Abstract
Base eversion is a fundamental process in the biochemistry of nucleic acids, allowing proteins engaged in DNA repair and epigenetic modifications to access target bases in DNA. Crystal structures reveal endpoints of these processes, but not the pathways involved in the dynamic process of base recognition. To elucidate the pathway taken by 8-oxoguanine during base excision repair by Fpg, we calculated free energy surfaces during eversion of the damaged base through the major and minor grooves. The minor groove pathway and free energy barrier (6–7 kcal/mol) are consistent with previously reported results.1 However, eversion of 8-oxoG through the major groove encounters a significantly lower barrier (3–4 kcal/mol) more consistent with experimentally determined2 rates of enzymatic sliding during lesion search. Major groove eversion has been suggested for other glycosylases, suggesting that in addition to function, dynamics of base eversion may also be conserved.
Keywords: 8-oxoguanine, Fpg, MutM, BER, base flipping, DNA repair
8-oxoguanine (8-oxoG), one of the most abundant products of oxidative DNA damage in cells, arises from oxidation of guanine by endogenous and exogenous reactive oxygen species.3 8-oxoG readily pairs with adenine in a Hoogsteen orientation; during DNA replication, this conformation results in a transversion mutation.4 Base excision repair (BER) pathways in eukaryote and prokaryote cells excise 8-oxoG to preserve genomic integrity. In prokaryotes, formamidopyrimidine-DNA glycosylase (Fpg), also known as MutM, excises 8-oxoG when the oxidized base is paired to cytosine.5 Human oxoguanine glycosylase (hOGG1) is a functional homologue of Fpg, but the two enzymes share little structural homology. 6
The base eversion mechanism is not limited to positioning damaged bases for enzymatic catalysis, and also plays a role in determining selectivity of substrates. Eversion of bases from the helix facilitates recognition of substrates. For example, studies of uracil DNA glycosylase (UDG) using imino proton exchange NMR indicate that UDG has a passive role in trapping spontaneously opened bases, the rates of which are increased for the substrate.7 However, the relatively low barrier to sliding of hOGG1 and Fpg (about 2 kcal/mol per base pair), as determined by single-molecule fluorescence studies2, suggests these glycosylases do not extrude every base into an extrahelical site, and that substrate recognition takes place at an earlier stage.
The low energy states of BER DNA glycosylases in general, and Fpg and hOGG1 in particular, have been characterized, primarily by X-ray crystallography. Catalytically inactive Fpg mutants allow trapping of substrates containing 8-oxoguanine in the active site.8 Intermolecular crosslinking has been used to generate structures of putative intermediates in the catalytic pathway9,10 as well as an intrahelical structure, interpreted as initial binding of Fpg to 8-oxoG damaged1 and undamaged11 DNA. Figure 1 illustrates the initial intrahelical and final extrahelical endpoints of 8-oxoG in the flipping pathway. Visual inspection suggests the minor groove pathway as the more direct route. However, crystal structures of hOGG1 complexed to DNA have been reported with bases in the major groove at putative intermediate locations along the eversion pathway.12,13 UDG crystallography also points to a major groove pathway to avoid unfavorable steric clashes with protein side chains in the minor groove.14
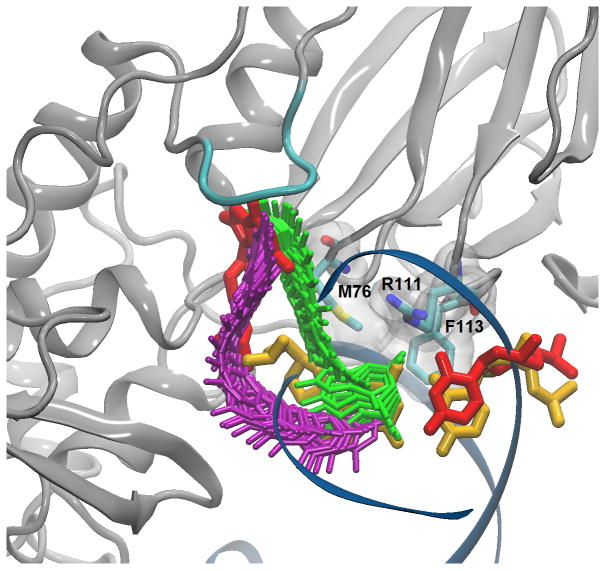
Figure 1.
Base eversion paths from NEB calculations. Paths are indicated by snapshots, with major groove shown in purple and minor groove in green. Intrahelical 8-oxoG:C base pair is shown in yellow, extrahelical 8-oxoG:C base pair, in red, catalytic loop, in cyan.
Molecular dynamics (MD) provides an unparalleled ability to simultaneously connect structure, dynamics and energy for conformational changes. To generate a minimum potential energy path (MEP) from the intrahelical to the extrahelical endpoint, the partial nudged elastic band (PNEB)15 implementation in Amber1016 was used with all-atom simulations in explicit water. Both paths begin and end with the same intrahelical and extrahelical coordinates, and are continuous in all solute degrees of freedom. The optimized, time-independent paths are shown in Figure 1 (Population histograms along each PNEB path are provided in Supporting Information, Figures S1 and S2). The minor groove path rotates around the glycosidic torsion early in eversion to minimize unfavorable steric clashes and form favorable electrostatic interactions with the intercalating triad (M76/R111/F113, see Fig. 1). The major groove path shows no steric hindrance for initial breaking of the intrahelical hydrogen bonds.
Two- dimensional umbrella sampling was performed to obtain free energy profiles along these paths, using two previously reported reaction coordinates that measure base eversion and the glycosidic torsion rotation.17,18 The resulting potential of mean force (PMF), shown in Figure 2 (with independent runs shown in Fig. S3), reveals free energy profiles for both pathways. Eversion through the minor groove encounters an energetic barrier of 6–7 kcal/mol; this observation is consistent with the ~10 kcal overall barrier to extrusion into Fpg’s active site reported in Qi et al. (who studied only the minor groove path).1 Although a small energy penalty is incurred upon breaking intrahelical hydrogen bonds, the main barrier to entering the active site corresponds to initial rotation of the glycosidic torsion.
Figure 2.
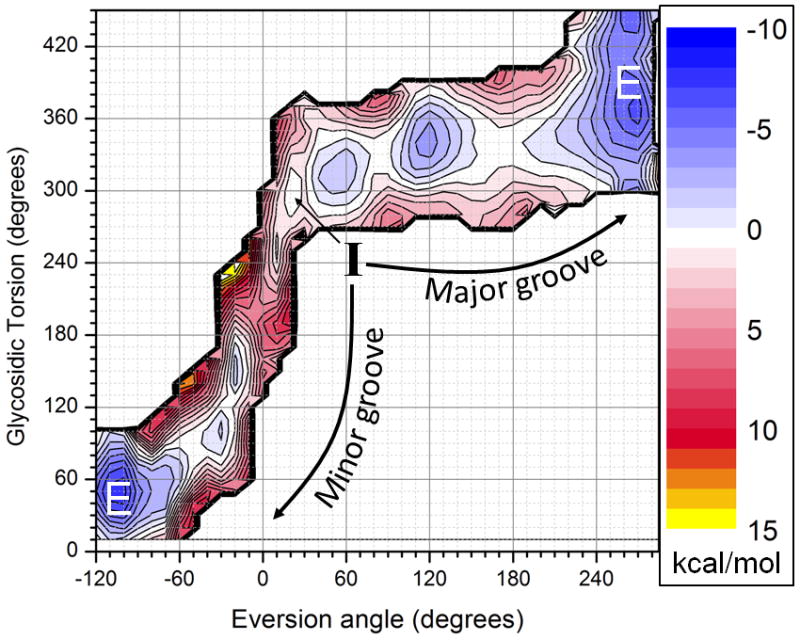
Free energy pathways of base eversion via the major (top) and minor (bottom left) grooves. The initial intrahelical state I is in the middle, and the final extrahelical state E is reached on both the right and left sides of the graph. Free energies are given in kcal/mol.
The major groove PMF indicates a maximum overall barrier of 3–4 kcal/mol, significantly less than through the minor groove. As suggested14 for UDG, the major groove pathway avoids steric clashes, and instead the smaller barrier arises from eversion past the β-hairpin of the zinc finger motif. 8-oxoG partially rotates to form a stable intermediate (eversion angle = 120°) that may have an important role in substrate recognition. Crystal structures of hOGG1 indicate a similar exo site where undamaged guanine is trapped to avoid processing.19 The major groove intermediate may provide a similar readout mechanism by Fpg, allowing 8-oxoG to continue along the path to the active site, while discriminating against G. This will be discussed in detail elsewhere.
In summary, we have used MD simulations to show the complete free energy profile of base eversion for Fpg with its 8-oxoG substrate. Our minor groove PMF indicates a free energy barrier to flipping into the active site that is significantly lower for major groove eversion (corresponding to ~2–3 orders of magnitude faster eversion rate), indicating that this is the preferred path for lesion processing in Fpg. Interestingly, the human functional analog hOGG1 is also believed to involve base eversion via a major groove pathway, reinforcing the functional conservation between the prokaryotic and eukaryotic enzymes.19,20 Our data suggest that Fpg not only shares functional similarity to hOGG1, but convergent evolution is also present in dynamic aspects of base recognition by DNA glycosylases.
Supplementary Material
Acknowledgments
This work was supported by the NIH Health, grant number CA17395 to CS & APG; the NSF, grant number 0549370 to CB; the NSF through TeraGrid under grant number MCA02N028 to CS; and the DOE under grant number DE-AC-02-98CH10886, and the State of New York.
Footnotes
Supporting Information Available: Detailed methods, histograms, convergence analysis and complete ref. 16 are available. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Qi Y, Spong MC, Nam K, Banerjee A, Jiralerspong S, Karplus M, Verdine GL. Nature. 2009;462:762. doi: 10.1038/nature08561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blainey PC, van Oijent AM, Banerjee A, Verdine GL, Xie XS. P Natl Acad Sci USA. 2006;103:5752. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pryor WA. Ann Rev Physiol. 1986;48:657. doi: 10.1146/annurev.ph.48.030186.003301. [DOI] [PubMed] [Google Scholar]
- 4.Grollman AP, Moriya M. Trends in Genetics. 1993;9:246. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 5.Tchou J, Grollman AP. Mutat Res. 1993;299:277. doi: 10.1016/0165-1218(93)90104-l. [DOI] [PubMed] [Google Scholar]
- 6.Bjoras M, Seeberg E, Luna L, Pearl LH, Barrett TE. Journal of Molecular Biology. 2002;317:171. doi: 10.1006/jmbi.2002.5400. [DOI] [PubMed] [Google Scholar]
- 7.Cao CY, Jiang YL, Stivers JT, Song FH. Nat Struct Mol Biol. 2004;11:1230. doi: 10.1038/nsmb864. [DOI] [PubMed] [Google Scholar]
- 8.Fromme JC, Verdine GL. J Biol Chem. 2003;278:51543. doi: 10.1074/jbc.M307768200. [DOI] [PubMed] [Google Scholar]
- 9.Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G. J Biol Chem. 2002;277:19811. doi: 10.1074/jbc.M202058200. [DOI] [PubMed] [Google Scholar]
- 10.Tchou J, Grollman AP. J Biol Chem. 1995;270:11671. doi: 10.1074/jbc.270.19.11671. [DOI] [PubMed] [Google Scholar]
- 11.Banerjee A, Santos WL, Verdine GL. Science. 2006;311:1153. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- 12.Norman DPG, Chung SJ, Verdine GL. Biochemistry. 2003;42:1564. doi: 10.1021/bi026823d. [DOI] [PubMed] [Google Scholar]
- 13.Radom CT, Banerjee A, Verdine GL. J Biol Chem. 2007;282:9182. doi: 10.1074/jbc.M608989200. [DOI] [PubMed] [Google Scholar]
- 14.Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. Nature. 1996;384:87. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- 15.Bergonzo C, Campbell AJ, Walker RC, Simmerling C. Int J Quantum Chem. 2009;109:3781. doi: 10.1002/qua.22405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Case DA, et al. University of California. San Francisco: 2008. [Google Scholar]
- 17.Song K, Campbell AJ, Bergonzo C, de los Santos C, Grollman AP, Simmerling C. J Chem Theory Comput. 2009;5:3105. doi: 10.1021/ct9001575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song K, Hornak V, de Los Santos C, Grollman AP, Simmerling C. Biochemistry. 2006;45:10886. doi: 10.1021/bi060380m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee A, Yang W, Karplus M, Verdine GL. Nature. 2005;434:612. doi: 10.1038/nature03458. [DOI] [PubMed] [Google Scholar]
- 20.Banerjee A, Verdline GL. P Natl Acad Sci USA. 2006;103:15020. doi: 10.1073/pnas.0603644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.