are currently focused on understanding how some antibiotics may affect, either positively or negatively, cell-cell communication systems, and the physiological responses that are affected as a result. In some cases, natural products can influence the ability of bacteria to transition from a planktonic state to complex multicellular aggregates attached to surfaces known as biofilms. Cells in bioflms are encased in an extracellular matrix which can serve as a barrier for antibiotics 10. One example is the biofilm produced by the gram-negative pathogen, Pseudomonas aeruginosa, which is resistant to antibiotics produced by gram-positive competitors 11. Studies on many model microorganisms from the genera Bacillus, Streptomyces, and Pseudomonas are shedding new light on the fascinating world of cell signaling and communication in microbial world 5,12,13.
We will discuss in this review several examples of the dual functions of some well-known antibiotics, including those that when used at sub-inhibitory concentrations (SIC) promote an interesting response in bacterial populations and the ecological implications of such varied responses. Also, the role of other naturally synthesized antibiotics will be discussed in the context of cell communication in natural environments, including one of the most exploited environmental niches for antibiotic discovery, the soil.
3. Antibiotics
The discovery of streptomycin in 1944 stimulated the exploration of new venues to find new metabolites with antimicrobial activity 14,15. The nature and structure of these molecules are remarkably varied, and they can be synthesized by humans, or produced by microorganisms, such as fungi or bacteria. New chemical approaches are contributing enormously to the development of a new generation of antibiotics termed ‘semi-synthetic.’ These drugs are naturally occurring antibiotics that are subsequently chemically modified to overcome deficiencies in terms of efficacy, stability, physiological target, etc., found in the originals 16–18. Moreover, studies in diverse systems have contributed to a better understanding of the pathways involved in natural product biosynthesis, and the conditions and signals that trigger their production 19.
3.1. Antibiotics produced in secondary metabolism
As mentioned above, soil has been the most explored environment in the search for natural products with antimicrobial activity. The variety of microorganisms living in soil is seemingly endless, with streptomycetes, bacilli, and myxococci, among the best studied as antimicrobial producers 19,20. Despite differences in activity or structure, most antibiotics are commonly produced as secondary metabolites, i.e. they are produced by secondary pathways dispensable in many growth conditions 17. When bacterial are grown under experimental conditions, the dynamics of cell growth clearly show different stages indicative of metabolic activity. When nutrients are abundant, machinery for production of nucleic acid, proteins and other macromolecules necessary for sustained growth is fully active and allows for exponential growth of the population. However, when nutrients become limiting cells arrest their growth and enter stationary phase. Nutrient limitation results in the turnover of enzymes, and often the opening of metabolic routes leading to production of secondary metabolites 17. The role of these antibiotics may range from killing or inhibiting competitors to controlling cell growth or modulating colony morphology.
3.2. Regulation of antibiotic production: Quorum sensing
The production of antibiotics is under strict genetic control. From studies on diverse bacterial systems, we know of the existence of complex signaling routes that allow communication between cells, including within and between species 21–23. Such signaling necessitates a transduction system that integrates this external information, resulting in the induction of antibiotic production, at the right time, amount, and by the right percentage or subpopulation of cells 23 (Figure 1). Studies of diverse antibiotics have led to a foundational understanding of the regulation of antibiotic production, signal molecules and gene clusters involved, and how these circuits are integrated into global regulatory pathways 24–27.
Figure 1.
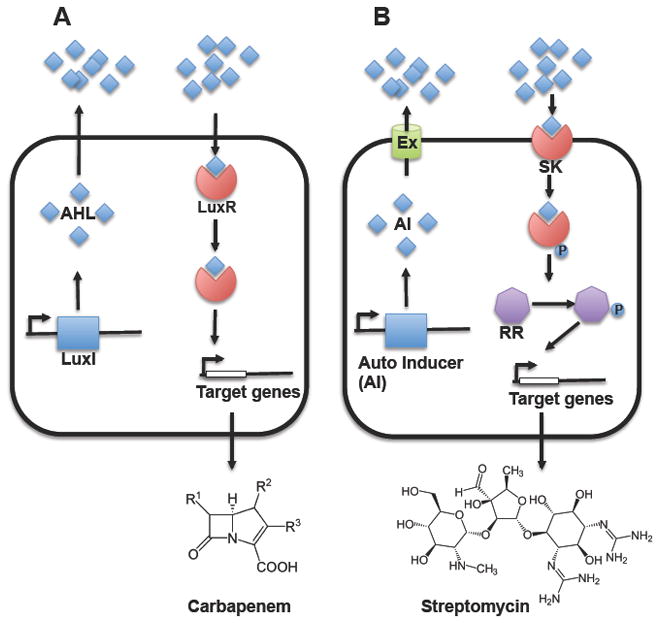
Quorum sensing communication systems in bacteria. A) In gram-negative bacteria, QS is regulated by the generally called LuxI-LuxR system. The signal molecule homoserine lactone (HL) is synthesized by LuxI. When the HL reaches a threshold, it binds to the transcription regulator LuxR that regulates the expression of the target genes. B) In gram-positive bacteria, the regulation is driven by a two-component signal-transduction system. The signal molecules are usually small peptides either post-translationally modified or not, which are secreted through ABC exporter system (Ex). The molecule secreted binds to the receptor, a sensor kinase (SK) that autophosphorilate a histidine residue. Then, the phosphate group is transferred to the response regulator (RR) protein that may be a transcription factor therefore regulating the expression of the target genes. Production of antibiotics are under control of QS system, for example C) the β-lactam antibiotic carbapenem by the gram negative pathogen Erwinia carotovora, and D) the aminoglycoside antibiotic streptomycin by Streptomyces species.
As mentioned above, production of antibiotics commonly occurs when cells reach stationary phase, and often, relatively high cell densities are required for robust antibiotic production. Bacteria control this process by literally ‘counting’ cell members in the culture. Bacteria count population members in a quorum-sensing process by releasing self-produced signaling molecules or autoinducers (AI) that accumulate externally. When the autoinducing molecules concentrate in the medium above a certain threshold, they trigger the quorum-sensing response. The nature of the molecules defines specificity for the receptor, and ensures the proper recognition and the expected genetic answer in the population 13. The word “quorum” is a legal term strictly denoting the minimum number of members from a board that is required to make a decision. Microbiologists however, use the meaning of this term to exemplify the decision-making process used by bacteria to coordinate their gene expression according to the density of their population. One rationale for limiting antibiotic production to situations in which a ‘quorum’ has been reached is that it might ensure that the antibiotic will be produced in an amount sufficient to impact the surrounding microbial community.
In gram-negative bacteria, acyl homoserine lactones (AHLs) are an important class of autoinducers, which contain structural differences in their aliphatic tails (Figure 1A). In P. aeruginosa two quorum-system AHLs are known, las and rhl, each of which posses their own AHL synthase (LasI and RhlI). Each AHL signal binds the correct cognate receptor, LasR or RhlR, and triggers the expression of a set of genes involved in different physiological process such as virulence or biofilm formation 28–30. As exemplified in Figure 1a, in Erwinia carotovora this signaling system regulates the expression of genes involved in the production of the β-lactam antibiotic carbapenem 31,32.
In gram-positive bacteria, quorum-sensing molecules are usually modified small peptides or amino acids, which are sensed by membrane-associated kinases (Figure 1B). In response, sensor kinases phosphorylate a cognate response regulator that finally turns on the expression of specific set of genes. Examples of signal molecules involved in cell-cell communication are the autoinducer peptide (AIP) in Staphylococcus aureus, ComX or CSF in Bacillus subtilis and CSP in Streptococcus pneumonie (reviewed in 33), and they control expression of genes involved in various processes including virulence, competence or sporulation. In the example of the pathogenic bacterium S. aureus, AIP is derived from the product of the agrD gene as a pre-peptide that is later processed into the mature form as a cyclic peptide with a thiolactone ring and exported from the cell. The mature form is externally sensed by a receptor that activates a regulator that finally regulates genes positively or negatively 34,35. An exception to this rule are the fatty acid derived γ-butyrolactones, the small molecules that regulate the induction of a large number of antibiotics and sometimes also differentiation into aerial hyphae in Streptomyces species 36. In Streptomyces griseus, the A-factor (γ-butyrolactone) is bound by the receptor, ArpA, which releases the repression of specific promoters involved, for example, in production of streptomycin (Figure 1B), and the formation of aerial hyphae 37. Similar γ-butyrolactone systems have been found to control the production of many natural products in an array of streptomycetes, including virginiamycin in S. virginiae, showdomycin in S. lavendulae, and actinorhodin and prodiginines in S. coelicolor 38.
Besides these QS mechanisms another fascinating communication system is the AI-2/Lux system, originally found in Vibrio harveyi, and now known to be a QS system widely distributed among both gram-positive and gram-negative bacteria. This system is proposed to be involved in many processes including virulence and biofilm formation. A notable feature of this system is that due to the prevalence of AI-2 signal [derived signals of dihydroxy-pentanedione (DPD)] receptors in a wide array of bacterial species, this molecule may facilitate both inter- and intra-species communication 39. There are examples in which AHLs signaling between two different species leads to a cooperative or competitive interaction. The most elegant examples of the significance of such communication may be found in multispecies biofilms formed by Pseudomonas aeruginosa and Bulkholderia cepacia. Both organisms coexist in lungs of patients with cystic fibrosis. It is proposed that sets of genes are expressed such that P. aerugionsa colonizes first and subsequently communicates with B. cepacia to facilitate its establishment in the host 40.
4. Bacterial biofilms
Cell signaling and cell density is especially important in the formation of tightly packed, spatially organized biofilms (Figure 2). These bacterial communities are often attached to surfaces and encased in an extracellular matrix normally produced by the bacteria themselves. Biofilm formation occurs in response to environmental cues, i.e. starvation, chemicals or antimicrobial insults. Though the signals and specific genes involved in the formation of biofilms vary among species, certain commonalities can be found 12. The formation of biofilms involves the transition of cells from a planktonic lifestyle to that of a sessile community attached to a surface, and the formation of an extracellular matrix that helps to form the characteristic architecture of biofilms and contributes to the movement of nutrients, oxygen, and other environmental signals 41.
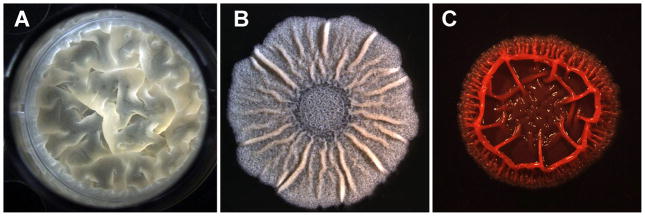
Figure 2.
Bacteria develop structurally complex communities called biofilms. Formation of biofilms responds to multiple factors and can be regulated by quorum sensing. These bacterial communities are formed by cells embedded within an extracellular matrix composed of proteins, exopolysaccharides and other molecules. This extracellular matrix confers stability and protect the cells from external aggressions as antibiotics. Biofilms of Bacillus subtilis grown in vitro develop: A) Top view of a pellicle in the interphase liquid-air in standing liquid culture and B) colony morphology in agarized medium and both characterized for the formation of typical wrinkles. C) A biofilm of Pseudomonas aeruginosa growth in agarized medium supplement with Congo Red appears as wrinkly colonies strongly stained red (Picture courtesy of Dr Liraz Chai).
Entry into stationary phase in response to nutrient limitation can trigger a cascade of signals, as explained in the previous section for QS system, that activates the expression of genes involved in biofilm formation 12,29,42. Some of these genes encode essential components of the extracellular matrix that provides stability and robustness to the biofilm. With all of this knowledge, it has been possible to develop in vitro biofilm assays that can identify mutants with no ability to build such communities. Two examples of in vitro bacterial biofilms are shown in Figure 2. The gram-positive bacterium B. subtilis forms pellicles in standing liquid cultures (Figure 2A) and colonies in agarized medium (Figure 2B) with typical features as wrinkles or aerial projections. In the case of the gram-negative Pseudomonas aeruginosa, biofilms can be visualized as wrinkly colonies that stain red in the presence of the dye Congo-Red (Figure 2C). Such ability is due to the production of the exopolysaccharide alginate, a component of the extracellular matrix. The extracellular matrix is not only necessary to build the backbone of robust biofilms it also affects the diffusion of molecules and becomes, as it will be presented later, a key element involved in the reduction of antibiotic efficiency.
Biofilms are widely distributed, and their effect may be beneficial or detrimental depending on the context. In clinical environments, biofilms of pathogenic bacteria represent a serious problem, where they serve as nidi of infection, and make eradication from clinical devices difficult due to increased antibiotic resistance 41,43–47. The resistance of biofilms to antibiotics is acquired by diverse mechanisms including the physico-chemical barrier provided by exopolysaccharides and other matrix components, and the degradation of the antimicrobial and/or development of resistant cell lines 11,48–50. It is known that efficacy of antibiotics depends on the dose, timing and mode of administration. Thus, some bacterial cells may only encounter SICs of the antibiotic used, which may induce biofilm formation, and ultimately result in increased resistance. Studies on biofilms, have shed light on bacterial physiology, gene regulation, and life cycle, and yielded information that is ultimately contributing to regulation of biofilms in a clinical context 51.
5. Antibiotics at sub-inhibitory concentrations
Taking in mind the original idea of hormesis 3, an increasing number of studies have used microarray technologies and libraries of transcriptional fusions to analyze the effect of sub-inhibitory concentrations of antibiotics on the physiology of target microbes 52–57. These studies have demonstrated changes in expression of genes involved in key biological process including transcription, translation of proteins, transport of exoproteins, general stress response, peptidoglycan synthesis, exopolysaccharide production, virulence, quorum-sensing, and biofilm formation (for a deeper consideration, see reviews 10). Many molecules have a pleiotropic effect, i.e. they affect multiple biological processes, or even induce variable responses depending on the bacterial species. Therefore, some antibiotics likely have multiple targets and, depending on the concentration used, bind preferentially to one or the other.
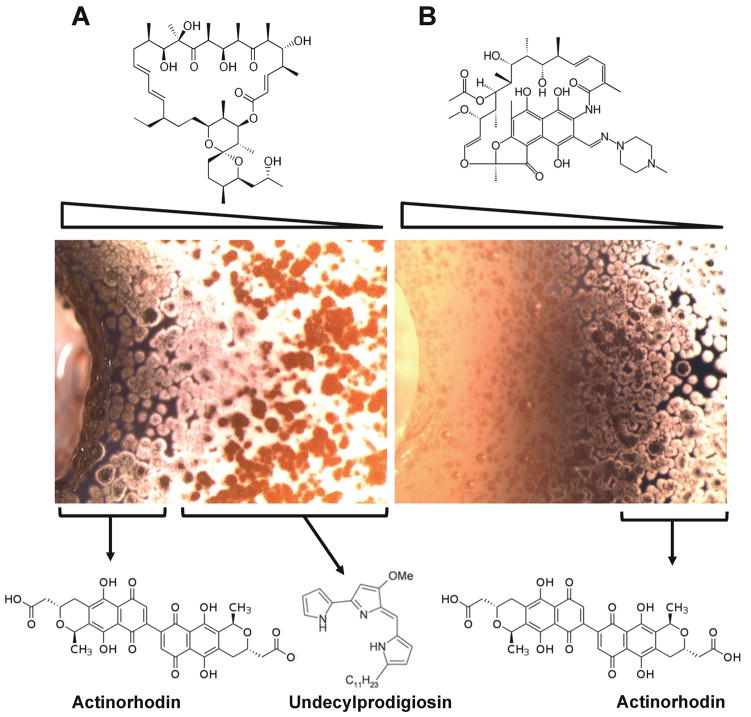
One example of how different concentrations of antibiotics may affect bacterial developmental process can be easily seen in the simplest agar diffusion tests (Figure 3). In this example, two antibiotics, Rifampicin (Figure 3A), a transcription inhibitor and Oligomycin A (Figure 3B), a mitochondrial ATP synthase inhibitor were tested against the gram positive bacterium Streptomyces coelicolor. This simple experiment illustrates how the diffusion gradient of each antibiotic induces differential phenotypes, for example, the acceleration or inhibition of aerial hyphae development, or the stimulation or inhibition of actinorhodin and undecylprodigiosin synthesis.
Figure 3.
Effect of rifampicin and oligomycin on development of S. coelicolor. Each well (edge visible at left) contains 60μl of a 20μg/ml solution of each drug. A) Rifampicin, a transcription inhibitor, causes a range of phenotypes across a diffusion gradient. This is visible as accelerated aerial hyphae and actinorhodin production near the well. B) The mitochondrial ATP synthase inhibitor oligomycin A causes a zone of growth inhibition, followed by an area of altered secondary metabolite production and aerial hyphe development. (Microscopy: Matt Traxler, Oligomycin A purified by Gavin Carr)
The observation that different antibiotics can induce similar bacterial responses is well illustrated by the β-lactams, a group of antibiotics that inhibit cell wall synthesis, and aminoglycosides, inhibitors of protein synthesis. Members of both groups have been demonstrated to induce the same response, formation of a biofilm, at sub-inhibitory concentrations 58,59. Indeed, a preliminary theoretical analysis showed that the similar response triggered by azithromycin (AZM) and other antibiotics like ciproflaxin and ceftazidine relied on AZM binding to specific receptors such as LasR, which, as stated above, is involved in cell-cell communication in P. aeruginosa 60. Some of these antibiotics are naturally produced by bacteria, and others are chemically modified. Therefore, structural similarities among synthetic and natural small molecules may contribute to similarities in their mode of action 3.
6. Clinically relevant antibiotics
It is intriguing that structurally divergent antibiotics with different modes of action can provoke similar responses in a bacterial population, i.e. biofilm formation. This observation exemplifies how a single behavioral response may prove advantageous in many stressful situations, including hostile interactions with microbes of diverse phylogeny. We have also seen how clinically useful antibiotics, when used at SIC, can provoke a global transcriptional response. The action of some of the most relevant clinical antibiotics has been studied in detail against important human pathogens, with the goal of deciphering the basis for antibiotic resistance and ultimately developing better chemotherapeutic programs. In the next section we consider some major classes of clinically used antibiotics and physiological responses they trigger in important human pathogens.
6.1. Aminoglycosides
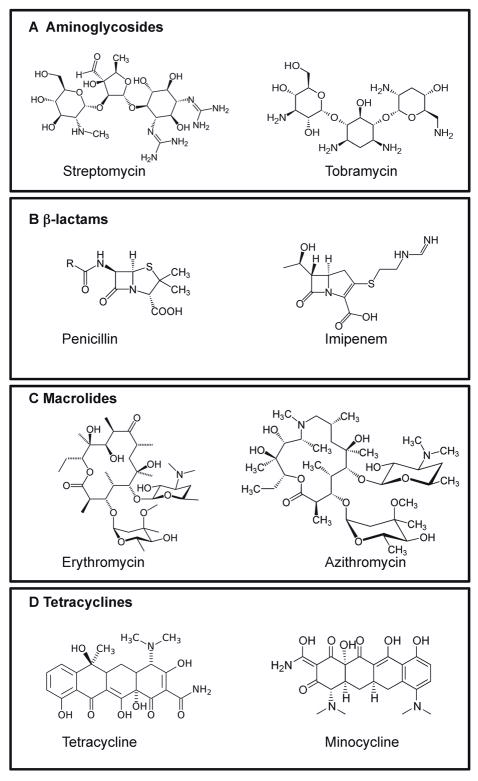
The starting point of this group of antibiotics can be dated to the mid-twentieth century with the discovery of streptomycin (Figure 4A). Later, many more compounds of this family were discovered, among them, kanamacin and tobramycin (Figure 4A). Their antibiotic activity is based on the ability to target the 30S ribosomal subunit, resulting in inhibition of protein synthesis 61. Tobramycin is produced by the soil bacterium Streptomyces tenebrarius, and it is used for treatment of chronic bacterial infections such as P. aeruginosa 62. The fact that both microorganisms coexist together in soil prompts an interesting hypothesis; that the development of resistance in P. aeruginosa to tobramicin would be an adaptive response of P. aeruginosa to the presence of this antibiotic in its natural environment and thus, the treatment of patients with tobramycin may ultimately fall short of curing infections caused by P. aeruginosa.
Figure 4.
Examples of clinically relevant antibiotics used for treatment of important human diseases. A) Aminoglycosides are inhibitors of protein synthesis by targeting the ribosomal subunit 30S, B) β-lactams target the bacterial peptido-glycan, C) Macrolides target the ribosomal subunit 50S and block protein synthesis and D) Tetracyclines block the entrance of t-RNAs in bacterial ribosomes.
This hypothesis led Hoffman and collaborators to investigate the effect of sub-lethal concentrations of tobramycin on P. aeruginosa and E. coli biofilm formation 58. They found that concentrations as low as 0.3 times the MIC induced biofilm formation to equivalent levels of a mutant with enhanced ability to form biofilms. Interestingly, this response was not related to an increase in the production of alginate, a major component of the extracellular matrix of P. aeruginosa biofilms, but rather it triggers an increase in the number of cells within the biofilm community. This response appears to be mediated by the arr gene (aminoglycoside response regulator), which encodes an inner membrane protein with an EAL domain (commonly found in phosphodiestherases involved in c-di-GMP degradation). In the model proposed, this antibiotic induces the EAL activity of the Arr protein. The mechanism by which tobramycin induces EAL activity is yet to be determined, but it is known that this induction causes a reduction in the levels of c-di-GMP. The ultimate consequence is the increase of biofilm formation. It is known that c-di-GMP regulates a myriad of genes involved in diverse physiological process, including motility and/or production of pili, both of which participate in the formation of biofilms. Thus, while the exact mechanism by which this antibiotic induces biofilm is unknown, it is clearly more complex than originally expected. More recently, a study with environmental isolates of P. aeruginosa has demonstrated that tobramycin may inhibit the RhlI/R system by reducing the production of the QS signal C4-HSL 62. In this case, tobramycin interacts with the 16S ribosomal RNA, blocking the peptide exit channel similarly to azithromycin, a macrolide antibiotic also used in the treatment of chronic infections. Surprisingly, this effect was not due to a decrease of rhlI transcription and required tobramycin-ribosome interaction.
These interesting observations highlight the fact that bacteria of the same species have diversified their genetic repertoire to respond efficiently to certain molecules, depending on the environment they occupy. These findings suggest the need for a better understanding of the signal transduction systems and physiological pathways involved in antimicrobial resistance.
6.2. β-lactams
β-lactams, are a group of antibiotics that target cell wall biosynthesis by blocking the transpeptidation reaction that enables the covalent cross-linking of peptides within strands of peptidoglycan. Within this family are found the penicillins (Figure 4B) and cephalosporins, which are produced by Penicillium chrysogenum and Acromonium chrysogenum respectively. Another β-lactam, is thienamycin, produced by Streptomyces flavogriseus. The clinical drug imipinem (Figure 4B), a variant produced from thienamycin, was designed to improve the features of the naturally occurring carbapanem backbone, and is used in the treatment of lung infections. However, there is great concern related to the stability of these antibiotics and development of resistance due to the expression of β-lactamases by some pathogenic bacteria. As an attempt to understand the molecular basis of the development of β-lactam resistance in the pathogen P. aeruginosa, a global transcriptome analysis was done with a SIC of imipinem 59. In this study, it was shown that, as expected, imipinem induced the expression of the gene ampC, which encodes a β-lactamase, and this correlated with antibiotic resistance. In addition, an increase in the expression of genes involved in the production of alginate was also observed. This resulted in increased production of extracellular matrix and thicker, more robust biofilms were produced. This is a particularly relevant observation since alginate has affinity for aminoglycoside antibiotics such as tobramycin. Thus an increase in alginate production likely reduces penetration of the antibiotics and enhances antibiotic resistance 11,63.
Recent experiments in the gram-positive bacterium Streptococcus pneumonie, have shown that exposure to penicillin at 50% of the minimal inhibitory concentration (MIC) also stimulates the formation of biofilms. Though the mechanism is unclear, it seems that this enhancement in biofilm formation is mediated by the up-regulation of luxS expression 54. Recently, Anmed and collaborators showed that sub-MICs of penicillin and other two structurally different drugs, ciprofloxacin (a fluoroquinolone), and tetracycline (a translation inhibitor) also induced the formation of biofilms in another gram-positive pathogen, Staphylococcus intermedieus. Even though the mode of action remains unclear, this induction seems to work through the LuxS/AI-2 signaling system 64.
6.3. Macrolides
The macrolides are a group of antibiotics that exert their bactericidal activity by targeting the 50S subunit of the ribosome and blocking polypeptide exit channel, thus inhibiting protein synthesis. Important antibiotics belonging to this group are erythromycin, produced by Saccharopolyspora erythraea, tylosin, produced by Streptomyces fradiae, and the semisynthetic drugs clarithromycin and azithromycin (Figure 4C) 65.
As discussed previously, some antibiotics may interfere with cell-cell communication systems and provoke a decrease in biofilm formation. While inhibiting biofilm production might seem like a desired effect, it may have the unintended consequence of enhancing the virulence of the pathogen, a consideration especially relevant in the treatment of chronic infections. This has been observed for the erythromycin derivative antibiotic azithromycin (AZM). At sub-inhibitory concentrations, this molecule inhibits the enzymatic activity of guanosine diphosphomannose dehydrogenase in the alginate biosynthetic pathway of mucoid P. aeruginosa strains. It has been proposed however, that the mode of action for AZM might be inhibition of cell-cell communication by interfering with the QS signaling system 66. In a recent study, the transcriptional profile of P. aeruginosa in reponse to a SIC of AZM demonstrated the decreased expression of raphnolipids and the adhesin protein LecA, both of which are involved in biofilm formation, as might be expected from the interference with the QS system 60,67. However, an increase in the level of expression of type three secretion system (T3S) genes was also observed. T3S is used by bacterial pathogens to deliver effectors into the cytoplasm of the host cell, and therefore plays a direct role in bacterial virulence and cytotoxicity 68. In a separate study, it was observed that treatment with AZM and the related macrolides, erythromycin and calrithromycin, resulted in increased mortality of mice after artificial infection with P. aeruginosa 69. Thus, the timing of antibiotic administration could have relevance for disease treatment, i.e. a variable response might be achieved in prophylactic vs. curative regimes. Such findings suggest that more research is required to better characterize the impact of small molecules on bacterial communication systems and virulence and the relevance of such changes from evolutionary and ecological perspectives.
6.4. Tetracyclines
Tetracyclines are a family of antibiotics that were discovered in the mid-twentieth century and include chlortetracycline, and tetracycline both produced by Streptomyces aureofaciens, or oxytetracycline isolated from S. rimosus, and the semi-synthetic drugs doxycycline and minocycline (Figure 4D). These antibiotics target the 30S ribosomal subunit by blocking the entry of aminoacyl-tRNAs to the A site.
As described previously for AZM, studies of transcriptional profiling of P. aeruginosa after exposure of SIC of tetracycline, showed an increase of the expression of genes of the T3S and in consequence a higher virulence of P. aerugionsa 70. Similarly to what has been described for penicillin, sub-inhibitory concentrations of tetracycline or quinupristin-dalfopristin have been observed to induce formation of biofilms. Cultures of Streptococci treated with these compounds at SIC enhanced the expression of the genes responsible for exopolysaccharide production 11-fold 71.
7. Signaling by natural small-molecules
As we have pointed out before, the majority of antibiotics used in treatment of human infections are based on naturally produced molecules. Problems of bacterial resistance, and stability or shelf life has driven development of variants that overcome these limitations, thus improving the efficiency of disease management. These molecules must be made in the natural habitat that the producers occupy. Therefore, the varied responses to sub-inhibitory concentrations of natural products (i.e. stimulation of biofilm formation, or increased virulence) observed in vitro should be considered in an ecological context. In addition to these antibiotics, one can imagine a universe of small molecules that might participate in diverse routes of communication, thus generating a wide array of responses.
7.1. Phenazines
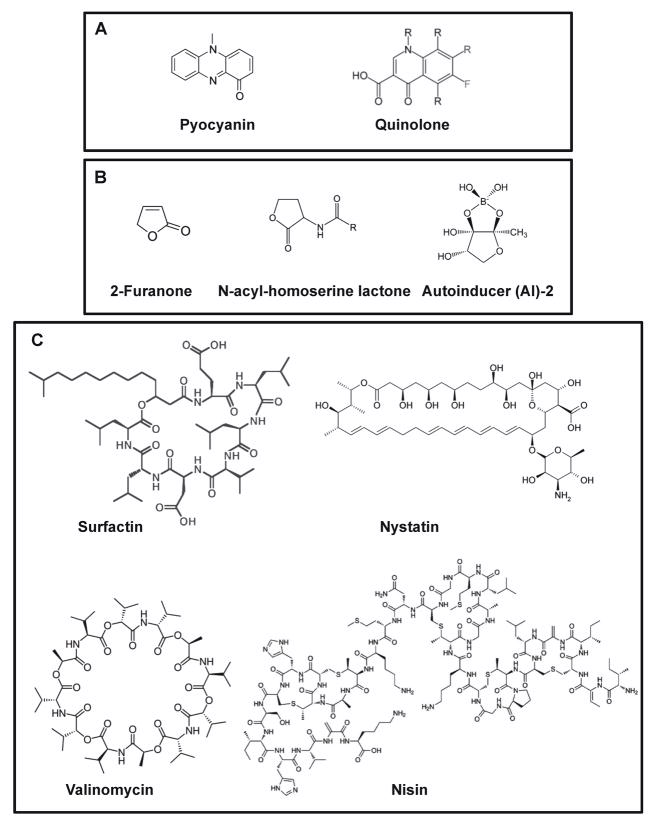
P. aeruginosa is known to produce a redox-active family of pigments called phenazines, which were initially found to posses antimicrobial activity 72. Interestingly, phenazines are remarkably similar in structure to quinones, a group of molecules used to transfer electrons in bacterial electron transport chains (Figure 5A). Newman and collaborators demonstrated phenazines to have a role in extracellular electron transfer to generate energy for growth 73,74. In the context of intraspecies interactions, phenazines may be considered as players in cell signaling. Work by Dietrich and collaborators showed that a mutant unable to produce phenazines produced a more robust biofilm as judged by a much more wrinkled colony morphology than a wild-type strain 74. This differential phenotype occurs by the induction of SoxR-regulated genes and other genes like pel, responsible for exopolysaccharide production. Interestingly, SoxR was previously thought to control gene expression in response to oxidative stresses.. A similar response to self-produced pigments was also observed in the unrelated bacterium S. coelicolor, suggesting a more complex, but widely conserved, role for redox-reactive pigments in regulating community behavior and cell-cell communication.
Figure 5.
Natural small molecules produced by different microorganisms. A) Pyocyanin, the phenazin produces by P. aeruginosa, and the structural similar antibiotic quinolone with a role in the formation of wrinkled biofilms. B) Furanones (2-furanone) produced by different plants are structurally similar to QS signal molecules N-acyl-homoserine lactone or autorinducer-2, and therefore may interfere with the communication among bacteria. C) Surfactin, a lipopeptide produced by B. subtilis acts a signal molecule in the formation of stable biofilms of B. subtilis. Other structurally divergent molecules produced by other organisms, as nystatin, valinomycin or nisin induces similar response via the KinC sensor kinase.
7.2. Furanones
Furanones (Figure 5B) are a group of molecules purified from the marine algae Delisea pulchra. This molecule is produced and secreted, ultimately reducing bacterial colonization by interfering with the QS system in both Gram-negatives (Acyl-homoserine lactone system) and Gram-postive pathogens (AI-2 signalling sytem) 75,76. For this reason it is considered a mechanism developed by algae to defend against colonization of bacteria. In a recent study, different furanones were evaluated for their activity at SIC against S. auerus 77. Interestingly, it was demonstrated that furanones interfered with the QS system, and as result provoked an increase in the formation of biofilm, an effect similarly stimulated by other structurally different molecules such as tetracycline or quinupristin-dalfopristin. This effect is due to an induction in the expression of ica genes responsible for production of the Polysaccharide Intercellular Adhesin (PIA) required for formation of robust biofilms. Though it needs to be clarified, initial data suggest that this overexpression of PIA production is due to a down-regulation of luxS genes that form part of the Lux Quorum Sensing communication system.
7.3. Surfactin
Though several antibiotics have been shown to induce biofilm formation, the signaling that underlies this effect remains unclear. Quite compelling in this regard are new insights gained from studies with the nonpathogenic bacteria B. subtilis. Studies with this organism, which is well-known as a model for bacterial cell differentiation, have demonstrated that biofilms are composed of different cell types, and most importantly, these cell types are maintained over time 78,79. Indeed, the clear spatial distribution of these cell types makes possible the formation of architecturally ordered biofilms 78. This knowledge gives us the ability to define the anatomy of biofilms, and is relevant in order to understand how diverse molecules may affect their formation and development.
More recently, Lopez and collaborators demonstrated that a natural small molecule, surfactin, plays a role as signal molecule that activates the pathway involved in biofilm formation 80. Surfactin is a lipopeptide originally described to have antifungal activity (Figure 5C). Like surfactin in B. subtilis, other lipopeptides are produced in diverse bacterial species and some of them have been exploited for clinical use, because of their surfactant and antifungal activities 81. This family of compounds was seen as an intriguing new family of antimicrobials, given their mode of action. Most of them form pores in membranes, eliciting the disaggregation of the membrane and eventually cell death 82–84. Another function attributed to lipopeptides is to reduce aqueous surface tension. However, the most striking and unexpected role of this compound is that of a key player in the quorum-sensing system that stimulates the activation of the regulatory pathway involved in biofilm formation in B. subtilis 80.
In Lopez et al. 2009, it was proposed that the formation of pores triggers a signaling cascade, starting with potassium leakage from the cytoplasm of the cells, which provokes a physiological stress that is finally sensed by a membrane-associated sensor kinase, KinC. KinC in turn induces the expression of the genes involved in extracellular matrix production via its cognate response regulator 80. Given that the system recognizes the function of the signaling molecule instead of its molecular structure, biofilm formation could be stimulated by a variety of natural products that share the ability to make pores in the membrane and cause potassium leakage from the cytoplasm. Among these molecules are the macrolide polyenes nystatin and amphotericin as well as the peptide antibiotics gramidicin and valinomycin, all of which are produced by soil-dwelling bacteria (Figure 5C). Because all the inducing small molecules are produced by soil-dwelling bacteria, it is reasonable to think that, B. subtilis uses this mechanism in nature to broadly sense and respond to the presence of different bacteria.
The study of B. subtilis has therefore accelerated acquisition of knowledge about the genetic pathways involved in the responses induced by these pore-forming molecules. For a more detailed description of this process, the reader is directed to a recent review 79. It is however interesting to note that rather than the whole population, only a subpopulation of specialized B. subtilis cells synthesizes surfactin. Thus, more studies are required to elucidate how differentiation of the surfactin producing cells is controlled. It is suggested that the regulator ComA~P, which directly activates the expression of the surfactin operon, might respond likewise to Spo0A in a bistable manner 85.
The surfactin signaling pathway has been reported more recently to exhibit another level of complexity, illustrating the sophistication of inter-cellular signaling in bacterial cells. It has been demonstrated that surfactin, once produced by a specific subpopulation of cells, can be uni-directionally sensed by another unique subpopulation. This situation differs dramatically from the architecture of archetypal QS systems, wherein all cells produce and sense the signal molecule 86. Therefore typical QS signaling systems can be described as autocrine, in clear contrast to that observed for surfactin, which could be considered an example of paracrine signaling since the producing cells do not detect the signal molecule 87.
7.4. Microcins
7.4.1. Cannibalism antimicrobial peptides
There are other small molecules that can trigger biofilm formation, though their original role was found to be totally different. This is the case for a number of toxins with antibiotic properties that are involved in cell differentiation. In B. subtilis for example, one differentiated subpopulation of specialized cells secretes two toxins which lyse a fraction of their sensitive siblings. This process is termed cannibalism because lysed cells release nutrients that are subsequently used by the community 88. The subpopulation of cannibal cells is the same subpopulation that produces the extracellular matrix required for biofilm formation. Cannibalism and matrix production are both triggered in the same subpopulation in response to the signaling molecule surfactin. Subsequently, matrix-producing cells use nutrients released by cannibalized cells, as it is the only subpopulation expressing resistance to these toxins and other similar antimicrobials. As a result, this subpopulation increases and matrix production is enhanced either when cannibalism toxins are produced or in the presence of other antimicrobials. This may constitute a defense mechanism to protect B. subtilis from other antibiotics present in natural settings 85. More recently, a study elucidated the structure of the two cannibalism toxins and dissected their bactericidal activity. In these experiments, the toxins were added externally, and it was demonstrated that one of the toxins, Sdp toxin (a 42-amino acid peptide), rather than the second toxin Skf (a 26-amino acid peptide) had the ability to inhibit the growth diverse pathogens 89.
Examples of cannibal toxins with a different role can be found in the gram-positive bacterium Streptococcus pneumoniae. In this case, a subpopulation of competent cells produce and secrete an antimicrobial peptide that kills a fraction of their sensitive siblings while growing in exponential phase 90,91. Another example is Myxococcus xanthus, a Gram-negative bacterium with the ability to sporulate, that has been used as a model for studies of cell differentiation. Under starvation conditions this bacterium is capable of obtaining nutrients by preying on other bacteria 92. This predatory activity is achieved through usage of a battery of secondary metabolites which not only posses antimicrobial activity but also have the ability to induce the production of DKxanthenes, a family of pigments related to sporulation 93.
7.4.2. Bacteriocins. Lantibiotics
Another interesting group of microcins are those produced by enteric bacteria, which have been proposed to play a key role in controlling the gastrointestinal bacterial population, supposedly by means of antibiosis. Microcins are typically ribosomally synthesized peptides, with variable lengths and features. These peptides have different antimicrobial spectrums and modes of action, with some targeting both cell walls and membranes, and others only membranes. Therefore they can be classified in different sub-families or groups 94–96. One of the most valuable features of these peptides is that they are used preferentially in the preservation of prepackaged food against undesirable contamination. AS-48 is one of the preferred bacteriocins used for such a purpose. This peptide is produced by Enteroccocus faecalis, and its structure has been elucidated 97. The bactericidal activity of this molecule depends on its ability to form pores in other bacterial membranes, but in contrast to other microcins such as nisin, it does not interact with the Lipid-II. However, as previously observed for other molecules, an adaptation to this enterocin has been observed in the food-borne pathogens Lysteria monocytogenes and Bacillus cereus, after continuous exposure to the molecule. The recent study of Grande-Brugos et al. addresses this question by analyzing the effect of sub-inhibitory concentrations of AS-48 against B. cereus. By using a combination of global transcriptome analysis and real-time PCR, they observed an up-regulation of the expression of genes encoding for membrane proteins, some of them under control of the regulator PadR, which is known to modulate important physiological process related to multidrug resistance, virulence and/or detoxification 98. Using a similar experimental procedure, another bacteriocin, Lcn972 from Lactoccocus lactis, was found to up-regulate the expression of 26 genes in L. lactis, some of them encoding for unknown membrane proteins and the two component system CesSR involved in cell-envelope stress response 99.
Nisin represents a fascinating example of a microcin with a role other than antibiosis. Nisin is a peptide antibiotic of 34 amino acids produced by Lactococcus lactis (Figure 5C). This bacteriocin has a broad-spectrum activity and is also used to preserve processed food from contamination with Gram-positive and Gram-negative bacteria. The mode of action of nisin is inhibition of peptidoglycan synthesis by interacting with Lipid-II, resulting in the formation of pores in the bacterial membrane. This molecule, similar to other Bacillus toxins, has been shown to promote the development of matrix producer cells in B. subtilis communities, therefore inducing the formation of biofilms 80. A possible explanation for this response is the existence of similarity in their structures and mode of action.
Elucidating the effects of these microcins at sub-lethal concentrations will help to understand whether or not a similar response is globally or rather specifically triggered in diverse bacterial species that occupy the same environmental niche. Identification of the molecules, receptors and genes targeted, might aid in the design of more efficient strategies aimed at enhancing the establishment of beneficial microbiota or interfering with the establishment of pathogenic communities.
8. Antibiotics-driven interspecies interactions
Most of the antibiotics used clinically are naturally produced by microbes, or modified from originals found in nature. We have seen how some of them may promote a differential response rather than antibiosis when used at SIC levels. The fact that some natural products unexpectedly trigger biofilm formation suggests that this might be a mechanism of defense from competitors. However, as described above, different bacterial species use different communication systems; so how signals from different origins can be sensed by diverse species is a question that requires deeper investigation.
The relatively common AI-2 communication system has already been discussed here, but in other many cases, the signaling pathways involved in interspecies communication have not been clearly determined. An exciting example of such communication has been demonstrated between Stenotrophomonas maltofilia and P. aeruginosa. These two organisms coexist in different environments ranging from soil to the Cystic fibrosis lung. In a recent work by Ryan et al. (2008), it was been beautifully shown how a diffusible signal factor (DSF), from S. maltofilia (initially identified in Xanthomonas campestris and characterized as the unsaturated fatty acid cis-11-methyl-2-dodecenoic acid), can be intercepted by P. aeruginosa 100. The reception of this molecule in P. aeruginosa relies on the sensor kinase PA1396 that triggers important physiological changes including bioifilm formation and increased tolerance to the antimicrobial peptide polymixin. Another interesting interspecies interaction is observed for the duo of P. aeruginosa and the yeast Candida albicans 101. In this interaction, C. albicans survives the attack of P. aeruginosa by combining two strategies. P. aeruginosa produces a homoserine lactone molecule that is perceived by C. albicans, stimulating a switch from filamentous growth to a more resistant yeast form. Interestingly, C. albicans produces an isoterpenoid molecule, farnesol, that interferes with the P. aeruginosa quinolone signaling cascade, which subsequently results in a reduction of piocyanine production and virulence.
Beyond the clinical context, soil represents a fascinating environment for exploring microbial interactions, and molecules with a role in cell-cell communication. One such interaction is that between Streptomyces coelicolor and the fungi Schizophyllum commune. S coelicolor is a filamentous bacterium that undergoes a complex developmental cycle in which spore chains are formed on the ends of aerial hyphae in response to nutrient limitation. S. coelicolor is also model organism for the genus Streptomyces, which are prodigious producers of many clinically-useful antibiotics. The genetic tractability of S. coelicolor has made it a valuable system for studying biosynthesis of secondary metabolites, cell differentiation, and signaling. The formation of aerial hyphae requires two operons, ramCSAB, which encodes for a lantibiotic-like surfactant peptide SapB, and chpA-H which encodes for the chaplin family of proteins that are involved in the formation of a hydrophobic spore coat 102–104. In separate studies, by using pair-wise interactions with different microorganisms, it was demonstrated that the cellular differentiation pathways in S. coelicolor can be altered in different ways. For example, B. subtilis produces two molecules, bacillaene, and surfactin, the latter of which, as stated above, has a role as a signal molecule in the developmental process of B. subtilis. Straight and collaborators beautifully demonstrated how these two molecules inhibited the formation of aerial hyphae of S. coelicolor. Though the exact mode of interaction requires more research, it was suggested to be mediated by interference with production of SapB and chaplins 105. In a separate study it was observed that the surfactant SC3, produced by the fungus Schizophyllum commune, could replace the native SapB surfactant in the formation of aerial hyphae 106.
Given the vast variety of secondary metabolites produced by Streptomyces species and the existence of similar developmental signaling pathways, it is possible that molecules involved in differentiation or the initiation of secondary metabolite production in one species may influence a similar behavior in other Streptomyces species. An example of such concept is illustrated in Figure 6. In these experiments, S. coelicolor was tested against a collection of different Streptomyces isolates. In a first round, bacterial suspensions were spotted close to each other, and after an incubation period, alterations on the morphology of S. coelicolor were visualized. The fact that variations with S. coelicolor were observed even without any physical contact, suggests that secreted molecules are responsible for the observed phenotypic changes. In the second round, extracts from cell cultures were obtained and tested. The extracts showed various activities against S. coelicolor. An extract from S. californicus caused growth inhibition near the well, followed by an area of aerial hyphae (white) inhibition. Within the ‘bald’ zone, separate areas of secondary metabolite production by S. coelicolor are observed; from none (nearest the well), undecylprodigiosin (red), and actinorhodin (blue). An extract from S. griseolus caused a sharp area of growth inhibition followed by an area of accelerated development (visible as a ring of white aerial hyphae around the well in column 2). Finally, an extract from S. griseoaurantiacus triggered enhanced actinorhodin production while simultaneously inhibiting the development of aerial hyphae.
Figure 6.
Screening for isolates that cause developmental/morphological changes in S. coelicolor. First column: 5 ul of dense spore solutions of WT Streptomyces coelicolor M145 (alone in row 1) and various wild Streptomyces isolates were spotted 1 cm apart on R2YE agar. Isolates were screened for their ability to influence S. coelicolor morphology. Second column: Wild isolates were grown as lawns on agar, and extracted with ethanol. Extracts were then tested for morphological activity against a lawn of S. coelicolor (pictured) after 48 hours. Third column: Close-ups of the same wells shown in column 2 after 72 hours. Row one shows a control colony of S. coelicolor and a well with extract from an uninoculated plate. The extracts showed various activities against S. coelicolor (Figure provided by Matt Traxler).
A good reported example of molecules produced by one Streptomyces species affecting the developmental programs of others, is pamamycin-607, a macrolide isolated from S. alboniger 107. This molecule has been reported to induce an imbalance in the levels of Ca2+ within cells that regulate the formation of aerial hyphae. It has been shown in vitro that external addition of this macrolide was able to restore the formation of aerial structures in different Streptomyces species, only some of which were producers of pamamycin-607. These findings suggest that though this molecule can function as a cell-differentiation regulator, it is not utilized as a global signal for aerial structure formation in all Streptomyces species.
The recent utilization of two-dimensional MALDI-TOF mass spectrometry analysis to study classical pair-wise interactions represents a powerful new way to define with more accuracy the signals exchanged among different bacteria. The strength and beauty of the technique relies on the fact that the spatial distribution of molecules involved in communication can be detected around microorganism colonies in situ. It also provides an opportunity to define the landscape of signals that fluctuate among different species when grown together. Using this technique Yang and collaborators investigated the interaction of two soil microorganisms, B. subtilis and S. coelicolor 108 They found that surfactin, produced by B. subtilis, not only affected the developmental program of S. coelicolor, but also decreased the production of calcium dependent antibiotic (CDA), a compound with activity against Gram-positive organisms. They provided a list of molecules affected during this interaction and how their production evolved over time, highlighting the complexity of the signals involved in interspecies communication. In other studies, 2D MALDI-TOF has been used to study how cannibalistic factors of B. subtilis are involved in killing of vulnerable siblings 89. In this study, interesting and unexpected results were observed, as exemplified by the fact that the domesticated strain of B. subtilis PY79, produced surfactin. In contrast, the two cannibalistic peptides (SKF and SDF) did not spread into the medium, as did surfactin and subtilosin. These experiments provided the basis for further purification protocols of the cannibalism factors, and their structures were elucidated.
9. Summary and perspectives
In the recent past, antibiotics have been widely employed for their ability to cure many human infectious diseases. The increasingly frequent failure of some of these molecules to control many previously treatable infections has spurred investigation of the mechanisms responsible for such resistance. A uniting feature among the results obtained in several pioneering studies has been the observation that many well-known antibiotics induce differential bacterial responses when applied at sub-inhibitory concentrations. In all of these studies, purified compounds were analyzed for their ability to effect global responses using transcriptomic analysis. Later, these results were confirmed by using other genetic tools, such as RT-PCR or transcriptional promoter fusions to reporter proteins.
Attempts to identify the targets affected by sub-inhibitory doses of these antibiotics have demonstrated that many of them interfere with cell-cell communication systems, and depending on the bacterial species, may affect virulence, stress response factors, or cell differentiation and morphological changes. More studies are necessary in order to clarify how this differential response is achieved and which players are involved. Hopefully, the knowledge gained by such studies will aid in the development of therapeutic regimes to promote or inhibit a specific bacterial response, for example, promotion of biofilm formation of beneficial bacteria to shield against invading pathogens or the specific inhibition of virulence processes.
Traditionally unculturable microorganisms may also represent a remarkable example of necessary microbial cooperation. Currently, we lack the understanding required to grow most microbes in a laboratory setting. One explanation might be that they require one or more microbial partners to grow. In such a framework, one bacterial species might provide the other with growth factors necessary to support their survival. As articulated by Lewis et al., 2010, these unculturable microorganisms may represent another interesting source of novel secondary metabolites 109. In this study, it was found that siderophores (molecules that bind to and make accessible the iron present in the medium) from one organism promoted the growth of another, previously uncultured partner microorganism. Another interesting example was observed for two marine bacteria, Micrococcus lotus and Micrococcus polysiphoniae 110. The growth of the first was supported by the latter, and interestingly in a distance dependent manner, the closer they were cultured, the better the growth. The production of five different siderophores was detected in further fractionation analysis. In this example, siderophores, used for competition in some environments, are proven to be useful for establishment of a mutualistic interaction. What other factors are involved, and how other secondary metabolites may be produced and participate in communication between species, will contribute to our understanding of the complexity of interspecies communication.
Now with a greater range of techniques are available, we are poised to make dramatic advances in our knowledge of bacterial communication. It is also likely that these studies will yield novel molecules that can be analyzed in detail for their therapeutic applications, including the modulation of bacterial consortia and cell differentiation.
Acknowledgments
Diego Romero is the recipient of a MEC/Fulbright post-doctoral fellowship from Secretaría General de Estado de Universidades e Investigación del Ministerio de Educación y Ciencia (Spain) and Matt Traxler is funded by NRSA F32GM089044-01 from the NIH. Work on this subject in R.K.’s laboratory is funded by grants from the NIH (GM58213 and GM82137)
Biographies
Dr. Diego Romero obtained his Bachelor’s degree in Biological Sciences at the University of Málaga (Spain). He received his Ph.D. in Microbiology from the University of Málaga with Dr. Antonio de Vicente and Dr. Alejandro Pérez, where his work focused on biotechnology applied to the control of plant diseases. After completing his doctoral studies, he joined Dr. Roberto Kolter’s laboratory at Harvard Medical School for his post-doctoral training. His recent research is focused on deciphering how amyloid proteins function in the extracellular matrix of Bacillis subtilis biofilms.

Dr. Matthew Traxler received both his Bachelor’s and Ph.D. degrees in Microbiology from the University of Oklahoma where he studied in the lab of Dr. Tyrrell Conway. His doctoral work focused on the role of the alarmone ppGpp in controlling global transcriptional responses in E. coli. He is currently a postdoctoral fellow in the lab of Roberto Kolter at Harvard Medical School. His research interests include investigating the role of natural products in mediating interspecies interactions between soil-dwelling microbes, with an emphasis on molecules that alter developmental processes in Streptomycetes.

Dr. Daniel Lopez received his Bachelor’s degree in Biochemical Sciences from University of Murcia (Spain) and earned hir Ph.D. in Biochemistry with Dr. Antonio Sanchez-Amat and Dr. Francisco Solano at the same university. After receiving hir doctorate, he trained as a postdoctoral fellow with Dr. Roberto Kolter at Harvard Medical School (USA), where he became interested in cell-cell communication in bacteria and small molecules involved in this process. He recently joined the Institute for Molecular Infection Biology IMIB at Würzburg University as Junior Research Group Leader, where he is building a research group that focuses on cell differentiation in infective bacteria and the molecular mechanisms involved in this process.

Dr. Roberto Kolter is Professor of Microbiology and Molecular Genetics at Harvard Medical School, where he has been a faculty member since 1983 and Co-Director of Harvard’s University-wide Microbial Sciences Initiative since 2003. At Harvard, he has worked on antibiotic synthesis, bacterial starvation physiology, experimental evolution, bacterial biofilms, and chemical communication in the microbial world. Dr. Kolter has been involved in science teaching and policy worldwide. He recently served as the President of the American Society for Microbiology and now serves as Chair of its Public and Scientific Affairs Board. A native of Guatemala, Dr. Kolter earned his B.S. at Carnegie-Mellon University, his Ph.D. at University of California, San Diego and carried out post-doctoral training at Stanford University.

References
- 1.Davies J. Science. 1994;264:375. doi: 10.1126/science.8153624. [DOI] [PubMed] [Google Scholar]
- 2.Davies J. Can J Infect Dis Med Microbiol. 2006;17:287. doi: 10.1155/2006/707296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yim G, Wang HH, Davies J. Philos Trans R Soc Lond B Biol Sci. 2007;362:1195. doi: 10.1098/rstb.2007.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies JJ. Ind Microbiol Biotechnol. 2006;33:496. doi: 10.1007/s10295-006-0112-5. [DOI] [PubMed] [Google Scholar]
- 5.Monds RD, O’Toole GA. Trends Microbiol. 2009;17:73. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Science. 2005;308:1635. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kent AD, Triplett EW. Annu Rev Microbiol. 2002;56:211. doi: 10.1146/annurev.micro.56.012302.161120. [DOI] [PubMed] [Google Scholar]
- 8.Ward DM, Ferris MJ, Nold SC, Bateson MM. Microbiol Mol Biol Rev. 1998;62:1353. doi: 10.1128/mmbr.62.4.1353-1370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Straight PD, Kolter R. Annu Rev Microbiol. 2009;63:99. doi: 10.1146/annurev.micro.091208.073248. [DOI] [PubMed] [Google Scholar]
- 10.Davies J, Spiegelman GB, Yim G. Curr Opin Microbiol. 2006;9:445. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MRJ. Bacteriol. 2001;183:5395. doi: 10.1128/JB.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez D, Vlamakis H, Kolter R. Cold Spring Harb Perspect Biol. 2010;2:a000398. doi: 10.1101/cshperspect.a000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keller L, Surette MG. Nat Rev Microbiol. 2006;4:249. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 14.Jones D, Metzger HJ, Schatz A, Waksman SA. Science. 1944;100:103. doi: 10.1126/science.100.2588.103. [DOI] [PubMed] [Google Scholar]
- 15.Davies J. Can J Infect Dis Med Microbiol. 2006;17:287. doi: 10.1155/2006/707296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osbourn A. Trends Genet. 2010;26:449. doi: 10.1016/j.tig.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Ruiz B, Chavez A, Forero A, Garcia−Huante Y, Romero A, Sanchez M, Rocha D, Sanchez B, Rodriguez−Sanoja R, Sanchez S, Langley E. Crit Rev Microbiol. 2010;36:146. doi: 10.3109/10408410903489576. [DOI] [PubMed] [Google Scholar]
- 18.Fischbach MA, Walsh CT. Science. 2009;325:1089. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clardy J, Fischbach MA, Walsh CT. Nat Biotechnol. 2006;24:1541. doi: 10.1038/nbt1266. [DOI] [PubMed] [Google Scholar]
- 20.Weissman KJ, Muller R. Nat Prod Rep. 2010;27:1276. doi: 10.1039/c001260m. [DOI] [PubMed] [Google Scholar]
- 21.Atkinson S, Williams PJR. Soc Interface. 2009;6:959. doi: 10.1098/rsif.2009.0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryan RP, Dow JM. Microbiology. 2008;154:1845. doi: 10.1099/mic.0.2008/017871-0. [DOI] [PubMed] [Google Scholar]
- 23.Horswill AR, Stoodley P, Stewart PS, Parsek MR. Anal Bioanal Chem. 2007;387:371. doi: 10.1007/s00216-006-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bibb M, Hesketh A. Methods Enzymol. 2009;458:93. doi: 10.1016/S0076-6879(09)04804-6. [DOI] [PubMed] [Google Scholar]
- 25.Rawlings BJ. Nat Prod Rep. 1999;16:425. doi: 10.1039/a900566h. [DOI] [PubMed] [Google Scholar]
- 26.Konz D, Marahiel MA. Chem Biol. 1999;6:R39. doi: 10.1016/S1074-5521(99)80002-7. [DOI] [PubMed] [Google Scholar]
- 27.Zhang W, Ostash B, Walsh CT. Proc Natl Acad Sci U S A. 2010;107:16828. doi: 10.1073/pnas.1011557107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandoz KM, Mitzimberg SM, Schuster M. Proc Natl Acad Sci U S A. 2007;104:15876. doi: 10.1073/pnas.0705653104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, Greenberg EP. Science. 1998;280:295. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 30.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. Nature. 2001;413:860. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 31.McGowan S, Sebaihia M, Jones S, Yu B, Bainton N, Chan PF, Bycroft B, Stewart GS, Williams P, Salmond GP. Microbiology. 1995;141(Pt 3):541. doi: 10.1099/13500872-141-3-541. [DOI] [PubMed] [Google Scholar]
- 32.McGowan SJ, Sebaihia M, O’Leary S, Hardie KR, Williams P, Stewart GS, Bycroft BW, Salmond GP. Mol Microbiol. 1997;26:545. doi: 10.1046/j.1365-2958.1997.6001974.x. [DOI] [PubMed] [Google Scholar]
- 33.Waters CM, Bassler BL. Annu Rev Cell Dev Biol. 2005;21:319. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 34.O’Gara JP. FEMS Microbiol Lett. 2007;270:179. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- 35.Yarwood JM, Bartels DJ, Volper EM, Greenberg EPJ. Bacteriol. 2004;186:1838. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin JF, Liras P. Curr Opin Microbiol. 2010;13:263. doi: 10.1016/j.mib.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 37.Horinouchi S, Ohnishi Y, Kang DKJ. Ind Microbiol Biotechnol. 2001;27:177. doi: 10.1038/sj.jim.7000068. [DOI] [PubMed] [Google Scholar]
- 38.Takano E. Curr Opin Microbiol. 2006;9:287. doi: 10.1016/j.mib.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Schauder S, Bassler BL. Genes Dev. 2001;15:1468. doi: 10.1101/gad.899601. [DOI] [PubMed] [Google Scholar]
- 40.Lewenza S, Conway B, Greenberg EP, Sokol PAJ. Bacteriol. 1999;181:748. doi: 10.1128/jb.181.3.748-756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stewart PS, Franklin MJ. Nat Rev Microbiol. 2008;6:199. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 42.Dickschat JS. Nat Prod Rep. 2010;27:343. doi: 10.1039/b804469b. [DOI] [PubMed] [Google Scholar]
- 43.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Nature. 2000;407:762. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 44.Donlan RM. Emerg Infect Dis. 2001;7:277. doi: 10.3201/eid0702.010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mack D, Davies AP, Harris LG, Rohde H, Horstkotte MA, Knobloch JK. Anal Bioanal Chem. 2007;387:399. doi: 10.1007/s00216-006-0745-2. [DOI] [PubMed] [Google Scholar]
- 46.Kreth J, Zhang Y, Herzberg MCJ. Bacteriol. 2008;190:4632. doi: 10.1128/JB.00276-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Carvalho CC. Recent Pat Biotechnol. 2007;1:49. doi: 10.2174/187220807779813965. [DOI] [PubMed] [Google Scholar]
- 48.Drenkard E. Microbes Infect. 2003;5:1213. doi: 10.1016/j.micinf.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 49.Walters MC, 3rd, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. Antimicrob Agents Chemother. 2003;47:317. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Folkesson A, Haagensen JA, Zampaloni C, Sternberg C, Molin S. PLoS One. 2008;3:e1891. doi: 10.1371/journal.pone.0001891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richards JJ, Melander C. Chembiochem. 2009;10:2287. doi: 10.1002/cbic.200900317. [DOI] [PubMed] [Google Scholar]
- 52.Goh EB, Yim G, Tsui W, McClure J, Surette MG, Davies J. Proc Natl Acad Sci U S A. 2002;99:17025. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mesak LR, Miao V, Davies J. Antimicrob Agents Chemother. 2008;52:3394. doi: 10.1128/AAC.01599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogers PD, Liu TT, Barker KS, Hilliard GM, English BK, Thornton J, Swiatlo E, McDaniel LSJ. Antimicrob Chemother. 2007;59:616. doi: 10.1093/jac/dkl560. [DOI] [PubMed] [Google Scholar]
- 55.Gmuender H, Kuratli K, Di Padova K, Gray CP, Keck W, Evers S. Genome Res. 2001;11:28. doi: 10.1101/gr.157701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ng WL, Kazmierczak KM, Robertson GT, Gilmour R, Winkler MEJ. Bacteriol. 2003;185:359. doi: 10.1128/JB.185.1.359-370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yim G, de la Cruz F, Spiegelman GB, Davies JJ. Bacteriol. 2006;188:7988. doi: 10.1128/JB.00791-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoffman LR, D’Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. Nature. 2005;436:1171. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 59.Bagge N, Schuster M, Hentzer M, Ciofu O, Givskov M, Greenberg EP, Hoiby N. Antimicrob Agents Chemother. 2004;48:1175. doi: 10.1128/AAC.48.4.1175-1187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skindersoe ME, Alhede M, Phipps R, Yang L, Jensen PO, Rasmussen TB, Bjarnsholt T, Tolker−Nielsen T, Hoiby N, Givskov M. Antimicrob Agents Chemother. 2008;52:3648. doi: 10.1128/AAC.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH. Nat Struct Mol Biol. 2007;14:727. doi: 10.1038/nsmb1271. [DOI] [PubMed] [Google Scholar]
- 62.Babic F, Venturi V, Maravic−Vlahovicek G. BMC Infect Dis. 2010;10:148. doi: 10.1186/1471-2334-10-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hodges NA, Gordon CA. Antimicrob Agents Chemother. 1991;35:2450. doi: 10.1128/aac.35.11.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ahmed NA, Petersen FC, Scheie AA. Antimicrob Agents Chemother. 2009;53:4258. doi: 10.1128/AAC.00546-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kobayashi H. Am J Med. 1995;99:26S. doi: 10.1016/s0002-9343(99)80282-4. [DOI] [PubMed] [Google Scholar]
- 66.Ichimiya T, Takeoka K, Hiramatsu K, Hirai K, Yamasaki T, Nasu M. Chemotherapy. 1996;42:186. doi: 10.1159/000239440. [DOI] [PubMed] [Google Scholar]
- 67.Nalca Y, Jansch L, Bredenbruch F, Geffers R, Buer J, Haussler S. Antimicrob Agents Chemother. 2006;50:1680. doi: 10.1128/AAC.50.5.1680-1688.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ader F, Le Berre R, Faure K, Gosset P, Epaulard O, Toussaint B, Polack B, Nowak E, Viget NB, Kipnis E, Guery BP. Infect Immun. 2005;73:4263. doi: 10.1128/IAI.73.7.4263-4271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kobayashi T, Tateda K, Matsumoto T, Miyazaki S, Watanabe A, Nukiwa T, Yamaguchi KJ. Antimicrob Chemother. 2002;50:59. doi: 10.1093/jac/dkf048. [DOI] [PubMed] [Google Scholar]
- 70.Linares JF, Gustafsson I, Baquero F, Martinez JL. Proc Natl Acad Sci U S A. 2006;103:19484. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rachid S, Ohlsen K, Witte W, Hacker J, Ziebuhr W. Antimicrob Agents Chemother. 2000;44:3357. doi: 10.1128/aac.44.12.3357-3363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Price−Whelan A, Dietrich LE, Newman DK. Nat Chem Biol. 2006;2:71. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- 73.Dubern JF, Diggle SP. Mol Biosyst. 2008;4:882. doi: 10.1039/b803796p. [DOI] [PubMed] [Google Scholar]
- 74.Dietrich LE, Teal TK, Price−Whelan A, Newman DK. Science. 2008;321:1203. doi: 10.1126/science.1160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Manefield M, de Nys R, Kumar N, Read R, Givskov M, Steinberg P, Kjelleberg S. Microbiology. 1999;145(Pt 2):283. doi: 10.1099/13500872-145-2-283. [DOI] [PubMed] [Google Scholar]
- 76.Ren D, Bedzyk LA, Ye RW, Thomas SM, Wood TK. Biotechnol Bioeng. 2004;88:630. doi: 10.1002/bit.20259. [DOI] [PubMed] [Google Scholar]
- 77.Kuehl R, Al−Bataineh S, Gordon O, Luginbuehl R, Otto M, Textor M, Landmann R. Antimicrob Agents Chemother. 2009;53:4159. doi: 10.1128/AAC.01704-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vlamakis H, Aguilar C, Losick R, Kolter R. Genes Dev. 2008;22:945. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lopez D, Vlamakis H, Kolter R. FEMS Microbiol Rev. 2009;33:152. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- 80.Lopez D, Fischbach MA, Chu F, Losick R, Kolter R. Proc Natl Acad Sci U S A. 2009;106:280. doi: 10.1073/pnas.0810940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chooi YH, Tang Y. Chem Biol. 2010;17:791. doi: 10.1016/j.chembiol.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 82.Sen R. Adv Exp Med Biol. 2010;672:316. doi: 10.1007/978-1-4419-5979-9_24. [DOI] [PubMed] [Google Scholar]
- 83.Canton R, Ruiz−Garbajosa P, Chaves RL, Johnson APJ. Antimicrob Chemother. 2011;65:1126. doi: 10.1093/jac/dkq087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Laverty G, McLaughlin M, Shaw C, Gorman SP, Gilmore BF. Chem Biol Drug Des. 2010;75:563. doi: 10.1111/j.1747-0285.2010.00973.x. [DOI] [PubMed] [Google Scholar]
- 85.Lopez D, Vlamakis H, Losick R, Kolter R. Mol Microbiol. 2009;74:609. doi: 10.1111/j.1365-2958.2009.06882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Camilli A, Bassler BL. Science. 2006;311:1113. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lopez D, Vlamakis H, Losick R, Kolter R. Genes Dev. 2009;23:1631. doi: 10.1101/gad.1813709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gonzalez−Pastor JE, Hobbs EC, Losick R. Science. 2003;301:510. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- 89.Liu WT, Yang YL, Xu Y, Lamsa A, Haste NM, Yang JY, Ng J, Gonzalez D, Ellermeier CD, Straight PD, Pevzner PA, Pogliano J, Nizet V, Pogliano K, Dorrestein PC. Proc Natl Acad Sci U S A. 2010;107:16286. doi: 10.1073/pnas.1008368107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Havarstein LS, Gaustad P, Nes IF, Morrison DA. Mol Microbiol. 1996;21:863. doi: 10.1046/j.1365-2958.1996.521416.x. [DOI] [PubMed] [Google Scholar]
- 91.Guiral S, Mitchell TJ, Martin B, Claverys JP. Proc Natl Acad Sci U S A. 2005;102:8710. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reichenbach H. Environ Microbiol. 1999;1:15. doi: 10.1046/j.1462-2920.1999.00016.x. [DOI] [PubMed] [Google Scholar]
- 93.Meiser P, Bode HB, Muller R. Proc Natl Acad Sci U S A. 2006;103:19128. doi: 10.1073/pnas.0606039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nes IF, Diep DB, Havarstein LS, Brurberg MB, Eijsink V, Holo H. Antonie Van Leeuwenhoek. 1996;70:113. doi: 10.1007/BF00395929. [DOI] [PubMed] [Google Scholar]
- 95.Abriouel H, Franz CM, Omar NB, Galvez A. FEMS Microbiol Rev. 2011;35:2,01. doi: 10.1111/j.1574-6976.2010.00244.x. [DOI] [PubMed] [Google Scholar]
- 96.Franz CM, van Belkum MJ, Holzapfel WH, Abriouel H, Galvez A. FEMS Microbiol Rev. 2007;31:293. doi: 10.1111/j.1574-6976.2007.00064.x. [DOI] [PubMed] [Google Scholar]
- 97.Sanchez−Barrena MJ, Martinez−Ripoll M, Galvez A, Valdivia E, Maqueda M, Cruz V, Albert A. J Mol Biol. 2003;334:541. doi: 10.1016/j.jmb.2003.09.060. [DOI] [PubMed] [Google Scholar]
- 98.Grande Burgos MJ, Kovacs AT, Mironczuk AM, Abriouel H, Galvez A, Kuipers OP. BMC Microbiol. 2009;9:227. doi: 10.1186/1471-2180-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martinez B, Zomer AL, Rodriguez A, Kok J, Kuipers OP. Mol Microbiol. 2007;64:473. doi: 10.1111/j.1365-2958.2007.05668.x. [DOI] [PubMed] [Google Scholar]
- 100.Ryan RP, Fouhy Y, Garcia BF, Watt SA, Niehaus K, Yang L, Tolker−Nielsen T, Dow JM. Mol Microbiol. 2008;68:75. doi: 10.1111/j.1365-2958.2008.06132.x. [DOI] [PubMed] [Google Scholar]
- 101.Hogan DA, Kolter R. Science. 2002;296:2229. doi: 10.1126/science.1070784. [DOI] [PubMed] [Google Scholar]
- 102.Nguyen KT, Willey JM, Nguyen LD, Nguyen LT, Viollier PH, Thompson CJ. Mol Microbiol. 2002;46:1223. doi: 10.1046/j.1365-2958.2002.03255.x. [DOI] [PubMed] [Google Scholar]
- 103.Kodani S, Hudson ME, Durrant MC, Buttner MJ, Nodwell JR, Willey JM. Proc Natl Acad Sci U S A. 2004;101:11448. doi: 10.1073/pnas.0404220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Elliot MA, Karoonuthaisiri N, Huang J, Bibb MJ, Cohen SN, Kao CM, Buttner MJ. Genes Dev. 2003;17:1727. doi: 10.1101/gad.264403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Straight PD, Willey JM, Kolter RJ. Bacteriol. 2006;188:4918. doi: 10.1128/JB.00162-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tillotson RD, Wosten HA, Richter M, Willey JM. Mol Microbiol. 1998;30:595. doi: 10.1046/j.1365-2958.1998.01093.x. [DOI] [PubMed] [Google Scholar]
- 107.Hashimoto M, Kondo T, Kozone I, Kawaide H, Abe H, Natsume M. Biosci Biotechnol Biochem. 2003;67:803. doi: 10.1271/bbb.67.803. [DOI] [PubMed] [Google Scholar]
- 108.Yang YL, Xu Y, Straight P, Dorrestein PC. Nat Chem Biol. 2009;5:885. doi: 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lewis K, Epstein S, D’Onofrio A, Ling LLJ. Antibiot (Tokyo) 2010;63:468. doi: 10.1038/ja.2010.87. [DOI] [PubMed] [Google Scholar]
- 110.D’Onofrio A, Crawford JM, Stewart EJ, Witt K, Gavrish E, Epstein S, Clardy J, Lewis K. Chem Biol. 2010;17:254. doi: 10.1016/j.chembiol.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]